Summary
Although intestinal bacteria live deep within the body, they are topographically on the exterior surface and thus outside the host. According to the classic notion that the immune system targets non-self rather than self, these intestinal bacteria should be considered foreign and therefore attacked and eliminated. While this appears to be true for some commensal bacterial species, recent data suggests that the immune system actively becomes tolerant to many bacterial organisms. The induction or activation of regulatory T (Treg) cells that inhibit, rather than promote, inflammatory responses to commensal bacteria appears to be a central component of mucosal tolerance. Loss of this mechanism can lead to inappropriate immune reactivity toward commensal organisms, perhaps contributing to mucosal inflammation characteristic of disorders such as inflammatory bowel disease.
Keywords: T cell, inflammatory bowel disease, T-cell receptors, mucosa
Introduction
The trillions of commensal bacteria that populate the gastrointestinal tract are of immense benefit to mammalian hosts. Commensal organisms are necessary for the proper digestion of dietary fibers, allowing for maximum nutrient extraction from food and the maintenance of host health (1–3). Thus, it is clear that we have co-evolved with commensal bacteria, incorporating them within our body to serve important biological functions.
Even though these bacteria are present deep within us, these microbes exist topographically on the exterior surface of the body. In the lumen of the gut, the commensal bacteria are separated from the interior of the body by a mucous layer fortified by anti-bacterial peptides that overlay the epithelial layer (4). These mechanical barriers limit the penetration of bacteria into the body that could trigger an immune response (5). Moreover, these barriers prevent bacterial antigens from accessing the specialized tissues that instruct self/non-self-recognition to adaptive immune cells during their development (6). Thus, bacteria that breach the mucosal barrier would be considered non-self and targeted for elimination by the immune system.
Recent reports demonstrate, however, that many commensal bacteria may not trigger an immune attack but rather elicit antigen specific tolerance. In particular, it appears that commensal bacteria induce regulatory subsets of CD4+ T cells, which may act to inhibit effector responses to those bacterial species. Here, we review the data supporting the notion that the immune system is educated to become tolerant of its own commensal bacteria and the molecular and cellular mechanisms by which bacteria influence this process. We also discuss the implications of these data, which suggest that the immune system, in addition to discrimination of self versus non-self, requires discrimination of commensal versus pathogenic ‘non-self’ bacteria to prevent inflammatory bowel disease while remaining responsive to enteric infections.
Microbial influences on T cells
To the best of our knowledge, it has been universally observed in murine models that pathologic inflammation in the gut is dependent on the presence of commensal bacteria. For example, mice with IL-10 or IL-2 deficiency develop colitis under conventional housing conditions where commensal bacteria are present but not under germ-free conditions (7–9). Similarly, adoptive transfer of naive T cells induces colitis in conventionally housed lymphopenic mice but not those kept germ-free (10, 11). Moreover, recent studies have shown that the presence of commensal microbiota or certain bacterial species contribute to autoimmune responses such as experimental allergic encephalitis (12) and a murine model of rheumatoid arthritis (13). These data therefore suggest that commensal bacteria are important initiators of effector T-cell responses that promote inflammation. In addition this supports the notion that the immune system sees commensal antigens as non-self, both because the bacterial antigens are not likely present during thymic T-cell selection and because bacteria display a variety of ligands for immune pattern recognition receptors (14).
It has become clear that commensal bacteria also elicit T cells that downregulate inflammation to maintain intestinal tolerance. An early indication came from studies by Powrie et al. (15), which demonstrated that transfer of the naive CD45RBhi subset of CD4+ T cells, but not the entire CD4+ population, induced colitis in lymphopenic hosts. This seminal observation was one line of evidence suggesting the existence of an inhibitory CD4+ T cell subset, now known to be Foxp3+ regulatory T (Treg) cells, which are required to maintain intestinal homeostasis and prevent colitis (16–18). This is further supported by the observation that mice deficient in Treg cell generation and function due to mutations in the IL-2 pathway also develop spontaneous colitis (19). Moreover, humans with genetic deficiencies of Foxp3, a transcription factor required for Treg cell development and function, suffer from IPEX (immune dysregulation polyendocrinopathy, enteropathy, X-linked) syndrome which includes intestinal issues and diarrhea amongst its manifestations (20). Thus, a large body of data supports an essential role for Treg cells in maintaining immune homeostasis in the gut and preventing effector cells from causing immunopathology in response to commensal bacteria.
Induction of Treg cells by commensal bacteria
Although it was clear that Treg cells were important for gut tolerance, it remained to be shown whether commensal bacteria directly influenced the generation or function of intestinal Treg cells. Seminal work by Sakaguchi et al. (21) showed that the thymus was an important site of Treg cell development required to prevent autoimmunity. Thymic Treg cell development begins very early during ontogeny, within a few days after birth in mice (22), and appears to be driven by T cell self-reactivity (23, 24). It was therefore possible that Treg cells generated in the thymus to self-antigens may also prevent gut inflammation as well as autoimmunity without previous exposure to commensal bacteria. This was supported by the observation that Treg cells could be easily found in the intestines of germ-free mice (25–27), demonstrating that commensal bacteria are not required for Treg cells to be present in the gut. In addition, Treg cells from germ-free mice are protective in the Powrie transfer model of colitis, although they are not as efficient as those from conventionally housed mice (27, 28). Taken together, these early reports suggested that commensal bacteria were not essential for Treg cell generation or function at mucosal sites.
Recent data have demonstrated that commensal bacteria have a major impact on colonic Treg cell generation and function, even if the bacteria are per se not strictly essential. While a number of groups found that commensal bacteria did not affect the percentage of colonic Treg cells (25, 29–31), other groups observed that the presence of commensal bacteria increased the frequency of colonic Treg cells (32–35). These disparate results were hypothesized to result from differences in the microbiota of the conventionally housed mice, implying that at least some microbial species affect Treg cell numbers in the colon.
The observation that commensal bacteria in conventionally housed specific pathogen-free (SPF) mice could increase the frequency of colonic Treg cells prompted a detailed analysis of Treg cells in germ-free mice with defined bacterial species. Altered Schaedler flora (ASF), which is comprised of only 8 commensal species, was sufficient to significantly increase the frequency of Treg cells, although interestingly, the magnitude of the increase was dependent on the genetic background of the mouse (32). An extensive study of a variety of commensals, including Lactobacillus, Bacteroides, and Clostridium species, demonstrated that clusters IV and XIVa Clostridium were primarily responsible for the increased frequency of colonic Treg cells in response to murine (33) and human (36) commensal microbiota.
The impact of commensals on Treg cells was further supported by the identification of a microbial product from a specific bacterial species that affects Treg cell function. Polysaccharide A (PSA) from Bacteroides fragilis was found to activate TLR2 expressed on Treg cells, inducing the production of IL-10 (31). This enhancement in Treg cell function facilitated the persistence of B. fragilis, suggesting that there is a mutualistic relationship between the host and this species.
Several groups have reported a ‘universal’ mechanism driving Treg cell expansion that is mediated by bacterially derived short-chain fatty acids (SCFAs) produced through the metabolism of dietary fiber (34, 37, 38). These reports show that orally administered SCFAs increase Treg cells numbers and enhance Treg cell function via the induction of IL-10. SCFAs may also induce Treg cell expression of GPR15 (34), a G coupled protein receptor important for Treg cell homing to the colon (39). Thus, these data demonstrate that some microbial products are sensed by the intestinal immune system to facilitate homeostasis and tolerance instead of inflammation, consistent with the notion of an evolutionary mutualistic relationship between commensal bacteria and the host (31, 32).
Many colonic Treg cells respond to commensal bacteria in an antigen-specific manner
As discussed above, Treg cells appear to recognize the presence of certain bacteria through non-antigen specific germ-line encoded receptors to bacteria-derived molecules. However, it was unclear for some time whether Treg cells could also recognize bacteria via their antigen-specific T-cell receptor. The fact that Treg cells are easily found in the colons of germ-free mice (25–27) might suggest that they develop and function due to recognition of self-antigen in the thymus and colon (24). Although Treg cells from germ-free mice were less effective at preventing colitis in the adoptive transfer model (27, 28), it remained possible that this resulted from a lack of prior exposure to non-TCR dependent bacterial signals such as SCFAs or PSA.
Data from the past several years have provided direct support for the notion that Treg cells can recognize antigens from commensal bacteria through their TCR. Using a fixed TCRβ chain model to permit analysis of TCR repertoires at the level of individual T cells, our group (40) observed that the TCR usage by peripheral Treg cells varied by anatomic location. This is consistent with prior reports suggesting that lymph nodes preferentially contain Treg cells that recognize antigens draining from local tissues (41–44). When these studies were extended to the gut, we also observed that colonic Treg cells utilize a unique set of TCRs, suggesting that they recognize antigens found only in this tissue, including both colon-specific self-antigens and antigens derived from commensal bacteria (29).
To test the hypothesis that colonic Treg cells recognize bacteria antigens, we asked whether common colonic Treg TCRs could be stimulated by fecal material in vitro (29). We were surprised to observe that approximately ½ of the TCRs tested could recognize antigens in the fecal material from conventional mice, but not in that from germ-free mice or in food. Importantly, fecal material from mice purchased from Jackson Labs was unable to stimulate these TCRs in vitro and in vivo unless these mice were first co-housed with mice from our colony, suggesting a transmissible agent. Two of the identified colonic Treg cell TCRs were stimulated by cultured bacterial isolates derived from our colony, supporting the notion that these TCRs directly recognize bacterial antigens. The observation that many colonic Treg TCRs could recognize fecal antigens was recently confirmed by an independent study using a different limited TCR repertoire model (45). Moreover, Cebula et al. (45) also found that the colonic Treg TCR repertoire was markedly changed after mice were exposed to broad spectrum antibiotics, consistent with Treg cell recognition of commensal bacteria. Similarly, Treg cells isolated from mice exposed to a mixture of Clostridium species were more suppressive in vitro in the presence of fecal material containing Clostridium, suggestive of antigen recognition (36). Together, this collection of data demonstrates that a substantial portion, and quite possibly the majority, of colonic Treg cells recognize antigens from commensal bacteria.
Origination of colonic Treg cells from peripheral versus thymic Treg cell differentiation
Colonic Treg cells that recognize foreign antigens from commensal bacteria may arise in the thymus, denoted tTreg cells (46), perhaps due to TCR reactivity with both self and foreign antigen (23, 47). Alternatively, colonic Treg cells may be comprised of peripherally derived Treg (pTreg) cells, which arise from the induction of Foxp3 during naive T-cell differentiation (48). While it appears clear that Treg cells derived from both thymic and peripheral development are present in the colon, the relative contribution of each developmental pathway to the total pool of mucosal Treg cells is controversial.
There are several lines of data supporting the notion that, unlike Treg cells in most other anatomical locations, colonic Treg cells arise primarily from peripheral T-cell differentiation. First, adoptive transfer experiments using naïve T cells demonstrated preferential accumulation of donor derived pTreg cells in the intestines of host mice (49). Another line of evidence came from studies of Foxp3-deficient mice, in which adoptive transfer of wildtype Treg cells was unable to fully complement the host’s Treg cell deficiency and restore the animal to full health and fecundity (50). Interestingly, co-transfer of Foxp3− cells, of which 50% developed into pTreg cells, along with Foxp3+ cells was required to fully complement Foxp3 deficiency. If these pTreg cells were selectively depleted in vivo, the previously healthy host mice developed inflammatory infiltrates in the liver, small intestine, colon, and lungs. In conjunction with TCR repertoire studies, they concluded that pTreg cells derived from naive Foxp3− cells are required to provide additional antigen specificities distinct from those of tTregs to generate tolerance particularly at mucosal sites.
Genetic deletion of the conserved non-coding sequence-1 (CNS1) region of the Foxp3 markedly decreases the efficiency of peripheral Treg cell differentiation (51). In these mice, Zheng et al. (51) observed that the frequency of Treg cells in the gut is reduced by approximately 50%. Similar, but milder, defects in gut Treg cells were also observed upon specific deletion within CNS1 of the SMAD binding site, which is involved in TGFβ signaling (52). Subsequent studies showed that the defect in pTreg selection in CNS1-knockout mice led to the eventual development of a Th2-mediated colitis (53). Thus, data from CNS1-deficient mice suggests that peripheral Treg cell differentiation is important for generating the bulk of the gut Treg cell population.
A fourth line of evidence comes from the use of markers for tTreg cells such as Helios, an Ikaros family transcription factor, which is reported to be highly expressed on Treg cells of thymic, but not peripheral, origin (54). In germ-free mice, ~80% of colonic Treg cells are Helioshi suggesting that most of these Treg cells are of thymic origin (29, 33). By contrast, only 20% of Treg cells are Helioshi in conventionally housed SPF mice, suggesting that the colonic Treg cell population is derived mostly from naive T cells that undergo pTreg cell differentiation in response to commensal bacteria. Similar results were reported for neuropilin-1, another putative marker of thymic Treg cells (55, 56). Although pTregs are capable of upregulating Helios under some inflammatory conditions (50, 57), low expression of Helios by tTreg cells has not been reported. While additional studies are necessary to validate these markers, the available data support the notion that the majority of colonic Treg cells arise from the peripherally derived Treg cell pool.
Additional work from our group has assessed the ability of colonic Treg TCRs to facilitate thymic Treg cell differentiation. As mentioned above, these TCRs were identified using the fixed TCRβ model, and were preferentially found in the colon but not elsewhere in the body, including the thymus. Unlike Treg TCRs isolated from peripheral lymph nodes (23, 29, 58), colonic Treg TCRs did not facilitate thymic Treg cell differentiation when retrovirally expressed on thymocytes (29). Using retroviral bone marrow chimeras to further study two colonic Treg TCRs, we observed peripheral Treg cell generation only when the purchased host mice were first co-housed with mice from our colony (29). Subsequent studies using adoptive transfer of naive T cells from colonic Treg TCR transgenic mice into congenically marked normal hosts have confirmed the ability of these TCRs to facilitate peripheral Treg cell differentiation (authors’ unpublished data).
There has been a recent challenge to the notion that most colonic Treg cells are of peripheral origin, as Cebula et al. (45) suggested that colonic Treg cells are primarily of thymic origin due to significant similarities in the TCR usage between colonic and thymic Treg cells in their model. Several factors may account for the discrepant results observed between this study and previous studies. First, although this group as well as ours analyzed mice with limited TCR repertoires, different TCRβ chains were used to restrict TCR diversity. Second, we utilized retroviral expression of TCRs in thymocytes to assess whether colonic Treg TCRs induce tTreg selection as discussed above (29). It was suggested by Cebula et al. (45) that intraclonal competition in the thymus precluded our ability to visualize thymic Treg cell development, a phenomenon previously described by our group and others (58, 59). However, this issue was examined in our manuscript (29). Third, Cebula et al. (45) assigned TCRs to be of thymic Treg origin based on repertoire studies. However, it may be difficult to draw conclusions regarding precursor:product relationships using TCR repertoire studies alone, as multiple factors may result in the detection of a peripherally derived Treg TCR amongst the tTreg TCR pool. Peripherally derived Treg cells can recirculate to the thymus where their TCRs may be erroneously counted as thymically derived Treg TCRs (40, 60). Impurities in FACS purification and errors in sequencing of the TCRs and barcodes may be compounded using next-gen platforms which generate many-fold more data. This can result in a greater perceived repertoire overlap if the primary criterion is a qualitative assessment of ‘presence’ or ‘absence’ of the TCR sequence in each subset. However, the currently available data suggests that TCR-driven thymic selection is a quantitative, rather than qualitative, process meaning that each individual TCR varies in its efficiency for promoting thymic Treg cell selection (23, 58) that likely depends on the amount of ligand in the thymus (24). Thus, the observation that a TCR sequence is detected in the thymic Treg cell sequences does not mean that all of the Treg cells in the periphery with that TCR were generated by thymic Treg cell differentiation.
Like the TCR repertoire data, studies of SCFAs have conflicted whether they affect pTreg cell differentiation versus tTreg cell expansion. One group suggested that SCFAs primarily acts to expand Treg cell numbers via GPR43 and histone deactylases (HDAC) inhibition (34), supporting the notion of tTreg cell expansion (45). However, other groups reported that the administration of SCFAs in the form of a butyrylated-diet lead to a marked increase in Nrp-1lo Treg cells, consistent with expansion or differentiation of pTreg cells (38). Moreover, SCFA-induced Treg cell expansion did not occur in CNS1−/− mice that are defective in peripheral Treg cell generation (37). Thus, while the mechanism by which SCFAs increase the number of colonic Treg cells remains uncertain, peripheral induction of Foxp3+ cells seems likely to be involved in this process.
It has not been controversial that both thymic and peripheral Treg cells are present in the colonic Treg cell population. The existence of Treg cells of both origins in the colon are suggested by the distribution of Helios and Nrp-1 markers for tTreg cells, and by the observation that CNS1-deficient mice still contain colonic Treg cells. Moreover, TCRs that facilitate thymic Treg cell selection and react with foreign antigen has been observed (23, 47). Thus, the unresolved issue remains the degree to which thymic versus peripheral Treg cell differentiation contributes to the overall colonic Treg cell population.
Effector T-cell differentiation in response to commensal bacteria
Although the precise ratio of thymic to peripheral Treg cells in the colon remains to be determined, the intestines are widely regarded as environments that favor peripheral Treg cell differentiation (61, 62). Interestingly, commensal bacteria also appear to elicit effector cell differentiation, as much higher numbers of Th1 and Th17 cells are observed in conventionally housed mice compared to those raised in germ-free conditions (63, 64). Note that these observations specifically refer to effector T-cell differentiation in normal mice under homeostatic conditions without evidence of inflammation or infection. Consistent with this observation, memory T cells from conventionally housed mice are much more efficient at inducing colitis after transfer into lymphopenic hosts than T cells from germ-free mice (64), suggesting that mucosal effector T cells, in addition to Treg cells, also develop in response to the commensal microbiota.
While relatively little is known about the specific bacterial species that induce Th1 cell responses under homeostatic conditions, the dominant bacterial species involved in eliciting Th17 responses in mice has been identified. The discovery of segmented filamentous bacteria (SFB) resulted from a serendipitous observation that mice from different vendors had different levels of Th17 cells in the gut lamina propria (65). SFB, a commensal microbe found in mice from Taconic, is unique as it is primarily found in the terminal ileum of the small intestine and makes tight attachments to the epithelial cells (66). The geographical location of SFB results in Th17 induction primarily in the small intestine, and it remains to be determined whether SFB is the dominant bacteria responsible for colonic Th17 cells. Another question is whether SFB represents a class of commensal bacteria that are more related to pathogenic species, i.e. a ‘pathobiont’, requiring effector immune responses to maintain homeostasis. Nonetheless, these data provide clear evidence that effector T-cell responses to specific commensal bacterial species occurs in normal healthy mice.
Treg cells express effector cell transcription factors during intestinal homeostasis
The above data suggest that naive T cells encountering commensal bacterial antigens can differentiate into Treg or effector cell lineages. In the classic paradigm of CD4+ T-cell differentiation, expression of lineage specific transcription factors in recently activated naive T cells leads to the unambiguous generation of canonical T-helper cell phenotypes such as Th1, Th2, Th17, and pTreg (67–71). Mutually exclusive lineage definition results in part from cross-inhibition between the transcription factors that define each T-cell phenotype. For example, co-expression of the Th17 transcription factor RORγt with Foxp3 was first noted with in vitro cultures of naive T cells with TGFβ (72). Antagonism between the two transcription factors results in either Th17 or Treg cells expressing only one transcription factor.
It has become evident that this may not strictly apply to Treg cells, which have been observed to co-express transcription factors associated with the various T-effector lineages. For example, Treg cell-specific deletion of IRF4, a transcription factor involved in Th2 development, results in spontaneous Th2-mediated autoimmunity (73). In contrast, Treg cell-specific deletion of GATA-3, another transcription factor required for Th2 development, exhibits spontaneous autoimmunity characterized by the production of multiple effector cytokines including IFNγ, IL-17, and IL-4 (74, 75). In another example, the Th1 transcription factor T-bet is expressed on a subset of Treg cells and is required for suppression of Th1 responses (76). While Treg cell-specific deletion of the canonical Th17 transcription factor RORγt has not yet been reported, deletion of another important factor for Th17 cells, STAT3, in Treg cells showed spontaneous colitis characterized by increased Th17 cells (77). Thus, these data have led to a model where Treg cells that co-express effector transcription factors gain the ability to specifically suppress the corresponding effector cells (17, 78).
An important question is the mechanism by which expression of an effector lineage transcription factor allows a Treg cell to specifically suppress that effector lineage. One proposed mechanism is exemplified by the Th1 factor T-bet, which induces the expression of the chemokine receptor CXCR3 on Treg cells. IFNγ induces the local expression of several CXCR3 ligands, thus allowing T-bet+ Treg cells to traffic to sites of Th1 activation and limit inflammation (76).
Another question is the mechanism by which Treg cells expressing effector transcription factors are generated. One suggestion is that pre-existing Treg cells exposed to lineage specifying cytokines upregulate the corresponding transcription factor, as observed for IL-12 and T-bet (79). Whether the expression of effector transcription factors is a transient response to environmental cues, or represents a stable developmental sublineage of Treg cells, remains to be established.
The relationship between Th17 and Treg cells deserves special consideration in gut immunity for several reasons. First, there is a strong developmental synergy between these two peripheral differentiation pathways, as TGFβ is an important factor for the development of both subsets in vitro, albeit with different dose optimum (72). Second, the colon, but not other locations in the body, contains a high frequency of RORγt+Foxp3+ cells (72, 80). In fact, the majority of RORγt+ cells in the colon co-express Foxp3, and conversely, nearly half of Foxp3+ cells co-express RORγt (72). Interestingly, expression of RORγt on Foxp3+ cells, just like induction of RORγt on effector cells, appears to be dependent on the microbiota (81). Despite their expression of RORγt, these Foxp3+ cells express high levels of classical Treg cell factors, such as IL-10, CTLA-4, and ICOS, rather than the cytokines associated with Th17 cells (80), and are protective against autoimmunity in a NOD-scid model of diabetes (82).
These observations regarding RORγt+Foxp3+ cells raise several intriguing questions regarding mucosal interactions with the gut microbiota. One question is to determine why the ratio of Th17 to RORγt+Foxp3+ cells is much higher in the small intestine and reversed in the colon. It could be due to different bacterial species, as SFB is found predominantly in the ileum, whereas microbes that induce RORγt in the colon are unknown and may have different properties. Alternatively, colonic microbes may induce RORγt in a manner similar to SFB, but other environmental cues such as SCFAs may hijack the development of Th17 cells, diverting them into Foxp3+ cells. Another puzzle is the developmental relationship between RORγt and Foxp3 expression. Do RORγt+Foxp3+ cell develop from Treg or Th17 cells? If they develop from Treg cells, is RORγt expression transient or stable, and does RORγt impart a special function on Treg cells? In summary, the questions regarding the generation of RORγt+Foxp3+ cells represent a microcosm of the issues regarding the mechanisms by which commensal bacteria induce the selective differentiation of effector and Treg cells.
How does the immune system determine effector versus Treg cell selection?
Although the gut is widely recognized to be an important site for Treg cell differentiation, it appears that these global signals favoring Treg cell selection do not override the process of effector T-cell lineage selection to commensal bacteria. Treg versus effector T-cell selection appears to be dependent on TCR specificity, as it was observed that the TCR repertoires are quite different between CD44hi effector T cells and Foxp3+ Treg cells (29, 40). One possible explanation is that molecular properties of the TCR interaction with antigen, such as affinity, direct T-lineage specification. However, this is unlikely to be a primary factor, as there is no obvious way in which this would distinguish bacteria that should elicit Treg versus effector cell responses. Another possibility is temporal compartmentalization, in which there are periods of effector cell differentiation and periods of Treg cell selection. This could be imagined as a process that would allow effector cell response to pathogens during periods of infection and Treg cell development to commensals during periods of homeostasis. While this may be an important mechanism for peripheral tissues that rarely encounter foreign antigens, it may not explain effector cell development in the intestines of specific pathogen free mice, which do not have pathology and should not be routinely exposed to new bacteria. We therefore favor the notion of spatial compartmentalization of effector and Treg cell responses, in which TCR specificity dictates the site of antigen encounter (Fig. 1). The cytokine milieu present during TCR activation would then direct T-cell lineage commitment. Thus, both the macroenvironment of the gut and the microenvironments within the mucosal immune tissues may play important roles in peripheral T-cell differentiation.
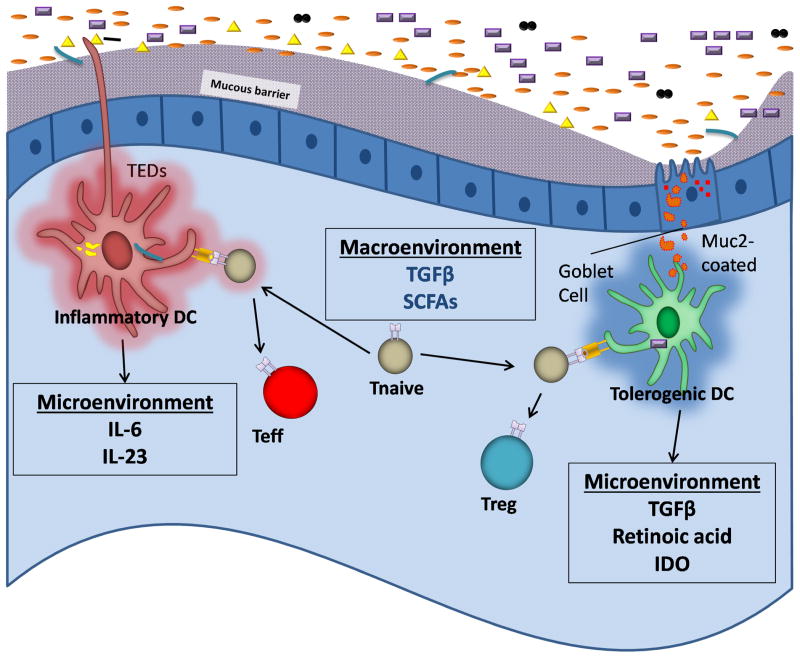
Fig. 1. Different microenvironments in the gut allow for antagonistic T-cell differentiation despite a tolerogenic macroenvironment.
The macroenvironment of the intestines is enriched with SCFAs which have been reported to induce tolerogenic factors including TGFβ. Antigens may be delivered to CD103+ DCs in the lamina propria via GAPs and muc2, or they may reside within the epithelium and acquire bacteria via intraepithelial dendrites. Muc2 and SCFAs may stimulate these DCs to secrete TGFβ, retinoic acid, and IDO, creating a microenvironment that directs pTreg cell differentiation. CX3CR1+CD103−DCs may acquire antigens via transepithelial dendrites during inflammation, and secrete cytokines enriched with pro-inflammatory factors such as IL-6. Thus, the microenvironment in which a naive T-cell encounters their antigen may direct whether they adopt a tolerogenic or effector fate. Note that although this figure illustrates naive T-cell encounter with APCs in the intestinal lamina propria, this phenomenon may first occur in the mesenteric lymph node and involve migratory DCs (90–93, 138).
In terms of the gut macroenvironment, it has been suggested that there are at least two important signals which globally enhance Treg cell induction in the gut. TGFβ has long been recognized as enriched in the gut (83, 84) and is a cytokine crucial for peripheral Treg cell differentiation (85) and survival (86, 87). The in vivo role for TGFβ was supported by the observation that deletion of the Smad3 TGFβ-dependent transcription factor binding site CNS1 in the Foxp3 locus resulted in decreased numbers of Treg cells in the gut (52). Commensal bacteria can also enhance the production of TGFβ (33). The other factor appears to be SCFAs produced by bacterial fermentation as discussed above (34, 37, 38). Thus, the presence of SCFAs and TGFβ appears to generate a macroenvironment that globally favors Treg cell selection or expansion in the gut.
DC subsets may provide selective microenvironments for T-cell differentiation
The observation that commensal bacteria makes a marked impact on the colonic Treg TCR repertoire during normal gut homeostasis (29, 45) suggests that bacteria antigens in the lumen routinely breach the mucosal barrier and are presented to the immune system. Accumulating data suggest that sampling of the lumen is a routine function for certain APC subsets. However, many current studies on antigen presentation focus on the small intestine rather than the colon, which have different mucosal, microbial, and immune physiology (88, 89). As the field of APCs in the intestines is complex and continually evolving (90–93), we will limit our discussion to consider the possibility that differential antigen uptake by DCs may result in spatial compartmentalization of T-cell selection (Fig. 1).
The presence of transepithelial dendrites from lamina propria CX3CR1+CD103−CD11c+ cells was observed to extend into the lumen of the small intestine and capture Salmonella (94–96). Recently, CX3CR1−CD103+ DCs in the epithelium were reported to preferentially sample luminal bacteria, rather than soluble antigen (97). Finally, an alternative mechanism of antigen sampling of luminal contents was proposed in which goblet cell associated passages (GAPs) allow luminal contents to pass into the lamina propria and preferentially transfer to CX3CR1− CD103+ DCs (98). While debate continues regarding the relative importance of these antigen uptake mechanisms in the small and large intestine and in infection versus homeostasis, these data support the hypothesis that the route of bacterial antigen entry through the mucosal barrier may dictate the APC subset presenting the antigen to T cells.
The observation that GAPs facilitate antigen transfer to CD103+ DCs synergizes with another report showing that a molecule in mucous secreted by goblet cells, muc2, signals CD103+ DCs to facilitate Treg cell differentiation (99). It is tempting to speculate that GAPs transfer a package of bacterial antigens coated with muc2 which both stimulates antigen uptake and tolerogenic cytokine production by CD103+ DCs, resulting in a microenvironment that directs Treg cell differentiation of commensal bacteria-specific naive T cells.
In vitro, CD103+ DCs have been reported for some time to facilitate Treg cell differentiation from naive T cells (100), and appear to do so via multiple mechanisms. One of the first described was the increased production by CD103+ DCs of TGFβ and retinoic acid (RA) (49, 101–103). RA also induces the expression of CCR9 involved in gut homing (104). The mechanism of RA may be indirect by inhibiting T-effector cell differentiation, rather than directly promoting the expression of Foxp3 itself (105), although a role for RA in effector cell generation has also been proposed (106). Another mechanism is that CD103+ DCs produce higher levels of IDO (107), which catalyzes the oxidative catabolism of tryptophan and induce a metabolic stress response leading to the inhibition of mammalian target of rapamycin (mTOR). The Akt-mTOR pathway is an antagonist of Treg cell differentiation (108–110) potentially via the transcription factor HIF-1α (111), although a recent report suggests paradoxically that mTOR facilitates Treg function (112). Consistent with this body of work favoring a role of CD103+ DCs in peripheral Treg cell differentiation, it was recently reported that in vivo ablation of CD103+ DCs resulted in a decrease in gut Treg cell numbers (113).
By contrast, it has been reported that Th17 (83) or Th1 (114) differentiation is facilitated by CD11b+ DCs. In vivo depletion of CD11b+CD103+, but not CD11b−CD103+, DCs revealed a decrease in Th17 effector cell numbers (113) or Th17-profile cytokines (115), despite the observation that each subset of CD103+ DCs individually is sufficient to sustaining Treg cell numbers (113). How this occurs is unclear. One possibility is that each DC subset can perform multiple roles (116). Another possibility is that DCs are plastic (114, 117), such that they respond to local signals from TLR agonists, SCFAs, apoptotic cells (118), and so forth, with a different array of cytokines. Third, the role of DC subsets has typically been assessed on bulk Treg and Th17 cell numbers. For Treg cells, compensatory maintenance of thymically educated Treg cells could obscure important defects in peripheral Treg cell selection. Thus, it will be important in future experiments to trace the impact of specific bacterial species, its uptake by DC subsets, and its recognition by antigen-specific T cells and the subsequent acquisition of Treg and/or effector T-cell lineage transcription factors (Fig. 2).
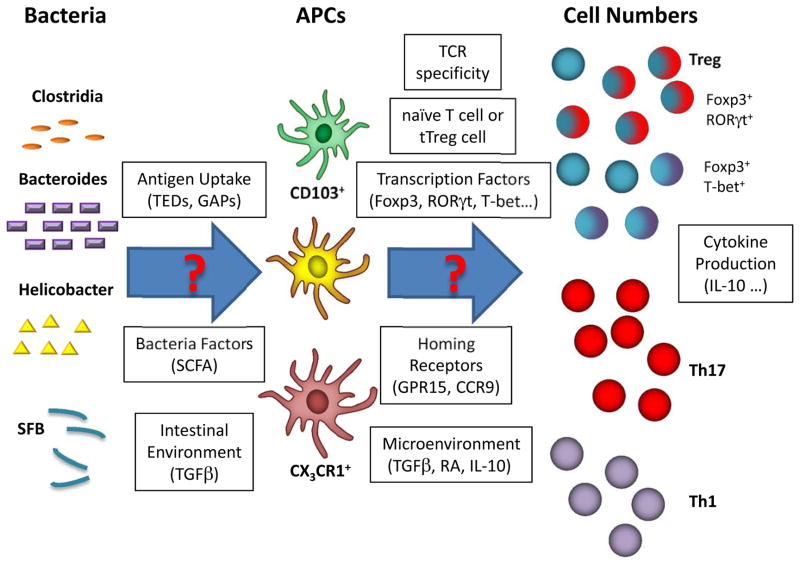
Fig. 2. The myriad of immune responses to commensal bacteria.
Recent studies have demonstrated a clear relationship between specific commensal species, APCs, and T-cell fates. Many studies report the total numbers of Treg and effector cells that arise from interaction with commensal bacteria, but the mechanisms behind these interactions remain unclear. For example, which commensal bacterial species elicit Treg, Th1, and Th17 cells? How is the antigen taken up, and by which APCs? Do they induce subsets of Treg cells expressing effector transcription factors like RORγt? Interestingly, like Treg cells, Th17 cells have recently been suggested to be comprised of subsets with different pathogenicity and developmental requirements (139, 140). Could these subsets of Th17 cells be related to Treg cells by transcription factors or TCR specificities? Thus, this diagram illustrates the complex relationships that exist between commensal bacteria and the T cell immune system as well as the myriad of factors that should be investigated in future experiments.
A role for effector T-cell responses to commensals during homeostasis
In comparison with the formalized notion that the immune system exists to eliminate pathogens without causing autoimmunity, the intestinal immune system appears considerably more complex. Considerable evidence exists suggesting that concurrent generation of both tolerogenic and inflammatory cells occurs in response to commensal bacteria. Moreover, these responses appear to be exaggerated in the gut, which may be best exemplified by the massive enrichment of pTreg and Th17 cells in the intestines—phenomena that appears dependent on the presence of commensal bacteria. Why then, does such a complex system of effectors and regulators exist?
This question is underscored by the observation that mice deficient in T and B cells do not develop spontaneous intestinal immunopathology, breed reasonably well, and live long lifespans in SPF housing conditions. In fact, they are more susceptible to environmental opportunistic infections such as pneumocystis rather than intestinal pathology from commensal bacteria. However, lymphocyte deficient mice do develop spontaneous colitis when compounded by the innate immune cell deficiency seen in T-bet knockout mice, suggesting a role for innate immune cells in controlling certain commensal bacteria (119). This raises the possibility that, instead of functioning as a basic mechanism of immunity in mucosal tissues, effector T-cell responses to commensal bacteria are rather a necessary ‘evil’ associated with the unique mucosal requirement for adaptive immune responses to discriminate between pathogenic and commensal bacteria and viruses.
Adaptive immune cells must make some contributions to immunity to commensal bacteria, as the number of live bacteria in the lamina propria and mesenteric lymph nodes is increased in T- and/or B-cell-deficient mice (120). It has been difficult to address whether this immunity is due to effector cells as transfer of effector cells without Treg cells results in colitis (15). The reverse experiment of transferring Treg cells alone results in the loss of Foxp3 expression in a small number of Treg cells which then undergo massive expansion (121), which may convolute interpretations of Treg cell transfer experiments into lymphopenic mice (122, 123). Although these experimental issues preclude proof of this hypothesis, the logical presumption is that adaptive immune control of commensal bacteria in mucosal associated tissues is enforced by effector T cells.
T-cell responses to pathogenic and commensal bacteria
While the primary role of the adaptive immune system may be to respond to pathogenic rather than commensal bacteria, the pre-existing effector and Treg cell responses to commensal bacteria can impact the course of infection. Conversely, infection may fundamentally alter the balance between effector and Treg cell responses to commensal bacteria, resulting in disturbed immune homeostasis and potential immunopathology well after the infection has been resolved (Fig. 3).
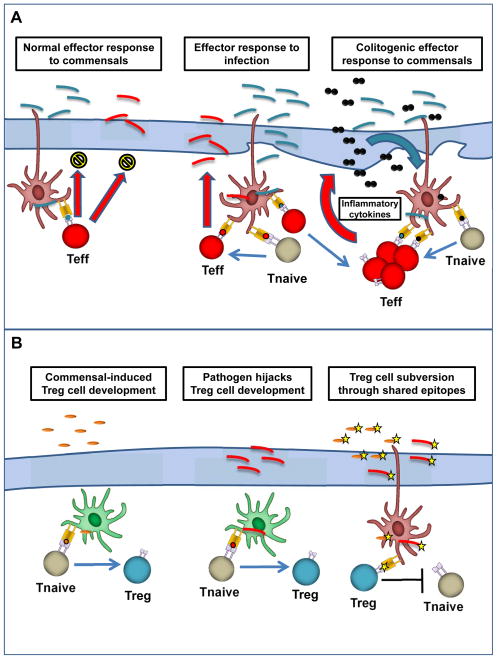
Fig. 3. Effector and Treg cell responses to pathogenic and commensal bacteria.
(A). Effector response to gut bacteria. Under homeostatic conditions, gut resident effector T cells (red) mediate sub-clinical immune response toward commensal bacteria (blue), limiting lamina propria invasion. This subclinical response may also provide protection against pathogenic bacteria (red). During infection, invasive pathogens promote a robust effector T-cell response, leading to their eventual clearance. Simultaneous presentation of commensal antigens during this inflammation can lead to the expansion of commensal-specific effector T cells. Sustained inflammation may lead to feed-forward loop (right) where commensal-T cells alone can maintain inflammation and promote immune response toward additional commensal bacteria and, eventually, overt colitis. (B). Regulatory response to gut bacteria. Under homeostatic conditions, commensal bacteria (orange) drive peripheral conversion of naïve T cells to a regulatory phenotype via the action of tolerogenic dendritic cells (DCs) (green). Pathogens (red) may be able to subvert this process to prevent their elimination by effector cells, potentially leading to the sustained inflammation described in (A). Pathogens that gain access to tolerogenic DCs may lead to generation of Treg rather than effector T cells. Pathogens that share epitopes (yellow star) with commensal bacteria may be protected by commensal-specific regulatory responses.
Commensal bacteria-mediated effector cell development may improve immune fitness by facilitating immune responses to new encounters with pathogenic bacteria. The presence of SFB confers resistance to infection by Citrobacter rodentium, an attaching and effacing bacterium that is a murine model of enteropathogenic E. coli, presumably due to the increased baseline expression of Th17 associated cytokines and antimicrobial defensins (66). It should be noted that SFB colonizes the small intestine and Citrobacter involves primarily the cecum, suggesting the possibility of cross-talk between the colonic and small intestinal adaptive immune systems. Thus, SFB induction of Th17-profile cytokines from innate lymphocytes and Th17 cells results in bystander protection against Citrobacter, much like the protection against bacterial infection conferred by the presence of latent virus (124).
Effector T cells that recognize commensal bacteria may be activated during infection due to an influx of antigens after mucosal injury, potentially exacerbating immunopathology. For example, GPR15−/− mice, which are deficient in colonic Treg cells due to impaired homing, are more susceptible to Citrobacter infection (39). This was hypothesized to result from insufficient numbers of Treg cells leading to impaired suppression of effector responses to commensals that infiltrate the lamina propria upon epithelial damage, and perhaps to Citrobacter itself. Thus, effector T cells that recognize commensal bacteria may enhance the inflammatory response during infection, requiring suppression by Treg cells to limit pathology.
In addition to activating pre-existing effector T cells responsive to commensal bacteria, infection can also introduce bacterial antigens that are not constitutively presented to the immune system. For example, CBir TCR transgenic T cells, which recognize flagellin found on certain commensal bacteria, typically possess a naïve phenotype in wildtype mice. However, in the context of mucosal injury due to Toxoplasma gondii infection or DSS administration, CBir T cells adopt an effector phenotype (125). Potentially, repeated injury could lead to the accumulation of these commensal-specific effector T cells sufficient to sustain an inflammatory response that prevents healing from epithelial injury, which allows more commensal bacterial antigens to be presented to effector T cells, generating a destructive feed-forward loop (125). Thus, effector T-cell responses to commensal antigens may improve immune fitness to infections, but potentially at the risk of incurring acute and chronic immunopathology.
Treg cell responses to commensal bacteria may increase susceptibility to pathogens. One possibility is that the host mechanisms that facilitate Treg cell responses to commensals, such as those involving SCFAs and TGFβ, could be co-opted by pathogens, resulting in blunted effector cell and enhanced Treg cell generation to the pathogen. Another interesting possibility is that commensal bacteria-specific Treg cells may block effector development to pathogens, particularly if the pathogenic bacteria share common antigenic epitopes with commensal species. This circumstance may be quite common with several known pathogenic variants of commensal species. For example, Shiga-toxin producing E. coli is an important cause of hemolytic uremic syndrome and is related to commensal E. coli (126). Similarly, enterotoxogenic B. fragilis is related to non-toxogenic B. fragilis, a normal commensal bacterial species (127). In these cases, pre-existing Treg cells to the commensal species may limit the effector response to pathogen. Although Treg cell control of inflammation is necessary for preventing effector cell-mediated immunopathology during resolution of infection and periods of homeostasis, this pathway may be co-opted by pathogens as an immune evasion strategy.
Systemic effects of T cells responses in the gut
The above discussion on T cell responses to commensal bacteria implies that these T cells are gut specific due to interactions with local antigens. Consistent with this, TCR repertoire studies suggest that the colon Treg TCR repertoire both varies with anatomic location and is markedly affected by antibiotics (29, 45). However, a number of studies have suggested that commensal bacteria may affect the immune system more globally and impact immune responses outside of the intestine. For example, the presence of commensal bacteria facilitates murine EAE, a model for human MS (12). Similarly, the murine K/B×N model for inflammatory arthritis was greatly attenuated in the absence of commensal bacteria (13). Interestingly, reconstitution of germ-free mice with a single bacteria strain, SFB, which is known for its ability to induce proinflammatory Th17 cells, was sufficient to generate arthritis with similar kinetics to conventionally housed SPF mice. As another example, Clostridium inoculated mice are resistant to colitis and systemic IgE responses (33). Thus, an increasing body of evidence suggests that microbiota can potentially both stimulate as well as inhibit immune responses beyond the intestinal compartment.
An important future goal involves defining the mechanism by which microbiota affect the ‘global’ immune system. Perhaps commensal organisms serve as a source of steady-state TLR ligands and other immunogenic bacterial products that stimulate effector or Treg cells either directly or indirectly via cytokines from innate cells. For instance, commensal-derived SCFAs have been shown to increase systemic Treg cell numbers when administered exogenously (37, 38). However, it is less clear whether microbiota produced SCFAs can cause global effects on Treg cell number or function. Interestingly, it was reported that spontaneous type 1 diabetes in the NOD model is inhibited by MyD88-deficiency, which is consistent with the notion that microbiota may stimulate the immune system through TLR signaling (128). However, re-derivation of MyD88−/− NOD mice under germ-free conditions restored susceptibility to diabetes, suggesting a complex pro- and anti-inflammatory role for the microbiota. Nonetheless, the idea is appealing that commensal bacteria provide factors which optimally condition the host immune system in its entirety as a byproduct of their co-evolution.
Alternatively, the microbiota might exert systemic effects by influencing the activation or differentiation of local T cells, which subsequently traffic to other anatomical locations. This may occur in response to ubiquitous self-antigens or tissue-restricted antigens that are also found in the intestine such as MBP (12). Encounter of self-antigen in the appropriate microbial microenvironment could result in the development of autoreactive Th17 cells. This may explain why the K/B×N model of arthritis is influenced by commensal microbiota (13) even though the TCR transgenic cells in this model recognize an antigen found in the serum (129). One question is whether a wildtype polyclonal T-cell population generates a different immune environment in the intestine in comparison with a TCR transgenic population. Polyclonal T cells would be able to generate Th17 effector responses specific to bacteria such as SFB and compete for a Th17 developmental or survival ‘niche’. Additionally, the generation of polyclonal Treg cells to commensal bacteria may provide a more tolerogenic environment. Thus, future studies are required to determine the mechanism by which the microbiota affects immune responses beyond the intestine.
T-cell selection to commensal bacteria and IBD
Both the Powrie transfer model and TNBS model suggest that excessive effector T-cell responses to commensal bacteria can induce chronic colitis that resembles human inflammatory bowel disease (IBD). As Treg cells can ameliorate colitis in these models, perturbation of the balance between effector and Treg cells that recognize commensal bacteria may contribute to IBD. Consistent with this notion, stool samples obtained from CD and UC patients were found to have deficiencies in members of the Firmicutes and Bacteroidetes phyla (130), which have been shown to impact Treg differentiation and function in murine studies (31, 36). Thus, a chronic lack of tolerogenic factors produced by bacteria such as SCFAs or PSA may tip the balance in favor of effector rather than Treg cell differentiation and expansion in response to commensal bacteria.
A primary defect in effector or Treg cells may not be an initiating factor for IBD. Genetic defects in Treg cell specific genes are most associated with IPEX syndrome (20). By contrast, genetic studies of IBD have revealed defects in genes associated with epithelial function such as autophagy (131), or genes associated with innate, rather than adaptive, immune components such as innate cytokine production and microbial sensing. For example, polymorphisms in the intracellular bacterial sensor NOD2 are clearly associated with IBD (132, 133). One hypothesis is that epithelial and innate immune defects may result in mild barrier breaches or slightly less efficient clearance of bacteria that penetrate the mucosa, leading to excess antigen presentation to adaptive immune cells. Over years, this may result in an imbalance between effector and Treg cells that results in inflammation sufficient to alter mucosal barrier function, eventually leading to a feed-forward loop where more and more commensal antigens are exposed to the immune system (125). Thus, defects in innate immune cells may result in immunopathology caused or perpetuated by an adaptive immune system with an imbalance between effector and Treg cells.
Although it may not be easy to reverse the genetic defects that promote IBD, it may be possible to alter the microbiota to eliminate or minimize the bacterial species that are particularly immunostimulatory. This could occur with antibiotic treatment, transplants of ‘designer microbiota’ (36), or direct administration of immunomodulatory factors such as SCFAs (34, 37, 38) or PSA (134). Alternative approaches could involve therapeutics aimed at increasing Treg cell numbers such as low dose IL-2 (135) or by ex vivo expansion of autologous Treg cells (136, 137). Of course, with any manipulation that promotes tolerance, susceptibility to infection may increase. However, historical experience using immunosuppression with IBD and other autoimmune diseases suggests that this may not be a prohibitive factor. In the future, it may be possible to expand bacterial antigen-specific Treg cells. While issues of cost, Treg cell antigen-specificity, persistence, phenotypic stability, and homing remain important challenges to personalized medicine with antigen-specific Treg cells, such an approach has the potential to provide prolonged periods of remission from IBD without systemic immunosuppression.
Acknowledgments
We thank Katherine Nutsch (Wash. U.) for critical reading of the manuscript. C.S.H. is supported by grants from the NIH (NIAID/NIDDK), CCFA, and the Burroughs Wellcome Fund.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106–114. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein L, Jovanovic K. Regulatory T cell lineage commitment in the thymus. Semin Immunol. 2011;23:401–409. doi: 10.1016/j.smim.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contractor NV, et al. Lymphoid hyperplasia, autoimmunity, and compromised intestinal intraepithelial lymphocyte development in colitis-free gnotobiotic IL-2-deficient mice. J Immunol. 1998;160:385–394. [PubMed] [Google Scholar]
- 9.Schultz M, et al. IL-2-deficient mice raised under germfree conditions develop delayed mild focal intestinal inflammation. AM J Physiol. 1999;276:G1461–1472. doi: 10.1152/ajpgi.1999.276.6.G1461. [DOI] [PubMed] [Google Scholar]
- 10.Aranda R, et al. Analysis of intestinal lymphocytes in mouse colitis mediated by transfer of CD4+, CD45RBhigh T cells to SCID recipients. J Immunol. 1997;158:3464–3473. [PubMed] [Google Scholar]
- 11.Powrie F, Mauze S, Coffman RL. CD4+ T-cells in the regulation of inflammatory responses in the intestine. Res Immunol. 1997;148:576–581. doi: 10.1016/s0923-2494(98)80152-1. [DOI] [PubMed] [Google Scholar]
- 12.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 13.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 15.Powrie F, Mason D. OX-22high CD4+ T cells induce wasting disease with multiple organ pathology: prevention by the OX-22low subset. J Exp Med. 1990;172:1701–1708. doi: 10.1084/jem.172.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 17.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol. 2010;11:7–13. doi: 10.1038/ni.1818. [DOI] [PubMed] [Google Scholar]
- 19.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 20.d’Hennezel E, et al. FOXP3 forkhead domain mutation and regulatory T cells in the IPEX syndrome. N Engl J Med. 2009;361:1710–1713. doi: 10.1056/NEJMc0907093. [DOI] [PubMed] [Google Scholar]
- 21.Itoh M, et al. Thymus and autoimmunity: Production of CD25+ CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunologic self-tolerance. J Immunol. 1999;162:5317–5326. [PubMed] [Google Scholar]
- 22.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A Broad Range of Self-Reactivity Drives Thymic Regulatory T Cell Selection to Limit Responses to Self. Immunity. 2012;37:475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan MS, et al. Thymic selection of CD4+ CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 25.Min B, et al. Gut flora antigens are not important in the maintenance of regulatory T cell heterogeneity and homeostasis. Eur J Immunol. 2007;37:1916–1923. doi: 10.1002/eji.200737236. [DOI] [PubMed] [Google Scholar]
- 26.Strauch UG, et al. Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut. 2005;54:1546–1552. doi: 10.1136/gut.2004.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36:2336–2346. doi: 10.1002/eji.200535244. [DOI] [PubMed] [Google Scholar]
- 28.Singh B, et al. Control of intestinal inflammation by regulatory T cells. Immunol Rev. 2001;182:190–200. doi: 10.1034/j.1600-065x.2001.1820115.x. [DOI] [PubMed] [Google Scholar]
- 29.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinen T, Volchkov PY, Chervonsky AV, Rudensky AY. A critical role for regulatory T cell-mediated control of inflammation in the absence of commensal microbiota. J Exp Med. 2010;207:2323–2330. doi: 10.1084/jem.20101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa H, et al. Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin Exp Immunol. 2008;153:127–135. doi: 10.1111/j.1365-2249.2008.03668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 37.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 39.Kim SV, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–1459. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008;205:3105–3117. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green EA, Choi Y, Flavell RA. Pancreatic lymph node-derived CD4+ CD25+ Treg Cells: Highly potent regulators of diabetes that require TRANCE-RANK signals. Immunity. 2002;16:183–191. doi: 10.1016/s1074-7613(02)00279-0. [DOI] [PubMed] [Google Scholar]
- 42.Seddon B, Mason D. Peripheral autoantigen induces regulatory T cells that prevent autoimmunity. J Exp Med. 1999;189:877–882. doi: 10.1084/jem.189.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taguchi O, Kontani K, Ikeda H, Kezuka T, Takeuchi M, Takahashi T. Tissue-specific suppressor T cells involved in self-tolerance are activated extrathymically by self-antigens. Immunology. 1994;82:365–369. [PMC free article] [PubMed] [Google Scholar]
- 44.Samy ET, Parker LA, Sharp CP, Tung KS. Continuous control of autoimmune disease by antigen-dependent polyclonal CD4+CD25+ regulatory T cells in the regional lymph node. J Exp Med. 2005;202:771–781. doi: 10.1084/jem.20041033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbas AK, et al. Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol. 2013;14:307–308. doi: 10.1038/ni.2554. [DOI] [PubMed] [Google Scholar]
- 47.Moon JJ, et al. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci USA. 2011;108:14602–14607. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lohr J, Knoechel B, Abbas AK. Regulatory T cells in the periphery. Immunol Rev. 2006;212:149–162. doi: 10.1111/j.0105-2896.2006.00414.x. [DOI] [PubMed] [Google Scholar]
- 49.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haribhai D, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011;35:109–122. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J Exp Med. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gottschalk RA, Corse E, Allison JP. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol. 2012;188:976–980. doi: 10.4049/jimmunol.1102964. [DOI] [PubMed] [Google Scholar]
- 58.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung MW, Shen S, Lafaille JJ. TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J Exp Med. 2009;206:2121–2130. doi: 10.1084/jem.20091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCaughtry TM, Wilken MS, Hogquist KA. Thymic emigration revisited. J Exp Med. 2007;204:2513–2520. doi: 10.1084/jem.20070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hand T, Belkaid Y. Microbial control of regulatory and effector T cell responses in the gut. Curr Opin Immunol. 2010;22:63–72. doi: 10.1016/j.coi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shale M, Schiering C, Powrie F. CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–182. doi: 10.1111/imr.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 64.Asseman C, Read S, Powrie F. Colitogenic Th1 cells are present in the antigen-experienced T cell pool in normal mice: control by CD4+ regulatory T cells and IL-10. J Immunol. 2003;171:971–978. doi: 10.4049/jimmunol.171.2.971. [DOI] [PubMed] [Google Scholar]
- 65.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 69.Wan YY, Flavell RA. How diverse--CD4 effector T cells and their functions. J Mol Cell Biol. 2009;1:20–36. doi: 10.1093/jmcb/mjp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wohlfert E, Belkaid Y. Plasticity of T reg at infected sites. Mucosal Immunol. 2010;3:213–215. doi: 10.1038/mi.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37:501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lochner M, et al. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 82.Tartar DM, et al. FoxP3+RORgammat+ T helper intermediates display suppressive function against autoimmune diabetes. J Immunol. 2010;184:3377–3385. doi: 10.4049/jimmunol.0903324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 84.Planchon SM, Martins CA, Guerrant RL, Roche JK. Regulation of intestinal epithelial barrier function by TGF-beta 1. Evidence for its role in abrogating the effect of a T cell cytokine. J Immunol. 1994;153:5730–5739. [PubMed] [Google Scholar]
- 85.Chen W, et al. Conversion of peripheral CD4+ CD25- naive T cells to CD4+ CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 87.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 2006;25:441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Macpherson AJ, McCoy KD. Stratification and compartmentalisation of immunoglobulin responses to commensal intestinal microbes. Semin Immunol. 2013;25:358–363. doi: 10.1016/j.smim.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 89.Denning TL, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farache J, Zigmond E, Shakhar G, Jung S. Contributions of dendritic cells and macrophages to intestinal homeostasis and immune defense. Immunol Cell Biol. 2013;91:232–239. doi: 10.1038/icb.2012.79. [DOI] [PubMed] [Google Scholar]
- 91.Knoop KA, Miller MJ, Newberry RD. Transepithelial antigen delivery in the small intestine: different paths, different outcomes. Curr Opin Gastroenterol. 2013;29:112–118. doi: 10.1097/MOG.0b013e32835cf1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mann ER, et al. Intestinal dendritic cells: their role in intestinal inflammation, manipulation by the gut microbiota and differences between mice and men. Immunol Lett. 2013;150:30–40. doi: 10.1016/j.imlet.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 93.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012;13:1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rescigno M, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 95.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 96.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farache J, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McDole JR, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shan M, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–453. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Annacker O, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 104.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 105.Hill JA, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hall JA, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matteoli G, et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595–604. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- 108.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 109.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shi LZ, et al. HIF1α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welty NE, Staley C, Ghilardi N, Sadowsky MJ, Igyarto BZ, Kaplan DH. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J Exp Med. 2013;210:2011–2024. doi: 10.1084/jem.20130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Satpathy AT, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Murphy KM. Transcriptional control of dendritic cell development. Adv Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
- 117.Zigmond E, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 118.Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458:78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 119.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci USA. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 123.Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197:111–119. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 125.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Buchholz U, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engle J Med. 2011;365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 127.Wick EC, Sears CL. Bacteroides spp. and diarrhea. Curr Opin Infect Dis. 2010;23:470–474. doi: 10.1097/QCO.0b013e32833da1eb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Matsumoto I, Staub A, Benoist C, Mathis D. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 130.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cadwell K, Stappenbeck TS, Virgin HW. Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol. 2009;335:141–167. doi: 10.1007/978-3-642-00302-8_7. [DOI] [PubMed] [Google Scholar]
- 132.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 133.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 134.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Webster KE, et al. In vivo expansion of T reg cells with IL-2-mAb complexes: induction of resistance to EAE and long-term acceptance of islet allografts without immunosuppression. J Exp Med. 2009;206:751–760. doi: 10.1084/jem.20082824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tang Q, et al. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J Exp Med. 2004;199:1455–1465. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tarbell KV, Yamazaki S, Olson K, Toy P, Steinman RM. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J Exp Med. 2004;199:1467–1477. doi: 10.1084/jem.20040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]