SUMMARY
Parkinson’s disease (PD) is characterized by loss of A9 dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc). An association has been reported between PD and exposure to mitochondrial toxins, including environmental pesticides paraquat, maneb, and rotenone. Here, using a robust, patient-derived stem cell model of PD allowing comparison of A53T α-synuclein (α-syn) mutant cells and isogenic mutation-corrected controls, we identify mitochondrial toxin-induced perturbations in A53T α-syn A9 DA neurons (hNs). We report a pathway whereby basal and toxin-induced nitrosative/oxidative stress results in S-nitrosylation of transcription factor MEF2C in A53T hNs compared to corrected controls. This redox reaction inhibits the MEF2C-PGC1α transcriptional network, contributing to mitochondrial dysfunction and apoptotic cell death. Our data provide mechanistic insight into gene-environmental interaction (GxE) in the pathogenesis of PD. Furthermore, using small-molecule high-throughput screening, we identify the MEF2C-PGC1α pathway as a therapeutic target to combat PD.
INTRODUCTION
Initial motor symptoms in Parkinson’s disease (PD) result from loss of A9-type dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc). Considerable damage occurs before onset of clinical symptoms, making identification of early events a challenge. Although the cause of sporadic PD is not fully understood, various factors, including environmental toxins, have been implicated. Mitochondrial toxins have been identified in epidemiological studies as contributing to “sporadic” PD, and mitochondrial-based toxin models gained attention following the discovery of MPTP-induced Parkinsonism (Langston et al., 1983). Paraquat (PQ; 1, 1′-dimethyl-4,4′-bipyridinium), a commonly used herbicide, shares structural similarity with MPP+, the active metabolite of MPTP. PQ crosses the blood-brain barrier, generates reactive oxygen and nitrogen species (ROS/RNS) and causes loss of SNpc DA neurons in animal models (Shimizu et al., 2001; Bonneh-Barkay et al., 2005; Morán et al., 2010). Additional pesticides, including the fungicide maneb (MB; manganese ethylnebisdithiocarbamate) and the insecticide rotenone, can induce neuronal death in PD models. Human epidemiological studies show association of PQ/MB exposure to development of PD (Costello et al., 2009), and this combination causes PD in animal models (Thiruchelvam et al., 2000). Though the contribution of pesticides to sporadic PD remains contentious, involvement of mitochondria is generally accepted. Thus, these toxins are used in disease models to induce mitochondrial electron transport chain dysfunction and related cell injury.
In contrast to sporadic PD, rare familial forms are causally linked to genetic mutations that are either dominant (PARK3, LRRK2, UCH-L1, PARK13, and SNCA [encoding α-syn]) or recessive (Parkin, PINK1, FBXO7, PLA2G6, and PARK9). Some gene products, including α-syn, contribute to aggregate formation in PD, appearing as intracellular inclusions (Lewy bodies and neurites). PD-associated gene mutations have been used to generate animal models to study the molecular basis of PD pathogenesis. Two categories of cellular events have been uncovered in these studies: (1) protein misfolding, aggregation, and aberrant proteostasis and (2) mitochondrial damage with oxidative/nitrosative stress (Dauer and Przedborski, 2003). Identifying pathways that link these events could provide information regarding events that lead to death of SNpc DA neurons. Recently, nitrosative/oxidative stress was reported to lead to tyrosine nitration and methionine oxidation on α-syn, thus contributing to aggregation (Giasson et al., 2000; Schildknecht et al., 2013; Chavarría and Souza, 2013). Considering the association of mitochondrial toxins with PD, another key question concerns interactions between environmental and genetic factors in PD. In fact, animal models suggest that PQ/MB renders mutant A53T α-syn transgenic mice particularly vulnerable to PD-like pathology (Norris et al., 2007). Cell-based studies on human tissue could potentially identify early molecular events linking environmental and genetic factors in PD.
Recent advances in human induced pluripotent stem cell (hiPSC) technology provide an opportunity to study these events in human context (Takahashi and Yamanaka 2006; Saha and Jaenisch 2009). Recently, this technology was used to generate hiPSC models of PD patients’ cells carrying disease-causing mutations (Nguyen et al., 2011; Cooper et al., 2012). These models recapitulate key features of PD and validate the feasibility of using hiPSCs to model the disease. However, lack of genetically matched nondiseased controls makes interpretation of observed phenotypes difficult to attribute solely to the disease-causing mutation. This confounding variable must be overcome, as differences in genetic background between cell types may critically influence data interpretation. Moreover, the relevance of findings from the above studies to PD is further undermined by limited representation of A9-type DA neurons following differentiation of hiPSCs to neurons.
Two recent events have allowed us to resolve the above issues. First, we generated isogenic stem cell models of PD (Soldner et al., 2011) in which we “correct” a SNCA-A53T mutation in patient-derived hiPSCs or “knock in” the same mutation in a human embryonic stem cell (hESC) line. This results in two pairs of isogenic stem cells that differ exclusively at the SNCA locus that can be used to study PD. Second, nearly pure populations of A9-type DA neurons can now be generated from pluripotent cells (Kriks et al., 2011). By combining these two methods, we tested a “multi-hit” scenario whereby mitochondrial toxins and cellular genetic factors interact in PD pathology. We report here a molecular pathway whereby increased basal and mitochondrial toxin-induced nitrosative stress results in inhibition of transcriptional activity of myocyte enhancer factor 2C (MEF2C) in A53T α-syn mutant A9 DA neurons (hNs) compared to corrected controls. We validate these findings using two distinct isogenic lines, representing both hiPSCs and hESCs, with disparate genetic backgrounds. Because MEF2C activity normally stimulates transcription of peroxisome proliferator-activated receptor-γ coactivator-1α (PGC1α), inhibition of MEF2C decreases this neuroprotective pathway. Disruption of the MEF2C-PGC1α pathway contributes to mitochondrial dysfunction and culminates in apoptotic cell death. Our results identify redox-mediated protein posttranslational modifications, including S-nitrosylation and sulfonation of a critical cysteine residue in MEF2, as an early event contributing to neuronal damage in PD. Screening for small molecules that rescue neurons from these mitochondrial toxins, we validate the MEF2C-PGC1α pathway as a new drug target for PD.
RESULTS
A9 Dopaminergic Neurons Derived from A53T Mutant hPSCs Display α-Syn Aggregation and Lewy Body/Neurite-like Pathology
Using hiPSCs that allow comparison of the A53T α-syn mutation (A53T) with isogenic-corrected controls (Corr) (Soldner et al., 2011), we characterized the lineage progression of hiPSCs to dopaminergic (DA) neurons. To determine the impact of the A53T α-syn mutation on cellular pathology in PD, it was critical to generate the specific cell type affected in PD, A9 dopaminergic (DA) neurons. Using the protocol of Kriks et al. (2011), we differentiated hiPSC into A9 DA neurons with high efficiency (~80% of total neurons) from both mutant A53T and corrected hiPSCs (Figures 1A–1E). Neurons progressed from hiPSCs to forkhead box A2 (FOXA2)+/LIM homeobox transcription factor 1α (LMX1A)+ or OTX2+ neural progenitor cells (NPCs). Next, upon terminal differentiation, they progressed to LMX1A+/tyrosine hydroxylase (TH)+, nuclear-receptor-related 1 protein (NURR1)+/TH+, or G-protein-regulated inward-rectifier potassium channel 2 (GIRK2)+/TH+ neurons (Figures 1A and 1B). Electrophysiological characterization of hNs revealed voltage-dependent sodium currents (Figure 1C) and evoked action potentials (Figure 1D). Moreover, spontaneous, rapid (2–7 Hz) spikes, a hallmark of the A9 DA neuronal phenotype and consistent with the presence of CaV1.3 channels (Kriks et al., 2011), were observed by day 35 of terminal differentiation in hiPSC-derived hNs (Figure 1E). Electrophysiological parameters are listed in Table S1 available online. Additionally, we confirmed these results in a second set of isogenic PSC-derived neurons of disparate genetic background obtained from wild-type (WT) and A53T-mutated hESCs (Soldner et al., 2011) (Figures S1A and S1B, available online). These hNs also displayed voltage-dependent sodium currents (Figure S1C) and spontaneous, rapid action potentials (Figure S1D) comparable to those of their hiPSC-derived counterparts.
Figure 1. Characterization of hiPSC Differentiation into A9-type DA Neurons.
(A) Micrographs of hiPSC differentiation into human DA neurons (hNs). NPCs (top) are both FOXA2/LMX1A double positive and OTX2/FOXA2 double positive at 11 days in vitro (DIV). Most β-III tubulin (TUJ1)+ neurons (lower-middle) stain for TH by DIV 25. TH+ neurons are also LMX1A+, NURR1+ (upper-middle) and GIRK2+ (bottom) at DIV 35. Blue, DAPI; scale bar, 50 µm.
(B) Quantification of TUJ1, TH/TUJ1, TH/LMX1A, and TH/GIRK2-positive cells relative to total cell number. Data from replicate experiments at DIV 35 are shown as mean + SEM; n = 6.
(C–E) Electrophysiological characterization of A53T hNs and corrected controls at DIV 35. INa and IK traces (n = 30, C). Cells were prepulsed to −90 mV for 150 ms and held at −60 mV with 10 mV voltage steps. Evoked action potential trains (Vh = −70 mV, n = 30, D). Spontaneous action potentials (n = 8, E). Mean resting potential, −39 ± 6 mV for A53T hN and −40 ± 3 mV (mean ± SEM) for corrected hN. Electrophysiological recordings were performed using K-gluconate internal solution.
(F) α-synuclein costaining with thioflavin T in hiPSC-derived corrected (Corr) and A53T hNs. A53T mutant shows increased thioflavin T. DIV, 35; blue, DAPI; scale bar, 10 µm.
(G) Quantification of thioflavin T fluorescence. Data from replicate experiments at DIV 35 shown as mean + SEM; n = 3.
(H and I) Immunostaining of corrected hNs (H) and A53T mutant hNs (I) shows PS-129 colocalization with ubiquitin in neurites and cell bodies of A53T hNs (arrows). DIV, 35; blue, DAPI; scale bar, 10 µm.
A53T mutation induces α-syn aggregation into oligomeric β sheets, which result in deposition in Lewy bodies/neurites. We thus assessed whether basal synuclein pathology could be detected in A53T mutant hNs. Costaining for thioflavin T and total α-syn showed a dramatic increase in thioflavin T fluorescence that colocalized with α-syn in A53T hNs relative to corrected hNs, indicating accumulation of oligomerized α-syn (Figures 1F and 1G). We next confirmed Lewy body/neurite-like α-syn deposition using an alternative antibody approach to detect phosphorylation of α-syn on serine 129 (Wang et al., 2012). Antigenic labeling of α-syn phosphoserine (PS)-129 and ubiquitin again revealed Lewy neurite/body-like deposition in A53T, but not corrected hNs (Figures 1H and 1I). Immunoblots also showed elevated α-syn PS129 levels in A53T relative to corrected hNs (Figure S1E). Whereas α-syn protein levels were higher in A53T compared to control hNs from both isogenic lines (Figures S1E and S1F), allele-specific PCR revealed no difference in SNCA mRNA expression (Figure S1G–S1I). Additionally, qRT-PCR of hiPSC-derived A53T and corrected hNs revealed no quantitative difference in SNCA mRNA expression (1.0 versus 1.1 arbitrary units). Taken together with prior work, these data suggest that synuclein pathology is associated with increased α-syn stability in A53T hNs, similar to A53T α-syn mouse models and human PD brain (Li et al., 2004; Wills et al., 2010).
A53T α-Syn Mutant hNs Manifest Alterations in Basal Mitochondrial Machinery and Increased Susceptibility to Mitochondrial Toxins
The presence of α-syn aggregates in A53T hNs under basal conditions led us to investigate other PD-relevant pathologies. For example, deficits in mitochondrial respiration are critically associated with neuronal dysfunction in PD. Therefore, we evaluated mitochondrial function in both A53T mutant and corrected hNs. Using the Seahorse platform to measure O2 consumption, we found a compromised maximal rate of mitochondrial respiration in A53T compared to corrected hNs (Figures 2A and 2B), resulting in deficiency in spare respiratory capacity (50% versus 190% for A53T and corrected hNs, respectively). We thus assessed the ability of A53T mutant versus corrected hNs to respond to stress induced by mitochondrial toxins known to affect neuronal viability.
Figure 2. A53T Mutant hNs Are Highly Susceptible to Mitochondrial Toxin-Induced Apoptosis.
(A and B) Mitochondrial O2 consumption monitored by Seahorse platform at DIV 35. After recording basal respiration, mitochondrial metabolic state 4 was induced with 1 µg/ml of the ATP synthase inhibitor oligomycin (Oligo), followed by induction of maximal respiration by the mitochondrial uncoupler FCCP in 1.5 µM increments. Maximal (Max) respiration was attained at a total FCCP concentration of 4.5 µM. Data from one of three replicate experiments are shown as mean ± SEM; n = 22 wells (A). Experimental data normalized to cell number per well (B). Neither basal respiration (reflecting cellular ATP demand) nor state 4 respiration (reflecting proton leak) differed between control and mutant cells. A53T mutant hNs displayed greatly reduced maximal respiratory capacity compared to corrected control.
(C–E) A53T mutant hNs are more susceptible to apoptosis than their genetically corrected counterparts following exposure to PQ alone (C), MB alone (D), or combined exposure (E). Data represent mean + SEM. **p < 0.01 by ANOVA with posthoc Tukey; n = 6–12; DIV, 35.
(F) Micrographs of TUNEL+ hNs following mitochondrial toxin exposure. DIV, 35; blue, DAPI; scale bar, 10 µm.
See also Figure S2.
Apoptosis was scored by TUNEL assay coupled with nuclear morphology or cleaved caspase-3 immunoreactivity. Choosing concentrations based on the EPA-reported lowest observable effect level (LOEL) (http://www.epa.gov/iris/subst/0183.htm), we determined the apoptotic index of the PD-associated pesticides PQ and MB alone and in combination, as well as that of rotenone in corrected and A53T mutant hNs (Figures 2C–2F, S2A, and S2B). These pesticides function in part by inhibiting mitochondrial respiratory chain complexes I (PQ and rotenone) or III (MB) (Gomez et al., 2007). A53T mutant hNs relative to corrected control manifested increased susceptibility to toxin mediated-apoptosis. Interestingly, combined exposure of A53T mutant hNs to PQ/MB produced synergistic effects, decreasing the concentration of each toxin required to evoke death (Figures 2E and 2F). At 2.8 µM PQ and 1 µM MB, differential susceptibility to toxin-evoked death was observed between A53T mutant and corrected hNs. This differential susceptibility was also observed in hESC-derived WT and A53T mutant hNs (Figure S2C and S2D). Although α-syn expression and phosphorylation on serine 129 were greater in A53T hNs, mitochondrial toxins had no additional effect on α-syn expression or aberrant phosphorylation. We next sought to understand this differential susceptibly to mitochondrial toxins by profiling changes in gene expression to identify signaling pathway(s) that are aberrantly regulated in A53T mutant hNs versus corrected controls.
Aberrant Modification of MEF2C by Oxidative/Nitrosative Stress in A53T Mutant Neurons
We used global gene expression analysis via microarray to identify biological processes specifically altered under basal conditions in A53T versus corrected hNs (Figure 3A). Enrichment analysis based on Gene Ontology (GO) revealed a decrease in expression of prosurvival genes and genes associated with metabolic function (Figure S3A). Next, we screened regulatory networks of multiple transcription factors against our microarray data set to identify factors whose target gene sets were significantly altered (p < 0.05) in A53T hNs relative to corrected. Based on GO analysis, we prioritized transcription factors with known functions related to apoptosis (Figure 3B), cell survival (Figure 3C), and DA neuron-specific transcriptional regulation (Figure 3D). We found that transcription factors whose targets are involved in DA neuron regulation were highly enriched among downregulated genes in A53T hNs, with the prosurvival/neurogenic transcription factor MEF2 (Leifer et al., 1993; Mao et al., 1999; Okamoto et al., 2000, 2002) being most enriched. MEF2 protein levels, however, showed little difference between A53T and corrected hNs (Figure S3B). We thus sought to determine how A53T SNCA mutation or mitochondrial stress might affect MEF2 transcriptional activation. We initially focused on MEF2C because it is the predominant isoform expressed in hPSCs developing into brainstem SNpc neurons (Lyons et al., 1995; Cho et al., 2011).
Figure 3. A53T Mutant hNs Manifest Altered Transcriptional Network Activity.
(A) Expression profiling of A53T mutant and corrected (Corr) hNs at DIV 35. Genes whose expression was altered ≥ 2-fold (p < 0.05 significance) were clustered based upon functional correlation using GenePattern (Broad Institute), and the resulting heatmap is depicted (red, up; blue, down; data from three replicate experiments). A53T mutant hNs had a strikingly different genetic profile from corrected counterparts.
(B–D) Pathway enrichment analysis was performed on all genes whose expression was altered (7,018 genes; p < 0.05 significance) in A53T hNs relative to corrected using NextBio and was filtered based on regulatory motifs using the Broad Institute Molecular Signatures Database (BroadMSigDB). Enrichment score was assigned by NextBio based on the method of Kupershmidt et al. (2010). Transcription factors were grouped based on their contribution to apoptosis (B), cell survival (C), or DA neuronal transcription (D). Transcription factor with the highest enrichment score was MEF2 (denoted in red). (Blue circle) Genes altered in A53T relative to corrected; (red circle) genes with transcription factor binding motifs within 2 kb of the transcriptional start site; (purple overlap) genes with transcription factor binding motifs within 2 kb of the transcriptional start site that are altered in A53T relative to corrected.
See also Figure S3.
One indication of mitochondrial stress, particularly after inhibition of complex I, is increased ROS production. Via MitoSOX fluorescence, we monitored superoxide anion, an indicator of ROS load, and found increased levels in A53T hiPSC-derived hNs relative to corrected both at baseline and following exposure to mitochondrial toxins (Figures 4A and 4B). We found a similar increase in basal MitoSOX fluorescence in A53T mutant human neurons derived from hESCs relative to isogenic WT controls (Figure S4A). ROS-induced posttranslational protein modifications can affect functional activity via reaction to a sulfonated adduct (Gu et al., 2002). To determine whether MEF2C can be so modified, we performed mass spectrometry (MS) on recombinant MEF2C following H2O2 exposure and detected a sulfenated (–SO−) derivative, indicated by a 16 Da shift in the MEF2C mass (Figure S4B). To identify the modified cysteine residue, we used DCP-Bio1 to detect sulfenated (–SOH) derivatives after exposure to H2O2. MS/MS on the DCP-Bio1 labeled-protein identified cysteine (Cys39) in the DNA-binding domain of MEF2C as the sulfenated site (Figure S4C). Next, we probed intact cells for SOH-MEF2C using the –SOH-specific detection technique of DCP-Bio1—assisted capture followed by immunoblot (Figures 4C and 4D) (Nelson et al., 2010). We found no substantial difference in SOH-MEF2C under basal conditions in A53T hNs relative to corrected controls but observed a dramatic induction after exposure to either H2O2 or PQ/MB. Additional MS experiments performed in the absence of DCP-Bio1 revealed higher oxidation states of the protein (representing –SO2/3H derivatization), often achieved in the presence of high ROS levels (Gu et al., 2002). Taken together, these experiments show that exposure to excessive ROS leads to the formation of SOxH-MEF2C (wherein x = 1–3).
Figure 4. Accumulation of ROS/RNS in A53T hNs Aberrantly Modifies the Neuronal Prosurvival Transcription Factor MEF2C.
(A) Representative micrographs of MitoSOX (red)-labeled A53T mutant and corrected hNs superimposed on phase-contrast images under basal conditions. DIV, 35; blue, DAPI; scale bar, 10 µm.
(B) Quantification of superoxide anion accumulation at DIV 35 in corrected and A53T mutant hNs measured via MitoSOX labeling following pesticide exposure. A53T mutant hNs displayed increased labeling relative to corrected (Corr) at baseline and following exposure to 200 nM rotenone or 2.8 µMPQ/1 µM MB. Values are mean + SEM. **p < 0.01 by t test; n = 6.
(C) Increased SOH-MEF2C detected at DIV 35 in A53T relative to corrected hNs after administration of mitochondrial toxins. SOH-protein capture using DCP-Bio1 and subsequent immunoblot for MEF2C show increased SOH-MEF2C upon exposure to H2O2 or PQ/MB.
(D) Quantification of SOH-MEF2C. Values are mean + SEM. *p < 0.05 by ANOVA with posthoc Tukey; n = 3.
(E and F) DAF-FM staining of NO levels shows higher basal values and more rapid accumulation of NO after pesticide exposure in A53T mutant relative to corrected hNs.
(G) Time course of DAF-FM signal in A53T and corrected hNs following exposure to PQ/MB normalized to respective baseline levels. Inset shows that A53T mutant hNs have a 17% higher level of DAF-FM fluorescence than corrected hNs at baseline.
(H) l-NAME pretreatment reduced both baseline and pesticide-evoked DAF-FM signal in A53T hNs. Pesticide exposure time, 6 hr; DIV, 35; blue, DAPI; scale bar, 10 µm. Data represent mean ± SEM. **p < 0.01 by two-way ANOVA with posthoc Tukey; n = 6.
(I and J) Biotin switch at DIV 35 shows that A53T mutant hNs had substantial basal levels of SNO-MEF2C compared to corrected hNs, pesticides increased SNO-MEF2C levels (I), and l-NAME inhibited basal SNO-MEF2C formation with negative control in the absence of ascorbate (–Asc) (J).
(K) Quantification of SNO-MEF2C. Values are mean + SEM. **p < 0.01, by ANOVA with posthoc Tukey; n = 3.
See also Figure S4.
In addition to ROS, RNS such as nitric oxide (NO) have been linked to mitochondrial stress and can react with critical cysteine residues to affect functional activity via protein S-nitrosylation (forming SNO proteins) (Lei et al., 1992; Lipton et al., 1993; Stamler et al., 1992, 1997). NO can also react rapidly with superoxide anion (O2−) to form peroxynitrite (ONOO−), causing tyrosine nitration and contributing to protein aggregation in the case of α-syn (Giasson et al., 2000). Using the NOsensitive dye DAF-FM, we found an increase in basal levels of NO in A53T mutant hNs derived from both PSC lines compared to their respective controls (Figures 4E and 4G, inset, and Figure S4D). Interestingly, low-dose mitochondrial toxins (2.8 µM PQ/1 µM MB) resulted in further accumulation of NO only in A53T mutant and not in corrected hNs (Figures 4F and 4G). Monitoring NO production via Griess reaction to measure the metabolite nitrite, we found that prolonged exposure to a higher dose of PQ/MB (28 µM/5 µM) also evoked NO accumulation in A53T-corrected hNs (Figure S4E). This increase in NO production both basally and following exposure to PQ/MB was blocked by pretreatment with the NO synthase (NOS) inhibitor l-NG-nitroarginine methyl ester (l-NAME), confirming that the increase in DAF-FM fluorescence was most likely due to NO (Figures 4H and S4F).
Next, we performed MS experiments to determine whether MEF2C can also be modified by S-nitrosylation. After exposing recombinant MEF2C to endogenous NO donor S-nitrosocysteine (SNOC), MS/MS revealed that MEF2C can indeed be S-nitrosylated, and this modification occurs on the same candidate cysteine (Cys39) as sulfonation (Figure S4G). Cys39 is located in the DNA-binding domain of MEF2C (Huang et al., 2000; Santelli and Richmond, 2000), suggesting that these posttranslational redox modifications might inhibit MEF2 transcriptional activity. Hence, we constructed a nonnitrosylatable/nonsulfonatable MEF2C mutant (MEF2C(C39A)), permitting us to test whether such redox reactions are causally linked to aberrant transcriptional activity and neuronal cell death.
To determine whether SNO-MEF2C is produced in neurons under basal conditions or following exposure to mitochondrial toxins, we probed A53T mutant and corrected hNs for S-nitrosylated MEF2C using the biotin switch assay. We found that A53T mutant hNs have high basal levels of SNO-MEF2C relative to corrected controls (Figures 4I–4K), suggesting that this was an aberrant modification existing under baseline conditions in A53T hNs. We found higher basal levels of SNO-MEF2C in our second set of isogenic A53T mutant hNs as well (Figure S4H). Additionally, high-dose PQ/MB exposure promoted SNOMEF2C formation in corrected hNs (Figures 4I and 4K). Pretreatment with l-NAME prevented this modification in both A53T hNs under basal conditions and in corrected hNs after exposure to pesticides (Figures 4J and S4I).
Formation of SNO-MEF2C Inhibits Transcription of the Mitochondrial Master Regulator PGC1α
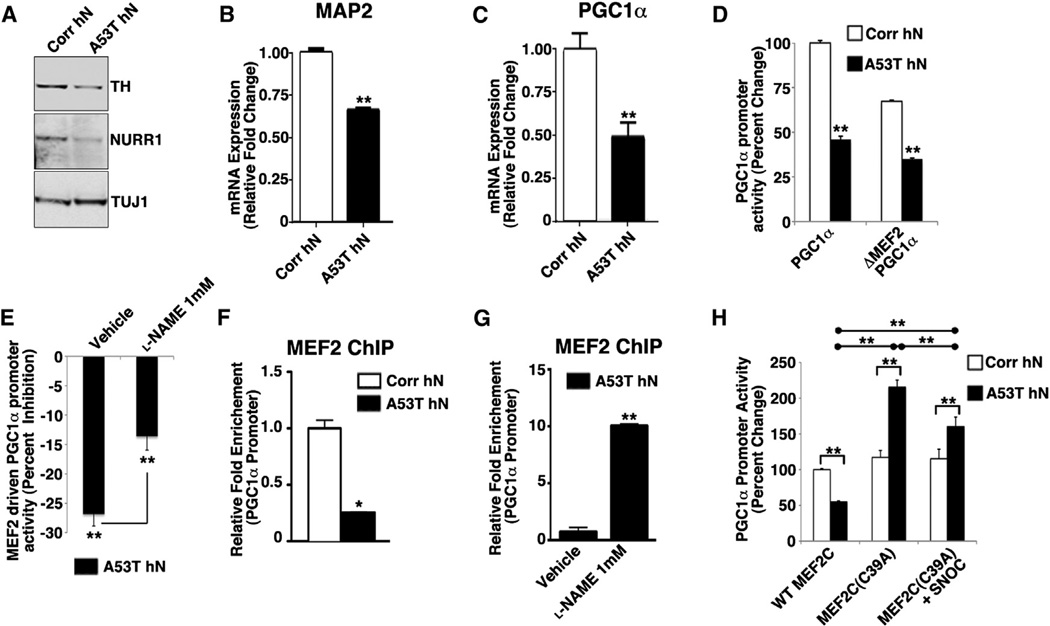
We next sought to determine whether redox modification of MEF2C could inhibit its transcriptional activity and, if so, to identify which transcriptional targets were altered in A53T hNs to account for increased neuronal susceptibility after mitochondrial stress. We performed both western blot and quantitative (q) real-time PCR analysis of MEF2C-regulated genes of importance to development or survival of A9 DA neurons. In A53T mutant hNs relative to control, we found decreased protein expression of the MEF2C transcriptional target NURR1 and the NURR1 transcriptional target TH, proteins highly enriched in A9 DA neurons (Figure 5A). Further analysis of mRNA levels showed decreased mRNA expression of MEF2-regulated genes, including microtubule-associated protein 2 (MAP2), encoding a neuronally restricted protein, and PGC1α, a transcriptional coactivator that is known to promote neuroprotection in PD models (Figures 5B and 5C) (Clark and Simon, 2009). Consistent with the microarray, we also found altered mRNA expression of the MEF2 transcriptional target GLDC (a mitochondrial glycine decarboxylase), whereas other markers of the A9 phenotype (GIRK2 and PITX3) were unchanged (Figure S5A). Importantly, treatment with l-NAME promoted mRNA expression of multiple MEF2 targets (Figure S5B). Although several of these targets are important for DA neurons, we identified PGC1α as a critical effector of MEF2, given its importance in mitochondria and in PD models (Jones et al., 2012).
Figure 5. MEF2C-Mediated Transcriptional Activation Is Inhibited in A53T hNs.
(A) Immunoblot analysis shows decreased TH and NURR1 protein expression in A53T mutant relative to corrected hNs at DIV 35.
(B and C) qRT-PCR shows significant reduction in MAP2 and PGC1α mRNA in A53T mutant relative to corrected hNs at DIV 35. Data represent mean + SEM. **p < 0.01 by t test; n = 3.
(D) Activation of native PGC1α promoter and PGC1α promoter lacking the MEF2-binding site (ΔMEF2) in A53T hNs relative to corrected hNs at DIV 35. Data represent mean + SEM. **p < 0.01 by t test; n = 4.
(E) MEF2-driven PGC1α promoter activity inhibited in A53T hNs relative to corrected hNs at DIV 35. l-NAME prevented 50% of this inhibition. PGC1α promoter activity attributable to MEF2 was derived from the following formula using the data in (D): (1 − (ΔMEF2/PGC1α) × 100). Data represent mean + SEM. **p < 0.01 by t test; n = 4.
(F) ChIP analysis on chromatin prepared from A53T mutant versus corrected (Corr) hNs at DIV 25 revealed 4-fold reduction in MEF2C bound to PGC1α promoter. Data represent mean + SEM. *p < 0.05 by t test; n = 3.
(G) l-NAME increases MEF2C binding to PGC1α promoter by ~10-fold. Data represent mean + SEM. **p < 0.01 by t test; n = 3.
(H) Effect of WT MEF2C or mutant MEF2C(C39A) on PGC1α promoter activity in the presence and absence of SNOC in A53T and corrected hNs at DIV 35. Data represent mean + SEM. *p < 0.05 by ANOVA with posthoc Tukey; n = 4.
See also Figure S5.
To assess involvement of MEF2C in PGC1α signal transduction, we first performed a reporter assay to determine whether MEF2C directly regulates PGC1α expression. We examined PGC1α promoter activity under normal conditions and after mutation of the MEF2-binding site (ΔMEF2-PGC1α) and found a significant decrease in normal WT-PGC1α promoter activity in A53T hNs relative to corrected. Whereas ΔMEF2-PGC1α reduced PGC1α promoter activity in corrected hNs, the effect was minimal in A53T cells, indicating that MEF2 was already inhibited (Figure 5D). l-NAME significantly prevented this inhibition on WT-PGC1α promoter (Figure 5E). Taken together, these findings suggest that decreased MEF2 activity contributes to inhibition of PGC1α expression, and NO is a critical factor in this inhibition.
To determine whether the basal level of redox-modified MEF2C in A53T mutant hNs interferes with MEF2C binding to the PGC1α promoter, we performed chromatin immuno-precipitation (ChIP) of MEF2C on the PGC1α promoter in both A53T hNs and corrected controls. We found a ~75% reduction in MEF2C bound to PGC1α promoter in A53T mutant hNs (Figure 5F). We confirmed that MEF2 binding to the PGC1α promoter was NO dependent by treating A53T mutant hNs with l-NAME, finding a 10-fold increase in bound MEF2C (Figure 5G). To show that the decrease in PGC1α promoter activity was due specifically to S-nitrosylation of MEF2C on Cys39, we overexpressed both WT and nonnitrosylatable MEF2 mutant (MEF2C(C39A)) in corrected and A53T mutant hNs. Overexpression of WT MEF2C in A53T mutant hNs was 50% less effective than in corrected hNs in driving PGC1α promoter activity, presumably due to the high basal levels of NO that are present in A53T hNs. Transfection with nonnitrosylatable MEF2C prevented the decrease in MEF2 transcriptional activity observed under basal conditions or following exposure to NO-generating agents (Figure 5H). Interestingly, MEF2C(C39A) expression resulted in higher basal levels of PGC1α promoter activity in A53T hNs than their corrected counterparts, presumably due to an effect on other MEF2 regulators such as HDAC9, which was downregulated on our microarray.
Next, to determine whether SNO-MEF2C-dependent loss of PGC1α expression could be observed in vivo, we administered intraperitoneal PQ/MB to adult WT and A53T mutant mice. A53T mutant mice manifested a decrease in basal levels of PGC1α protein expression relative to WT (Figure 6A). This loss of PGC1α expression was exacerbated in both WT and A53T mutant animals following exposure to PQ/MB. Moreover, PQ/MB resulted in substantial increase in SNO-MEF2C, detectable up to 4 weeks postadministration (Figures 4B and 4C). In addition, we found markedly reduced PGC1α protein levels in mouse brains from MEF2C global heterozygotes or Nestin-Cre-driven brain-specific knockout (Li et al., 2008a) (Figure 5E). Collectively, these data show that MEF2C can be S-nitrosylated or sulfonated by RNS/ROS at Cys39 and raise the possibility that this reaction can result in negative regulation of MEF2C transcriptional activity in the context of PD. Moreover, the results implicate loss of MEF2 transcriptional activity and thus failure to induce neuroprotective PGC1α expression as an effector of nitroxidant stress in A9 DA neurons.
Figure 6. MEF2C and Pesticides Regulate PGC1α Protein Expression In Vivo.
(A) PGC1α protein expression in WT and A53T mutant mouse brain in the presence and absence of PQ/MB exposure.
(B) PQ/MB increases SNO-MEF2C expression in WT mouse brain by biotin switch assay.
(C) Quantification of SNO-MEF2C in WT mouse brain. Values are mean + SEM. **p < 0.01, by t test; n = 3.
(D) PGC1α protein levels are reduced in MEF2C+/– and MEF2C conditional knockout mouse brains.
Preventing S-Nitrosylation and Promoting MEF2C-PGC1α Transcriptional Activity Rescue A53T Mutant hNs from Death
We assessed whether pretreatment with l-NAME to inhibit S-nitrosylation, with consequent enhancement of the MEF2C-PGC1α transcriptional pathway, would offer therapeutic benefit in A53T hNs exposed to mitochondrial toxins. Indeed, l-NAME partially rescued A53T mutant hNs derived from both PSC lines from pesticide-induced death (Figures 7A, S6A, and S6B). l-NAME also conferred partial protection to corrected hNs exposed to high-dose PQ/MB (Figure S6C). Because inhibiting NO synthesis conferred protection from cell death, we next sought to determine whether genetic rescue of MEF2C-PGC1α activity could promote cell survival. Although we found that overexpression of WT MEF2C was able to partially rescue (64%) from PQ/MB toxin-induced death in A53T mutant hNs, the nonnitrosylatable/nonsulfonatable MEF2C(C39A) provided significantly more neuroprotection (83%) (Figures 7B and 7C). Interestingly, this level of protection was similar to that observed with overexpression of WT MEF2C in conjunction with l-NAME treatment (81%) (Figure 7C). The nonredox-modifiable MEF2C mutant also rescued from toxin-induced death in our second set of isogenic PSCs, with greater efficacy than WT MEF2C (Figure S6D and S6E). The fact that mutant MEF2C(C39A) provided the greatest degree of neuroprotection from mitochondrial toxins and that this level of protection was similar to that seen with WT MEF2C plus l-NAME is consistent with the notion that S-nitrosylation of Cys39 by the toxins inhibited MEF2C activity, thus contributing to cell death. To gather further evidence for the importance of the MEF2C-PGC1α pathway in protecting hNs from mitochondrial stress, we next assessed whether PGC1α could directly rescue from PQ/MB-mediated apoptosis. In fact, we observed that overexpression of PGC1α nearly completely abrogated mitochondrial toxin-induced apoptosis (~93% reduction in death) (Figures 7B and 7C).
Figure 7. Activation of MEF2C Transcriptional Network Rescues A53T Mutant hNs from Mitochondrial Toxin-Evoked Death.
(A) l-NAME partially blocks PQ/MB-induced death in A53T mutant hiPSC-derived hNs. **p < 0.01 by ANOVA with posthoc Tukey; n = 6.
(B) TUNEL reactivity in A53T hNs exposed to mitochondrial toxins PQ and MB following overexpression of MEF2C (with or without 1 mM l-NAME) or PGC1α. WT MEF2C, mutant MEF2C(C39A), and PGC1α rescue from PQ/MB-induced apoptosis relative to empty vector (EV)-expressing cells. DIV, 25; blue, DAPI; scale bar, 10 µm.
(C) Quantification of data in (C). Values are mean + SEM. **p < 0.01, *p < 0.05 by ANOVA with posthoc Tukey or Fisher PLSD; n = 6.
(D) TUNEL reactivity in A53T hNs exposed to PQ and MB following pretreatment with the MEF2 activator Isoxazole (10 µM). Isoxazole rescued from rotenone- or PQ/MB-induced apoptosis. DIV, 35; blue, DAPI; scale bar, 10 µm.
(E) Quantification of data in (D). Values are mean + SEM. *p < 0.05 by t test; n = 3.
(F) In human A9-type DA neurons, MEF2C drives MEF2 target genes, including PGC1α expression, which in turn promotes mitochondrial biogenesis, mitochondrial respiration, and neuronal health (i). When SNCA is mutated (A53T) in familial PD, higher steady-state levels of RNS result in the S-nitrosylation of MEF2C under basal conditions. Resulting inhibition of MEF2C transcriptional activity decreases MEF2 target gene expression, including PGC1α (dashed line), with consequent impairment of mitochondrial respiration and disruption of neuronal function (ii). Combined effect of mitochondrial stress due to A53T SNCA and exposure to mitochondrial toxins produces a dramatic increase in RNS and ROS over basal conditions, resulting in increased S-nitrosylation and/or sulfonation of MEF2C and further decrease in MEF2C activity. The decreased MEF2C activity leads to further inhibition of MEF2 target gene transcription, including PGC1α, and consequently a more profound drop in mitochondrial biogenesis and respiration, culminating in neuronal cell death (width of red lines indicates magnitude of effect) (iii).
See also Figure S6.
In our final set of experiments, aimed at developing a therapeutic, we performed high-throughput screening (HTS) for small molecules capable of targeting the MEF2C-PGC1α pathway. We screened a chemical library of compounds for their ability to activate MEF2 transcription in the human neuronal context. One molecule identified was isoxazole (Figure S6F), consistent with previous results in rodent cells (Schneider et al., 2008). We thus assessed isoxazole as a potential therapeutic in A53T mutant hNs. Isoxazole effectively drove expression of both MEF2C and PGC1α in A53T hNs (Figure S6G). Moreover, isoxazole protected A53T hNs from apoptosis induced by the mitochondrial toxins rotenone or PQ/MB (Figures 7D and 7E). In summary, these data identify a pathway common to both genetic and environmental stress in PD centered on ROS/RNS-mediated repression of the MEF2C-PGC1α transcription pathway. This pathway can be therapeutically targeted to promote survival of human A9 DA neurons derived from PD patients (Figure 7F). Moreover, because several FDA-approved drugs contain isoxazole rings, our findings may have potential clinical implications for the repurposing of known drugs to treat PD.
DISCUSSION
SNpc A9-type DA neurons are known to possess a unique form of pacemaking activity that can produce Ca2+ overload, which has been implicated in both mitochondrial and endoplasmic reticulum (ER) stress; the associated increase in ROS/RNS may contribute to α-syn-dependent cell death in PD (Mosharov et al., 2009; Guzman et al., 2010). Given the high energy demands of neurons in general and the pacemaking activity of A9 neurons in particular, it is not surprising that these cells display increased dependence on mitochondrial respiration. Compromised mitochondrial function can thus be devastating to these neurons. Here, we show that hiPSC-derived A9 DA neurons bearing the A53T SNCA mutation exhibit diminished mitochondrial spare-respiratory capacity and produce increased basal levels of ROS/RNS compared to isogenic corrected controls. Moreover, we find that even brief exposure to mitochondrial toxins is sufficient to further increase ROS/RNS, contributing to neuronal damage. We observed that administration of commonly used pesticides at doses below EPA-accepted levels is sufficient to inflict oxidative/nitrosative stress and consequent death in these hiPSC-derived A9 neurons. Increased susceptibility of hiPSC-derived neurons with the A53T SNCA mutation likely reflects the fact that these mutant cells manifest increased basal levels of ROS/RNS compared to isogenic corrected cells. We show that ROS/RNS generated during toxin exposure leads to increased sulfonation/S-nitrosylation of the antiapoptotic transcription factor MEF2C and that NO contributes to cell death because inhibition of NOS with l-NAME attenuated the damage. Causality of SNO-MEF2 in this form of cell death is implicated by rescue of neurons with the nonnitrosylatable MEF2C(C39A) mutant construct.
A number of aberrantly S-nitrosylated proteins have been implicated in neurodegenerative disorders and may contribute to specific disease pathology (Nakamura et al., 2013). In support of this notion in PD, gene ontology clustering after genetic profiling of A53T mutant hNs relative to corrected cells showed aberrations in biological processes related to metabolism of nitrogen-containing compounds. Additionally, this analysis identified downregulation of a number of targets of the prosurvival/pro-DA neurogenic transcription factor MEF2 (Leifer et al., 1993; Mao et al., 1999; Okamoto et al., 2000, 2002; Li et al., 2008a,b; Cho et al., 2011), suggesting possible involvement of MEF2 in neuronal damage in PD. MEF2 isoforms localize in part to mitochondria and influence oxidative phosphorylation (OXPHOS) (She et al., 2011). Hence, MEF2 is an excellent candidate for a protein whose dysfunction might have a major impact on DA neuron homeostasis. Supporting this hypothesis, in the present study, we show that mitochondrial toxins such as pesticides induce damage in DA neuronal cells at least in part by inhibiting MEF2C activity via S-nitrosylation, and the nonoxidizable form of MEF2 rescues this damage.
Our bioinformatics analysis of potential MEF2-binding sites in the neuronal genome identified several potential targets whereby dysfunctional MEF2 might elicit mitochondrial damage and neuronal death. PGC1α was among the targets that we experimentally validated by ChIP-qPCR analyses. Interestingly, prior work had suggested that PGC1α and MEF2 were functionally linked (Czubryt et al., 2003; Handschin et al., 2003). As a master regulator of mitochondrial bioenergetics and oxidative metabolic programs, transgenic expression of PGC1α in DA neurons can protect them from MPTP-induced death (Mudò et al., 2012). Moreover, the Global PD Gene Expression consortium identified PGC1α as a therapeutic target for early intervention in PD (Zheng et al., 2010). Consistent with these findings, we show that posttranslational redox modification of MEF2C contributes to mitochondrial dysfunction and DA neuronal apoptosis by decreasing PGC1α expression. Given the important regulatory influence of MEF2C on PGC1α and the fact that even short exposures to mitochondrial toxins impair this effect, oxidative/nitrosative stress mediated by sulfonation/S-nitrosylation of MEF2C may well be an early, critical event in PD pathogenesis. Although these results identify disruption of the MEF2C-PGC1α pathway as a potential contributor to DA neuronal demise in PD, they do not exclude additional pathways either involving other MEF2 isoforms or independent of MEF2 altogether from being important.
An intriguing observation in the present study is the finding that human DA neurons carrying the A53T SNCA mutation display increased vulnerability to mitochondrial toxin-induced cell death. This result suggests that the mutant background may facilitate induction of PD by environmental toxins, supporting a “two-hit hypothesis.” In accord with our findings on human neurons, prior studies in rodents had suggested that aberrant expression of α-syn increases the vulnerability of SNpc neurons to the neurotoxic effects of PQ/MB (Thiruchelvam et al., 2004; Norris et al., 2007). Mutation of α-syn promotes oligomerization and aggregation, leading to formation of Lewy bodies/neurites (Lashuel et al., 2013), as observed in A53T hNs. Oligomerization or aggregation of several proteins, including α-syn, is associated with aberrant accumulation of RNS/ROS (Giasson et al., 2000; Gregersen and Bross 2010; Schildknecht et al., 2013). Thus, mutant α-syn neurons are exposed to oxidative/nitrosative stress under basal conditions. Accordingly, we observed an increase in the basal level of SNO-MEF2C in A53T hNs compared to corrected controls. These observations may thus help to explain the increased vulnerability of the A53T mutant neurons to nitrosative damage after mitochondrial toxin exposure compared to isogenic corrected controls.
In summary, in A53T SNCA mutant hNs, we identify the MEF2-PGC1α transcriptional pathway as a contributor to neuronal damage. Using hiPSC-derived A9 neurons and high-throughput screening studies, we show that this pathway represents a target for potential therapeutic intervention in PD. Finally, we provide a new molecular mechanism whereby genes and environment (GxE) can interact to contribute to the PD phenotype in human cells. Namely, mitochondrial toxins, represented by environmental pesticides, are shown to induce nitrosative/oxidative stress that inhibits MEF2C activity via S-nitrosylation (or possibly sulfonation) of Cys39 in the MEF2C DNA-binding domain. In turn, this attenuates PGC1α expression and thus the neuroprotective effect of this transcriptional coactivator. In the future, therapeutics aimed at counteracting the aberrant redox pathway that inhibits MEF2C transcriptional activity may prove effective in combating neuronal injury in PD.
EXPERIMENTAL PROCEDURES
hiPSC/hESC Culture and A9 DA Neuronal Differentiation
hiPSCs/hESCs were cultured as previously described, with details of this procedure and A9 DA neuronal differentiation provided in the Extended Experimental Procedures.
Measurements of Oxygen Consumption Rate
Oxygen consumption rate was analyzed in day 35 hNs using an XF24-3 Extra-cellular Flux Analyzer (Seahorse Biosciences), as described in the Extended Experimental Procedures.
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling, Active Caspase-3 Assessment, and Image Analysis
For evaluation of apoptosis, cells exposed to toxins or vehicle were subjected to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, immunocytochemistry for active (cleaved) caspase-3, and immunolabeling for neuronal markers. Details are provided in the Extended Experimental Procedures.
Global Gene Expression Analysis
Expression profiling was performed using the One-Color Microarray Gene Expression Platform with Illumina beadchip human-HT-12 v4. Transcription factor targets enriched in A53T relative to corrected hNs are listed in Table S2, and those not enriched are listed in Table S3. Details of data analysis are in the Extended Experimental Procedures. Data available from the Gene Expression Omnibus under accession number GSE46798.
Detection of ROS/RNS and Redox-Modified Proteins
MitoSOX detection of superoxide and DAF-FM assay of NO were performed as per manufacturer’s instructions (Invitrogen, CA). Biotin switch assay (Jaffrey et al., 2001; Uehara et al., 2006) was used to detect S-nitrosylated cysteine residues in MEF2C protein. Details, with Griess and DCP-Bio assays, are provided in the Extended Experimental Procedures.
Reporter Gene Assay
Cells were transfected via Lipofectamine 2000 (Invitrogen) with MEF2 or PGC1α luciferase reporter and Renilla luciferase control vector. Analysis used Dual-Glo luciferase assay kit (Promega). See the Extended Experimental Procedures.
Chromatin Immunoprecipitation
ChIP assays were performed using the ChIP-IT EXPRESS assay kit (Active Motif, CA). Immunoprecipitations were performed using anti-MEF2C antibody (Santa Cruz). Input and ChIP DNA were analyzed by qPCR using Stratagene Mx3000P (see the Extended Experimental Procedures).
Small-Molecule Screening and Testing of “Hit” Compounds
MEF2 luciferase reporter screen was performed in human neuronal context against a chemical library for MEF2 activators (see the Extended Experimental Procedures).
Statistical Analysis
Data represent at least three independent experiments, presented as mean ± SEM. Statistical significance was ascertained by t test or two-way ANOVA with appropriate posthoc testing; p < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by NIH grants P01 HD29587, P01 ES016738, and P30 NS076411 (to S.A.L.); R37 CA084198 (to R.J.); United Mitochondrial Disease Foundation grant (to R.A.); and a Parkinson Society of Canada Fellowship (to S.D.R.).
Footnotes
ACCESSION NUMBERS
mRNA expression data have been deposited in Gene Expression Omnibus under accession number GSE46798.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.11.009.
REFERENCES
- Bonneh-Barkay D, Langston WJ, Di Monte DA. Toxicity of redox cycling pesticides in primary mesencephalic cultures. Antioxid. Redox Signal. 2005;7:649–653. doi: 10.1089/ars.2005.7.649. [DOI] [PubMed] [Google Scholar]
- Chavarría C, Souza JM. Oxidation and nitration of α-synuclein and their implications in neurodegenerative diseases. Arch. Biochem. Biophys. 2013;533:25–32. doi: 10.1016/j.abb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Cho EG, Zaremba JD, McKercher SR, Talantova M, Tu S, Masliah E, Chan SF, Nakanishi N, Terskikh A, Lipton SA. MEF2C enhances dopaminergic neuron differentiation of human embryonic stem cells in a parkinsonian rat model. PLoS ONE. 2011;6:e24027. doi: 10.1371/journal.pone.0024027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Simon DK. Transcribe to survive: transcriptional control of antioxidant defense programs for neuroprotection in Parkinson’s disease. Antioxid. Redox Signal. 2009;11:509–528. doi: 10.1089/ars.2008.2241. [DOI] [PubMed] [Google Scholar]
- Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003985. 141ra90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S, Cockburn M, Bronstein J, Zhang X, Ritz B. Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 2009;169:919–926. doi: 10.1093/aje/kwp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 α (PGC-1 α) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective α-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- Gomez C, Bandez MJ, Navarro A. Pesticides and impairment of mitochondrial function in relation with the parkinsonian syndrome. Front. Biosci. 2007;12:1079–1093. doi: 10.2741/2128. [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P. Protein misfolding and cellular stress: an overview. Methods Mol. Biol. 2010;648:3–23. doi: 10.1007/978-1-60761-756-3_1. [DOI] [PubMed] [Google Scholar]
- Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pace-making in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 2000;19:2615–2628. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jones AW, Yao Z, Vicencio JM, Karkucinska-Wieckowska A, Szabadkai G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012;12:86–99. doi: 10.1016/j.mito.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS ONE. 2010;5:e13066. doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Overk CR, Oueslati A, Masliah E. The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013;14:38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8:1087–1099. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc. Natl. Acad. Sci. USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lesuisse C, Xu Y, Troncoso JC, Price DL, Lee MK. Stabilization of α-synuclein protein with aging and familial parkinson’s disease-linked A53T mutation. J. Neurosci. 2004;24:7400–7409. doi: 10.1523/JNEUROSCI.1370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. USA. 2008a;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, McKercher SR, Cui J, Nie Z, Soussou W, Roberts AJ, Sallmen T, Lipton JH, Talantova M, Okamoto SI, Lipton SA. Myocyte enhancer factor 2C as a neurogenic and antiapoptotic transcription factor in murine embryonic stem cells. J. Neurosci. 2008b;28:6557–6568. doi: 10.1523/JNEUROSCI.0134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Morán JM, Ortiz-Ortiz MA, Ruiz-Mesa LM, Fuentes JM. Nitric oxide in paraquat-mediated toxicity: A review. J. Biochem. Mol. Toxicol. 2010;24:402–409. doi: 10.1002/jbt.20348. [DOI] [PubMed] [Google Scholar]
- Mosharov EV, Larsen KE, Kanter E, Phillips KA, Wilson K, Schmitz Y, Krantz DE, Kobayashi K, Edwards RH, Sulzer D. Interplay between cytosolic dopamine, calcium, and α-synuclein causes selective death of substantia nigra neurons. Neuron. 2009;62:218–229. doi: 10.1016/j.neuron.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudò G, Mäkelä J, Di Liberto V, Tselykh TV, Olivieri M, Piepponen P, Eriksson O, Mälkiä A, Bonomo A, Kairisalo M, et al. Transgenic expression and activation of PGC-1α protect dopaminergic neurons in the MPTP mouse model of Parkinson’s disease. Cell. Mol. Life Sci. 2012;69:1153–1165. doi: 10.1007/s00018-011-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CCL, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S-i, Lipton SA. Aberrant protein s-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris EH, Uryu K, Leight S, Giasson BI, Trojanowski JQ, Lee VM-Y. Pesticide exposure exacerbates α-synucleinopathy in an A53T transgenic mouse model. Am. J. Pathol. 2007;170:658–666. doi: 10.2353/ajpath.2007.060359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Krainc D, Sherman K, Lipton SA. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2000;97:7561–7566. doi: 10.1073/pnas.130502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Li Z, Ju C, Scholzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc. Natl. Acad. Sci. USA. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Gerding HR, Karreman C, Drescher M, Lashuel HA, Outeiro TF, Di Monte DA, Leist M. Oxidative and nitrative α-synuclein modifications and proteostatic stress: implications for disease mechanisms and interventions in synucleinopathies. J. Neurochem. 2013;125:491–511. doi: 10.1111/jnc.12226. [DOI] [PubMed] [Google Scholar]
- Schneider JW, Gao Z, Li S, Farooqi M, Tang TS, Bezprozvanny I, Frantz DE, Hsieh J. Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J. Clin. Invest. 2011;121:930–940. doi: 10.1172/JCI43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Ohtaki K, Matsubara K, Aoyama K, Uezono T, Saito O, Suno M, Ogawa K, Hayase N, Kimura K, Shiono H. Carrier-mediated processes in blood—brain barrier penetration and neural uptake of paraquat. Brain Res. 2001;906:135–142. doi: 10.1016/s0006-8993(01)02577-x. [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Singel DJ, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thiruchelvam MJ, Richfield EK, Baggs RB, Tank AW, Cory-Slechta DA. The nigrostriatal dopaminergic system as a preferential target of repeated exposures to combined paraquat and maneb: implications for Parkinson’s disease. J. Neurosci. 2000;20:9207–9214. doi: 10.1523/JNEUROSCI.20-24-09207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruchelvam MJ, Powers JM, Cory-Slechta DA, Richfield EK. Risk factors for dopaminergic neuron loss in human α-synuclein transgenic mice. Eur. J. Neurosci. 2004;19:845–854. doi: 10.1111/j.0953-816x.2004.03139.x. [DOI] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi Z-Q, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shi M, Chung KA, Zabetian CP, Leverenz JB, Berg D, Srulijes K, Trojanowski JQ, Lee VM, Siderowf AD, et al. Phosphorylated α-synuclein in Parkinson’s disease. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3002566. 121ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J, Jones J, Haggerty T, Duka V, Joyce JN, Sidhu A. Elevated tauopathy and α-synuclein pathology in postmortem Parkinson’s disease brains with and without dementia. Exp. Neurol. 2010;225:210–218. doi: 10.1016/j.expneurol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. Global PD Gene Expression (GPEX) Consortium. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3001059. 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.