Abstract
When produced at physiological levels reactive oxygen species (ROS) can act as signaling molecules to regulate normal vascular function. Produced under pathological conditions, ROS can contribute to oxidative damage of cellular components (e.g., DNA and proteins) and trigger cell death. Moreover, the reaction of superoxide with nitric oxide (NO) produces the strong oxidant peroxynitrite and decreases NO bioavailability, both of which may contribute to activation of cell death pathways. The effects of ROS generated from the 1,4-naphthoquinones alone and in combination with NO on the activation status of poly(ADP-ribose) polymerase (PARP) and cell viability were examined. Treatment with redox cycling quinones activates PARP, and this stimulatory effect is attenuated in the presence of NO. Mitochondria play a central role in cell death signaling pathways and are a target of oxidants. We show that simultaneous exposure of endothelial cells to NO and ROS results in mitochondrial dysfunction, ATP and NAD+ depletion, and cell death. Alone, NO and ROS have only minor effects on cellular bioenergetics. Further, PARP inhibition does not attenuate reduced cell viability or mitochondrial dysfunction. These results show that concomitant exposure to NO and ROS impairs energy metabolism and triggers PARP-independent cell death. While superoxide-mediated PARP activation is attenuated in the presence of NO, PARP inhibition does not modify the loss of mitochondrial function or adenine and pyridine nucleotide pools and subsequent bioenergetic dysfunction. These findings suggest that the mechanisms by which ROS and NO induce endothelial cell death is closely linked to maintenance of mitochondrial function and not overactivation of PARP.
Keywords: Mitochondria, oxidative stress, mitochondrial bioenergetics
Endothelial dysfunction contributes to the pathogenesis of several diseases including diabetes, atherosclerosis, cardiovascular, and pulmonary diseases.1 Endothelium-derived nitric oxide (NO) is essential for normal vessel function as it regulates arterial pressure by causing vascular smooth muscle relaxation.2 Reactive oxygen species (ROS), such as superoxide anion and hydrogen peroxide, are produced in the endothelium during cellular metabolism by various enzymes including NADPH oxidase, xanthine oxidase, uncoupled endothelial NO synthase, and mitochondrial respiratory chain complexes.3, 4 In addition to cellular sources of ROS, naphthoquinone compounds (e.g., menadione, DMNQ) represent a group of environmental toxins present in the air as products of fuel combustions and tobacco smoke. Quinones interact with biological systems to cause oxidative stress by intracellular one- and two-electron redox cycling to generate superoxide and hydrogen peroxide,5, 6 and in some cases modification of cellular nucleophiles.7 Quinones have been used to generate ROS to study oxidative stress and redox signaling.8–11 For example, menadione has been shown to activate redox-dependent gene expression at low levels8 and cause mitochondrial DNA damage and cell death at high levels.12, 13 DMNQ exerts its prooxidative effect via redox cycling,14 while menadione, in addition to ROS generation, can modify protein thiols, which are strong nucleophiles.7
When produced at physiological levels, ROS can act as signaling molecules that regulate normal vascular function,15 however, at pathophysiological levels, ROS can decrease NO bioavailability due to the reaction between NO and superoxide, yielding the strong oxidant peroxynitrite and the subsequent post-translational modifications of proteins (e.g., tyrosine nitration).16, 17 Further, there is a complex interplay between NO- and ROS-dependent signaling, as NO has been shown to inhibit hydrogen peroxide-induced death,18 and interactions between NO and superoxide limit NO signaling.19
ROS produced by redox cycling quinones, i.e., superoxide and hydrogen peroxide, can damage proteins and DNA.20 Poly(ADP)-ribose polymerase (PARP) is activated by DNA damage,21, 22 and is responsible for menadione-stimulated cell death.11 PARP catalyzes the transfer of ADP-ribose from NAD+ to itself and other acceptors (e.g., histones) to facilitate changes in chromatin structure and recruitment of DNA repair machinery. PARP-1 is responsible for ~90% of poly (ADP-ribosy)lation in most cells.21 High levels of DNA damage and prolonged activation of PARP can lead to depletion of cellular NAD+ and subsequent loss of ATP, which results in cell death.23, 24 PARP activation has been linked to the pathogeneses of multiple vascular diseases including endothelial dysfunction associated with diabetes,25 hemorrhagic shock,26 stroke,27 and myocardial28, 29 and cerebral ischemia.30 In each of these examples, oxidative stress was proposed as the underlying mechanism responsible for DNA damage and subsequent activation of PARP. While superoxide and/or peroxynitrite were indicated as mediators of DNA damage that triggered PARP, an investigation of systems generating superoxide and nitric oxide under a defined set of conditions has not been performed.
In this study, the mechanisms of action of ROS and NO on endothelial metabolic function and viability were examined. We show that actions of ROS on bovine aortic endothelial cells (BAEC) are reversible, associated with PARP-1 activation and the inhibition in mitochondrial function, but do not lead to subsequent cell death. Alone, NO transiently inhibits mitochondrial respiration without activating PARP; however, in combination, NO donor and redox cyclers activate PARP-1 and decrease cellular ATP and NAD+ pools, and these effects result in the complete inhibition of oxidative metabolism. We conclude that under these conditions cell death occurs due to inability to maintain metabolic functions and not due to overactivation of PARP.
Experimental procedures
Materials
All chemicals were of analytical grade and purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Deta/NO (2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethyl-1H-imidazolyl-1-oxy-3-oxide) was purchased from Cayman Chemical (Ann Arbor, MI). DMNQ was obtained from Enzo Life Sciences (Farmingdale, NY). Hydroethidine was purchased from Molecular Probes (Grand Island, NY).
Cell culture
BAEC were obtained from Lonza (Walkersville, MD) and grown in high glucose (25 mM) DMEM media supplemented with 10 % FBS (Invitrogen, Carlsbad, CA) as described previously.31 Cells were exposed to NO donor and quinones in assay media (Dulbecco’s phosphate buffered saline, DPBS, supplemented with 5.5 mM glucose and 1 mM pyruvate). Cells were grown in 6-well cluster plates unless otherwise noted. Experiments using extracellular flux technology were performed in specialized Seahorse Bioscience microplates with 40,000 cells/well. BAEC were seeded on 48-well cluster plates for MTT assay. Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Cells were used between passages 4 and 10 for all experiments and at 90% confluence at the beginning of each assay.
Detection of superoxide
Superoxide formation was assessed by measuring oxidation of hydroethidine (HE) to 2-hydroxyethidium (2-OH-E+) as described previously.32 Briefly, cells were treated with indicated compounds for 4 h and HE (10 µM) was added for the last hour of incubation. After the wash with ice cold DPBS, cells were scraped into 1 mL of DPBS and pellets (1000 × g for 1 min) were snap frozen in liquid nitrogen. Pellets were syringe-lysed in 120 µL DPBS containing 0.1 % Triton-X100. Then 80 µL of lysate was mixed with 80 µL of 0.2 M HClO4 in methanol to extract oxidation products of HE. Samples were incubated on ice for 1 h, followed by centrifugation (16000 × g for 15 min). Resulting supernatant (120 µL) was mixed with 1 M potassium phosphate, pH 2.6 (40 µL). Samples were centrifuged and supernatants were analyzed by HPLC using Shimadzu Prominence system equipped with UV/Vis and fluorescent detectors. The analysis was performed on Kromasil C18 column (100 × 4.6 mm, 2.6 µm, Phenomenex, Torrance, CA) using solvent A (0.1 % trifluoroacetic acid in water, v/v) and solvent B (0.1 % trifluoroacetic acid in acetonitrile, v/v) with a flow rate of 1 ml/min. The column was equilibrated with 80 % solvent A and 20 % solvent B, followed by linear increase in solvent B to 40 % over the next 10 min. The fraction of solvent B was increased to 100 % over the next 2 min and maintained at this level for 2.5 min. 2-OH-E+ in the sample was detected with fluorescent detector (ex. 490nm, em. 567 nm) and quantified using the known amounts of standard.
Lactate dehydrogenase release
After the treatment with NO donor and quinones and a 12 h incubation in complete medium, aliquots of the medium were taken, and cells were harvested by scraping into lysis buffer (PBS with 0.1% Triton X-100). Cell lysates were centrifuged (14,000 × g for 10 min), and LDH (lactate dehydrogenase) activity in the supernatant and medium samples was measured by monitoring the oxidation of NADH (0.3 mM) at 340 nm.33
MTT assay
MTT assay was performed as described previously.34 After the treatment, the media were replaced with complete culture medium containing 0.4 mg/mL thiazolyl blue tetrazolium, and the cells were incubated at 37°C for 2 h. The medium was then removed, and the resulting formazan crystals were solubilized in DMSO. The absorbance was read at 590 nm with background reference at 620 nm.
Determination of protein poly(ADP-ribosyl)ation
Cell lysate proteins were resolved using reducing SDS/PAGE and transferred on to nitrocellulose membranes (Bio-Rad, Hercules, CA). Equivalent amounts of protein were loaded, and protein-attached poly(ADP-ribose) polymers were detected using specific antibody from Trevigen (Gaithersburg, MD) and visualized with enhanced chemiluminescence. The levels of protein poly(ADP-ribosyl)ation were normalized to β-actin levels (Sigma).
Extracellular flux technology
Seahorse Bioscience XF24 Extracellular Flux Analyzer (North Billerica, MA) was used to measure mitochondrial function in intact BAEC.35–37 This instrument allows for the sensitive measurement of oxygen consumption rates (OCRs) from adherent, intact cultured cells. The mitochondrial function assay employed in the present study used sequential injection of oligomycin and carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) to define the mitochondrial function parameters basal OCR, maximal OCR, and reserve respiratory capacity as described previously.38, 39 After 5 h treatment with NO donor and quinones, cells were washed and switched to assay media 1 h prior to the beginning of the assay and maintained at 37°C. Values were normalized to the total protein per well after the completion of the extracellular flux assay. Protein levels were assessed by the Bradford protein assay (Bio-Rad, Hercules, CA). When examining the recovery of mitochondrial function with extracellular flux analyzer, the treatment medium was replaced with unbuffered DMEM, supplemented with 10% FBS, 25 mM glucose, 1 mM pyruvate and adjusted to pH 7.4, and oxygen consumption and extracellular acidification were followed for 12 hours.
Nucleotide measurements
Adenine (ATP, ADP, and AMP) and pyridine nucleotides (NAD+, NADH, NADP+, and NADPH) were analyzed using HPLC following acidic or alkaline extraction based on previously published methods.40–42 Adenine and oxidized forms of pyridine nucleotides were extracted using perchloric acid precipitation as described previously.42 Solvent A (75 µL; 0.1 M potassium phosphate, 4 mM tetrabutylammonium bisulfate, pH 6.0, diluted v/v in water 64:36) was added to supernatants. Protein concentration was determined using the Bradford assay in protein pellets resuspended in 0.5 N NaOH (200 µL). For NADH and NADPH measurements, cells were harvested under alkaline conditions (0.5 M KOH/Hank’s balanced salt solution, 3:1). The pH of lysates was adjusted to ~8 using 6 M HCl and ammonium acetate (1 M, pH 4.7).40, 41 All samples were filtered prior to HPLC analysis. HPLC analysis of nucleotides was performed on Kinetex C-18 column (2.6 µm, 100 mm × 4.6 mm internal diameter) using solvent A and solvent B (0.1 M potassium phosphate, 4 mM tetrabutylammonium bisulfate, pH 6.0, diluted v/v in methanol 64:36) with a flow rate of 1 mL/min. The column was equilibrated with solvent A and compounds were eluted during a linear increase in solvent B to 50% between 1 min and 5 min, followed by increase to 65% over next 6 min. Solvent B was maintained at that level for 1.5 min. ATP, ADP, AMP, NAD+, NADH, NADP+, and NADPH peaks were measured for each sample, compared with the standards, and expressed in nanomoles per milligram of protein.
Statistical analysis
Results are reported as means ± SE for n ≥ 3, as indicated in the figure legends. Statistical significance was evaluated by Student's t-test. The minimum level of significance was set at p < 0.05.
Results
Effects of NO and ROS on poly(ADP-ribose) polymerase activation
Menadione and DMNQ generate superoxide intracellularly via redox cycling, and superoxide production in BAEC was confirmed by monitoring the oxidation of HE to 2-OH-E+.32 Within 4 h, the levels of 2-OH-E+ increased from 12 ± 1 pmol/mg protein in controls (mean ± SE, n=3) to 629 ± 14 pmol/mg protein in cells treated with menadione (20 µM) and to 345 ± 9 pmol/mg protein in cells exposed to DMNQ (20 µM). Similar amounts of 2-OH-E+ were detected in macrophages stimulated with phorbol ester,43 hence superoxide generation by quinones corresponded to levels of superoxide produced by NADPH oxidase in immune cells. The nitric oxide donor Deta/NO has a half-life of 20 h under cell culture conditions (37°C, pH 7.4)44 liberating NO in the extracellular space with an initial rate of about 10 nM/s. The levels of NO liberated by this donor (at 500 µM) are comparable to levels of NO generated by iNOS.45 The calculated steady state concentration of NO in solution is ~ 2 µM (considering consumption only by oxygen).
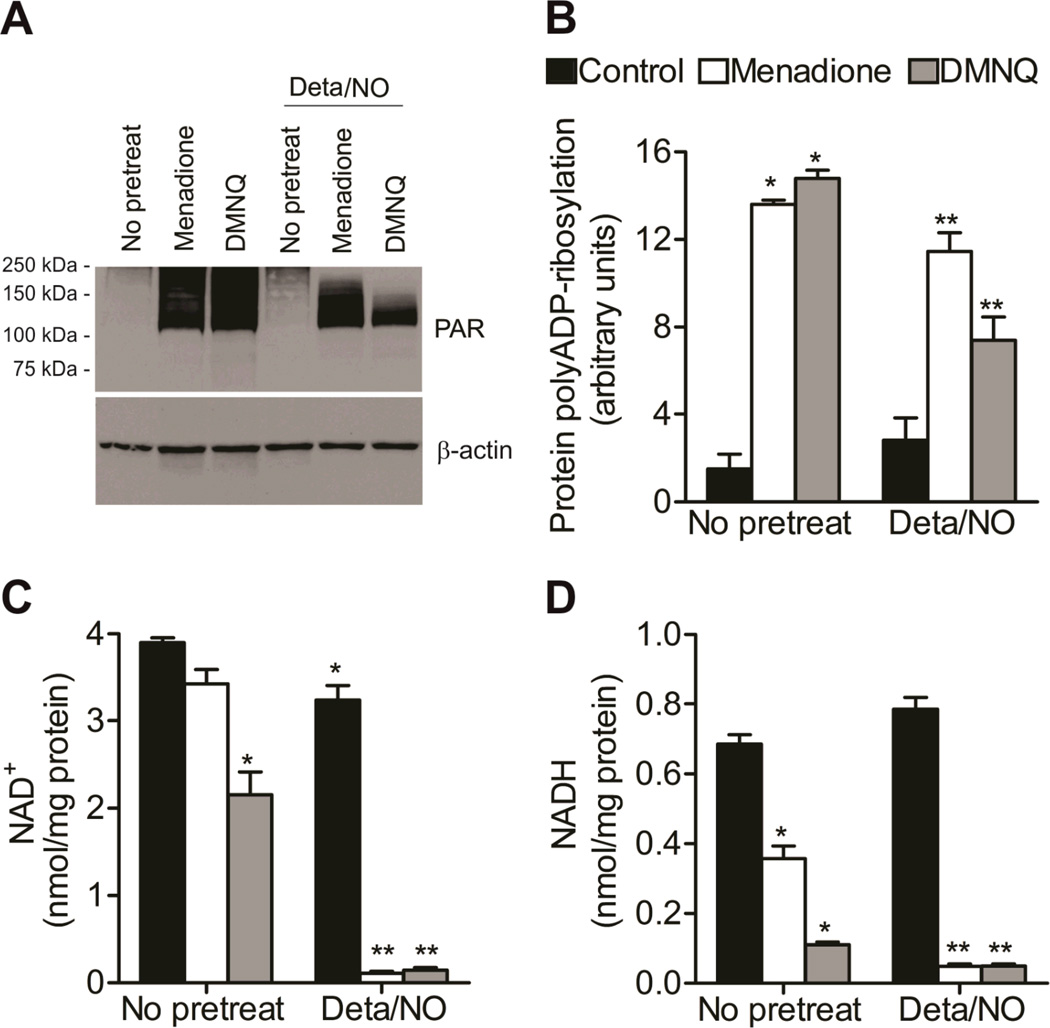
We first measured formation of poly(ADP-ribose) (PAR) polymers, indicative of PARP activation, after the treatment with NO donor (500 µM) and quinones alone and in combination. As expected, alone menadione and DMNQ caused significant protein poly(ADP-ribosyl)ation indicative of DNA damage (Figure 1A and B). Consistent with previous reports,46 NO alone did not activate PARP. Surprisingly, protein poly(ADP-ribosyl)ation was decreased when NO was administered in the presence of redox cyclers as compared to redox cyclers alone. This suggests that NO inhibits ROS-dependent PARP activation.
Figure 1. NO abrogates quinone-dependent PARP activation.
BAEC were exposed to Deta/NO (500 µM) for 1 h prior to treatment with menadione (20 µM) or DMNQ (20 µM) for an additional 4 h. (A) Protein poly(ADP-ribosyl)ation was measured by Western blotting using antibody against poly(ADP-ribose). (B) Densitometric analysis of protein PAR normalized to β-actin levels. NAD+ (C) and NADH (D) levels were measured by HPLC and normalized to total protein. Control (black bars), menadione-treated (white bars), and DMNQ-treated (grey bars) groups are shown. Values represent means ± SE; n = 3. * p < 0.05 compared to untreated control, ** p < 0.05 compared to samples without Deta/NO.
Since PARP-1, once activated by DNA strand breaks, is a major NAD+-consuming enzyme,21, 22 we next measured the levels of NAD+ and NADH. Treatment with menadione, DMNQ, or Deta/NO resulted in modest decreases in NAD+ levels (by 12%, 44.7%, and 16.9%, respectively; Figure 1C). However, when the NO donor was added in the presence of the quinone, less than 5% of the NAD+ remained. NADH levels followed the same trend as NAD+, with combined treatment of NO and quinone, resulting in over 90% depletion of the pool (Figure 1D). Moreover, combined exposure to NO and redox cycler significantly decreased intracellular NADPH, but did not modify NADP+ levels (Figure S1), indicating that the observed loss of NAD+ and NADH pools was not due to conversion to the NADP(H) pool of nucleotides. Cells exposed to NO and menadione maintained the plasma membrane integrity compared to untreated controls and there was about 25% trypan blue positive staining in cells treated with NO and DMNQ (Figure S2). Thus, loss of membrane integrity was not responsible for the depletion of pyridine nucleotides. Taken together, these results demonstrate a striking lack of correlation between activation of PARP and NAD+ depletion, and further studies sought to understand this mechanistically.
The effects of NO and quinone on cell viability
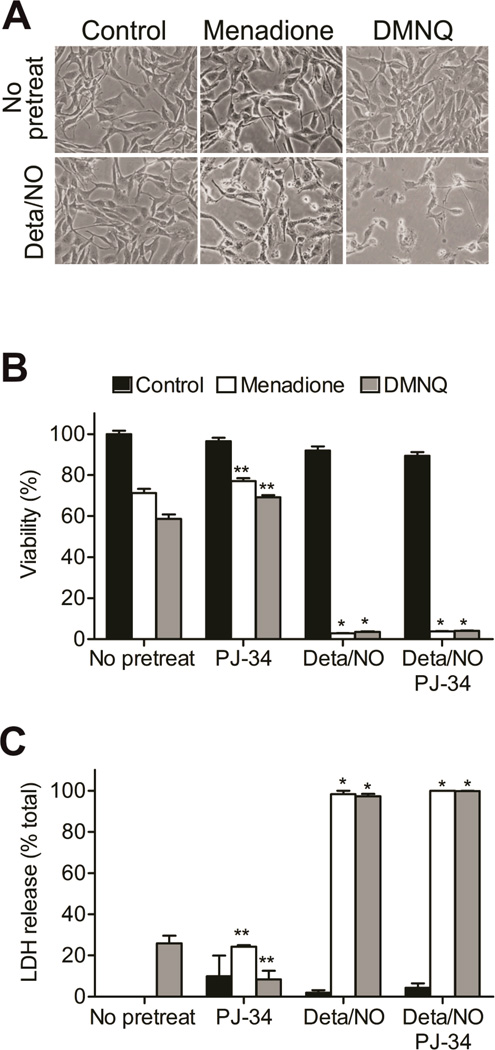
BAEC were exposed to the NO donor Deta/NO (500 µM) or quinone (DMNQ, 20 µM, or menadione, 20 µM) alone or in combination for 5 h. The effects of NO and ROS (generated from menadione or DMNQ) on cellular viability were assessed. Simultaneous treatment with the NO donor and DMNQ or menadione led to marked changes in cell morphology, that include the loss of projections, detachment from the plate and cell shrinking. Alone these agents did not modify cell morphology (Figure 2A). Viability measurements using MTT reduction demonstrated menadione and DMNQ decreased BAEC viability by 30 to 40%, and addition of PARP-1 inhibitor, PJ-34, afforded only a small protection (Figure 2B). Alone, the NO donor did not reduce BAEC viability, but in the presence of menadione or DMNQ, NO caused a greater than 95% loss in cell viability. Importantly, co-treatment with PJ-34 under the same conditions did not exert any protective effects. Together, these studies were extended using LDH release as a second measure of cell death, and similar results were found. The combined treatment of NO donor and menadione or DMNQ caused significant LDH release, and this process was not affected by addition of PJ-34 (Figure 2C). These findings indicate that NO and quinones act synergistically to induce cell death in BAEC which is insensitive to PARP-1 inhibition. This implies that PARP-1 overactivation does not mediate cell death under conditions of concomitant NO and ROS production, a finding inconsistent with other literature reports. Additional studies were aimed at identifying alternative mechanisms of cell death.
Figure 2. NO and quinone synergistically contribute to cell death.
BAEC were exposed to Deta/NO (500 µM) for 1 h prior to treatment with menadione (20 µM) or DMNQ (20 µM) for an additional 4h. (A) Light micrographs were taken at 5 h and show morphological changes in cells exposed to both quinone and NO donor. (B) Viability was assessed by MTT in cells incubated in the absence or presence of PJ-34 (10 µM). (C) Cells were washed and incubated in full medium for an additional 12 h. LDH release was measured to assess the cell viability. Control (black bars), menadione-treated (white bars), and DMNQ-treated (grey bars) groups are shown. Values represent means ± SE; n = 3. * p < 0.05 compared to samples without Deta/NO, **p < 0.05 compared to samples without PJ-34.
NAD+ levels in response to PARP-1 inhibition
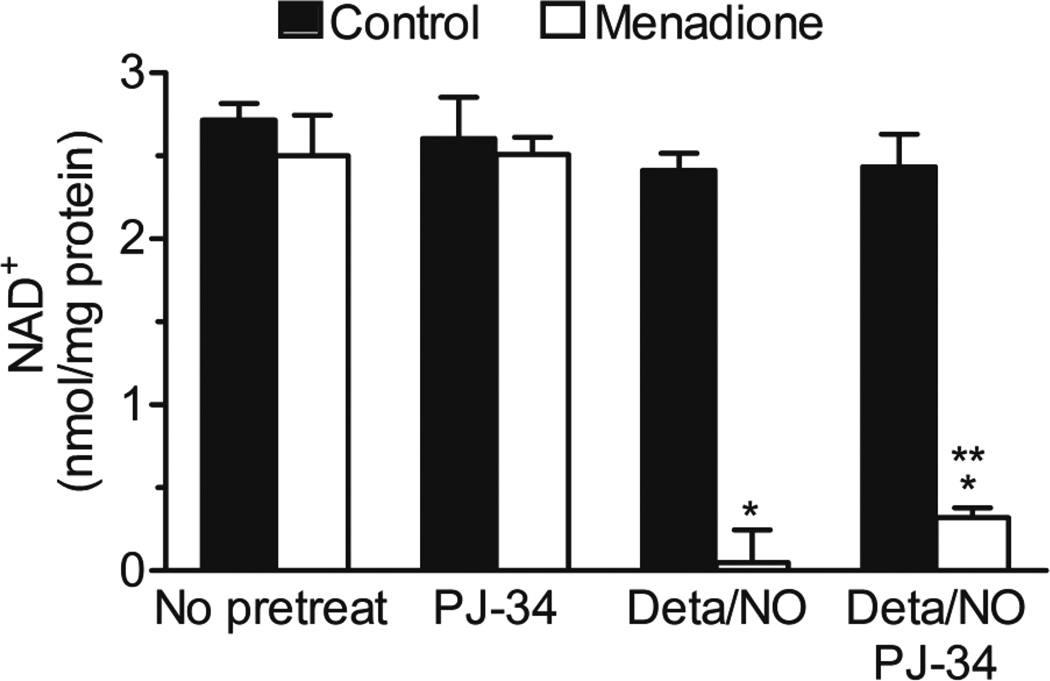
PARP-1 is a major NAD+-consuming enzyme that is activated by DNA strand breaks21, 22 and we have shown that in response to concomitant generation of NO and ROS, there is dramatic loss of the cellular NAD+ pool. Thus, we investigated whether inhibition of PARP-1 protects this nucleotide pool. After treatment with NO donor and menadione, in the presence of PJ-34, the NAD+ level was significantly higher than in its absence, however this increase is less than 12 % of the total pool of NAD+ (Figure 3). These data demonstrate that while the addition of PJ-34 is able to protect from complete consumption of NAD+, PARP-dependent NAD+ depletion represents only a small component of the loss in response to NO and superoxide/hydrogen peroxide exposure.
Figure 3. The effect of PARP inhibitor on NAD+ depletion.
BAEC were exposed to Deta/NO (500 µM) for 1 h prior to incubation without (black bars) and with menadione (20 µM, white bars) for an additional 4 h in the presence and absence of PARP-1 inhibitor, PJ-34 (10 µM). NAD+ levels were measured by HPLC and normalized to total protein. Values represent means ± SE; n = 3. * p < 0.05 compared to samples without Deta/NO, **p < 0.05 compared to samples without PJ-34.
Adenine nucleotide levels after Deta/NO and quinone administration
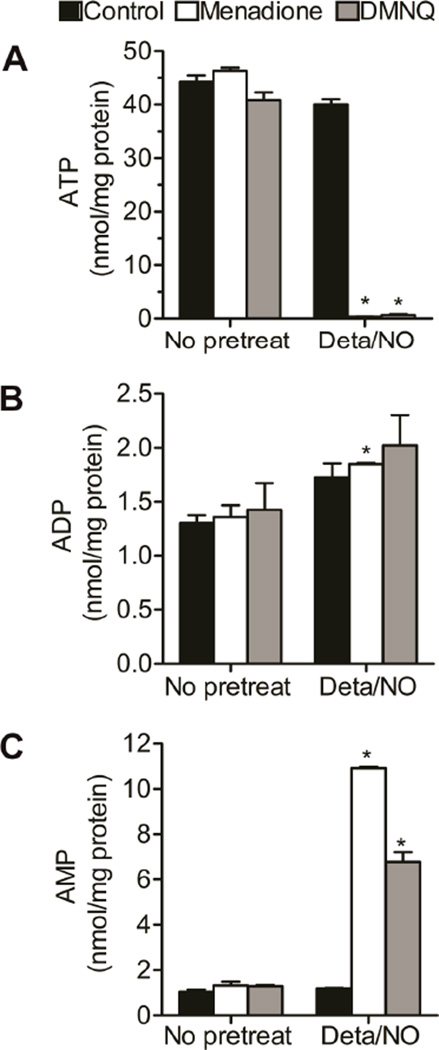
To investigate the mechanism mediating the NAD+/NADH depletion and toxicity after NO and ROS exposure we examined the effects of these compounds on the levels of ATP, critical substrates for de novo and salvage pathways of NAD+ biosynthesis.47 Alone, the NO donor, menadione, or DMNQ did not affect ATP, ADP, and AMP levels. In contrast, simultaneous exposure to NO and ROS led to ATP depletion in BAEC (Figure 4A). A nearly complete loss of ATP was associated with 50% elevation in ADP, while AMP levels were increased 8- to 11-fold (Figure 4B and C). Interestingly, the increase in ADP and AMP did not account for the entire loss in ATP, suggesting there is further metabolism of adenosine phosphates. Inhibition of PARP-1 with PJ-34 did not prevent ATP depletion or changes in ADP and AMP levels in response to combined Deta/NO and menadione treatment (Figure S3).
Figure 4. Changes in adenine nucleotides in response to combination of NO and quinone.
BAEC were exposed to Deta/NO (500 µM) for 1 h prior to treatment with menadione (20 µM, white bars) or DMNQ (20 µM, grey bars) for an additional 4 h. Black bars represent control treatment. ATP (A), ADP (B) and AMP (C) levels were measured by HPLC and normalized to total protein. Values represent means ± SE; n = 3. * p < 0.05 compared to samples without Deta/NO.
Mitochondrial function in response to NO and quinone
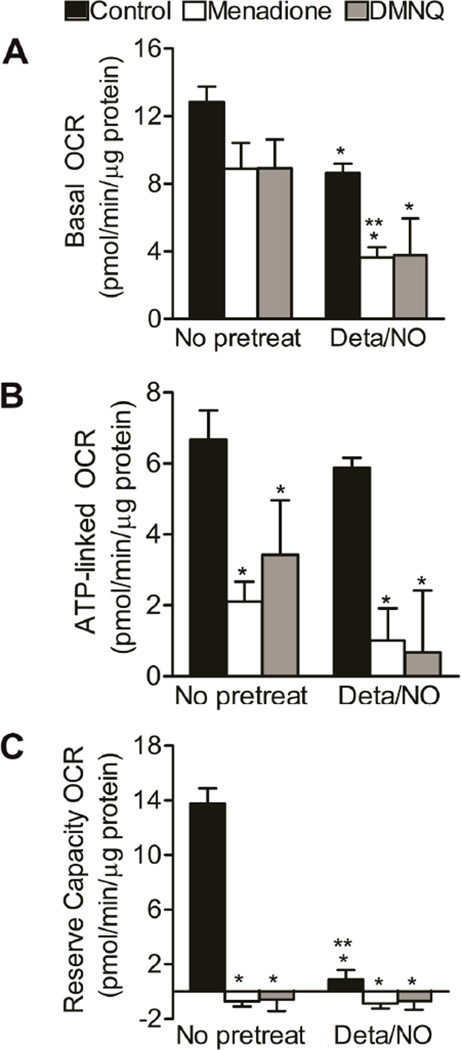
As shown above, combined NO and ROS treatment dramatically decreased ATP and NAD+ levels. Yet, PARP-1 overacitvation was insufficient to explain this depletion, as PARP was activated by redox cyclers both in the presence and absence of NO. An alternative mechanism for deregulation of adenine and pyridine nucleotides homeostasis is through bioenergetic dysfunction, and mitochondria represent a critical hub for nucleotide catabolism and anabolism. Bioenergetic function was assessed under conditions of NO and superoxide/hydrogen peroxide formation using extracellular flux technology. There was a ~30% decrease in basal oxygen consumption rate (OCR) of BAEC upon treatment with NO donor, menadione or DMNQ (Figure 5A). Administration of redox cycler in the presence of Deta/NO reduced OCR by 66%, a finding that is indicative of loss of mitochondrial function. Mitochondrial function was further probed by examining the effects of sequential administration of electron transport chain inhibitors to assess multiple mitochondrial function parameters.38 A schematic representation of this mitochondrial function assay and the calculation of these parameters, including basal OCR, ATP-linked OCR, proton leak, reserve capacity and oxygen consumption that occurs independent of Complex IV (non-mitochondrial), is presented in Supplemental Figure 4 along with time-resolved data obtained after the treatment with quinones in the presence and absence of Deta/NO. ATP-linked respiration was inhibited after treatment with menadione and DMNQ, but consistent with previous reports,48 it was not affected by NO (Figure 5B). In response to simultaneous administration of a redox cycler and Deta/NO there was a further decrease in ATP-linked respiration as compared to the redox cycler alone. Reserve capacity was abolished by all compounds added alone or combination (Figure 5C). Non-mitochondrial oxygen consumption and proton leak were not significantly affected by any of the treatments (Figure S4). In sum, these results indicate that simultaneous exposure to NO and ROS results in severe mitochondrial dysfunction in BAEC, a likely cause of adenine and pyridine nucleotides depletion.
Figure 5. Mitochondrial function is decreased in response to combination of NO and quinone.
BAEC were exposed to Deta/NO (500 µM) for 1 h prior to treatment with menadione (20 µM, white bars) or DMNQ (20 µM, grey bars) for an additional 4 h. Black bars represent control treatment. (A) Basal oxygen consumption rate (OCR), (B) ATP-linked OCR and (C) reserve capacity OCR were measured using extracellular flux technology. OCRs were normalized to total protein per well after completion of assay. Values represent means ± SE; n = 3 to 4. * p < 0.05 compared to control, ** p < 0.05 compared to samples without Deta/NO.
Discussion
In response to DNA damage caused by oxidative stress, PARP is activated and participates in the repair of DNA and cell survival. When overactivated, PARP can deplete cellular NAD+ and ATP23 and this process has been linked to several vascular pathologies.25–28, 30 Since overactivation of PARP-1 and oxidative stress have been associated with vascular disease, the effects of NO and ROS on PARP-1 activation status and endothelial cells function/survival were examined. After quinone treatment, NAD+ and ATP levels as well as basal oxygen OCR were decreased about 30%, and this effect was associated with PARP-1 activation. Interestingly, the depletion of NAD+ and ATP and decline in mitochondrial function after combined treatment with NO and ROS occurred under conditions when PARP activity was diminished as compared to ROS alone, demonstrating PARP activation is attenuated, not stimulated in response to the combination of ROS and nitric oxide. Although the mechanism is not clear, it is likely that hydrogen peroxide is the major genotoxic oxidant in this system. NO could diminish hydrogen peroxide by directly scavenging superoxide and diverting it away from dismutation, or alternatively, NO could act down-stream of hydrogen peroxide formation on the PARP activation signaling pathway. Although inhibition of PARP-1 has been shown to protect from hydrogen peroxide- and peroxynitrite-induced NAD+ depletion in human umbilical vein endothelial cells,49 in the case of BAEC, the inhibition of PARP-1 did not prevent the loss of cellular levels of NAD+ and ATP, or the inhibition of mitochondrial function after treatment with Deta/NO and quinones. These findings suggest PARP-1 overactivation is not responsible for the depletion of NAD+ and ATP. The differences observed between the two cell types may arise from the different oxidants produced in each experimental model or the use of a single dose (bolus addition) in the study with HUVEC as compared to the use of redox cycling quinones and Deta/NO to deliver ROS and NO in this study. There was a significant induction of PARP-1 activity after menadione and DMNQ treatment in BAEC, suggesting that the production of ROS by these redox cyclers causes DNA damage to levels sufficient to activate PARP. Nevertheless, the cellular NAD+ and ATP levels remained intact, suggesting that PARP-1 is not overactivated. This quinone-induced protein poly(ADP-ribosyl)ation was also inhibited in the presence of NO donor, implicating NO as a suppressor of this response to genotoxic stress. In combination, the inhibition of mitochondrial function by NO appears to synergize with the genotoxic effects of the quinones resulting in irreversible damage and cell death. While our studies are in agreement with previous reports indicating that superoxide/hydrogen peroxide and NO are together far more toxic than any of these compounds alone,17, 50, 51 we provide new evidence that ROS-stimulated PARP-1 activation is attenuated in the presence of NO indicating that targets in addition to PARP-1 and DNA damage contribute to the demise of endothelial cells producing ROS and NO.
Superoxide-producing compounds, such as menadione induce cell death by apoptosis in endothelial cells52 or PARP-dependent necrosis in cardiomyocytes.11 We show that superoxide and hydrogen peroxide production in response to menadione and DMNQ treatment (20 µM) does not reduce cell viability and does not affect cellular morphology. Much like superoxide and hydrogen peroxide, the administration of NO also does not change cell morphology; however, in combination, in response to Deta/NO and superoxide producing quinones (menadione or DMNQ) cellular morphology is affected as evidenced by cell shrinking and loss of projections and cell detachment from plates (Fig 2A). While there are morphological changes associated with the loss of cell viability, there does not appear to be a breach in membrane integrity as cells exclude Trypan Blue and LDH is not released into the supernatant following 5 h incubation. Continuing the incubation leads to the loss of cell viability including membrane integrity as nearly all of the cellular LDH is released from the cells following a 12 h incubation (Fig 2C). These results indicate that the mechanisms responsible for cell death in response to NO and superoxide-generating compounds are not rapid and are associated with loss of membrane integrity, suggesting endothelial cell death by necrotic mechanisms.
Because PARP-1 overactivation does not appear to contribute to BAEC death in response to NO and ROS, but the loss of viability is associated with reductions in ATP and NAD+, the role of mitochondrial oxidative capacity was examined using the extracellular flux analyzer. Reserve capacity is defined as the bioenergetic reserve that cells can mobilize to respond to stress, and its loss has been associated with the subsequent cell death and end organ dysfunction.39, 53, 54 We show that NO and ROS generated from quinones can acutely decrease the reserve capacity of BAEC (Fig 5C), but when these compounds are removed and cells are allowed to recover in complete culture medium for up to 12 h, the cells remain viable (absence of LDH release, Fig 2C). While the stressors can negatively affect cellular bioenergetics, alone they do not cause cell death. These data are consistent with previous reports showing that NO generated from Deta/NO has relatively little effect on basal and ATP-linked OCR, but decreases reserve capacity.39 In contrast, simultaneous administration of NO donor and menadione or DMNQ leads to complete inhibition of mitochondrial respiration resulting in irreversible inhibition of cellular metabolism and cell death. Similar results have been obtained with BAEC exposed to S-nitrosocysteine (nitrosative stress) with DMNQ (oxidative stress) where these agents were not toxic individually but in combination they cause bioenergetic dysfunction, depletion of ATP and NAD+, and cell death.55 The decreases in NADH levels observed following treatment with menadione and DMNQ are likely due to the reduction of oxygen that occurs during the transfer of electron(s) from flavoenzymes to molecular oxygen by the redox cycling agents.56 However, NAD+ levels were also diminished under these conditions indicating that the mechanism is more complex than the simple oxidation of NADH. Consistent with the complexity, NO, in the presence of quinone, caused almost complete loss of NADH and NAD+ indicating large perturbations in pyridine nucleotide homeostasis under these conditions. Although NAPDH was depleted after combined treatment with Deta/NO and redox cycler, the levels of NADP+ were maintained. This finding is consistent with studies by Circu et al. who reported increased NADPH consumption in colon epithelial cells after exposure to menadione that was associated with enhanced NAD+ to NADP+ conversion to compensate for the NAPDH depletion.57 We have observed a similar scenario in endothelial cells exposed to quinones. Despite the decline in NADH and NAD+ levels, BAEC were able to maintain an intact NADPH pool. However, when NO was added as an additional stressor, loss of NADPH was observed. The disruption of pyridine nucleotide homeostasis, combined with the loss of mitochondrial function, likely contributes to disruption of cellular bioenergetics.
ATP depletion, observed after combined treatment with Deta/NO and quinone, was associated with only small increase in ADP levels and several-fold increase in cellular AMP pool (Fig 4). This is an important observation, as the energetic status of the cell is often expressed as the ratio of adenine nucleotides (e.g., ATP/ADP). Our findings suggest that the total amount of these high-energy substrates is equally important. Further, a direct consequence of elevated levels of AMP is the activation of AMP-activated protein kinase (AMPK), and AMPK has been attributed to providing protection of cells to NO-mediated damage.58 In combination NO and ROS treatment causes a net decrease in the total pool of adenine nucleotides across mono-, di-, and triphosphate. For example, combined treatment with Deta/NO and menadione resulted in a decrease of ~44 nmol of ATP per mg of protein while only 1.1 % and 22.7 % of adenine could be accounted for in the ADP and AMP pools, respectively. This indicates that under severe stress conditions, further metabolism and/or degradation of these nucleotides takes place.
Our study provides an interesting association between PARP-1 activation state and mitochondrial function. The mechanism responsible for cell death in BAEC treated with NO and superoxide involves mitochondrial dysfunction that leads to bioenergetic failure and subsequent cell death. Under these conditions DNA repair process appears to be attenuated. It is becoming apparent that mitochondrial metabolism and adequate supply of high-energy substrates are crucial for proper DNA repair.59, 60 What specifically triggers bioenergetic failure will require further investigation; however, several possibilities exist and include decrease in biosynthetic pathways and/or increased catabolism. Inhibition of oxidative phosphorylation will result in decreased levels of ATP and subsequently NAD+ due to ATP requirement for NAD+ synthesis.47 Similarly, in the absence of NAD+, glycolysis and citric acid cycle function will be impaired since NAD+ is a critical cofactor for several enzymes in these pathways, and this may inhibit ATP formation. PARP-1 overactivation can be one of the mechanisms responsible for increased catabolism of NAD+ in cells, but our data exclude this possibility in the case of BAEC exposed to NO and ROS. Alternatively, increased demand on NADP+/NADPH redox couple can deplete NAD+ - a substrate for synthesis of NADP+ and NADPH through conversion of NAD+ to NADP+ by NAD kinase.61 It is also possible that these events occur in combination and together result in severe impairment of cellular bioenergetics.
Overall, we show that NO and ROS synergize, likely through formation of peroxynitrite, to cause irreversible inhibition of mitochondrial function, disruption of nucleotide homeostasis, and cell death. It is apparent that oxidants control not only the redox state of the NADH/NAD+ couple, but also the absolute levels of pyridine and adenine nucleotides, and the latter may be more important in determining cell fate. Endothelium is a site of nitric oxide production and, under inflammatory conditions, may be exposed to ROS from several sources. Thus these findings could contribute to better understanding of events during the development and progression of vascular pathologies.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIG grants: R01-GM-55792 (N.H.), R01-AI-044458 and R01-DK-052194 (J.A.C) and Interdisciplinary Cancer Research Post-Doctoral Fellowship from the Cancer Center of the Medical College of Wisconsin (A.R.D.).
ABBREVIATIONS
- BAEC
bovine aortic endothelial cells
- Deta/NO
diethylenetriamine NONOate
- DMEM
Dulbecco’s Modified Eagle Medium
- DMNQ
2,3-dimethoxy-1,4-naphthoquinone
- ECAR
extracellular acidification rate
- FCCP
carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- LDH
lactate dehydrogenase
- MTT, NO
nitric oxide
- OCR
oxygen consumption rate
- PAR
poly(ADP-ribose)
- PARP
poly(ADP-ribose) polymerase, PJ-34 N-(6-Oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride
- ROS
reactive oxygen species
- SE
standard error
Footnotes
Supporting Information
The levels of adenine nucleotides probed in the presence of PARP inhibitor, the levels of NADP+/NADPH and time resolved XF data reported here. This material is available free of charge via the Internet at http://pubs.acs.org.
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1.Li JM, Shah AM. Endothelial cell superoxide generation: regulation and relevance for cardiovascular pathophysiology. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1014–R1030. doi: 10.1152/ajpregu.00124.2004. [DOI] [PubMed] [Google Scholar]
- 2.Waldman SA, Murad F. Biochemical mechanisms underlying vascular smooth muscle relaxation: the guanylate cyclase-cyclic GMP system. J Cardiovasc Pharmacol. 1988;12(Suppl 5):S115–S118. [PubMed] [Google Scholar]
- 3.Paravicini TM, Touyz RM. NADPH oxidases, reactive oxygen species, and hypertension: clinical implications and therapeutic possibilities. Diabetes Care. 2008;31(Suppl 2):S170–S180. doi: 10.2337/dc08-s247. [DOI] [PubMed] [Google Scholar]
- 4.Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BS, Karoui H, Tordo P, Pritchard KA., Jr Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors. Proc Natl Acad Sci U S A. 1998;95:9220–9225. doi: 10.1073/pnas.95.16.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadenas E. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem Pharmacol. 1995;49:127–140. doi: 10.1016/s0006-2952(94)00333-5. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe N, Forman HJ. Autoxidation of extracellular hydroquinones is a causative event for the cytotoxicity of menadione and DMNQ in A549-S cells. Arch Biochem Biophys. 2003;411:145–157. doi: 10.1016/s0003-9861(02)00716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunmark A, Cadenas E. Redox and addition chemistry of quinoid compounds and its biological implications. Free Radic Biol Med. 1989;7:435–477. doi: 10.1016/0891-5849(89)90126-3. [DOI] [PubMed] [Google Scholar]
- 8.Chuang YY, Chen Y, Gadisetti, Chandramouli VR, Cook JA, Coffin D, Tsai MH, DeGraff W, Yan H, Zhao S, Russo A, Liu ET, Mitchell JB. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246–6254. [PubMed] [Google Scholar]
- 9.Gerasimenko JV, Gerasimenko OV, Palejwala A, Tepikin AV, Petersen OH, Watson AJ. Menadione-induced apoptosis: roles of cytosolic Ca(2+) elevations and the mitochondrial permeability transition pore. J Cell Sci. 2002;115:485–497. doi: 10.1242/jcs.115.3.485. [DOI] [PubMed] [Google Scholar]
- 10.Kim KA, Lee JY, Park KS, Kim MJ, Chung JH. Mechanism of menadione-induced cytotoxicity in rat platelets. Toxicol Appl Pharmacol. 1996;138:12–19. doi: 10.1006/taap.1996.0092. [DOI] [PubMed] [Google Scholar]
- 11.Loor G, Kondapalli J, Schriewer JM, Chandel NS, Vanden Hoek TL, Schumacker PT. Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic Biol Med. 2010;49:1925–1936. doi: 10.1016/j.freeradbiomed.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1300–L1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- 13.Luo X, Pitkanen S, Kassovska-Bratinova S, Robinson BH, Lehotay DC. Excessive formation of hydroxyl radicals and aldehydic lipid peroxidation products in cultured skin fibroblasts from patients with complex I deficiency. J Clin Invest. 1997;99:2877–2882. doi: 10.1172/JCI119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gant TW, Rao DN, Mason RP, Cohen GM. Redox cycling and sulphydryl arylation; their relative importance in the mechanism of quinone cytotoxicity to isolated hepatocytes. Chem Biol Interact. 1988;65:157–173. doi: 10.1016/0009-2797(88)90052-x. [DOI] [PubMed] [Google Scholar]
- 15.Lee MY, Griendling KK. Redox signaling, vascular function, and hypertension. Antioxid Redox Signal. 2008;10:1045–1059. doi: 10.1089/ars.2007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 17.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotamraju S, Tampo Y, Keszler A, Chitambar CR, Joseph J, Haas AL, Kalyanaraman B. Nitric oxide inhibits H2O2-induced transferrin receptor-dependent apoptosis in endothelial cells: Role of ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 2003;100:10653–10658. doi: 10.1073/pnas.1933581100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas DD, Ridnour LA, Espey MG, Donzelli S, Ambs S, Hussain SP, Harris CC, DeGraff W, Roberts DD, Mitchell JB, Wink DA. Superoxide fluxes limit nitric oxide-induced signaling. J Biol Chem. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 20.Morgan WA. DNA single-strand breakage in mammalian cells induced by redox cycling quinones in the absence of oxidative stress. J Biochem Toxicol. 1995;10:227–232. doi: 10.1002/jbt.2570100407. [DOI] [PubMed] [Google Scholar]
- 21.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 23.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia Soriano F, Virag L, Jagtap P, Szabo E, Mabley JG, Liaudet L, Marton A, Hoyt DG, Murthy KG, Salzman AL, Southan GJ, Szabo C. Diabetic endothelial dysfunction: the role of poly(ADP-ribose) polymerase activation. Nat Med. 2001;7:108–113. doi: 10.1038/83241. [DOI] [PubMed] [Google Scholar]
- 26.Liaudet L, Soriano FG, Szabo E, Virag L, Mabley JG, Salzman AL, Szabo C. Protection against hemorrhagic shock in mice genetically deficient in poly(ADP-ribose)polymerase. Proc Natl Acad Sci U S A. 2000;97:10203–10208. doi: 10.1073/pnas.170226797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endres M, Scott G, Namura S, Salzman AL, Huang PL, Moskowitz MA, Szabo C. Role of peroxynitrite and neuronal nitric oxide synthase in the activation of poly(ADP-ribose) synthetase in a murine model of cerebral ischemia-reperfusion. Neurosci Lett. 1998;248:41–44. doi: 10.1016/s0304-3940(98)00224-9. [DOI] [PubMed] [Google Scholar]
- 28.Faro R, Toyoda Y, McCully JD, Jagtap P, Szabo E, Virag L, Bianchi C, Levitsky S, Szabo C, Sellke FW. Myocardial protection by PJ34, a novel potent poly (ADP-ribose) synthetase inhibitor. Ann Thorac Surg. 2002;73:575–581. doi: 10.1016/s0003-4975(01)03329-x. [DOI] [PubMed] [Google Scholar]
- 29.Schriewer JM, Peek CB, Bass J, Schumacker PT. ROS-Mediated PARP Activity Undermines Mitochondrial Function After Permeability Transition Pore Opening During Myocardial Ischemia-Reperfusion. J Am Heart Assoc. 2013;2:e000159. doi: 10.1161/JAHA.113.000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, Snyder SH, Dawson VL. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 31.Broniowska KA, Hogg N. Differential mechanisms of inhibition of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosothiols and NO in cellular and cell-free conditions. Am J Physiol Heart Circ Physiol. 2010;299:H1212–H1219. doi: 10.1152/ajpheart.00472.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zielonka J, Hardy M, Kalyanaraman B. HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic Biol Med. 2009;46:329–338. doi: 10.1016/j.freeradbiomed.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diers AR, Higdon AN, Ricart KC, Johnson MS, Agarwal A, Kalyanaraman B, Landar A, Darley-Usmar VM. Mitochondrial targeting of the electrophilic lipid 15-deoxy-Delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochem J. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, Ferrick DA, Nicholls DG, Brand MD. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med. 2010;48:905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merker MP, Bongard RD, Kettenhofen NJ, Okamoto Y, Dawson CA. Intracellular redox status affects transplasma membrane electron transport in pulmonary arterial endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L36–L43. doi: 10.1152/ajplung.00283.2001. [DOI] [PubMed] [Google Scholar]
- 41.Stocchi V, Cucchiarini L, Canestrari F, Piacentini MP, Fornaini G. A very fast ion-pair reversed-phase HPLC method for the separation of the most significant nucleotides and their degradation products in human red blood cells. Anal Biochem. 1987;167:181–190. doi: 10.1016/0003-2697(87)90150-3. [DOI] [PubMed] [Google Scholar]
- 42.Perez J, Hill BG, Benavides GA, Dranka BP, Darley-Usmar VM. Role of cellular bioenergetics in smooth muscle cell proliferation induced by platelet-derived growth factor. Biochem J. 2010;428:255–267. doi: 10.1042/BJ20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, Kalyanaraman B. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem. 2012;287:2984–2995. doi: 10.1074/jbc.M111.309062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keefer LK, Nims RW, Davies KM, Wink DA. "NONOates" (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 45.Orsi A, Beltran B, Clementi E, Hallen K, Feelisch M, Moncada S. Continuous exposure to high concentrations of nitric oxide leads to persistent inhibition of oxygen consumption by J774 cells as well as extraction of oxygen by the extracellular medium. Biochem J. 2000;346(Pt 2):407–412. [PMC free article] [PubMed] [Google Scholar]
- 46.Andreone T, Meares GP, Hughes KJ, Hansen PA, Corbett JA. Cytokine-mediated beta-cell damage in PARP-1-deficient islets. Am J Physiol Endocrinol Metab. 2012;303:E172–E179. doi: 10.1152/ajpendo.00055.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diers AR, Broniowska KA, Darley-Usmar VM, Hogg N. Differential regulation of metabolism by nitric oxide and S-nitrosothiols in endothelial cells. Am J Physiol Heart Circ Physiol. 2011;301:H803–H812. doi: 10.1152/ajpheart.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathews MT, Berk BC. PARP-1 inhibition prevents oxidative and nitrosative stress-induced endothelial cell death via transactivation of the VEGF receptor 2. Arterioscler Thromb Vasc Biol. 2008;28:711–717. doi: 10.1161/ATVBAHA.107.156406. [DOI] [PubMed] [Google Scholar]
- 50.Pryor WA, Squadrito GL. The chemistry of peroxynitrite: a product from the reaction of nitric oxide with superoxide. Am J Physiol. 1995;268:L699–L722. doi: 10.1152/ajplung.1995.268.5.L699. [DOI] [PubMed] [Google Scholar]
- 51.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 52.Warren MC, Bump EA, Medeiros D, Braunhut SJ. Oxidative stress-induced apoptosis of endothelial cells. Free Radic Biol Med. 2000;29:537–547. doi: 10.1016/s0891-5849(00)00353-1. [DOI] [PubMed] [Google Scholar]
- 53.Gong G, Liu J, Liang P, Guo T, Hu Q, Ochiai K, Hou M, Ye Y, Wu X, Mansoor A, From AH, Ugurbil K, Bache RJ, Zhang J. Oxidative capacity in failing hearts. Am J Physiol Heart Circ Physiol. 2003;285:H541–H548. doi: 10.1152/ajpheart.01142.2002. [DOI] [PubMed] [Google Scholar]
- 54.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diers A, Broniowska K, Hogg N. Nitrosative stress and redox-cycling agents synergize to cause mitochondrial dysfunction and cell death in endothelial cells. Redox Biology. 2013;1:1–7. doi: 10.1016/j.redox.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe N, Dickinson DA, Liu RM, Forman HJ. Quinones and glutathione metabolism. Methods Enzymol. 2004;378:319–340. doi: 10.1016/S0076-6879(04)78024-6. [DOI] [PubMed] [Google Scholar]
- 57.Circu ML, Maloney RE, Aw TY. Disruption of pyridine nucleotide redox status during oxidative challenge at normal and low-glucose states: implications for cellular adenosine triphosphate, mitochondrial respiratory activity, and reducing capacity in colon epithelial cells. Antioxid Redox Signal. 2011;14:2151–2162. doi: 10.1089/ars.2010.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meares GP, Hughes KJ, Naatz A, Papa FR, Urano F, Hansen PA, Benveniste EN, Corbett JA. IRE1-dependent activation of AMPK in response to nitric oxide. Mol Cell Biol. 2011;31:4286–4297. doi: 10.1128/MCB.05668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng J, Yang GY, Ying W, Kelly M, Hirai K, James TL, Swanson RA, Litt L. Pyruvate improves recovery after PARP-1-associated energy failure induced by oxidative stress in neonatal rat cerebrocortical slices. J Cereb Blood Flow Metab. 2007;27:304–315. doi: 10.1038/sj.jcbfm.9600335. [DOI] [PubMed] [Google Scholar]
- 60.Mercer JR, Cheng KK, Figg N, Gorenne I, Mahmoudi M, Griffin J, Vidal-Puig A, Logan A, Murphy MP, Bennett M. DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ Res. 2010;107:1021–1031. doi: 10.1161/CIRCRESAHA.110.218966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.