Abstract
We previously identified short synthetic shRNAs (sshRNAs) that target a conserved hepatitis C virus (HCV) sequence within the internal ribosome entry site (IRES) of HCV and potently inhibit HCV IRES-linked gene expression. To assess in vivo liver delivery and activity, the HCV-directed sshRNA SG220 was formulated into lipid nanoparticles (LNP) and injected i.v. into mice whose livers supported stable HCV IRES-luciferase expression from a liver-specific promoter. After a single injection, RNase protection assays for the sshRNA and 3H labeling of a lipid component of the nanoparticles showed efficient liver uptake of both components and long-lasting survival of a significant fraction of the sshRNA in the liver. In vivo imaging showed a dose-dependent inhibition of luciferase expression (>90% 1 day after injection of 2.5 mg/kg sshRNA) with t1/2 for recovery of about 3 weeks. These results demonstrate the ability of moderate levels of i.v.-injected, LNP-formulated sshRNAs to be taken up by liver hepatocytes at a level sufficient to substantially suppress gene expression. Suppression is rapid and durable, suggesting that sshRNAs may have promise as therapeutic agents for liver indications.
Keywords: HCV, lipid nanoparticles, PK, RNAi, shRNA, sshRNA
Introduction
Hepatitis C virus (HCV) is a worldwide health problem, and treatment and prevention of HCV infection remain a major challenge.1,2,3 The HCV virion contains a single, positive-strand RNA of 9.4 kb that acts as both the viral genome and the message encoding the viral proteins. The high error rate of its RNA-dependent RNA polymerase results in the generation of a huge number of sequence variants among the viral species present in an infected individual. Therefore, creating an effective vaccine has been very challenging, and any effective therapy is likely to require multiple antiviral agents in order to block all likely sequence variants. The ability of RNA interference (RNAi) to target multiple sequences, including noncoding regions, of the viral genome makes it an attractive therapeutic option for this purpose. RNAi-induced cleavage of the genomic RNA can abolish both replication and translation of the virus. Several groups have reported the ability of RNAi approaches to target HCV in cell-culture models, including cell-based reporter gene systems, HCV subgenomic replicon systems, and cell culture-based infectious HCV systems.4,5,6,7,8
We previously reported the identification of short synthetic shRNAs (sshRNAs) targeting the HCV internal ribosome entry site (IRES) that are highly potent inhibitors of HCV IRES-dependent gene expression in cell-based assays and have minimal immune stimulatory effects.9,10,11,12,13 Members of this novel class of inhibitors induce cleavage of target RNAs by a dicer-independent RNAi mechanism.11,14 A recent study indicated that sshRNAs are predominantly loaded as intact molecules into Argonaute (Ago)-containing complexes without prior processing by Dicer and are activated by Ago2-mediated cleavage of the passenger arm of the hairpin.14
The liver, the site of infection by HCV, is also the organ most accessible to delivery of therapeutic RNA when formulated into lipid nanoparticles (LNP) and administered systemically.15,16,17,18,19,20,21,22,23,24 Because of the limited availability of robust animal models of HCV infection,25,26 we have performed initial in vivo evaluation of LNP-formulated sshRNAs against HCV in a reporter mouse in which a plasmid introduced by hydrodynamic injection provides long-term expression in the liver of firefly luciferase (fLuc) under the control of the HCV IRES. We demonstrate the presence in the liver of significant levels of SG220 after a single intravenous injection of the formulated sshRNA, and its ability to potently and durably knock-down HCV-IRES-dependent gene expression.
Results
In vitro efficacy of sshRNA inhibitors targeting the HCV IRES
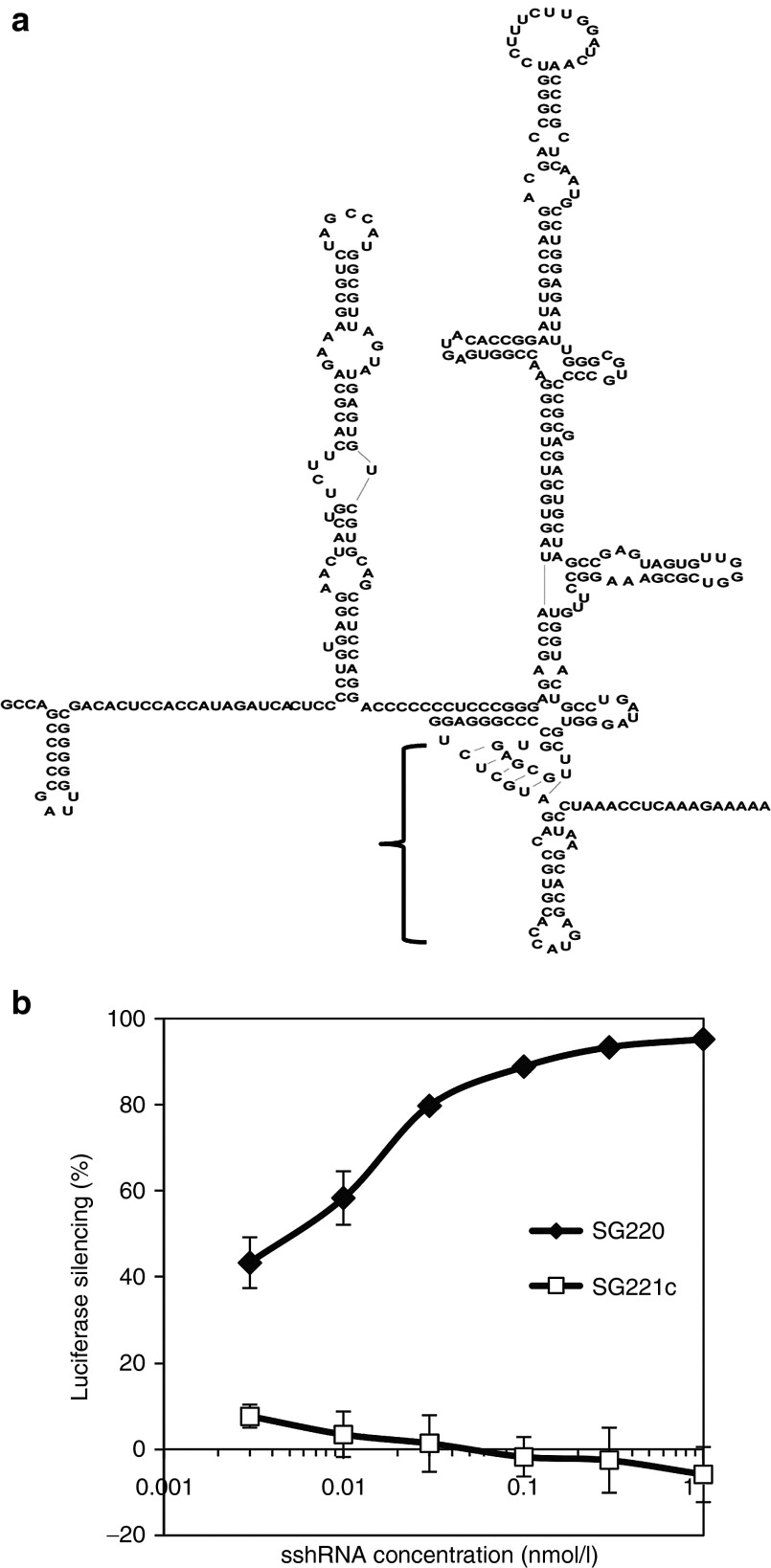
In previous studies, we found that introduction of 2′-O-methyl (2′-OMe) modifications at certain residues of 19-bp sshRNAs allows retention of RNA interfering potency while providing improved serum stability and diminished immune stimulatory potential.10 One such sshRNA, SG220, targets a conserved region within the IRES of the HCV genome (Figure 1a). In a transient transfection assay in 293FT cells, SG220 inhibited IRES-directed firefly luciferase (fLuc) reporter gene activity with a median 50% effective concentration (EC50)of ~5 pmol/l (Figure 1b).
Figure 1.
Gene silencing activity of HCV sshRNAs in vitro. (a) Secondary structure of the HCV IRES with the region targeted by sshRNA SG220 indicated by the bracket. (b) Potency of SG220 in suppressing luciferase gene expression driven by the HCV-IRES in 293FT cells. SG220 (filled diamonds); SG221C (scrambled control) (squares). The results shown are the mean values ± SEM from triplicate transfections.
Lack of immune stimulatory activity
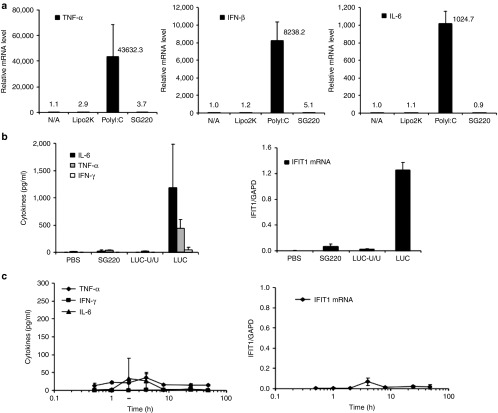
Certain RNA oligonucleotides, including a variety of RNAi constructs, are known to activate the mammalian innate immune response, and the incorporation of 2′-OMe nucleotides can avoid recognition by this antiviral defense mechanism.27 As anticipated based on their 2′-OMe modifications, SG220 did not show measurable induction of inflammatory cytokines in vitro when transfected into the human fetal lung fibroblast cell line MRC-5 (Figure 2a). These cells can be activated by cytosolic RNA through retinoic acid inducible gene-I (RIG-I)-like receptors. To determine whether the sshRNA constructs possessed significant immune stimulatory activity in vivo, SG220 was formulated with the LNP intended for use in subsequent in vivo efficacy studies and injected intravenously into immunocompetent mice. LNP-formulated SG220 caused minimal change in serum inflammatory cytokines and no induction of IFIT1 within the liver throughout the 48-hour period postinjection (Figure 2b,c). This response to SG220 was comparable with that of a canonical 2′-OMe-modified siRNA that was previously characterized as having minimal immune stimulatory potential.28
Figure 2.
Lack of immune stimulatory effect of SG220 in vitro and in vivo. (a) In vitro analysis. Twenty nmol/l sshRNAs SG220 or SG273 were transfected into human MRC-5 cells in triplicate. Untransfected (N/A) cells and cells transfected with Lipofectamine 2000 alone (Lipo2K) served as negative controls. The numbers shown are the mean and standard deviations of the mean of the indicated cytokine mRNAs relative to the untransfected control and normalized to GAPDH. (b, c) In vivo analysis. CD1 ICR mice were administered 2.5 mg/kg LNP-formulated sshRNA (SG220) or LNP-formulated control siRNAs (LUC-U/U, LUC) by intravenous injection. At the designated time point, blood was collected by cardiac puncture and processed as plasma for cytokine determination, and liver was excised and placed in RNAlater (Sigma–Aldrich) for IFIT1 mRNA analysis. The values shown are mean and standard deviation of measurements from each group of four mice. (b) Plasma cytokines (left) and liver IFIT mRNA (right), determined 4 hours after administration. (c) Time course of SG220-mediated plasma cytokine (left) and liver IFIT1 mRNA (right) induction. LNP, lipid nanoparticles.
Efficient and durable liver uptake of LNP-formulated HCV sshRNAs in mice
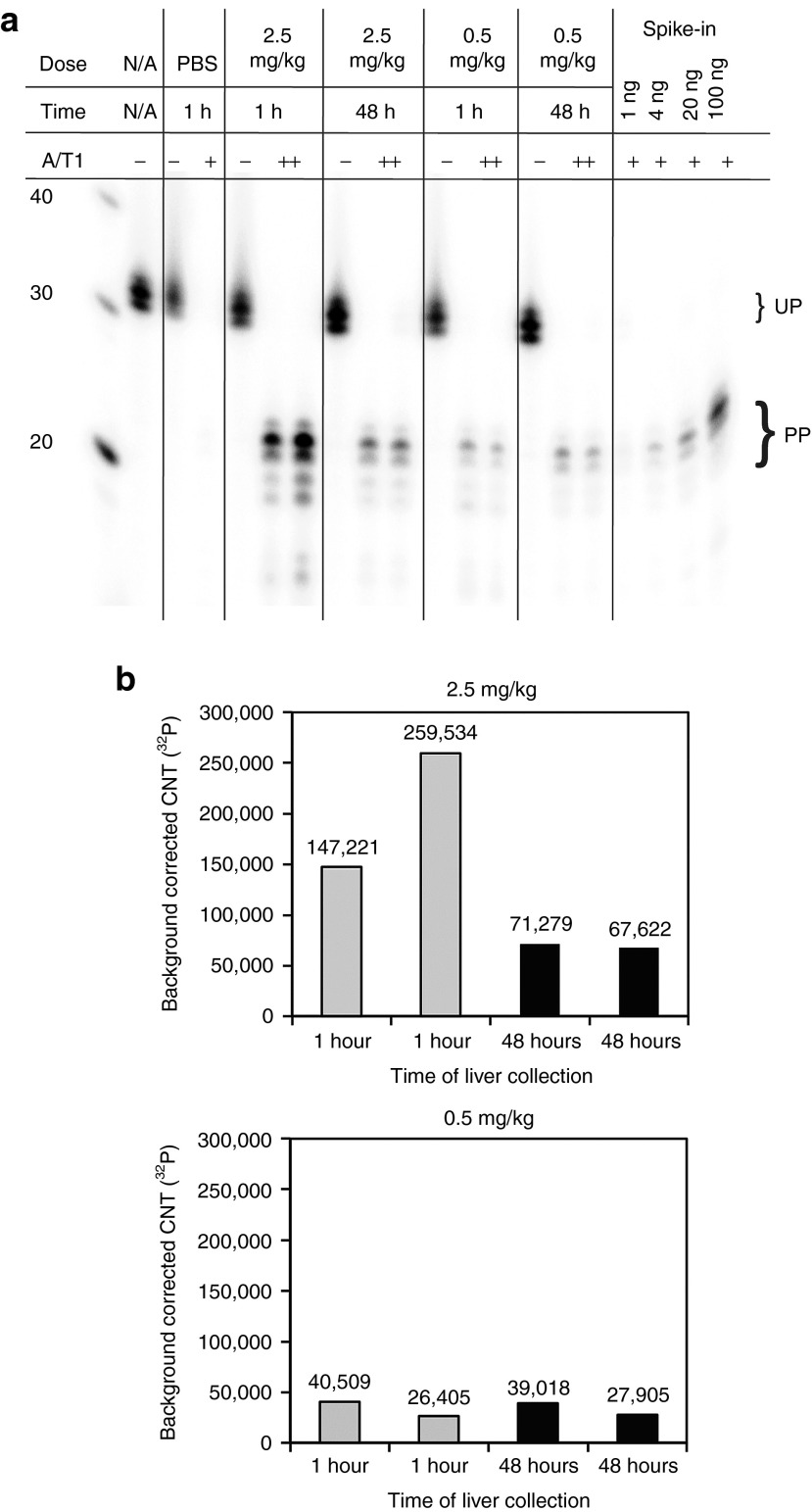
To evaluate the efficacy of sshRNAs in mice, we first examined the uptake of the LNP-formulated sshRNA SG220 in liver. Mice received single doses of formulated SG220 in 2 dosing groups (0.5 or 2.5 mg/kg) via low-pressure injection into the tail vein. Liver samples were collected either 1 or 48 hour after dosing (two mice for each time point) and levels of SG220 in 50 µg of total liver RNA were determined by a ribonuclease protection assay (RPA). The specificity of the RPA for SG220 was demonstrated by the lack of protection of the probe by total liver RNA from control mice treated with phosphate-buffered saline (PBS: 150 mmol/lNaCl, 10 mmol/l phosphate, pH 7.4) (Figure 3a, PBS). Comparing the band intensities of the protected probe at the different time points and doses (Figure 3b), we observed a 50–75% loss of protected signal at the 2.5 mg/kg dose between 1 and 48 hour after injection, whereas at 0.5 mg/kg the protected signals remained comparatively steady. This difference suggests that the higher dose may be saturating one or more sinks (e.g., RISC) for binding of SG220 and that the excess is degraded or otherwise lost over this time period. To follow the liver uptake of lipid as well as RNA components of LNP-formulated SG220 in more detail with a larger cohort size (four mice per time point), the experiment was repeated with quantification of SG220 in both blood and liver using the RPA (Figure 4a,b and Supplementary Figure S1, online) and measurement of tritiated cholesteryl oleyl ether (3H-CHE) in those same compartments (Figure 4c). In this experiment, a single dose of LNP-formulated SG220 (2.5 mg/kg) was delivered by ordinary intravenous injection. The results show that the lipid component CHE moves rapidly and nearly quantitatively from blood to liver (Figure 4c). The sshRNA initially follows similar pharmacokinetics, but after 2 hours the liver levels decline until about 10-hour post injection, after which they level out (Figure 4a, b). This pattern confirms what was seen in the first experiments and is again consistent with the dual process of liver uptake accompanied by (or followed by) degradation of sshRNA molecules that are not protected by protein binding; the final plateau may represent the fraction that is stably bound to RISC.24 It should be noted that the RPA probe used in this analysis cannot distinguish between the full-length and Ago2-processed forms of SG220, as the sequences protected by the probe are common to both forms and the processed form of SG220 is detected with ~tenfold greater sensitivity than full-length because of a lesser degree of competing secondary structure (Supplementary Figure S2, online). In addition, the radioactivity assay does not distinguish between intact and degraded CHE. By comparing the band intensities of the protected probe to those of SG220 standards (Figure 3a, Spike-in), the concentrations of SG220 in the 50 µg total liver RNA were calculated. From this number we determined that 5.7 ± 2.0% (mean ± SD) of the total injected SG220 remained in the mouse liver 48 hours after treatment. The presence of abundant sshRNA in the liver 48 hours after treatment suggests that the hepatocytes in the mouse liver can efficiently take up large macromolecular complexes such as LNP-formulated double-stranded nucleic acids and that the chemically modified sshRNA in these nanoparticles is sufficiently stable in the bloodstream to reach hepatocytes in intact form.
Figure 3.
Liver uptake of LNP-SG220 in CD1 mice. (a) Denaturing 10% PAGE analysis of RNase protection assay to monitor SG220 present in livers of CD1 mice at the indicated doses and time points. A mouse injected with PBS only was used as a control (PBS). Bands corresponding to undigested probe and probe protected after RNase A/T1 digestion are labeled. For quantification, the amounts of SG220 indicated in the last four lanes were spiked into 50 µg of total liver RNA from untreated mice to generate a calibration curve for the quantification of the liver uptake of SG220. PP, protected probe; UP, undigested full-length probe. (b) Plots of background corrected counts (32P) for individual mice for dosing groups 0.5 and 2.5 mg/kg. LNP, lipid nanoparticles.
Figure 4.
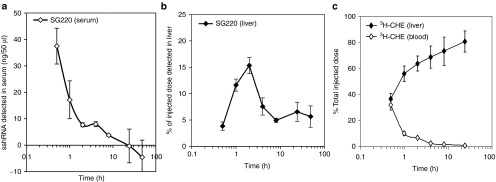
Kinetics of uptake of sshRNA and lipid components of LNP-formulated SG220 into mouse liver. CD1 ICR mice were administered one intravenous injection of LNP-formulated SG220 at 2.5 mg/kg (n = 4 for each time point). Levels of sshRNA and CHE in the serum and liver at various times after dosing were quantified as described in “Materials and Methods.” Liver uptake is presented as a percentage of the initial dose. The error bars show mean ± SEM from four mice. LNP, lipid nanoparticles.
In vivo activity in a reporter mouse model
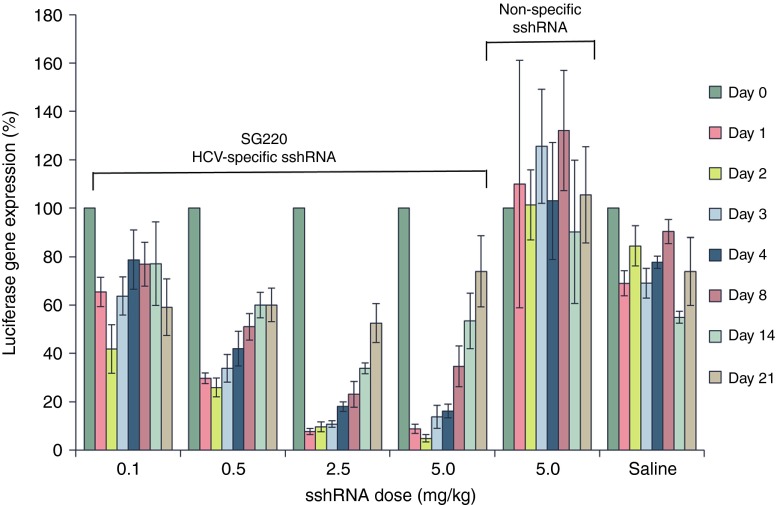
The ability of the sshRNAs to inhibit HCV IRES activity in vivo was assessed using an HCV IRES-dependent bioluminescence mouse model. Balb/C mice were subjected to hydrodynamic intravenous injection of a plasmid (pSG231) containing the targeted region of the HCV IRES linked to firefly luciferase (fLuc) expressed through the liver-specific mouse transthyretin (mTTR) promoter. Bioluminescence measurements showed stable, durable (>30 days) expression of HCV IRES-luciferase in the livers of these mice. Seven days after injection of pSG231, the mice received a single (low pressure) intravenous injection of SG220 formulated in LNP at 0.1, 0.5, 2.5, or 5.0 mg/kg, or an LNP-formulated irrelevant sshRNA at 5.0 mg/kg, or a buffer control (PBS). HCV IRES-directed luciferase expression in the livers of the mice was monitored by in vivo bioluminescence with imaging performed at various time points after sshRNA injection, and the signals were compared with those prior to sshRNA administration. A single administration of SG220 resulted in a dose-dependent reduction of luciferase activity in the livers of the mouse, reaching more than 90% knock-down with 2.5 or 5.0 mg/kg SG220 (Figure 5). This knock-down was prolonged, with over 50% luciferase reduction maintained at least 14 days after treatment with 2.5 or 5.0 mg/kg LNP-formulated sshRNA. The knock-down was also sshRNA-specific, as neither the irrelevant sshRNA nor the PBS control led to significant luciferase reduction at any time point analyzed.
Figure 5.
Inhibition of HCV IRES-dependent luciferase gene expression in mouse liver by HCV sshRNA SG220. Seven days after the mice were hydrodynamically injected with the HCV-IRES-fLucplasmid pSG231, either SG220 or an irrelevant (sequence-scrambled) sshRNA (irr sshRNA) formulated with lipid nanoparticles at the indicated doses, or PBS, was injected intravenously at low pressure. At the indicated time points post-sshRNA injection, luciferin was injected intraperitoneally and luciferase gene expression in the mouse liver was detected by in vivo bioluminescence imaging. The luciferase gene expression at the indicated time points is presented as a percentage of that prior to the sshRNA injection.
Discussion
In this study, we have investigated the in vivo efficacy of LNP-formulated sshRNAs targeting the HCV IRES. Our results showed that, consistent with their potent in vitro inhibition of HCV IRES activity, LNP-formulated HCV-targeting sshRNAs are efficiently taken up from the bloodstream of mice into the liver and induce potent and rapid inhibition of HCV IRES-dependent reporter gene expression in that organ. These studies are, to the best of our knowledge, the first report of potent and durable inhibition of an HCV target by synthetic RNAs acting through an RNAi mechanism. Importantly, the RNAi effect of LNP-formulated sshRNA treatment was very long-lasting, suggesting that such compounds could be effective even with infrequent dosing. This indicates a long effective half-life of the sshRNAs in hepatocytes and suggests that the chemical modifications (2′-O-methyl) within the sshRNAs have successfully protected them against RNases, as we reported previously,10 or that the sshRNAs were sequestered in subcellular compartments that protect them from nuclease digestion. We observed a good correlation between in vitro and in vivo efficacy for SG220, suggesting that initial screening for lead candidate sequences may be performed by in vitro assay.
Therapeutic RNAs can be sensed by the cellular innate immune system, leading to activation of cellular antiviral responses including the type I interferon pathway. Such a process could lead to indirect inhibition of HCV replication.29 The fact that inflammatory cytokines, interferon, and Ifit1 mRNA are not induced by the SG220 either in vitro or in vivo, together with the lack of activity of the scrambled control RNA, indicate that the LNP-formulated sshRNAs characterized here achieve in vivo efficacy through a sequence-specific mechanism30,31,32 rather than a generalized innate immune response.
Several lines of evidence demonstrated convincingly that our sshRNAs act on the HCV RNA through an RNAi mechanism. First, HCV inhibition is sequence-specific, with little or no immune stimulatory component. Second, the RNA target is cleaved at the site expected for an RNAi mechanism in cells treated with these sshRNAs.11,14 Finally, the sshRNAs can be immunoprecipitated (in both full-length and “sliced” form) by Ago2.14 The potent and prolonged knock-down observed in this mouse reporter model suggests that LNP-formulated sshRNAs may be effective in a true HCV infection (H. Ma, A. Dallas, H. Ilves, K. Klumpp, I. MacLachlan, and B.H. Johnston, Gastroenterology, in press). With recent improvements in lipid nanoparticle formulation potency improving delivery to liver,33 efficacy of sshRNAs at doses well below those seen here seems probable.
Materials and methods
Preparation of sshRNAs. sshRNAs were chemically synthesized and HPLC-purified by Integrated DNA Technologies (IDT, Coralville, IA, USA). The sequences of sshRNAs used in this study are as follows: SG220 5′- UGAGGUUUAGGAUUCGUGCUUGCACGAAUCCUAAACCUCA -3′ and SG221c 5′- CGUGCUUAGGAUUUGGAGUUUACUCCAAAUCCUAAGCACG -3′. The positions of 2′OMe RNA bases are underlined. sshRNAs were dissolved either in RNase- and pyrogen-free buffer containing 20 mmol/l KCl, 6 mmol/l HEPES-KOH (pH 7.5), and 0.2 mmol/l MgCl2 (Thermo Fisher Scientific, Dharmacon Products, Lafayette, CO, USA) for in vitro cell-culture assays, or in sterile RNAse-free H2O for subsequent formulation with LNP and in vivo experiments. To convert sshRNAs from a heterogeneous mixture of monomers, dimers, and higher-order multimers to a homogeneous monomeric hairpin conformation, sshRNAs were heated to 95 °C for 4 minutes and then transferred immediately to an ice-water bath, where they remained for 10–20 minutes before further use. RNAs were confirmed to be in monomeric hairpin form by nondenaturing PAGE and staining with SYBR Gold (Invitrogen, Carlsbad, CA).
Transfection and in vitro assays. The human kidney cell line 293FT (Invitrogen) was maintained in DMEM (Cambrex, Walkersville, IN) with 10% fetal bovine serum (Hyclone, Logan, UT), supplemented with 2 mmol/l l-glutamine and 1 mmol/l sodium pyruvate. One day prior to transfection, cells were seeded at 23,000 cells/well in a 96-well plate, resulting in ~80% cell confluency at the time of transfection. Transfections were performed using Lipofectamine™ 2000 (Invitrogen) following the manufacturer's instructions. Thirteen nanograms of pIRES/fLuc (an IRES-linked firefly luciferase reporter construct), 20 ng of pSEAP2-control plasmid (BD Biosciences Clontech, San Jose, CA) as a transfection control, and the indicated amounts of sshRNAs were cotransfected into 293FT cells. Forty-eight hours later, the cells were lysed and luciferase activity was measured in a MicroLumat LB 96P luminometer (Berthold Technologies, Bad Wildbad, Germany). All sshRNA samples were tested in triplicate. Percent silencing was calculated relative to transfections with the reporter plasmid in the absence of sshRNA inhibitors. Interferon and cytokine assays were performed as described previously.11
Formulation of sshRNA into LNP. Monomeric sshRNAs and control siRNAs were formulated into LNP by the process of step-wise ethanol dilution and spontaneous vesicle formation as previously described.34,35 LNP were comprised of synthetic cholesterol (Sigma), dipalmitoylphosphatidylcholine (Avanti Polar Lipids, Alabaster, AL), the PEG-lipid PEG-C-DMA (3-N-[(ω-methoxy poly(ethylene glycol)2000)carbamoyl]- 1,2-dimyrestyloxy-propylamine), and the cationic lipid DLinDMA (1,2-dilinoleyloxy-3-N,N-dimethylaminopropane) in the molar ratio of 34.3:7.1:1.4:57.1. LNP were dialyzed against PBS and filter sterilized through a 0.2 μm filter before use. Mean particle sizes were 85–90 nm with polydispersity values <0.1 by dynamic light scattering, and 92–98% of the siRNA was encapsulated within the lipid particles. The final lipid-to-nucleic acid ratio in formulations used in this study was approximately 6.5:1 (wt:wt).
Reporter plasmids. For in vitro cell-culture experiments, a dual luciferase expression plasmid (IRES/fLuc) was used in which the sequence encoding the HCV IRES of genotype 1b is placed between the coding sequences for Renilla and firefly luciferase (fLuc) such that fLuc expression is dependent on the IRES. For in vivo experiments, a luciferase reporter under control of the liver-specific promoter mTTR was constructed. The Ig kappa MAR sequence36,37 from pCG419 (generously provided by H. Ilves, Cell Genesys, San Francisco, CA) was inserted upstream of HCV IRES-Fluc reporter resulting in a plasmid capable of producing durable, liver-specific expression of luciferase (pSG231).
Proinflammatory cytokine detection in vitro. Cells of the human fetal lung fibroblast line MRC-5 were seeded in 24-well plates at 6 × 104 cells per well with MEM containing 10% fetal calf serum. Transfections were performed using Lipofectamine 2000 following the manufacturer's instructions. Twenty nmol/l sshRNAs or an equivalent amount of positive control (polyI:C) was transfected in triplicate. Untransfected cells and cells receiving Lipofectamine 2000 alone were used as negative controls. Twenty-four hours later, the cells were lysed in Trizol (Invitrogen) and total RNA was extracted according to the manufacturer's instructions. Quantitative RT-PCR was performed using high-capacity cDNA Reverse Transcription Kits with the TaqMan Universal PCR Master Mix, IFN-β (Hs01077958_s1), TNFα (Hs99999043_m1), and GAPDH (Hs99999905_m1) TaqMan probes and a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA) following the manufacturer's protocol.
In vivo immune stimulation assays. Mouse immune response studies were performed at Tekmira Pharmaceuticals in accordance with Canadian Council on Animal Care guidelines and following protocol approval by the Institutional Animal Care and Use Committee of Tekmira. Six to eight week-old CD1 ICR mice were obtained from Harlan and subjected to a 2-week acclimation period prior to use. Mice were administered LNP-formulated sshRNA or siRNAs (2.5 mg/kg RNA) in PBS via standard i.v. injection in the lateral tail vein. In this study, LNP-formulated native LUC siRNA was used as a positive control for immune stimulation.28 A previously described 2′-OMe modified form of the same siRNA sequence targeting LUC (LUC-U/U) was used as an additional negative control.28 Sequences of siRNAs are as follows as reported in ref. 28: LUC (sense) GAUUAUGUCCGGUUAUGUAUU + LUC (antisense) UACAUAACCGGACAUAAUCUU; LUC-U/U(sense) GAUUAUGUCCGGUUAUGUAUU + LUC-U/U(antisense) UACAUAACCGGACAUAAUCUU. 2′-OMe-modified residues are underlined. Cohorts of n = 4 mice were sacrificed at t = 0.5, 1, 2, 4, 8, 24, and 48 hours postinjection. Blood was collected by cardiac puncture and processed as plasma for cytokine analysis. Liver was collected into RNAlater (Sigma–Aldrich) for IFIT1 mRNA analysis. Plasma cytokines TNF-α, IL-6, IFN-γ, IL-12 p70, IL-10, and MCP-1 were assayed by Cytometric Bead Array (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. The six-cytokine bead populations were resolved in the red channel with a 488 nm laser capable of detecting and distinguishing fluorescence emissions at 576 and 670 nm using a FACSCanto II flow cytometer equipped with FACSDiva data capture software version 6.0. Data analysis was performed using Tree Star FlowJo software version 7.6.1. Cytokine concentrations in each plasma sample were calculated from a standard curve of the mouse recombinant cytokine provided with the assay kit. IFIT1 mRNA was quantified in mouse liver homogenates by branched DNA (bDNA) assay as previously described.28 The IFIT1 probe set was specific to mouse IFIT1 mRNA (positions 4–499, GenBank accession number NM_008331), and the GAPDH probe set was specific to mouse GAPDH mRNA (positions 9–319, GenBank accession number NM_008084). Data are shown as the ratio of IFIT1 RLU to GAPDH RLU.
Determination of liver uptake of LNP-sshRNA in mice. Liver uptake experiments were performed at Stanford University, and mice were treated according to NIH and Stanford guidelines for animal care. LNP-SG220 was delivered via hydrodynamic injection into the tail vein of CD1 mice in two dosing groups (0.5 and 2.5 mg/kg). Cohorts of n = 2 mice were sacrificed at 1 and 48 hours and livers were collected and sliced, and the slices were immersed in RNAlater solution (Ambion, cat#AM7024) overnight at 4 °C. The RNAlater solution was then removed and liver slices were stored at −80 °C until analysis.
RNA isolation from mouse liver. Total RNA was isolated from mouse livers using a FastPrep instrument (FastPrep-24, FP24, MP Biomedicals, Solon, OH) to disrupt tissue. Approximately 100 mg of preserved liver tissue from each mouse was added to 1.2 g of lysing Matrix “D” beads (MP Biomedicals) and 1 mL of QIAZOL (Qiagen). Samples were processed in the FP24 homogenizer twice for 60 seconds at a setting of 6 ms−1, with the samples kept on ice for 5 minutes in between homogenization steps. Cellular debris was removed by centrifugation at 12,000×g at ambient temperature. The liver homogenate was extracted with chloroform, and the aqueous layer was precipitated with isopropanol. The resulting RNA pellet was washed with 70% ethanol, resuspended in 100 μl of RNAse-free H2O, and quantified by A260 determination.
RNase protection assay. The following DNA oligonucleotides (from IDT, Coralville, IA) were annealed to form a duplex T7 RNA polymerase transcription template encoding a probe specific for SG220 for the RNAse protection assay: 5′-CTAATACGACTCACTATAGGCAAGCACGAATCCTAAACCTCATTTTTT-3′ and 5′-AAAAAATGAGGTTTAGGATTCGTGCTTGCCTATAGTGAGTCGTATTAG-3′. The 32P-labeled probe specific for the detection of SG220 was prepared by in vitro transcription using T7 RNA polymerase (Promega) in the presence of [α32P]CTP (Perkin-Elmer, Boston, MA). Following transcription, the probe was purified by a G-50 spin column (Amersham/GE Healthcare, Piscataway, NJ, USA) and 10% denaturing PAGE. The probe (sequence 5′-GGCAAGCACGAAUCCUAAACCUCAUUUUUU-3′) is complementary to the guide sequence, loop, and two nucleotides of the passenger sequence. For each sample, ~100,000 cpm of 32P-labeled probe was coprecipitated with 50 μg of total RNA isolated from each mouse liver. Hybridization was performed at 42 °C for 12 hours, followed by cooling to 38 °C for 2 hours and finally to 32 °C for an additional 2 hours (Robbins Scientific Model 400 hybridization oven). RNAse protection assays were performed using the RPA III kit (Ambion) according to the manufacturer's instructions with nondigested controls. Samples were analyzed by 10% denaturing PAGE (8 mol/l urea). Protected bands were visualized and quantified by a Molecular Imager FX (Bio-Rad, Hercules, CA, USA). In parallel, a standard curve was generated by analyzing spiked-in synthetic SG220 to final amounts of 1, 4, 20, 100 ng/50 μg total RNA. Quantification of SG220 sshRNA was performed by comparing the intensities of the protected bands to those of the spiked-in standards to estimate the percent of SG220 present in the mouse livers 48 hours after injection relative to the total amount dosed.
In vivo rodent PK experiment. Plasma clearance and tissue distribution studies were performed essentially as described in ref. 23. Radiolabeled LNP was prepared by incorporation of the nonexchangeable lipid label 3H-CHE at 2.7 μCi/mg total lipid.38 LNP-formulated sshRNA was administered at a dose of 2.5 mg RNA/kg via lateral tail vein injection in 8-week old female BALB/c mice (Harlan Labs, IN) and blood was collected via tail vein nick over a 24-hour period. At 24 hours after injection, mice were euthanized and harvested tissues were homogenized in FastPrepLysing Matrix Tubes (MP Biomedicals) containing distilled water. Tissue homogenates were assayed for radioactivity by liquid scintillation counting with Picofluor 40 and whole blood was assayed using Picofluor 15 (Perkin-Elmer).
Inhibition of HCV IRES-mediated luciferase gene expression in mice. Stable, IRES-dependent expression of firefly luciferase from a liver-specific promoter (mTTR) in balb/c mice was established by hydrodynamic injection of plasmid pSG231. Seven days later, on day 0, a single low-pressure tail vein injection of LNP-formulated SG220 was administered at the following doses: 0.1, 0.5, 2.5, and 5.0 mg/kg (day 0, 5 mice/dose group). A nonspecific control group received LNP-formulated SG221C (irrelevant control) at 5.0 mg/kg. Two additional groups, injected with saline or untreated, were also included in the study. The expression of luciferase was monitored by in vivo imaging with a Xenogen/Caliper IVIS-50 camera at days 0, 1, 2, 3, 4, 8, 14, and 21. For day 0, imaging was done immediately prior to administration of sshRNA. For each measurement, mice were anesthetized with 1–2% isofluorane, 300 µl d-luciferin (15 mg/ml in PBS) was injected i.p., and 10 minutes later images were collected with an integration time of 1 minute.
Conflict of Interest
B.H.J., A.D., and H.I. are current employees of SomaGenics, Inc., and J.S. is a former employee. I.M. is a current employee of Tekmira Pharmaceuticals, Inc., and A.J. is a former employee of Tekmira.
SUPPLEMENTARY MATERIAL Figure S1. Time course of liver uptake of LNP-formulated sshRNA in mice analyzed by RNase protection assay (RPA) on 10% denaturing PAGE. Figure S2. Schematic of RPA probe hybridization.
Acknowledgments
The authors thank Sergei A. Kazakov (SomaGenics) for advice in development of the sshRNA detection assay. Supported by National Institutes of Health (NIH) [R44AI056611, R44AI074256 and R43AI074214 to B.H.J.]
Supplementary Material
Time course of liver uptake of LNP-formulated sshRNA in mice analyzed by RNase protection assay (RPA) on 10% denaturing PAGE.
Schematic of RPA probe hybridization.
References
- GraaGHCGWHO http://www.who.int/csr/disease/hepatitis/whocdcsrlyo2003/3n/index4.htnl .
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–714. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo MY, Abrignani S, Houghton M, Han JH. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J Virol. 2003;77:810–812. doi: 10.1128/JVI.77.1.810-812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Steele R, Ray R, Ray RB. Small interfering RNA targeted to hepatitis C virus 5' nontranslated region exerts potent antiviral effect. J Virol. 2007;81:669–676. doi: 10.1128/JVI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov AV, Korba B, Farrar K, Mukerjee S, Seyhan AA, Ilves H, et al. shRNAs targeting hepatitis C: effects of sequence and structural features, and comparision with siRNA. Oligonucleotides. 2007;17:223–236. doi: 10.1089/oli.2006.0069. [DOI] [PubMed] [Google Scholar]
- Ge Q, Dallas A, Ilves H, Shorenstein J, Behlke MA, Johnston BH. Effects of chemical modification on the potency, serum stability, and immunostimulatory properties of short shRNAs. RNA. 2010;16:118–130. doi: 10.1261/rna.1901810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Ilves H, Dallas A, Kumar P, Shorenstein J, Kazakov SA, et al. Minimal-length short hairpin RNAs: the relationship of structure and RNAi activity. RNA. 2010;16:106–117. doi: 10.1261/rna.1894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Ilves H, Kaspar RL, Wang Q, Seyhan AA, Vlassov AV, Contag CH, et al. Inhibition of hepatitis C IRES-mediated gene expression by small hairpin RNAs in human hepatocytes and mice. Ann N Y Acad Sci. 2006;1082:52–55. doi: 10.1196/annals.1348.060. [DOI] [PubMed] [Google Scholar]
- Dallas A, Ilves H, Ge Q, Kumar P, Shorenstein J, Kazakov SA, et al. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012;40:9255–9271. doi: 10.1093/nar/gks662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner A. Cellular delivery in vivo of siRNA-based therapeutics. Curr Pharm Des. 2008;14:3603–3619. doi: 10.2174/138161208786898815. [DOI] [PubMed] [Google Scholar]
- de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther. 2008;19:125–132. doi: 10.1089/hum.2008.928. [DOI] [PubMed] [Google Scholar]
- Li L, Shen Y. Overcoming obstacles to develop effective and safe siRNA therapeutics. Expert Opin Biol Ther. 2009;9:609–619. doi: 10.1517/14712590902911420. [DOI] [PubMed] [Google Scholar]
- Li SD, Huang L. Targeted delivery of siRNA by nonviral vectors: lessons learned from recent advances. Curr Opin Investig Drugs. 2008;9:1317–1323. [PubMed] [Google Scholar]
- Novobrantseva TI, Akinc A, Borodovsky A, de Fougerolles A. Delivering silence: advancements in developing siRNA therapeutics. Curr Opin Drug Discov Devel. 2008;11:217–224. [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, McMillan NA. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11:639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Hoerter JA, Krishnan V, Lionberger TA, Walter NG. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PLoS ONE. 2011;6:e20359. doi: 10.1371/journal.pone.0020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Kim M, Shin D, Kim SI, Park M. Inhibition of hepatitis C virus gene expression by small interfering RNAs using a tri-cistronic full-length viral replicon and a transient mouse model. Virus Res. 2006;122:1–10. doi: 10.1016/j.virusres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT, et al. Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L, et al. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- Bouchie A. Companies in footrace to deliver RNAi. Nat Biotechnol. 2012;30:1154–1157. doi: 10.1038/nbt1212-1154. [DOI] [PubMed] [Google Scholar]
- Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S, MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- Yi M, Wu P, Trevorrow KW, Claflin L, Garrard WT. Evidence that the Igkappa gene MAR regulates the probability of premature V-J joining and somatic hypermutation. J Immunol. 1999;162:6029–6039. [PubMed] [Google Scholar]
- Xu M, Hammer RE, Blasquez VC, Jones SL, Garrard WT. Immunoglobulin kappa gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J Biol Chem. 1989;264:21190–21195. [PubMed] [Google Scholar]
- Stein Y, Halperin G, Stein O. Biological stability of [3H]cholesteryl oleyl ether in cultured fibroblasts and intact rat. FEBS Lett. 1980;111:104–106. doi: 10.1016/0014-5793(80)80771-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time course of liver uptake of LNP-formulated sshRNA in mice analyzed by RNase protection assay (RPA) on 10% denaturing PAGE.
Schematic of RPA probe hybridization.