Abstract
Mutation in presenilin 1 (PS1) is one of the leading causes of familial Alzheimer’s disease (fAD). PS1 mutation exacerbates the autophagic and lysosomal pathology in AD patients, leading to accumulation of partially degraded material in bloated lysosomes and autophagosomes – a pathology that bears some resemblance to other diseases characterized by elevated lysosomal pH, like age-related macular degeneration. In this study, we examined the effect of the PS1-fAD mutation A246E on lysosomal pH and lysosomal function, and asked whether restoration of lysosomal pH could reverse some of these changes. Lysosomal pH was elevated by 0.2-0.3 pH units in human fibroblasts with the PS1-fAD mutation. The lysosomal alkalization in PS1-fAD fibroblasts was supported by a reduction in the pH-dependent cleavage of cathepsin D and by a reduction in binding of BODIPY FL-pepstatin A to the cathepsin D active site. PS1-fAD cells had increased LC3B-II/-I ratios and p62 levels, consistent with impaired lysosomal degradation and analogous to changes induced by lysosomal alkalinization with chloroquine. PS1-fAD fibroblasts had increased expression of ATP6V1B2, ATG5, BECN1 TFEB mRNA, and of ATP6V1B2, ATG5 and beclin at the protein level, consistent with chronic impairment of autophagic and lysosomal functions in the mutant cells. Critically, cAMP treatment reacidified lysosomal pH in mutant PS1-fAD; cAMP also increased the availability of active cathepsin D and lowered the LC3B-II/-I ratio. These results confirm a small elevation in the lysosomal pH of human PS1-fAD fibroblasts, demonstrate that this lysosomal alkalization is associated with chronic changes in autophagy and degradation, and suggest that treatment to reacidify the lysosomes with cAMP can reverse these changes.
Keywords: Alzheimer’s disease, Presenilin 1, Autophagy, Lysosomal pH, Therapeutic strategy, Age-related Macular Degeneration (AMD), Cathepsin D
1. Intracellular waste products, damaged organelles and other targets of bulk cellular degradation reach the lysosomes via the process known as macroautophagy (henceforward, simply “autophagy”). The efficient clearance of this material is of particular importance in post-mitotic cells such as cortical neurons (Boland et al., 2008). This degradation is highly dependent on lysosomal pH (pHL): activity of lysosomal enzymes is optimal over a narrow range of acidic levels. Substantial shifts in pHL, such as those induced by drugs like chloroquine or bafilomycin, can severely disrupt degradative enzyme activity and block fusion of autophagosomes with lysosomes (Yamamoto et al., 1998, Klionsky et al., 2008). However, even an increase of only a few tenths of a unit is sufficient to depress the activity of key lysosomal proteases and lipases (Barrett, 1970, 1972, 1973, Schwartz and Bird, 1977, Ameis et al., 1994). These moderate elevations of pHL can perturb the clearance of cellular waste and lead to a backup of the autophagic pathway, resulting in a slow accumulation of waste with time.
Though the pH dependence of lysosomal enzyme activity has been recognized for decades, a role for impaired degradation has only been implicated in neurodegenerative diseases more recently (Pacheco et al., 2007, Cheung and Ip, 2009, Martinez-Vicente et al., 2010, Winslow et al., 2010, Elrick and Lieberman, 2013). Alzheimer’s disease (AD) is of importance in this regard, as fundamental defects in autophagy and autophagic degradation have been observed (Cataldo and Nixon, 1990, Cataldo et al., 1996, Cataldo et al., 2004, Nixon, 2005, Nixon et al., 2005, Nixon and Cataldo, 2006, Khurana et al., 2010, Lipinski et al., 2010). Although the canonical pathologies of Alzheimer’s disease include tau and amyloid-β deposition, the disease is also associated with the pathological build-up of partially degraded protein in bloated lysosomes and autophagosomes (Nixon et al., 2005). While this so-called “autophagic pathology” is observed in multiple forms of the disease, it is accentuated by mutations in the transmembrane protein presenilin 1 (PS1), the catalytically active component of the ı-secretase complex (Cataldo et al., 2004). As PS1 mutation is a common cause of early-onset, inherited, familial Alzheimer’s disease (fAD), these autophagic defects may impact disease progression. The missense mutation A246E, one of the first PS1 mutations to be identified (Sherrington et al., 1995), is of particular relevance. While the protein with this point mutation is still capable cleaving amyloid precursor protein, it is associated both with elevated Aı42/40 ratio (Scheuner et al., 1996, Qian et al., 1998) and autophagic pathology (Lee et al., 2010). In addition, mice expressing the human A246E transgene show increased amyloid beta in the absence of plaques, as well as reduced performance (Lalonde et al., 2003). However, the mechanistic links between the mutation and these pathologies remain unclear.
The potential contribution of lysosomal alkalization to this impaired degradation is currently a matter of considerable interest. It has been suggested that the A246E mutation disrupts the trafficking of a v-(H+)ATPase subunit to lysosomes and that lysosomal pH is elevated in these mutant cells (Lee et al., 2010). However, others have been unable to detect a significant change in lysosomal pH using a variety of approaches (Neely et al., 2011, Coen et al., 2012, Zhang et al., 2012) or confirm a role for defective lysosomal pH in disease (Bezprozvanny, 2012). Given that accurate measurement of lysosomal pH is technically challenging, this discrepancy is understandable. However, we have spent the past decade developing a protocol that can accurately detect small changes in lysosomal pH. We have demonstrated the effects of elevated lysosomal pH in retinal pigmented epithelial (RPE) cells associated with Age-related Macular Degeneration (AMD) and have screened to identify treatments that can reacidify damaged lysosomes and reverse the accumulation of waste material (Liu et al., 2008, Baltazar et al., 2012, Guha et al., 2012, Liu et al., 2012, Guha et al., 2013). Alzheimer’s disease has many parallels with AMD, including the slow accumulation of incompletely degraded material in the lysosome and in lysosome-associated organelles of aging post-mitotic cells (Isas et al., 2010, Ding et al., 2011, Kaarniranta et al., 2011, Ohno-Matsui, 2011, Sivak, 2013). We thus applied our technique for accurate detection of lysosomal pH to skin fibroblasts from humans with the PS1-fAD mutation and found an elevation in lysosomal pH, a decrease in pH-dependent processing of cathepsin D and changes in molecular and protein markers.
2. Experimental procedures
2.1. Culture of human skin fibroblast cells
This study used two distinct sets of control (CTRL) and PS1-fAD (A246E) human skin fibroblasts from the NIA Aging Cell Culture Repository (Coriell, Camden, NJ): cell numbers AG6840 and AG08170 were from two different PS1-fAD donors and termed “PS1-fAD” cells, while numbers AG07621 and AG07623 were the control cells from unaffected spouses of AD patients (termed “CTRL” cells). Cells were grown to confluence in 25cm2 primary culture flasks in minimum essential Eagle’s medium (Sigma-Aldrich, St. Louis, MO) with 2mM GlutaMAX™, 100U/mL penicillin and 100μg/ml streptomycin and 15% fetal bovine serum (all Life Technologies Corp., Grand Island, NY). Cells were incubated at 37°C in 5.5% CO2 and sub-cultured by room temperature incubation in 0.53mM EDTA in HBSS (−Mg2+/−Ca2+), followed by incubation with 0.05% trypsin/0.48mM EDTA (Life Technologies) at 37°C. Culture protocols followed in accordance with those provided by the cell supplier. Data presented from PS1-fAD fibroblasts represent composite data from both AG08640 and AG08170 lineages; data for CTRL fibroblasts represent composite data from both AG07621 and AG07623 lineages. No clear differences were detected between cells from different lineages but the same PS1 genotype.
2.2. Measurement of lysosomal pH from fibroblast cells
Lysosomal pH was measured as described using the dye LysoSensor Yellow/Blue DND-160 (Liu et al., 2008, Baltazar et al., 2012, Guha et al., 2012, Liu et al., 2012). The use of LysoSensor Yellow/Blue (Life Technologies, Inc.) to measure lysosomal pH has the advantage, common to ratiometric dyes, that the readout is independent of concentration. Because the dye is membrane-permeable, readout is also representative of a broader range of lysosomes than those reached by endocytosis of a dextran-tagged probe. Extensive preliminary trials have optimized key experimental parameters including incubation time, dye concentration, etc. to minimize variation and give the best signal-to-noise ratio with the lowest concentration of dye (Liu et al., 2008). For example, all readings used for a given experimental set were performed simultaneously in 96-well plates. Incubation times were monitored precisely, and all measurements were taken within 12-14 min of washing off the dye. Calibrations were performed simultaneously in adjacent wells to ensure the relevance of the measures.
In brief, CTRL and PS1-fAD fibroblasts from unaffected spouse donors were grown to at least 80% confluence in black 96-well plates. Cells were assayed after a minimum of 6 days in culture. The two cell types were plated in alternating rows to control for any signal variation across the plate. To begin the assay, culture medium was removed and cells were incubated for 3 min with 2μM LysoSensor Yellow/Blue in isotonic solution (NaCl, 105mM; KCl, 5mM; HEPES-Acid, 6mM; Na-HEPES, 4mM; NaHCO3, 5mM; mannitol, 60mM; glucose, 5mM; MgCl2, 0.5mM; CaCl2, 1.3mM; pH, adjusted to 7.4; osmolality, 300mOsm). Dye loading and incubation steps were carried out at room temperature and in the dark. After 3 min, cells were rinsed 3× with isotonic solution and incubated with additional isotonic solution, with a relevant drug, or with pH calibration buffers. All final drug applications (200nM bafilomycin A1, 20mM NH4Cl, 30μM tamoxifen, cAMP cocktail (500 μM cpt-cAMP, 100μM IBMX, 10μM forskolin (forskolin from LC Laboratories, Woburn, MA)) were made up in isotonic solution. After 10 min, fluorescence was measured with a Fluoroskan Ascent Microplate Fluorometer and recorded using the ASCENT software package (both Thermo Scientific, Waltham, MA). Lysosomal pH was determined from the ratio of light excited at 340nm over 380nm (F340nm/F380nm, 527nm emission). Mean light levels at both emission wavelengths were integrated over 60 msec and recorded for each well in sequence; this course was repeated every 60 seconds for 11 iterations. pH data are the mean levels from 3-7 measurements taken 12-14 min after removal of the dye. In certain recordings, absolute pH levels were obtained by calibrating the lysosomal pH against standards; calibration wells were incubated with 15μM monensin and 30μM nigericin, each a proton-cation ionophore that permeabilizes the lysosomal membrane to Na+ and K+, respectively. These ionophores were added in a solution of 20mM MES (2-(N-Morpholino)ethanesulfonic acid), 110mM KCl and 20mM NaCl, at pH values of 4.0, 4.5, 5.0, 5.5 and 6.0. It should be noted that while the use of ionophores to calibrate fluorescent ratios into absolute values is relatively precise when working with a cytoplasmic readout such as with fura-2 (Grynkiewicz et al., 1985), the complexities of permeating both the plasma membrane and vesicular membranes can lead to small variations in the absolute levels. However, our approach of measuring output simultaneously from cells grown on a single plate, and then calibrating and normalizing to the mean control level for each plate, ensures that the differences observed between cell types and between treatment conditions are repeatable and reliable.
2.3. Confocal microscopy
CTRL and PS1-fAD fibroblasts were grown to near-confluence on glass coverslips. For LysoSensor/LysoTracker staining, coverslips were first rinsed 3× in warm isotonic solution, then co-incubated in fresh growth medium with 2μM LysoSensor Yellow/Blue and 50nM LysoTracker DND-99 for just over 30 min at 37°C. For BODIPY FL-pepstatin A staining, coverslips were rinsed and then co-incubated in fresh growth medium with 1ug/ml μM BODIPY FL-pepstatin A (Life Technologies) for 1h. Following incubation, coverslips were rinsed 3× times and then imaged on a warm stage using a Nikon A1R Laser Scanning Confocal Microscope and the University of Pennsylvania Live Cell Imaging Core. LysoSensor signal was excited at 407nm using the microscope’s 405nm Diode laser; emission was read at 450nm. The LysoTracker signal was excited at 562nm using the microscope’s DPSS laser; emission was read at 595nm. The BODIPY FL-pepstatin A signal was excited at 488nm using the microscope’s Argon laser; emission was read at 525nm. Image capture and analysis were performed using Nikon Elements Advanced Research Software package 3.2. Image capture on live cells was completed within 15min of completing dye incubation for LysoSensor/LysoTracker staining, and within 30min of completing dye incubation for BODIPY FL-pepstatin A.
2.4. Immunoblots
CTRL and PS1-fAD fibroblasts were grown to confluence in adjacent wells of 6-well plates, and assayed after a minimum of 6 days in culture. On the day of protein collection, existing medium was replaced with one of three media solutions: fresh medium (components as described above: minimum essential Eagle’s medium, fetal bovine serum, GlutaMAX, penicillin/streptomycin), fresh medium plus 10μM chloroquine (CTRL fibroblasts only), or fresh medium plus cAMP cocktail. Fibroblast cultures were incubated in these solutions for 6 hours at 37°C. Cells were rinsed 3× in DPBS (+Ca2+/+Mg2+)(Life Technologies). Total protein was extracted from fibroblasts using RIPA buffer (Sigma-Aldrich OR made in-house) plus Complete Mini Protease Inhibitor Cocktail Tablets (Roche Applied Science, Indianapolis, IN) according to standard protocols. Protein concentration in cellular extracts was quantified on the BioPhotometer spectrophotometer (Eppendorf AG, Hamburg, Germany) using the Pierce® BCA Protein Assay Kit (ThermoFisher Scientific).
Protein samples were prepared for agarose gel electrophoresis according to standard protocols and run at 130V on a 4-15% Mini-PROTEAN® TGX™ Gel with 50μL well depth (Bio-Rad, Hercules, CA). Gel-bound samples were then transferred to a PVDF membrane (EMD Millipore, Billerica, MA) for immunoblotting. A solution of 5% non-fat milk (Bio-Rad) was used for membrane blocking. To detect p62, the mouse mAb p62/SQSTM1 (D-3) was used (#sc-28359, Santa Cruz Biotechnology, Santa Cruz, CA). To detect LC3B, the rabbit mAb LC3B (D11) XP® was used (#3868S, Cell Signaling Technology, Beverly, MA). To detect vATPaseB2, the rabbit pAb Anti-ATP6V1B2 was used (#ab73404, Abcam, Cambridge, MA). To detect Atg5, the rabbit mAb Atg5 (D1G9) was used (#8540, Cell Signaling Technology). To detect Beclin-1, the rabbit mAb Beclin-1 (D40C5) was used (#3495P, Cell Signaling Technology). To detect GAPDH, the rabbit mAb GAPDH (14C10) was used (#2118S, Cell Signaling Technology). The antibody used to detect cathepsin D was a generous gift from Kathleen Boesze-Battaglia. All primary Abs were used at a dilution of 1:1000. Secondary Abs were used at a dilution of 1:3000 and consisted of either sheep anti-mouse (GE Healthcare Life Sciences, Amersham, UK) or donkey anti-rabbit (GE Healthcare Life Sciences) IgG as HRP-linked whole Ab for ECL chemiluminescence. Visualization and band quantification were performed on an ImageQuant LAS 4000 biomolecular imaging system (GE Healthcare Life Sciences, Pittsburgh, PA).
2.5. qPCR
Total RNA was isolated from cultured human skin fibroblast cells using the TRIzol (Life Technologies) extraction protocol, with glycogen (Life Technologies) added to improve yield. RNA yield was determined by absorbance at 260nm, and purity confirmed by measurement of 260nm/280nm ratio on Eppendorf Biophotometer Spectrophotometer. 1μg of total RNA was converted into cDNA using the High Capacity RNA-to-cDNA first strand synthesis kit (Applied Biosystems). Human primers were purchased from Life Technologies/Sigma-Aldrich and constructed according to specifications given in Table 1. qPCR was performed using the Power SYBR Green detector (Life Technologies) on a Life Technologies 7300 Real Time PCR System. Final primer concentration in each well was 1μM sense and 1μM antisense primer. The thermal cycling profile was as follows: Stage 1, 50°C for 2min (1 cycle); Stage 2, 95°C for 10 min (1 cycle); Stage 3, 95°C for 15s followed by 60°C for 1min (40 cycles). Expression levels of genes of interest were normalized internally to each sample’s expression of the housekeeping gene ACTB (β-actin). ACTB expression did not differ between CTRL and PS1-fAD fibroblasts across all experimental trials. All runs included a final dissociation stage to confirm amplification of only desired products. To control for genomic DNA contamination, PCR was also performed on samples from reverse transcriptase reactions in which the enzyme was omitted. No products were observed from these samples, indicating that no genomic DNA contaminated our experimental samples.
Table 1. qPCR Primers.
| Target | NCBI code | Primer Pair | Tm (C°) |
Product
Length (bp) |
|---|---|---|---|---|
| ACTB | NM_01101.3 |
Sense: AGAAAATCTGGCACCACACC Anti-Sense: GGGGTGTTGAAGGTCTCAAA |
59.97 59.94 |
142 |
| ATP6V1B2 | NM_001693.3 |
Sense: GAGGGGCAGATCTATGTGGA Anti-Sense: GCATGATCCTTCCTGGTCAT |
60.00 59.89 |
128 |
| ATG5 | NM_004849.2 |
Sense: GCAAGCCAGACAGGAAAAAG Anti-Sense: GACCTTCAGTGGTCCGGTAA |
59.99 59.97 |
137 |
| BECN1 | NM_003766.3 |
Sense: AGGTTGAGAAAGGCGAGACA Anti-Sense: GCIIIIGTCCACTGCTCCTC |
59.99 60.00 |
139 |
| TFEB | NM_001167827.2 |
Sense: GTCCGAGACCTATGGGAACA Anti-Sense: CGTCCAGACGCATAATGTTG |
58.52 57.28 |
218 |
2.6. Data analysis
All data are given as mean ± standard error of the mean. Significance was defined as p<0.05 and was determined using a one-way ANOVA followed by an appropriate post hoc test using SigmaPlot statistics software (v11.0, Systat Software, Inc., San Jose, CA) unless otherwise noted. When data were not normally distributed, ANOVAs were performed on ranks.
All chemicals and compounds used were from Sigma-Aldrich Corp., St. Louis, MO unless otherwise indicated.
3. Results
3.1. Lysosomal pH is elevated in PS1-fAD fibroblasts
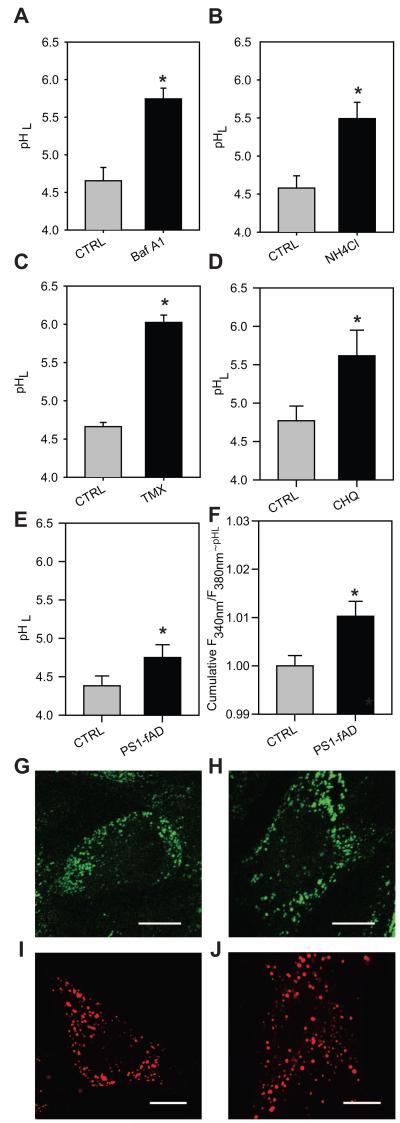
Initial experiments established the feasibility of performing reliable measurements of lysosomal pH from fibroblasts using our protocols. Fibroblasts derived from normal subjects (CTRL) were plated into black-walled 96-well plates and briefly incubated with different compounds known to increase pHL through varied mechanisms. These compounds included: 200nM bafilomycin A1, a specific inhibitor of v-(H+)ATPase (Bowman et al., 1988); 20mM NH4Cl, a lysosomotropic weak base (Ohkuma and Poole, 1978); 30μM tamoxifen, a tertiary amine whose pHL-elevating activity is distinct from its estrogen-like activity (Altan et al., 1999); and 10μM chloroquine, a lysosomotropic agent that becomes “trapped” in the lysosome following protonation (de Duve et al., 1974, Chung, 1986). A representative pH measurement is shown to illustrate the strong effect of each compound upon pHL (Fig. 1, panels A-D). Bafilomycin A1 (Fig. 1A), NH4Cl (Fig. 1B), tamoxifen (Fig. 1C), and chloroquine (Fig. 1D) led to a significant rise in lysosomal pH levels, as expected. While absolute level of baseline pHL varied somewhat, the baselines recorded were comparable to lysosomal pH levels in other cell types, and were well within the usual acidic range of the lysosomal enzymes. Importantly, the differences found within the same experimental trial were constant. These findings verified our ability to effectively measure pHL in these fibroblasts, and showed that the pHL could be manipulated using standard procedures and compounds.
Figure 1. PS1-fAD mutation yields elevated pHL in human skin fibroblasts.
A-D, Fifteen minute treatment with pHL-elevating compounds significantly increases lysosomal pH in fibroblasts from controls. A, Bafilomycin A1 (Baf A1) 200nM. B, NH4Cl 20mM. C, Tamoxifen (Tmx) 30μM. D, Chloroquine (CHQ) 10μM; for each n=6-14 wells. E, Lysosomal pH levels in CTRL vs. PS1-fAD fibroblasts, n= 10. F, Summary of baseline lysosomal pH levels across 26 separate experiments, expressed as uncalibrated ratios of fluorescence excited at 340 vs. 380 nm - ~ pHL (lysosomal pH). Each experiment from 7-10 wells of each genotype, total 222 CNTL and 224 PS1-fAD. All values normalized to the mean control for each experimental set. G-H, Confocal images of 2μM LysoSensor staining at ex/em 405/450nm for CTRL, G, and PS1-fAD fibroblasts, H. I-J, Confocal images of 50nM LysoTracker staining at ex/em 405/450nm for CTRL, I, and PS1-fAD fibroblasts, J. Scale bars = 10μm. Throughout, “ * ” signifies p <0.05 and CTRL = untreated control fibroblasts.
Subsequent experiments compared the baseline lysosomal pH levels between CTRL and PS1-fAD fibroblasts. The fibroblasts examined had the PS1-fAD missense mutation A246E, which is linked to both perturbed autophagy (Lee et al., 2010) and elevated Aı42/40 ratio (Scheuner et al., 1996, Qian et al., 1998). Fig. 1E illustrates a typical experiment, with CTRL fibroblasts at a pH of from 4.38 ± 0.13 and PS1-fAD fibroblasts at 4.75 ± 0.17. This small but significant elevation in lysosomal pH was confirmed in numerous trials from both sets of donor pairs. In a total of 26 individual trials, a significant elevation in lysosomal pH was detected in the PS1-fAD cells, as indicated by an increased emission ratio (Fig. 1F, the ratios were more reliable than calibration across double membranes - see Methods). This rise corresponds to an increase of approximately 0.2 pH units. No substantive difference was observed in autofluorescence between CTRL and PS1-fAD fibroblasts at wavelengths used for the pHL assay.
While fluorescent readings from the plate reader provided the most accurate comparison of lysosomal pH, it was important to examine the lysosomes microscopically. No major difference in overall LysoSensor staining patterns or fluorescence output from the two cell types was observed by confocal imaging (Figs. 1G-H). Similarly, no substantial differences were observed between CTRL and PS1-fAD cells using LysoTracker dye (Figs. 1I-J). With both dyes, the number and distribution of fluorescing organelles varied on a cell-to-cell basis. Bafilomycin treatment completely eliminated LysoTracker fluorescence for both cell types, confirming the readout.
3.2. Availability of active cathepsin D is reduced in PS1-fAD fibroblasts
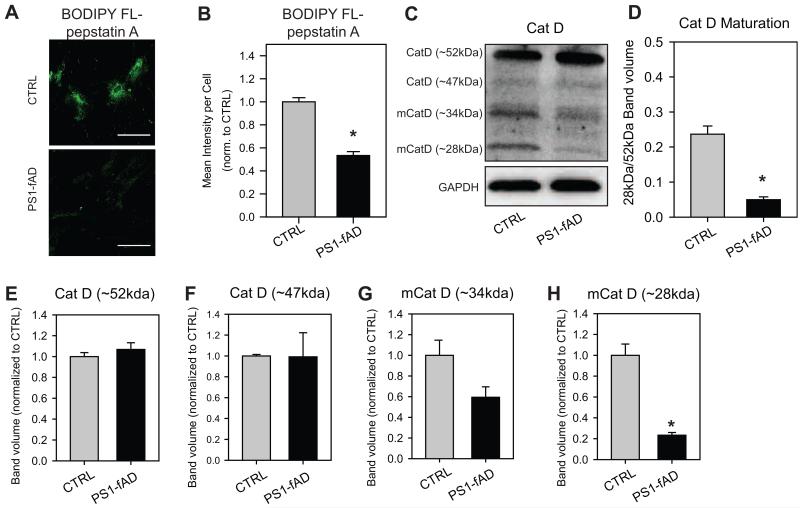
Cathepsin D, the primary aspartyl protease of the lysosome, has a particularly acidic pH optimum (Barrett, 1970) as well a pH-dependent maturation (Rosenfeld, 1982). It was therefore reasoned that the enzyme’s sharp tuning would make its activity and availability particularly sensitive to pH shifts over the range observed in the PS1-fAD fibroblasts. To this end, both CTRL and PS1-fAD fibroblasts were incubated with BODIPY FL-pepstatin A, which selectively binds to active cathepsin D (Chen et al., 2000), and examined under a confocal microscope. PS1-fAD cells exhibited markedly reduced BODIPY FL-pepstatin A fluorescence when compared against CTRL fibroblasts (Fig. 2A). When mean fluorescence intensity per cell was calculated for both CTRL and PS1-fAD fibroblasts, BODIPY FL-pepstatin A fluorescence was significantly reduced by approximately 50%, indicating a substantial loss in the availability of active cathepsin D with PS1-fAD mutation (Fig. 2B).
Figure 2. Cathepsin D active site availability and maturation reduced in PS1-fAD fibroblasts.
A, BODIPY FL-pepstatin A fluorescence is markedly reduced in PS1-fAD fibroblasts when compared to CTRL. Scale bar = 100μM. B, Mean intensity per cell analysis of BODIPY FL-pepstatin A fluorescence reveals a significant loss in fluorescence; n = 42 CTRL and 40 PS1-fAD cells. C, A sample Western blot showing reduction of mature cathepsin D in PS1-fAD fibroblasts. D, The ratio of 28kDa/52kDa cathepsin D is reduced in PS1-fAD fibroblasts. E. The level of 52kDa or, in F, 47kDa cathepsin D, is the same in PS1-fAD and control fibroblasts. The reduction in the mature 34kDa band approaches significance in G at p=0.07, while the mature 28kDa band is significantly reduced in PS1-fAD cells, H. For D-H, n = 3. Throughout, “ * ” signifies p <0.05 and CTRL = untreated control fibroblasts.
Having observed a significant reduction in active site availability by BODIPY FL-pepstatin A, cathepsin D levels in both CTRL and PS1-fAD fibroblasts were next examined by Western blot. The antibody detected four primary bands: one each at 52 and 47kDa, corresponding with immature/intermediate forms of cathepsin D (Gieselmann et al., 1983), and one each at 34 and 28kDa, corresponding with processed, mature forms of cathepsin D (Fig. 2C) (Erickson et al., 1981). The ratio of 28kDa/52kDa cathepsin D was found to be significantly reduced in PS1-fAD fibroblasts, indicating impaired maturation of cathepsin D in mutant cells (Fig. 2D). Importantly, while no significant changes were observed in the relative levels of either the 52 or 47kDa forms of cathepsin D (Fig. 2E,F), the reduction in levels of mature cathepsin D approached significance at 34kDa (Fig. 2G) and was significant at 28kDa when compared against CTRL fibroblasts (Fig. 2H). This indicated that the PS1-fAD cells have reduced maturation, and not initial production, of the protease.
Together, these data support the premise that even a small pH elevation has real consequences for the functionality of a major lysosomal enzyme, and suggest that PS1-fAD cells may have impaired degradation as a consequence of the loss of cathepsin D.
3.3 Altered levels of key proteins support elevated pHL
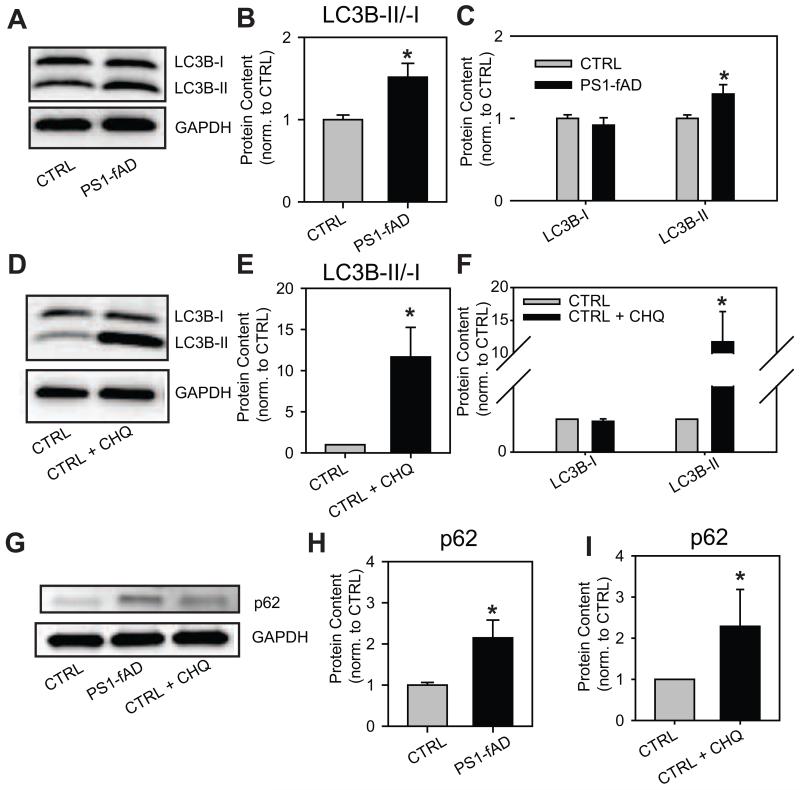
The finding that cathepsin D availability and maturation is markedly reduced in PS1-fAD fibroblasts (Fig. 2) suggested that the autophagic degradative system may be perturbed as a result of pH elevation, and prompted an examination of autophagic markers. To provide a general index of autophagic activity, the levels of the proteins LC3B and p62 were determined in control and mutant cell types. LC3B is an intracellular protein, the accumulation of which is often used to identify shifts in autophagic flux (Klionsky et al., 2012). Upon initiation of autophagy the cytosolic LC3B-I form of the protein is cleaved and conjugated to phosphatidylethanolamine to form LC3B-II, which associates with autophagosomal membranes. Elevation of lysosomal pH can lead to a backlog of autophagy and can raise the ratio of LC3B-II to LC3B-I, as previously shown in LS174 cells exposed to bafilomycin A1 (Bellot et al., 2009). The ratio of LC3B-II/-I was also elevated in PS1-fAD fibroblasts compared to CTRL (Fig. 3A, B). This increase reflected a significant elevation in the relative level of LC3B-II in PS1-fAD fibroblasts (Fig. 3,A, C). Chloroquine also induced a rise in LC3B-II/-I in CTRL fibroblasts (Fig. 3D, E) which, as with the PS1-fAD fibroblasts, reflected a significant change in LC3B-II (Fig. 3F). While qualitatively similar, the magnitude of this increase in LC3BII/I with chloroquine was considerably greater than that observed with PS1-fAD cells, consistent with the relative magnitude of lysosomal alkalinization under the two conditions.
Figure 3. Elevated pHL impairs autophagic degradation through the lysosomes.
A, A sample Western blot showing elevation of LC3B-II in PS1-fAD fibroblasts. B, LC3B-II/-I is elevated in PS1-fAD fibroblasts. C, LC3B-I is unchanged in PS1-fAD fibroblasts, while LC3B-II is significantly elevated. For B, C, n =15 CTRL, 16 PS1-fAD, 9 blots total. D, A sample Western blot showing elevation of LC3B-II in CTRL cells treated with 10μM chloroquine (CHQ) for 6 hours. E, LC3B-II/-I is elevated in CTRL fibroblasts treated with CHQ. F, LC3B-I is unchanged in CHQ-treated fibroblasts, while LC3B-II is significantly elevated. For E, F, n =5 CTRL, 5 CTRL + CHQ, 5 blots total. G, A sample Western blot showing elevation of p62 in PS1-fAD fibroblasts and in CTRL cells treated with 10μM CHQ for 6 hrs. H, p62 is elevated in PS1-fAD fibroblasts, (n =13 CTRL, 14 PS1-fAD, 8 blots total). I, p62 is elevated following 6h chloroquine incubation, n = 5 per condition, 5 blots total. Throughout, “ * ” signifies p <0.05 and CTRL = untreated control fibroblasts. Protein level of each protein was normalized to GAPDH as an internal control.
The increased lysosomal pH levels in PS1-fAD cells is also supported by changes in the levels of the autophagy-associated protein p62, also known as sequestosome-1. p62 itself is primarily degraded by autophagy (Bjorkoy et al., 2005), and therefore its elevation can be indicative of impaired autophagic degradation. Levels of p62 were increased more than two-fold in PS1-fAD cells as compared to control (Fig 3G,H). Chloroquine also triggered an increase in p62 levels in control fibroblasts (Fig. 3G,I). Together, elevation of the LC3B-II/-I ratio and of p62 provides support for an elevated lysosomal pH in PS1-fAD cells on a protein level, and implies functional consequences consistent with this rise, although the use of these two measures does not rule out other mechanisms by which autophagic degradation might be inhibited in PS1-fAD fibroblasts, such as through the mTOR signaling pathway.
3.4. PS1-fAD mutation results in altered expression of lysosome- and autophagy-associated genes
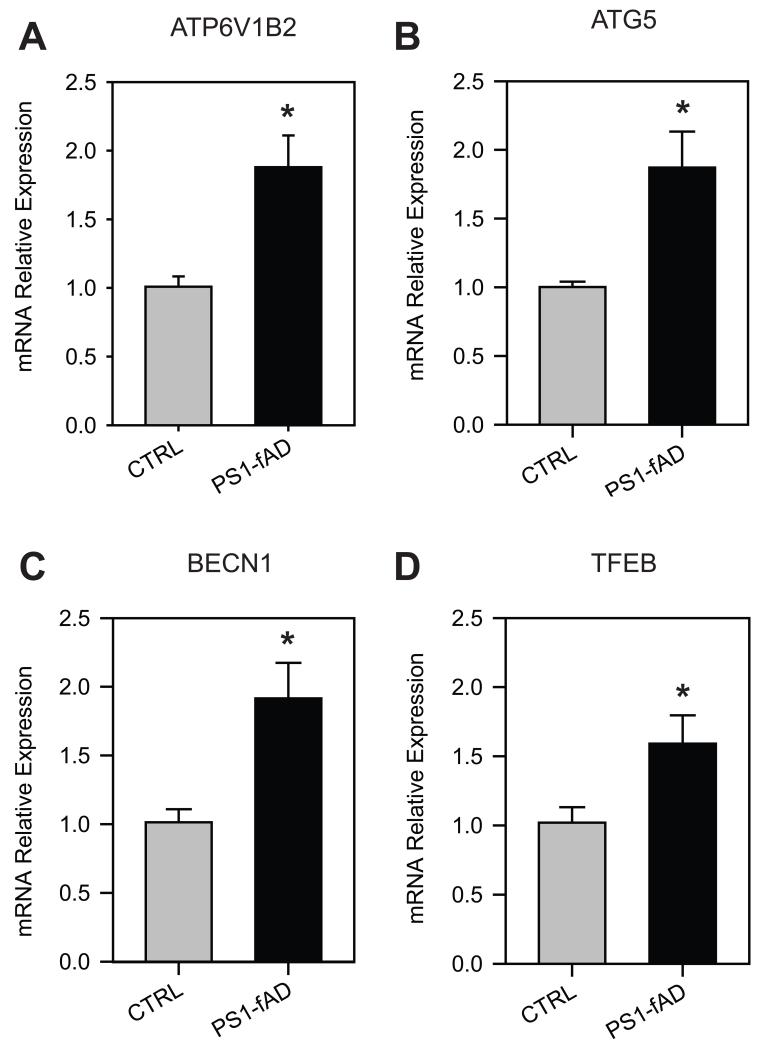
Additional support for a perturbed degradative system in PS1-fAD cells was sought at a molecular level by examining the expression of genes associated with lysosomal pH and autophagy. Lysosomal pH is regulated by a complex series of feedback systems, with pH elevation leading to alterations in mRNA message for certain key genes (Settembre et al., 2012). qPCR demonstrated that relative expression of the B2 subunit of the v-(H+)ATPase proton pump rose 89% in PS1-fAD fibroblasts (Fig. 4A). For ATG5, whose gene product is associated with autophagosome elongation, relative expression rose 87% in PS1-fAD fibroblasts (Fig. 4B). Similarly, expression of BCN1, whose product beclin is involved in the genesis of autophagosomes, was also increased by 92% in PS1-fAD fibroblasts (Fig. 4C). Finally, there was a significant increase in expression of TFEB, the transcription factor responsible for lysosomal biogenesis, which has also recently been identified as a link between lysosomal and autophagic processes (Settembre et al., 2011, Roczniak-Ferguson et al., 2012); relative expression rose 59% in PS1-fAD fibroblasts (Fig. 4D). All expression data were first normalized internally to the ubiquitously- and highly-expressed ı-actin (ACTB) message prior to comparison, as no difference in ACTB expression was detected between CTRL and PS1-fAD fibroblasts.
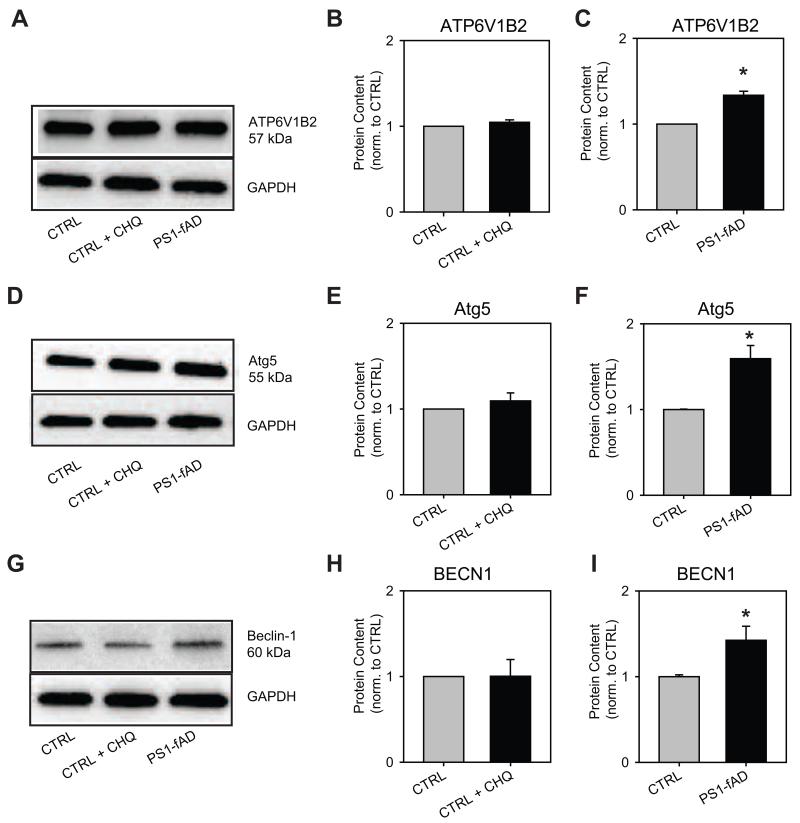
Figure 4. PS1-fAD fibroblasts exhibit altered gene expression profile.
Results from quantitative PCR experiments indicating the increased expression of genes in fibroblasts from the PS1-fAD cells as compared to unaffected controls. A, ATP6V1B2 expression elevated in PS1-fAD fibroblasts. B, ATG5 expression elevated in PS1-fAD fibroblasts. C, BECN1 expression elevated in PS1-fAD fibroblasts. D, TFEB expression elevated in PS1-fAD fibroblasts. n=4 independent trials for each. “ * ” signifies p <0.05 for entire figure. Expression level of each gene was first normalized to ACTB expression as an internal control.
3.5. Protein level shifts in PS1-fAD fibroblasts of lysosome- and autophagy-associated proteins mirror gene expression changes
While disruption of the lysosome- and autophagy-associated genes identified in Fig. 4 provided support for perturbed autophagy in PS1-fAD fibroblasts, validation at the protein level was desired to confirm the functional effect of the increased mRNA levels. To this end, the levels of vATPaseB2, Atg5, and beclin-1 were evaluated by Western blot. TFEB levels were not examined at this time, since the primary mechanism of TFEB’s action is through a nuclear translocation event (Settembre et al., 2012), and not purely through increased expression.
The protein level of vATPaseB2 in CTRL fibroblasts was found to be unaffected by 6h incubation with CHQ, but was significantly increased in PS1-fAD fibroblasts when compared against CTRL cells (Fig. 5A-C). Similar results were observed for the protein level of Atg5 (Fig. 5D-F) and for the level of beclin-1 (Fig. 5G-I): in these cases, as well, CHQ incubation proved insufficient to increase protein levels, but PS1-fAD mutation reliably produced this increase. Together, these data provide further support for a perturbation of the degradative system in PS1-fAD cells, while also highlighting a possible difference between the effect of acute and chronic lysosomal pH elevation upon that system.
Figure 5. PS1-fAD fibroblasts have increased levels of lysosome- and autophagy-associated proteins.
A, A sample Western blot showing elevation of vATPaseB2 (ATP6V1B2) in PS1-fAD fibroblasts, but not in CTRL cells treated with 10μM CHQ for 6 hrs. B, vATPaseB2 is not elevated following 6h CHQ incubation; n = 4 CTRL, 4 CHQ, 4 blots total. C, vATPaseB2 is elevated in PS1-fAD fibroblasts; n = 4 CTRL, 4 PS1-fAD, 4 blots total. D, A sample Western blot showing elevation of Atg5 in PS1-fAD fibroblasts, but not in CTRL cells treated with 10μM CHQ for 6 hrs. E, Atg5 is not elevated following 6h CHQ incubation; n = 5 CTRL, 5 CHQ, 5 blots total. F, Atg5 is elevated in PS1-fAD fibroblasts; n = 7 CTRL, 8 PS1-fAD, 6 blots total. G, A sample Western blot showing elevation of beclin-1 in PS1-fAD fibroblasts, but not in CTRL cells treated with 10μM CHQ for 6 hrs. H, Beclin-1 is not elevated following 6h CHQ incubation; n = 4 CTRL, 4 CHQ, 4 blots total. I, Beclin-1 is elevated in PS1-fAD fibroblasts; n = 8 CTRL, 8 PS1-fAD, 6 blots total. Throughout, “ * ” signifies p <0.05 and CTRL = untreated control fibroblasts. Protein level of each protein was normalized to GAPDH as an internal control.
3.6. Intracellular cAMP elevation re-acidifies lysosomes and reduces LC3B accumulation
Since pH measurement, protein level, and gene expression data all support the conclusion that the lysosomal pH is defective in the PS1-fAD fibroblasts, attempts were made to restore pH using the intracellular signaling molecule cAMP. Previous work from our laboratory has demonstrated that intracellular elevation of cAMP can partially restore pHL that has been elevated by either pathological or by pharmacological means (Liu et al., 2008, Guha et al., 2012). Importantly, cAMP re-acidified RPE lysosomes in cells from old mice whose lysosomes had been damaged for an extended time, suggesting the approach might also restore acidity to lysosomes in the PS1-fAD fibroblasts. cAMP also has been shown to promote acidification in a variety of contexts and across a range of cell types (Alzamora et al., 2010, Paunescu et al., 2010).
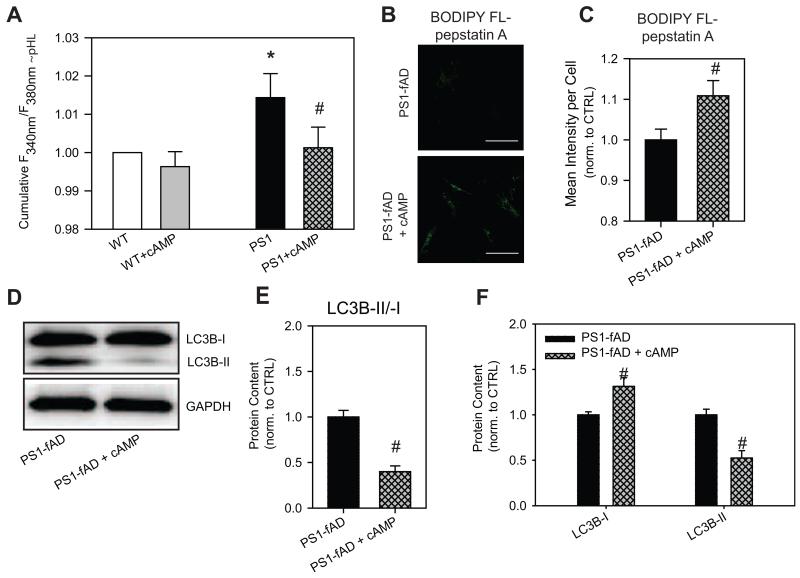
Increased intracellular cAMP levels reduced pHL in PS1-fAD fibroblasts (Fig. 6A). A cAMP-elevating cocktail, composed of cell-permeable cpt-cAMP, IBMX, and forskolin, consistently restored pHL in the mutant fibroblasts. The cAMP mix led to a small but insignificant drop in the lysosomal pH of control cells. Interestingly, preliminary data indicate that cAMP induced a larger acidification of lysosomes from control fibroblasts whose lysosomes had been alkalized either with NH4Cl or with tamoxifen. The re-acidifying effect of cAMP was also larger in epithelial cells with alkalized lysosomes (Liu et al., 2012), consistent with the findings in fibroblasts.
Figure 6. cAMP restores pHL and improves clearance in PS1-fAD fibroblasts.
A, Treatment of PS1-fAD cells with cAMP reduced lysosomal pH levels back to control levels. Treatment of control cells with the cocktail had a minimal effect. LysoSensor ratios normalized to mean control of each day’s experiment. N = 14 plates with 7-10 wells each. cAMP cocktail = 500μM cpt-cAMP + 100μM IBMX + 10μM forksolin. * = p <0.05 vs control, # = p <0.05 vs PS1-fAD alone. B, BODIPY FL-pepstatin A fluorescence is increased in PS1-fAD fibroblasts treated with cAMP cocktail. Scale bar = 100μM. C, Mean intensity per cell analysis of BODIPY FL-pepstatin A fluorescence reveals a significant increase in fluorescence; n = 60 PS1-fAD and 81 PS1-fAD cells. D, Western blot showing incubation with cAMP cocktail decreases LC3B-II/-I in PS1-fAD fibroblasts. E, Summary of 6 Western blots showing a significant decrease in LC3B-II/-I in PS1-fAD fibroblasts treated with the cAMP cocktail. F, LC3B-I is significantly increased in cAMP-treated fibroblasts, while LC3B-II is significantly reduced. For E, F, n = 11 PS1-fAD, 8 PS1-fAD + cAMP, 6 blots total. # = p < 0.05 for the entire figure.
To validate the functional implications of lysosomal acidification of PS1-fAD fibroblasts by cAMP, the consequences of cAMP-induced pHL restoration on both cathepsin D active site availability and on autophagy were examined. PS1-fAD cells treated with cAMP cocktail had greater BODIPY FL-pepstatin A fluorescence when compared against untreated PS1-fAD fibroblasts (Fig. 6B). When mean fluorescence intensity per cell was calculated for both treated and untreated PS1-fAD fibroblasts, BODIPY FL-pepstatin A fluorescence was significantly increased by about 10% in treated fibroblasts, indicating that 6h cAMP treatment and pH restoration brings about at least partial recovery of active cathepsin D (Fig. 6C). Still, even partial restoration of enzyme activity proved sufficient to improve clearance through the autophagosomal degradation pathway; incubation with the cAMP cocktail reduced the LC3B-II/I ratio by 60% (Fig. 6D, E), and significantly decreased LC3B-II levels while increasing LC3B-I (Fig. 6D, F). As cAMP interferes with p62 independently through the ubiquitin-proteasome degradative pathway (Myeku et al., 2012), an examination of p62 levels was not pursued here.
4. Discussion
In this study, human skin fibroblasts containing the PS1-fAD mutation A246E were found to exhibit elevated lysosomal pH (Fig. 1), reduced availability of active cathepsin D and reduced cleavage to the mature form of the enzyme (Fig. 2), and also impaired degradation of autophagic substrates (Fig. 3) as compared to levels from control fibroblasts. Substantive increases in expression of genes associated with lysosomal and autophagic degradative machineries were also detected in PS1-fAD fibroblasts (Fig. 4), with increases mirrored by protein levels (Fig. 5). Finally, lysosomal acidification was restored by elevation of intracellular cAMP, leading to a partial restoration of cathepsin D active site availability, as well as reduction in the LC3B-II/-I ratio (Fig. 6), Together, these changes indicate a small but significant steady-state dysfunction in lysosomal pH and degradation as a consequence of the A246E PS1-fAD mutation, and offer some clues as to how to address this problem therapeutically.
4.1. Elevation of lysosomal pH
The concurrence of effects on pH, cathepsin D, protein and gene levels strongly support the conclusion that lysosomes in PS1-fAD fibroblasts are alkalized as compared to controls. Our protocol to measure lysosomal pH enables the accurate detection of small changes in this pH. While the elevation of lysosomal pH by 0.2 units may appear small, this change occurs across the sharpest part of the pH/activity curve for many lysosomal enzymes, where even a minimal rise in pHL is sufficient to depress enzymatic activity and slow down degradation of cellular materials (Barrett, 1970). A pHL elevation of only 0.2 units was recently shown to impair proteolysis in macrophage phagosomes (Jiang et al., 2012), confirming that modest alkalizations are capable of inducing pathological changes. The decreased cleavage of cathepsin D into the mature form and the reduction in binding of BODIPY FL-pepstatin A to the active site of this major lysosomal protease are consistent with functional consequences of this lysosomal alkalization. This deficit is likely small enough that most cellular processes can progress unhindered under normal circumstances. However, the gradual accumulation of improperly processed material may decrease efficient cellular function over time. This slow accumulation is predicted to be more severe in active post-mitotic cells like neurons, which rely upon high baseline levels of autophagic degradation as a primary clearance mechanism (Boland et al., 2008). Given the differences between acute modulations of autophagy and the chronic changes in PS1-fAD fibroblasts, however, the situation is likely to be complex, and differences in the magnitude of the LC3 response in PS1 cells compared to control and those treated with CHQ are not unreasonable. Of interest in this regard is a manuscript demonstrating that chronic treatment by chloroquine and NH4Cl, but not 3-MA, leads to increased p62 and LC3B-ll/LC3B-l in rat cortical neurons (Myeku and Figueiredo-Pereira, 2011). Whether that result reflects the short-term actions of 3-MA (Wu et al., 2010), or merely different feedback systems activated by lysosomal alkalization, is not clear.
While this modest alkalization may have functional consequences over time, the small magnitude of the change may explain why this difference has been so difficult to detect. Measuring lysosomal pH is a difficult task under ideal conditions, and single-wavelength assays such as LysoTracker Red may not be sensitive enough to detect such small changes (Neely et al., 2011, Coen et al., 2012). Similarly, while the microscopy-based assay for measuring pHL used by Zhang et al. (2012) may enable specific localization of acidic organelles, its readout of average vesicular pH apparently yields a value far higher, both in WT- and PS1k/PSdko-ES, than would be found in functional lysosomes, and therefore the utility of such an approach to address the question of lysosomal pH is rather unclear. Our assay has been designed to minimize variation and detect small differences in vesicular pH; the use of a membrane-permeable pH probe may also enable access to different compartments than dextran-conjugated probes, the readout of which is necessarily restricted to that lysosomal sub-population undergoing fusion with endosomes, and thus may not be indicative of compromised lysosomal subpopulations. In particular, the concordance of our direct pH measurement data with the more indirect BODIPY FL-pepstatin A data, as well as with the immunoblots showing impaired maturation of cathepsin D, provides strong support for the accuracy of our approach.
This study does not identify the ultimate source of pHL dysregulation in these fibroblast culture types. It may indeed result from improper trafficking of v-(H+)ATPase due to a defect in the v0a1 subunit, as the Nixon laboratory has suggested (Lee et al., 2010), or perhaps from some other underlying defect or defects in the autophagic or endo-lysosomal axes. Our qPCR results indicating an increase in message for the proton pump suggest a compensatory mechanism is being attempted. It is important to note that our qPCR probes were designed to amplify ATP6V1B2, the B2 subunit of the proton pump, which is the more ubiquitously expressed variant of the B subunit of v-(H+)ATPase (Puopolo et al., 1992, van Hille et al., 1994). No issues with this subunit have been identified thus far in AD.
The gene expression and protein data are generally consistent with feedback attempts to compensate for a perturbed lysosomal pH. The fact that a relatively short-term incubation with CHQ is insufficient to induce the protein increases observed in PS1-fAD suggests, too, that these fibroblasts may initially attempt to manage pH increases by some other means than increased autophagic initiation. A sustained, life-long increase in pH, however, may differentially affect the degradative system along the entirety of its axis. Interestingly, BCN1 expression is also increased in fibroblasts from patients with certain storage diseases (Pacheco et al., 2007), further substantiating the similarities observed between primary lysosomal disorders and Alzheimer’s disease (Nixon, 2004, Nixon et al., 2008), and suggesting that up-regulation of autophagy in response to accumulated material, while perhaps a conserved strategy, may not be effective in all cases. The small-but-significant up-regulation of TFEB in PS1-fAD fibroblasts is particularly intriguing, given that Zhang et al. (2012) reported no difference from wild type in the expression of TFEB while investigating gene expression profiles of PS1ko and PSdko-ES cells, as well as PScdko mice. The group also saw no change in the expression of ATG5 and BCN1 in any of these PSko models. These discrepancies suggest that cells containing mutated, but otherwise intact, PS1 exhibit a differential expression profile from cells that lack PS1 and/or PS2 all together; some caution should therefore be observed in conflating and comparing results obtained through the use of a variety of models and techniques. Together, the elevated expression and protein levels of these genes critical for autophagy and lysosomal acidification provide additional evidence for perturbed lysosomal pH and, moreover, may be indicative of a degradative system attempting to compensate for a chronic defect at the lysosomal pH level.
Regardless, the major difference between these control and PS1-fAD fibroblasts is the A246E mutation in presenilin 1, and as multiple groups have implicated autophagic disruption in Alzheimer’s disease, the evidence points toward a basic deficit in cells with the Alzheimer’s disease-associated PS1 mutation that predisposes the cells toward a phenotype of accelerated aging, including lysosomal failure and protein buildup.
4.2. Re-acidification by cAMP
Importantly, our data show that re-acidifying lysosomes via elevated intracellular cAMP can help to restore the lysosomal pH and autophagic turnover associated with the PS1-fAD (A246E) mutation. The mechanism by which cAMP may do this in these fibroblasts is currently unknown, though similar acidifying processes have been linked to protein kinase A (PKA) activity in other cells (Liu et al., 2008, Alzamora et al., 2010, Paunescu et al., 2010) and may involve the activation of a Cl− channel (Liu et al., 2012). Another intriguing mechanistic possibility for cAMP’s effect on pHL could involve PKA-mediated inhibition of glycogen synthase kinase 3 (GSK3) (Fang et al., 2000). While the activity of both the ı and ı isoforms of GSK3 has been associated with AD pathology (Cho and Johnson, 2003, Phiel et al., 2003), a number of recent studies have also identified links between GSK3 and lysosomal dysfunction, whether pharmacologically- or pathologically-induced (Dobrowolski et al., 2012, Parr et al., 2012, Avrahami et al., 2013). GSK3 inhibition reportedly restores lysosomal acidification defects in both the 5XFAD mouse model and in cells lacking PS1/2 (Avrahami et al., 2013) and also improves amyloid pathology (Parr et al., 2012, Avrahami et al., 2013). Interestingly, low PKA activity has been reported in brain tissue from AD patients (Liang et al., 2007), consistent with a reduction in regulated lysosomal acidification. Augmenting this system by increasing intracellular cAMP levels and re-acidifying lysosomes may help to enhance clearance in older neurons and prevent additional consequences of lysosomal defect, such as perturbed calcium signaling (Christensen et al., 2002). The ability of cAMP to reverse the rise in LC3B-II/-I ratio seen in the PS1-fAD mutant cells also supports the central role of lysosomal pH in controlling this ratio. In particular, the fact that cAMP simultaneously increases LC3B-I while decreasing LC3B-II points its considerable power to affect autophagy and autophagic clearance, perhaps through some more fundamental effect on the system than pH alone. Intriguingly, the effect of short-term cAMP treatment apparently reduces LC3B-II to a level below the control baseline; we suspect that this may be due to differences between chronic and acute effects on autophagic states. By contrast, the reduction of baseline pHL observed in control fibroblasts was minimal and not significant. Preliminary results suggest, however, that cAMP lowers lysosomal pH in control fibroblasts when that pH is artificially elevated, which is consistent with the idea that the cAMP cocktail proves most effective when pHL is perturbed from baseline.
Overall, this study suggests that small deficits in autophagic degradation, linked to improper maintenance of lysosomal pH, may contribute to pathologic build-up of protein in PS1-fAD fibroblasts, and that lysosomal re-acidification may offer a strategy by which this accumulation can be ameliorated. In other words: regardless of whether lysosomal failure is the precipitating event in disease, or simply a consequence of PS1-mutation-associated phenotypes, re-acidification could equalize the cellular playing field.
Highlights.
Fibroblasts with the Alzheimer’s-associated PS1 mutation A246E have elevated lysosomal pH
PS1-fAD fibroblasts with reduction in pH-dependent cathepsin D maturation and active site access
Increased lysosomal pH correlates with accumulation of proteins normally degraded by autophagy
PS1-fAD fibroblasts up-regulate expression of genes and proteins linked to lysosomal pH
Raising intracellular cAMP lowers lysosomal pH, reduces protein buildup in PS1-fAD fibroblasts
Acknowledgements
We thank Wennan Lu, Jason Lim, Ann O’Brien Jenkins and Gabriel Baltazar for training and support, and Kathleen Boesze-Battaglia for help with the cathepsin D immunoblots. These experiments were supported by the Hearst Foundation Fellowship (EEC), National Institutes of Health through grants EY013434, EY015537 Vision Research Core Grant EY001583 (CHM, AML), Research to Prevent Blindness (AML), and the Foundation Fighting Blindness (AML)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work has been previously presented in abstract form (Coffey et al., 2013).
References
- Altan N, Chen Y, Schindler M, Simon SM. Tamoxifen inhibits acidification in cells independent of the estrogen receptor. Proc Natl Acad Sci U S A. 1999;96:4432–4437. doi: 10.1073/pnas.96.8.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzamora R, Thali RF, Gong F, Smolak C, Li H, Baty CJ, Bertrand CA, Auchli Y, Brunisholz RA, Neumann D, Hallows KR, Pastor-Soler NM. PKA regulates vacuolar H+-ATPase localization and activity via direct phosphorylation of the a subunit in kidney cells. J Biol Chem. 2010;285:24676–24685. doi: 10.1074/jbc.M110.106278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameis D, Merkel M, Eckerskorn C, Greten H. Purification, characterization and molecular cloning of human hepatic lysosomal acid lipase. Eur J Biochem. 1994;219:905–914. doi: 10.1111/j.1432-1033.1994.tb18572.x. [DOI] [PubMed] [Google Scholar]
- Avrahami L, Farfara D, Shaham-Kol M, Vassar R, Frenkel D, Eldar-Finkelman H. Inhibition of glycogen synthase kinase-3 ameliorates beta-amyloid pathology and restores lysosomal acidification and mammalian target of rapamycin activity in the Alzheimer disease mouse model: in vivo and in vitro studies. J Biol Chem. 2013;288:1295–1306. doi: 10.1074/jbc.M112.409250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltazar GC, Guha S, Lu W, Lim J, Boesze-Battaglia K, Laties AM, Tyagi P, Kompella UB, Mitchell CH. Acidic nanoparticles are trafficked to lysosomes and restore an acidic lysosomal pH and degradative function to compromised ARPE-19 cells. PLoS One. 2012;7:e49635. doi: 10.1371/journal.pone.0049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ. Cathepsin D. Purification of isoenzymes from human and chicken liver. Biochem J. 1970;117:601–607. doi: 10.1042/bj1170601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ. A new assay for cathepsin B1 and other thiol proteinases. Anal Biochem. 1972;47:280–293. doi: 10.1016/0003-2697(72)90302-8. [DOI] [PubMed] [Google Scholar]
- Barrett AJ. Human cathepsin B1. Purification and some properties of the enzyme. Biochem J. 1973;131:809–822. doi: 10.1042/bj1310809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I. Presenilins: a novel link between intracellular calcium signaling and lysosomal function? J Cell Biol. 2012;198:7–10. doi: 10.1083/jcb.201206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Properties of the endosomal-lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer’s disease. J Neurosci. 1996;16:186–199. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Nixon RA. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci U S A. 1990;87:3861–3865. doi: 10.1073/pnas.87.10.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Schmidt SD, Terio NB, Duff K, Beard M, Mathews PM, Nixon RA. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol. 2004;63:821–830. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]
- Chen C-S, Chen W-NU, Zhou M, Arttamangkul S, Haugland RP. Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. Journal of Biochemical and Biophysical Methods. 2000;42:137–151. doi: 10.1016/s0165-022x(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Cheung ZH, Ip NY. The emerging role of autophagy in Parkinson’s disease. Mol Brain. 2009;2:29. doi: 10.1186/1756-6606-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Johnson GV. Glycogen synthase kinase 3beta phosphorylates tau at both primed and unprimed sites. Differential impact on microtubule binding. J Biol Chem. 2003;278:187–193. doi: 10.1074/jbc.M206236200. [DOI] [PubMed] [Google Scholar]
- Christensen KA, Myers JT, Swanson JA. pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci. 2002;115:599–607. doi: 10.1242/jcs.115.3.599. [DOI] [PubMed] [Google Scholar]
- Chung CH. pH-Dependent Inhibition of Cathepsin B1 by Chloroquine. Korean Biochem J. 1986;19:93–98. [Google Scholar]
- Coen K, Flannagan RS, Baron S, Carraro-Lacroix LR, Wang D, Vermeire W, Michiels C, Munck S, Baert V, Sugita S, Wuytack F, Hiesinger PR, Grinstein S, Annaert W. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J Cell Biol. 2012;198:23–35. doi: 10.1083/jcb.201201076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey EE, Beckel JM, Laties AM, Mitchell CH. Elevated lysosomal pH and autophagy dysfunction in human fibroblasts bearing the Alzheimer’s-associated presenilin 1 A246E mutation can be ameliorated with cAMP; Society for Neuroscience Annual Meeting 525.01; 2013; Abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- Ding JD, Johnson LV, Herrmann R, Farsiu S, Smith SG, Groelle M, Mace BE, Sullivan P, Jamison JA, Kelly U, Harrabi O, Bollini SS, Dilley J, Kobayashi D, Kuang B, Li W, Pons J, Lin JC, Bowes Rickman C. Anti-amyloid therapy protects against retinal pigmented epithelium damage and vision loss in a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2011;108:E279–287. doi: 10.1073/pnas.1100901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski R, Vick P, Ploper D, Gumper I, Snitkin H, Sabatini DD, De Robertis EM. Presenilin deficiency or lysosomal inhibition enhances Wnt signaling through relocalization of GSK3 to the late-endosomal compartment. Cell Rep. 2012;2:1316–1328. doi: 10.1016/j.celrep.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrick MJ, Lieberman AP. Autophagic dysfunction in a lysosomal storage disorder due to impaired proteolysis. Autophagy. 2013;9:234–235. doi: 10.4161/auto.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AH, Conner GE, Blobel G. Biosynthesis of a lysosomal enzyme. Partial structure of two transient and functionally distinct NH2-terminal sequences in cathepsin D. J Biol Chem. 1981;256:11224–11231. [PubMed] [Google Scholar]
- Fang X, Yu SX, Lu Y, Bast RC, Jr., Woodgett JR, Mills GB. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci U S A. 2000;97:11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann V, Pohlmann R, Hasilik A, Von Figura K. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J Cell Biol. 1983;97:1–5. doi: 10.1083/jcb.97.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Guha S, Baltazar GC, Coffey EE, Tu L-A, Lim JC, Beckel JM, Eysteinsson T, Lu W, O’Brien-Jenkins A, Patel S, Laties AM, Mitchell CH. Lysosomal alkalinization, lipid oxidation, impaired autophagy and reduced phagosome clearance triggered by P2X7 receptor activation in retinal pigmented epithelial cells. Faseb J. 2013;27:4500–4509. doi: 10.1096/fj.13-236166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Baltazar GC, Tu LA, Liu J, Lim JC, Lu W, Argall A, Boesze-Battaglia K, Laties AM, Mitchell CH. Stimulation of the D5 dopamine receptor acidifies the lysosomal pH of retinal pigmented epithelial cells and decreases accumulation of autofluorescent photoreceptor debris. J Neurochem. 2012;122:823–833. doi: 10.1111/j.1471-4159.2012.07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isas JM, Luibl V, Johnson LV, Kayed R, Wetzel R, Glabe CG, Langen R, Chen J. Soluble and mature amyloid fibrils in drusen deposits. Invest Ophthalmol Vis Sci. 2010;51:1304–1310. doi: 10.1167/iovs.09-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Salao K, Li H, Rybicka JM, Yates RM, Luo XW, Shi XX, Kuffner T, Tsai VW, Husaini Y, Wu L, Brown DA, Grewal T, Brown LJ, Curmi PM, Breit SN. Intracellular chloride channel protein CLIC1 regulates macrophage function through modulation of phagosomal acidification. J Cell Sci. 2012;125:5479–5488. doi: 10.1242/jcs.110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K, Salminen A, Haapasalo A, Soininen H, Hiltunen M. Age-related macular degeneration (AMD): Alzheimer’s disease in the eye? J Alzheimers Dis. 2011;24:615–631. doi: 10.3233/JAD-2011-101908. [DOI] [PubMed] [Google Scholar]
- Khurana V, Elson-Schwab I, Fulga TA, Sharp KA, Loewen CA, Mulkearns E, Tyynelä J, Scherzer CR, Feany MB. Lysosomal dysfunction promotes cleavage and neurotoxicity of tau in vivo. 2010;6:e1001026. doi: 10.1371/journal.pgen.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Lalonde R, Qian S, Strazielle C. Transgenic mice expressing the PS1-A246E mutation: effects on spatial learning, exploration, anxiety, and motor coordination. Behav Brain Res. 2003;138:71–79. doi: 10.1016/s0166-4328(02)00230-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Down-regulation of cAMP-dependent protein kinase by over-activated calpain in Alzheimer disease brain. J Neurochem. 2007;103:2462–2470. doi: 10.1111/j.1471-4159.2007.04942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lu W, Guha S, Baltazar GC, Coffey EE, Laties AM, Rubenstein RC, Reenstra WW, Mitchell CH. Cystic fibrosis transmembrane conductance regulator contributes to reacidification of alkalinized lysosomes in RPE cells. Am J Physiol Cell Physiol. 2012;303:C160–169. doi: 10.1152/ajpcell.00278.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lu W, Reigada D, Nguyen J, Laties AM, Mitchell CH. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(−/−) mice: pharmacologic approaches and functional recovery. Invest Ophthalmol Vis Sci. 2008;49:772–780. doi: 10.1167/iovs.07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N, Figueiredo-Pereira ME. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J Biol Chem. 2011;286:22426–22440. doi: 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N, Wang H, Figueiredo-Pereira ME. cAMP stimulates the ubiquitin/proteasome pathway in rat spinal cord neurons. Neurosci Lett. 2012;527:126–131. doi: 10.1016/j.neulet.2012.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KM, Green KN, LaFerla FM. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a gamma-secretase-independent manner. J Neurosci. 2011;31:2781–2791. doi: 10.1523/JNEUROSCI.5156-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. Niemann-Pick Type C disease and Alzheimer’s disease: the APP-endosome connection fattens up. Am J Pathol. 2004;164:757–761. doi: 10.1016/S0002-9440(10)63163-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Cataldo AM. Lysosomal system pathways: genes to neurodegeneration in Alzheimer’s disease. J Alzheimers Dis. 2006;9:277–289. doi: 10.3233/jad-2006-9s331. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog Retin Eye Res. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Pacheco CD, Kunkel R, Lieberman AP. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- Parr C, Carzaniga R, Gentleman SM, Van Leuven F, Walter J, Sastre M. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol Cell Biol. 2012;32:4410–4418. doi: 10.1128/MCB.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunescu TG, Ljubojevic M, Russo LM, Winter C, McLaughlin MM, Wagner CA, Breton S, Brown D. cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells. Am J Physiol Renal Physiol. 2010;298:F643–654. doi: 10.1152/ajprenal.00584.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phiel CJ, Wilson CA, Lee VM, Klein PS. GSK-3alpha regulates production of Alzheimer’s disease amyloid-beta peptides. Nature. 2003;423:435–439. doi: 10.1038/nature01640. [DOI] [PubMed] [Google Scholar]
- Puopolo K, Kumamoto C, Adachi I, Magner R, Forgac M. Differential expression of the “B” subunit of the vacuolar H(+)-ATPase in bovine tissues. J Biol Chem. 1992;267:3696–3706. [PubMed] [Google Scholar]
- Qian S, Jiang P, Guan XM, Singh G, Trumbauer ME, Yu H, Chen HY, Van de Ploeg LH, Zheng H. Mutant human presenilin 1 protects presenilin 1 null mouse against embryonic lethality and elevates Abeta1-42/43 expression. Neuron. 1998;20:611–617. doi: 10.1016/s0896-6273(00)80999-x. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M. Biosynthesis of lysosomal hydrolases: their synthesis in bound polysomes and the role of co- and post-translational processing in determining their subcellular distribution. The Journal of Cell Biology. 1982;93:135–143. doi: 10.1083/jcb.93.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- Schwartz W, Bird JW. Degradation of myofibrillar proteins by cathepsins B and D. Biochem J. 1977;167:811–820. doi: 10.1042/bj1670811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HA, Haines JL, Perkicak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Sivak JM. The aging eye: common degenerative mechanisms between the Alzheimer’s brain and retinal disease. Invest Ophthalmol Vis Sci. 2013;54:871–880. doi: 10.1167/iovs.12-10827. [DOI] [PubMed] [Google Scholar]
- van Hille B, Richener H, Schmid P, Puettner I, Green JR, Bilbe G. Heterogeneity of vacuolar H(+)-ATPase: differential expression of two human subunit B isoforms. Biochem J. 1994;303(Pt 1):191–198. doi: 10.1042/bj3030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, Brown S, O’Kane CJ, Rubinsztein DC. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garbett K, Veeraraghavalu K, Wilburn B, Gilmore R, Mirnics K, Sisodia SS. A role for presenilins in autophagy revisited: normal acidification of lysosomes in cells lacking PSEN1 and PSEN2. J Neurosci. 2012;32:8633–8648. doi: 10.1523/JNEUROSCI.0556-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]