Abstract
Noonan, LEOPARD and cardiofaciocutaneous syndromes (NS, LS and CFCS) are developmental disorders with overlapping features including distinctive facial dysmorphia, reduced growth, cardiac defects, skeletal and ectodermal anomalies, and variable cognitive deficits. Dysregulated RAS-mitogen-activated protein kinase (MAPK) signal traffic has been established to represent the molecular pathogenic cause underlying these conditions. To investigate the phenotypic spectrum and molecular diversity of germline mutations affecting BRAF, which encodes a serine/threonine kinase functioning as a RAS effector frequently mutated in CFCS, subjects with a diagnosis of NS (N= 270), LS (N= 6) and CFCS (N= 33), and no mutation in PTPN11, SOS1, KRAS, RAF1, MEK1 or MEK2, were screened for the entire coding sequence of the gene. Besides the expected high prevalence of mutations observed among CFCS patients (52%), a de novo heterozygous missense change was identified in one subject with LS (17%) and 5 individuals with NS (1.9%). Mutations mapped to multiple protein domains and largely did not overlap with cancer-associated defects. NS-causing mutations had not been documented in CFCS, suggesting that the phenotypes arising from germline BRAF defects might be allele specific. Selected mutant BRAF proteins promoted variable gain of function of the kinase, but appeared less activating compared than the recurrent cancer-associated p.Val600Glu mutant. Our findings provide evidence for a wide phenotypic diversity associated with mutations affecting BRAF, and occurrence of a clinical continuum associated with these molecular lesions.
Keywords: Noonan syndrome, LEOPARD syndrome, cardiofaciocutaneous syndrome, BRAF, CFCS, mutation analysis, genotype-phenotype correlation, functional studies
Introduction
In the last few years, mutations in genes coding for transducers participating in the RAS-mitogen activated protein kinase (MAPK) signaling pathway have been identified as the molecular cause underlying a group of clinically related developmental disorders with features including reduced postnatal growth, facial dysmorphia, cardiac defects, ectodermal anomalies, variable cognitive deficits and susceptibility to certain malignancies [Gelb and Tartaglia, 2006; Schubbert et al., 2007b; Aoki et al., 2008]. Noonan syndrome (NS; MIM# 163950), which is the most common condition among these Mendelian traits, has been demonstrated to be caused by heterozygous germline mutations in the PTPN11 (MIM# 176876), SOS1 (MIM# 182530), KRAS (MIM# 190070), RAF1 (MIM# 164760), and MEK1 (MIM# 176872) genes in approximately 65% of affected individuals [Tartaglia and Gelb, 2005, 2009]. SHP2 (encoded by PTPN11), SOS1, RAF1, and MEK1 positively contribute to RAS-MAPK signal traffic, and possess complex autoinhibitory mechanisms that can be impaired by mutations [Keilhack et al., 2005; Tartaglia et al., 2006, 2007; Pandit et al., 2007; Roberts et al., 2007; Martinelli et al., 2008]. Similarly, reduced GTPase activity or increased guanine nucleotide release underlies the aberrant signal flow through the MAPK cascade promoted by most NS-causing KRAS mutations [Schubbert et al., 2006, 2007a]. PTPN11 and RAF1 mutations also account for approximately 95% of LEOPARD syndrome (LS; MIM# 151100]) [Digilio et al., 2002; Legius et al., 2002; Pandit et al., 2007], a condition which resembles NS phenotypically but is characterized by multiple lentigines dispersed throughout the body, café-au-lait spots, and a higher prevalence of electrocardiographic conduction abnormalities, obstructive cardiomyopathy and sensorineural hearing deficits [Sarkozy et al., 2009]. In addition, enhanced RAS-MAPK signaling has been established in other clinically related conditions, including cardiofaciocutaneous syndrome (CFCS; MIM# 115150), which is caused by activating mutations in the KRAS, BRAF (MIM# 164757), MEK1, and MEK2 (MIM# 601263) genes in approximately 60–80% of affected subjects [Niihori et al., 2006; Rodriguez-Viciana et al., 2006; Narumi et al., 2007; Nava et al., 2007; Schulz et al., 2008], Costello syndrome (CS; MIM# 218040), which is caused by HRAS (MIM# 190020) missense mutations [Aoki et al., 2005; Kerr et al., 2006; Estep et al., 2006; Gripp et al., 2006; Zampino et al., 2007], and neurofibromatosis type 1 (NF1; MIM# 162200) and related phenotypes, which are caused by inactivating mutations or deletions of the NF1 (MIM# 162200) and SPRED1 (MIM# 609291) genes [De Luca et al., 2005; Brems et al., 2007].
While the clinical overlap occurring among these disorders and the shared pathogenesis with respect to RAS-MAPK signaling justifies their grouping within a disease family, the substantial phenotypic variation observed among and within these conditions as well as the documented phenotypic heterogeneity associated with individual disease genes provide evidence for the diverse potential of individual mutations to perturb developmental processes. Of note, the available data support the view that the phenotypic diversity occurring in NS, which is the most variable clinically among these disorders, can be ascribed, in part, to the gene mutated and even the specific molecular lesion [Tartaglia and Gelb, 2005, 2009]. For instance, pulmonic stenosis (PS) is more prevalent among individuals with a mutated PTPN11 allele, whereas RAF1 mutations are almost invariably associated with HCM. Similarly, SOS1 and KRAS mutations are associated with specific phenotypes, the former including ectodermal abnormalities, normal stature and absence of cognitive deficits, and the latter, a more severe condition approaching CFCS and CS and characterized by pronounced growth failure and mental retardation. The finding that mutations in the PTPN11, KRAS and BRAF genes occur as somatic events in human cancers and largely do not overlap with the germline transmitted defects underlying inherited disorders further supports this idea.
Based on these observations, we explored the possibility that a previously unrecognized class of mutations in BRAF might specifically occur in NS and/or LS. While this work was in progress, two groups reported on the identification of heterozygous BRAF mutations in two unrelated sporadic cases with phenotype fitting NS [Razzaque et al., 2007; Nystrom et al., 2008]. Here, we report that BRAF mutations also underlie a small fraction of LS, and provide a more extensive evaluation of BRAF mutation prevalence, diversity and associated phenotypic spectrum in NS and CFCS.
Materials and Methods
Clinical evaluation
Three cohorts including unrelated subjects with NS (N= 270), LS (N= 6) and CFCS (N= 33) were included in the study. Nearly all subjects were of European ancestry with the majority being Italian. Subjects were evaluated by clinical dysmorphologists experienced with these disorders (G.Z., M.C.D., A.S., L.M., M.C.S. and B.D). Clinical assessment included family history, physical, anthropometric, neurologic and cardiac evaluation, and accurate examination for craniofacial and other dysmorphism, ophthalmologic and otorhinolaryngologic defects, and ectodermal and musculoskeletal anomalies. Clinical features for the majority of subjects satisfied diagnostic criteria reported by van der Burgt et al. [1994] and Allanson [1987] (NS), Voron et al. [1976] and Sarkozy et al. [2009] (LS), and Roberts et al. [2006] (CFCS), but a few individuals who lacked sufficient features for a definitive diagnosis were also included. No subject harbored a mutation in the PTPN11, SOS, KRAS, HRAS, RAF1, MEK1 or MEK2 gene based on scanning of the coding exons by denaturing high-performance liquid chromatography (DHPLC) analysis and/or direct sequencing. Two individuals with the diagnosis of NFNS, based on the presence of features of neurofibromatosis type 1 [Stumpf et al., 1988; Gutmann et al., 1997] and NS, but without mutation in NF1, PTPN11 and SPRED1, were also included.
Molecular analyses
Genomic DNA specimens were collected under Institutional Review Board–approved protocols and with informed consent, and isolated from peripheral blood lymphocytes according to standard methods. The entire coding sequence, exon/intron boundaries and flanking intronic portions of the BRAF gene (GenBank accession number NM_004333.3) were screened for mutations using DHPLC (3100 and 3500HT WAVE DNA fragment analysis system, Transgenomic, Omaha, NE). Primer pair sequences as well as PCR and DHPLC analysis settings are available upon request. Amplimers having abnormal denaturing profiles were purified (Microcon PCR, Millipore, Billerica, MA) and sequenced bi-directionally using the ABI BigDye terminator Sequencing Kit v.1.1 (Applied Biosystems, Foster City, CA) and an ABI 3700 Capillary Array Sequencer or ABI Prism 310 Genetic Analyzer (Applied Biosystems). Positions of mutations were numbered with the A of the ATG-translation initiation codon in the reference cDNA sequence being 1 (NM_004333.3). Genotyping of parent-offspring trios was performed in all families with an affected individual with a BRAF mutation by using the AmpDESTER Profiler Plus kit (Applied Biosystems).
To explore whether BRAF gene copy number variation might underlies a fraction of NS, multiplex ligation-dependent probe amplification (MLPA) analysis was performed by using the SALSA P173-Gain3 kit (MRC-Holland, Amsterdam, The Netherlands) according to the manufacturer’s recommendations. Two BRAF-specific probes were complementary to exons 5 and 13 of the gene. Reaction products were analyzed using ABI PRISM 3100 automated sequencer (Applied Biosystems). MLPA data were collected using the Gene Mapper software (MRC-Holland) and exported to be analyzed by Coffalyzer software (MRC-Holland). Fifteen unrelated control DNA samples were included in the analysis as reference population. Based on the results obtained on the control group, observed values falling within the range of 0.7–1.3 were considered to have two copies of the gene.
Functional characterization of germline BRAF mutations
The single-base changes resulting in the p.Thr241Pro (LS), p.Glu275Lys (CFCS), p.Trp531Cys (NS), p.Leu597Val (NS), p.Thr599Arg (CFCS), p.Lys601Gln (CFCS) and p.Val600Glu (cancer-associated mutant) were introduced by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene, La Jolla, CA) into a full-length human Myc-tagged BRAF cDNA cloned in a pEFP vector. NIH-3T3 cell colony focus formation assay was assessed as described by Di Fiore et al. [1987]. Briefly, cells were grown in DMEM supplemented with 10% calf serum (Cambrex, Rockland, MA), and seeded at a density of 1.3 × 105 cells/10cm dish the day before the transfection. After 24 h, cells were shifted to 5% calf serum DMEM, and left at confluency for 15 days with medium changes every three days. Foci were visualized by crystal violet staining. To evaluate the effects exerted by each mutant on the MAPK cascade, HEK293 cells were seeded at a density of 1.5 × 106 cells/dish the day before transfection, and kept in DMEM supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Cells were transfected transiently with wild type or mutant BRAF expression constructs, and, after 24 h, cells were switched to serum-deprived medium for 12 h and lysed. Cell lysates (50–100 μg) were run on 10% SDS-PAGE, transferred onto nitrocellulose membrane, and examined for the basal phosphorylation levels of MEK and ERK proteins using the following antibodies: anti-pMEK1/2, #9121 (Cell Signalling, Danvers, MA); anti-MEK1/2, #9122 (Cell Signalling); anti-pERK1/2, #9102 (Cell Signalling); anti-ERK1/2, #SC-94 (Santa Cruz Biotechnology, Santa Cruz, CA).
Results
BRAF mutation analysis
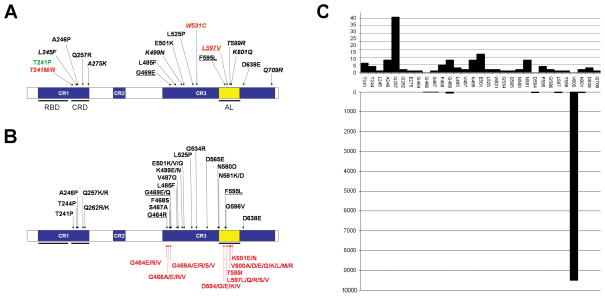
DHPLC screening of the entire BRAF coding sequence in peripheral blood leukocyte genomic DNA specimens allowed the identification of a heterozygous missense mutation in one of six subjects with LS, and five individuals with NS (1.9%) (Table 1 and Figure 1A). All cases were sporadic. Parental DNAs were available for five subjects, and no parent was identified as carrying the mutation, providing evidence for their de novo occurrence. Genotyping confirmed paternity in all tested families. To verify that the C-to-G transversion at position 722 (p.Thr241Arg), identified in one subject with NS for whom parental DNAs were not available, was not a gene variant occurring in the population, 150 population-matched control individuals were screened by DHPLC and sequencing, and none harbored that change. Mutations were observed in exons 6, 13 and 15, and were predicted to affect residues located in the cysteine-rich domain, within the conserved region 1 (CR1), and the kinase domain (Figure 1A). The p.Thr241Pro amino acid substitution identified in the subject with LS had previously been reported in three children with a phenotype apparently fitting CFCS [Nava et al., 2007; Schulz et al., 2008], but all NS-causing mutations were novel (Figure 1B).
TABLE 1.
List of BRAF gene mutations identified in Noonan syndrome, LEOPARD syndrome and cardiofaciocutaneous syndrome.
| Disorders and cases | Nucleotide changea | exon | Amino Acid change | Domain | Status |
|---|---|---|---|---|---|
| NS (N= 270) | |||||
| 1 | c.722C>T | 6 | p.Thr241Met | CRD (CR1) | de novo |
| 1 | c.722C>G | 6 | p.Thr241Arg | CRD (CR1) | controls |
| 2 | c.1593G>C | 13 | p.Trp531Cys | KD (CR3) | de novo |
| 1 | c.1789C>G | 15 | p.Leu597Val | KD (CR3) | de novo |
| LS (N= 6) | |||||
| 1 | c.721A>C | 6 | p.Thr241Pro | CRD (CR1) | de novo |
| CFCS (N= 33) | |||||
| 1 | c.735A>T | 6 | p.Leu245Phe | CRD (CR1) | controls |
| 1 | c.736G>C | 6 | p.Ala246Pro | CRD (CR1) | de novo |
| 1 | c.770A>G | 6 | p.Gln257Arg | CRD (CR1) | de novo |
| 1b | c.823G>A | 6 | p.Glu275Lys | CRD (CR1) | de novo |
| 1 | c.1406G>A | 11 | p.Gly469Glu | KD (CR3) | de novo |
| 1 | c.1455G>C | 12 | p.Leu485Phe | KD (CR3) | de novo |
| 1 | c.1497A>C | 12 | p.Lys499Asn | KD (CR3) | de novo |
| 1 | c.1501G>A | 12 | p.Glu501Lys | KD (CR3) | de novo |
| 1 | c.1574T>C | 13 | p.Leu525Pro | KD (CR3) | de novo |
| 2 | c.1785T>G | 15 | p.Phe595Leu | KD (CR3) | de novo |
| 1 | c.1796C>G | 15 | p.Thr599Arg | KD (CR3) | de novo |
| 2 | c.1801A>C | 15 | p.Lys601Gln | KD (CR3) | de novo |
| 1 | c.1914T>G | 16 | p.Asp638Glu | KD (CR3) | de novo |
| 1 | c.1914T>A | 16 | p.Asp638Glu | KD (CR3) | de novo |
| 1b | c.2126A>G | 17 | p.Gln709Arg | KD (CR3) | de novo |
Position referred to the A of the ATG-translation initiation codon in the reference cDNA sequence (NM_004333.3).
These patients exhibited severe neonatal feeding difficulties, typical facial dysmorphisms, ectodermal anomalies associated with normal growth and cognitive development. NS: Noonan syndrome; LS: LEOPARD syndrome; CFCS: cardiofaciocutaneous syndrome; CRD: cysteine-rich domain; KD: kinase domain; CR1: conserved region 1; CR3: conserved region 3.
FIGURE 1. BRAF domain structure and location of residues altered in Noonan, LEOPARD and cardiofaciocutaneous syndromes.
BRAF protein domains are indicated (CR, conserved region; RBD, RAS binding domain, CRD, cysteine-rich domain, AL, activation loop). (A) Mutations identified in the present study. Amino acid substitutions identified in Noonan syndrome, LEOPARD syndrome or cardiofaciocutaneous syndrome are indicated in red, green and black, respectively. Novel mutations are indicated in italics, while mutations documented to occur as somatic changes in human cancers are underlined. (B) Mutations previously identified in cardiofaciocutaneous syndrome (including individuals with a phenotype resembling Costello syndrome during infancy) (above the cartoon) or human cancers (under the cartoon) (prevalence ≥1.5%, according the COSMIC database, http://www.sanger.ac.uk/genetics/CGP/cosmic/). A heterozygous condition for the p.Lys499Glu and p.Glu501Lys substitutions has also been observed in two individuals diagnosed with NS [Razzaque et al., 2007; Nystrom et al., 2008]. (C) Histogram showing distribution and relative prevalence of germline (above panel) and somatic (lower panel) BRAF mutations in human disease (updated to June 2008).
Based on the recent evidence supporting the idea that a small fraction of NS might be caused by duplication of the PTPN11 gene [Shchelochkov et al., 2008], we also explored possible occurrence of BRAF gene copy number variation in NS. MLPA analysis was performed on 50 of the 265 BRAF mutation-negative patients included in this study by analyzing two probes that were complementary to exons 5 and 13 of the gene. The analysis revealed the presence of peak sizes within the normal range values for both exons in all subjects, excluding a major contribution of BRAF gene amplification in NS pathogenesis.
To characterize the molecular diversity of germline BRAF mutations and their phenotypic spectrum further, a cohort of 33 unrelated sporadic subjects with CFCS without mutations in known disease genes and two individuals with a diagnosis of NFNS without mutation in NF1, SPRED1 and PTPN11 were analyzed. BRAF mutation scanning identified 15 different missense nucleotide substitutions in 17 individuals (51.5% of cases), while no sequence variant was observed in the two subjects with NFNS (Table 1 and Figure 1A). Nine amino acid substitutions had been previously reported among subjects with a diagnosis of CFCS (Figure 1A) [Niihori et al., 2006; Rauen, 2006; Rodriguez-Viciana et al., 2006; Gripp et al., 2007; Makita et al., 2007; Narumi et al., 2007; Nava et al., 2007; Schulz et al., 2008], while five were novel. All cases were considered sporadic by the referring clinicians, and genotyping of parental DNAs performed in 16 families for which samples were available confirmed the de novo occurrence of the missense changes. The A-to-T transversion at position 735 (p.Leu245Phe), which we were not able to assess in that manner, was not observed among controls. Mutations primarily affected exons 6, 12, 15 and 16 (82% of identified changes) and altered affected residues located in the cysteine-rich and kinase domains. Three mutations (p.Phe595Leu, p.Lys601Gln and p.Asp638Glu) were recurrent.
BRAF is a member of a small family of threonine-serine protein kinases functioning as effectors of RAS and upstream components of the MAPK module [Wellbrock et al., 2004]. Different from the other two members of the family, RAF1 and ARAF, that are only occasionally mutated in human cancers, somatic BRAF mutations frequently occur in malignant melanomas and in thyroid, colorectal and ovarian cancers. Comparison of the spectra of germline transmitted and somatically acquired BRAF mutations indicated minimal overlap (Figure 1C), which is reminiscent of the available data collected for PTPN11 [Tartaglia et al., 2003, 2006] and KRAS [Schubbert et al., 2006; Niihori et al., 2006; Carta et al., 2006; Zenker et al., 2007] mutations. Three BRAF germline changes, c.1406G>A (p.Gly469Glu), c.1785T>G (p.Phe595Leu) and c.1789C>G (p.Leu597Val), had rarely been reported to be acquired somatically (pooled prevalence less then 3/10,000, according to the catalogue of somatic mutations in cancer [COSMIC] database at http://www.sanger.ac.uk/genetics/CGP/cosmic/), while the c.1796C>G (p.Thr599Arg) and c.1801A>C (p.Lys601Gln) transversions affected residues that are substituted by different amino acids in human cancers. None of the patients carrying these changes had developed any malignancy at the time of their last examination (mean age = 6.1 years) (Supp. Table S1).
Phenotypic spectrum of germline BRAF mutations
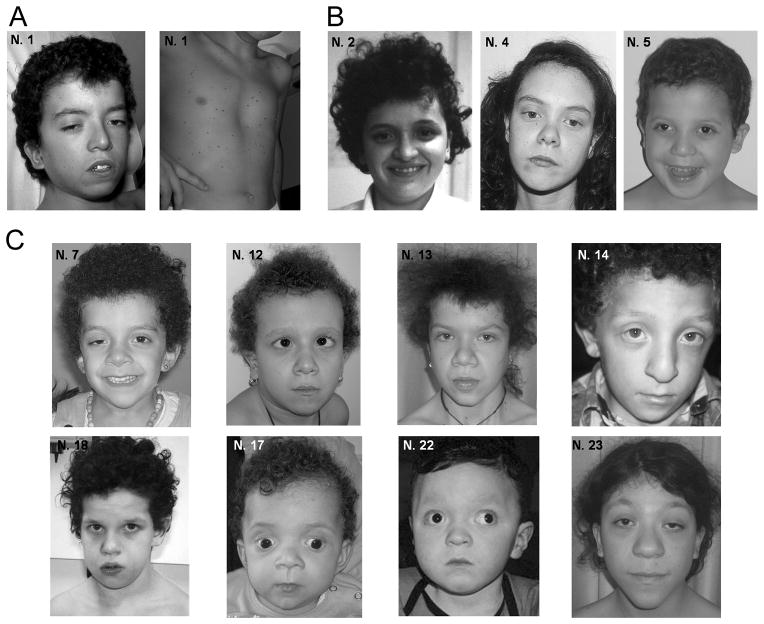
A detailed clinical characterization of BRAF mutation-positive NS patients indicated that these individuals exhibited as constant findings neonatal growth failure and variable feeding difficulties, short stature, dysmorphic facial features (dolicocephaly, prominent forehead, hypertelorism and low-set ears with thickened helix, most commonly), mild-to-moderate cognitive deficits, skeletal anomalies and hypotonia (Figure 2 and Supp. Table S1). Congenital cardiac defects were present in two subjects (PS and atrial septal defect), while hyperpigmented cutaneous lesions were observed in three. Of note, CFCS-related skin anomalies, polydramnios and HCM were absent in all the subjects. Clinical re-evaluation of the individual diagnosed with LS documented reduced growth, craniofacial anomalies, short and webbed neck, thorax defects, mitral and aortic valve dysplasia, delayed puberty, cognitive deficits, sensorineural deafness, seizures, neonatal hypotonia, generalized skeletal hypomineralization and fibrous cystic lesions of the pelvis, hyperkeratosis, cafè-au-lait spots, as well as multiple nevi and dark colored lentigines, which were spread on the whole body including some on the palms and soles.
FIGURE 2. Facial dysmorphia and other features of BRAF mutation-positive subjects.
(A) LEOPARD syndrome; (B) Noonan syndrome; (C) cardiofaciocutaneous syndrome.
Analysis of the clinical features of BRAF mutation-positive CFCS patients indicated a wide phenotypic variability, although all subjects displayed typical dysmorphic facies, cardiac defects, and skin and skeletal anomalies (Figure 2 and Supp. Table S1). The most common facial features included prominent forehead, bitemporal narrowing, hypertelorism, downslanting palpebral fissures, ptosis, thick palpebral lids, epicanthal folds, flat nasal bridge, thick lips and low set ears with thick helices and large lobes. PS and HCM were the most common cardiac defects, observed in approximately 90% and 55% of cases, respectively. Ectodermal anomalies included absent or hypoplastic eyebrows, curly and sparse hair, hyperkeratosis, and keratosis pilaris. Short stature (<3 centile) was present in 53% of the patients. Most subjects showed some degree of neonatal growth failure, poor sucking and/or swallowing, or required tube feeding. Ocular anomalies were detected in 82% of the patients, including strabismus in half of them. Hyperhidrosis appeared a common feature in CFCS patients with BRAF mutations (78% of cases). Moderate-to-severe mental retardation was observed in all but two individuals (87%), which was found to be associated with seizures or hypotonia in 9 and 13 individuals, respectively. The two subjects with normal cognitive development exhibited hypotonia at birth (N07) or deafness (N23) as the only neurological anomalies observed. These two individuals had originally been classified as affected by CFCS based on the presence of severe neonatal feeding difficulties, typical facial dysmorphism, ectodermal anomalies and hemangiomas (N07), and fetal macrosomia, distinctive facial dysmorphisms, and skin and ocular anomalies (N23). Pigmentary changes, such as café-au-lait spots, naevi or lentigines, were observed in 9/17 individuals, two of which (N13 and N23) exhibiting a considerably high number of lentigines.
Functional analysis
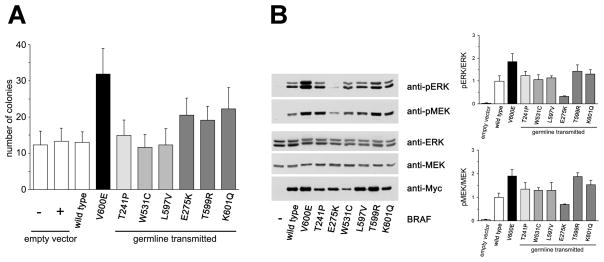
Consistent with the non-overlapping distribution of germline and somatic mutations, biochemical data indicate that the NS/CFCS-causing and cancer-associated PTPN11 and KRAS gene lesions have diverse abilities to perturb signaling traffic [Seeburg et al., 1984; Tartaglia et al., 2003, 2006; Chan et al., 2005; Mohi et al., 2005; Schubbert et al., 2005, 2006, 2007a], suggesting that cancer-associated lesions might severely affect embryonic or fetal development. In agreement with this idea, widespread expression of the p.Val600Glu BRAF change, which accounts for the large majority (> 90%) of total BRAF amino acid substitutions in human cancers, is embryonic lethal [Mercer et al., 2005]. Of note, this oncogenic lesion has been documented to confer a substantially higher transforming behavior to BRAF compared to other less common amino acid substitutions identified in cancer cell lines or primary tumors [Davies et al., 2002]. To further explore this issue, the activating strength of six selected BRAF mutants, p.Thr241Pro (LS), p.Trp531Cys and Leu597Val (NS), and p.Glu275Lys, p.Thr599Arg and p.Lys601Gln (CFCS), in promoting NIH-3T3 colony focus formation was assessed in vitro, and compared to the recurrent cancer-associated p.Val600Glu protein. Results from at least three independent experiments, each performed in duplicate, demonstrated that untransfected cells and cells transfected with empty vector or overexpressing wild type BRAF developed comparable numbers of foci, while those expressing the oncogenic p.Val600Glu change exhibited a statistically significantly higher number of foci (Figure 3A). CFCS-causing mutants appeared less activating compared with the p.Val600Glu mutant, while the LS- (p.Thr241Pro) and NS-causing (p.Trp531Cys and p.Leu597Val) changes did not confer enhanced transformation to cells. The effects exerted by each mutant on the MAPK cascade were evaluated by comparing the levels of MEK and ERK phosphorylation in transiently transfected HEK293 cells cultured in DMEM supplemented with 10% fetal bovine serum, and then switched to serum-deprived medium. Enhanced phosphorylation of MEK and ERK proteins was observed in cells overexpressing the oncogenic p.Val600Glu BRAF and, to a lesser extent, in those cells expressing the p.Thr599Arg and p.Lys601Gln mutants. A slight increase in pMEK level was also observed in cells expressing the p.Thr241Pro, p.Trp531Cys and p.Leu597Val mutants, while expression of the p.Glu275Lys mutant was associated with an impaired phosphorylation level of the MEK and ERK proteins (Figure 3B).
FIGURE 3. Functional characterization of selected germline transmitted BRAF mutations.
(A) Focus assays of NIH-3T3 cells transfected with each of the BRAF constructs coding for the p.Thr241Pro (LS), p.Glu275Lys (CFCS), p.Trp531Cys (NS), p.Leu597Val (NS), p.Thr599Arg (CFCS) and p.Lys601Gln (CFCS) mutants, the cancer-associated p.Val600Glu mutant, and the wild type (WT) protein. Data illustrate results of at least three experiments performed in duplicate. (B) MEK and ERK phosphorylation assays in cells transiently expressing one of the six selected BRAF mutants, the cancer-associated p.Val600Glu or wild type BRAF proteins. Blots are representative of three experiments performed (left), while phosphorylation ratios (right) are expressed as the mean of three replicates ± SD.
Discussion
In this report, we established that heterozygous missense mutations in the BRAF gene can underlie LS, and expanded the molecular diversity of BRAF lesions causing NS and CFCS. We also provided an estimate of the prevalence of BRAF mutations in NS as well as a more complete assessment of the phenotypic variation associated with these molecular lesions. Finally, we showed that a selected panel of mutations associated with LS, NS or CFCS have a reduced transforming capability compared with the recurrent oncogenic p.Val600Glu substitution, indicating less potency in deregulating the BRAF-mediated signal flow.
BRAF involvement in NS pathogenesis was suggested by Razzaque et al. [2007], who described the occurrence of the CFC-associated c.1501G>A missense change (p.Glu501Lys) in a sporadic case diagnosed with NS. More recently, Nystrom et al. [2008] reported on the identification of the c.1495A>G transition (p.Lys499Glu), which was also previously documented to recur in CFCS, in one subject with a phenotype fitting NS and characterized by severe cognitive impairment and retarded motor development. Here, we extended those initial observations in multiple respects. Our present analysis of a large cohort with sporadic and familial cases of NS estimated the BRAF mutation prevalence to be less than 2%, indicated that mutations preferentially occur in sporadic cases, and excluded a major contribution of BRAF gene amplification in NS pathogenesis. None of the NS-associated mutations identified in the present work have previously been observed in CFCS, suggesting a genotype-phenotype correlation. The detection of the p.Trp531Cys amino acid substitution in two of the five individuals with the disorder, as well as the clustering of mutations identified in three NS cases and the single LS case at the threonine residue 241, further supports the idea that the phenotype resulting from germline BRAF defects might be allele specific. While the p.Thr241Pro change identified in one individual with LS had previously been reported in three subjects with a phenotype apparently fitting CFCS [Nava et al., 2007; Schulz et al., 2008], the newborn age at diagnosis of two of these subjects and the unavailability of clinical data do not allow any conclusion regarding the phenotypic variation associated with this molecular lesion.
Together with the previously published records, 38 germline BRAF mutations have been identified so far. All are missense defects that are not randomly distributed, clustering in the cysteine-rich domain and in the amino-terminal portion and activation segment of the kinase domain. Most mutations are recurrent, with substitutions of residues Gln257 and Glu501 accounting for approximately 40% of total defects. While a high percentage of mutations affect exons 6 and 12 (approximately 70% of total changes), this study recognized a novel mutational hot spot located within exon 15. The distribution of BRAF mutations significantly differs from that observed for RAF1 gene lesions, for which the reason can be ascribed to the distinct regulatory mechanisms controlling the function of these two kinases [Wellbrock et al., 2004; Dhomen and Marais, 2007]. With the exception of changes affecting two homologous residues (Asp594 and Thr599 in BRAF, and Asp486 and Thr491 in RAF1) located within the activation segment of the kinase domain, mutations alter different domains of the two proteins. Most of the RAF1 amino acid substitutions affect the amino-terminal 14-3-3 consensus sequence (within CR2) and two residues located near the carboxy-terminal 14-3-3 binding site. On the other hand, BRAF changes cluster within the cysteine-rich region of CR1 and the amino-terminal portion of the kinase domain.
Consistent with the observation that cancers are uncommon among subjects carrying a BRAF mutation, germline BRAF mutations are rarely observed as somatic mutations contributing to oncogenesis. Such a differential spectrum of mutations is consistent with the present NIH-3T3 cell colony focus formation assay data indicating that CFCS-associated BRAF mutants (p.Glu275Lys, p.Thr599Arg and p.Lys601Gln) have a reduced transforming capability compared with the recurrent oncogenic p.Val600Glu BRAF protein, and that the NS- (p.Trp531Cys and p.Leu597Val) and LS-associated (p.Thr241Pro) BRAF mutants do not confer enhanced transformation to cells. While these results support the view that the germline transmitted mutations have less potency in deregulating BRAF function, they also suggest that NS- and LS-associated mutations are less activating compared to those associated with CFCS, which parallels the present observations indicating that phenotypes arising from germline BRAF defects might be allele specific. As previously documented by other groups [Rodriguez-Viciana et al., 2006; Dhomen and Marais, 2007], however, the present in vitro functional studies indicate that the selected amino acid changes can confer either increased or decreased activity upon the BRAF mutant as compared to the wild type protein. Even though additional studies are required to characterize more precisely the perturbing effects of the NS-, LS- and CFCS-associated mutations on BRAF activity and signal transduction, our data suggest that multiple alternative mechanisms are likely to be involved in the functional dysregulation of the kinase.
Based on the present and previously gathered data, BRAF gene mutations account for approximately 50–75% of subjects with a diagnosis of CFCS, including a number of cases reported to exhibit a phenotype resembling CS during infancy [Nava et al., 2007; Schulz et al., 2008]. In contrast, BRAF mutations appear to underlie only a small fraction of NS and LS (<2%). The striking finding of a causative association between BRAF mutations and NS/LS expands the phenotypic heterogeneity associated with these gene defects, and further points out that the large clinical variability in NS is closely related to the underlying gene involved. These subjects share neonatal growth failure and feeding difficulties, mild-to-moderate cognitive deficits, and hypotonia, and have higher prevalence of multiple nevi and dark colored lentigines. As adults, they display the full-blown phenotype of this disorder, which appears more severe compared to that associated with PTPN11 and SOS1 mutations. In these individuals, however, polyhydramnios, HCM, and CFCS-related skin features [Roberts et al., 2006; Armour and Allanson, 2008] are uncommon or absent, and cardiac defects, neurological impairment and feeding problems appear to be less severe compared to what is generally observed in CFCS. Similarly, the individual with LS who was heterozygous for the p.Thr241Pro change displayed a phenotype overlapping that of PTPN11 or RAF1 mutation-positive LS cases, although cognitive difficulties were more severe and the cardiac involvement milder. Of note, two previously unreported BRAF missense changes (p.Glu275Lys and p.Gln709Arg) were identified in two subjects (N07 and N23) originally diagnosed with CFCS on the basis of the severe neonatal growth and feeding difficulties, distinctive dysmorphic features and extensive ectodermal involvement who, however, showed normal growth and cognitive development. Case N23 also exhibited multiple lentigines and other features recurring in LS, which also were observed in patient N13. While the occurrence of distinct BRAF gene mutations in subjects with a phenotype undoubtedly fitting NS indicates that some features might be allele-specific, the observation of a subgroup of subjects in the present cohort with an “intermediate” phenotype suggests that a clinical continuum characterized by a differential combination and severity of features is associated with defects in the BRAF gene. These findings further emphasize the difficulty in identifying efficient clinical criteria to define CFCS, LS or NS nosologically, and make evident the usefulness of a molecular-based definition (individual gene involved and type of gene lesion) of these clinically overlapping conditions that might direct clinicians toward a more appropriate management of patients.
Overall, the present data indicate that patients heterozygous for BRAF mutations do not invariably have full-blown CFCS, and that molecular screening of this gene would be worthwhile in individuals with clinical features fitting NS or LS with moderate to severe cognitive deficits. While the analysis of a larger cohort of patients is required to characterize more precisely the phenotypic variability associated with these molecular lesions, consistent with recent data by other groups [Rauen, 2006; Nava et al., 2007; Nystrom et al., 2008; Schulz et al., 2008], the present report documents that germline BRAF mutations are associated with a wide phenotypic spectrum, further emphasizing the tight clinical and molecular relationships of this family of disorders. Like the previous discovery that different PTPN11 mutations cause NS and LS, our current findings suggest that the phenotypes arising from germline BRAF defects are likely to be also allele specific.
Supplementary Material
Acknowledgments
We are indebted to the patients and families who participated in the study and the physicians who referred the subjects. This work was supported by Telethon-Italy grant GGP07115 (M.T.), “Programma di Collaborazione Italia-USA/malattie rare 2007” grants (M.T. and A.S.), US National Institutes of Health Grants HL71207, HD01294, HL074728 (B.D.G.), Italian Ministry of Health Grant RC 2007, 2008 (B.D.), and Regione Piemonte Ricerca Sanitaria Finalizzata 2006–2007 (GBF and MCS). V.C. was supported by a fellowship from Associazione Genitori Oncologia Pediatrica ONLUS “Per un sorriso in più”, Lecce.
References
- Allanson JE. Noonan syndrome. J Med Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- Armour CM, Allanson JE. Further delineation of cardio-facio-cutaneous syndrome: clinical features of 38 individuals with proven mutations. J Med Genet. 2008;45:249–254. doi: 10.1136/jmg.2007.054460. [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, Cools J, Marynen P, Thomas G, Yoshimura A, Legius E. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Carta C, Pantaleoni F, Bocchinfuso G, Stella L, Vasta I, Sarkozy A, Digilio C, Palleschi A, Pizzuti A, Grammatico P, Zampino G, Dallapiccola B, Gelb BD, Tartaglia M. Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RJ, Leedy MB, Munugalavadla V, Voorhorst CS, Li Y, Yu M, Kapur R. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood. 2005;105:3737–3742. doi: 10.1182/blood-2004-10-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Bottillo I, Sarkozy A, Carta C, Neri C, Bellacchio E, Schirinzi A, Conti E, Zampino G, Battaglia A, Majore S, Rinaldi MM, Carella M, Marino B, Pizzuti A, Digilio MC, Tartaglia M, Dallapiccola B. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet. 2005;77:1092–1101. doi: 10.1086/498454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Kraus MH, Segatto O, King CR, Aaronson SA. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]
- Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, Pizzuti A, Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet. 2006;15:R220–R226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Lin AE, Stabley DL, Nicholson L, Scott CI, Jr, Doyle D, Aoki Y, Matsubara Y, Zackai EH, Lapunzina P, Gonzalez-Meneses A, Holbrook J, Agresta CA, Gonzalez I, Sol-Church K. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Lin AE, Nicholson L, Allen W, Cramer A, Jones KL, Kutz W, Peck D, Rebolledo MA, Wheeler PG, Wilson W, Al-Rahawan MM, Stabley DL, Sol-Church K. Further delineation of the phenotype resulting from BRAF or MEK1 germline mutations helps differentiate cardio-facio-cutaneous syndrome from Costello syndrome. Am J Med Genet A. 2007;143:1472–1480. doi: 10.1002/ajmg.a.31815. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubenstein A, Viskochil D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997;278:51–57. [PubMed] [Google Scholar]
- Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants: implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- Kerr B, Delrue MA, Sigaudy S, Perveen R, Marche M, Burgelin I, Stef M, Tang B, Eden OB, O’Sullivan J, De Sandre-Giovannoli A, Reardon W, Brewer C, Bennett C, Quarell O, M’Cann E, Donnai D, Stewart F, Hennekam R, Cavé H, Verloes A, Philip N, Lacombe D, Levy N, Arveiler B, Black G. Genotype-phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J Med Genet. 2006;43:401–405. doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legius E, Schrander-Stumpel C, Schollen E, Pulles-Heintzberger C, Gewillig M, Fryns JP. PTPN11 mutations in LEOPARD syndrome. J Med Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita Y, Narumi Y, Yoshida M, Niihori T, Kure S, Fujieda K, Matsubara Y, Aoki Y. Leukemia in Cardio-facio-cutaneous (CFC) syndrome: a patient with a germline mutation in BRAF proto-oncogene. J Pediatr Hematol Oncol. 2007;29:287–290. doi: 10.1097/MPH.0b013e3180547136. [DOI] [PubMed] [Google Scholar]
- Martinelli S, Torreri P, Tinti M, Stella L, Bocchinfuso G, Flex E, Grottesi A, Ceccarini M, Palleschi A, Cesareni G, Castagnoli L, Petrucci TC, Gelb BD, Tartaglia M. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum Mol Genet. 2008;17:2018–2029. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C. Expression of endogenous oncogenic V600E B-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohi MG, Williams IR, Dearolf CR, Chan G, Kutok JL, Cohen S, Morgan K, Boulton C, Shigematsu H, Keilhack H, Akashi K, Gilliland DG, Neel BG. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7:179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Narumi Y, Aoki Y, Niihori T, Neri G, Cave H, Verloes A, Nava C, Kavamura MI, Okamoto N, Kurosawa K, Hennekam RC, Wilson LC, Gillessen-Kaesbach G, Wieczorek D, Lapunzina P, Ohashi H, Makita Y, Kondo I, Tsuchiya S, Ito E, Sameshima K, Kato K, Kure S, Matsubara Y. Molecular and clinical characterization of cardio-facio-cutaneous (CFC) syndrome: overlapping clinical manifestations with Costello syndrome. Am J Med Genet A. 2007;143:799–807. doi: 10.1002/ajmg.a.31658. [DOI] [PubMed] [Google Scholar]
- Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, Aoki Y, Matsubara Y, Arveiler B, Lacombe D, Pasmant E, Parfait B, Baumann C, Heron D, Sigaudy S, Toutain A, Rio M, Goldenberg A, Leheup B, Verloes A, Cave H. Cardio-facio-cutaneous and Noonan syndromes due to mutations in RAS/MAPK signaling pathway: genotype/phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- Nyström AM, Ekvall S, Berglund E, Björkqvist M, Braathen G, Duchen K, Enell H, Holmberg E, Holmlund U, Olsson-Engman M, Annerén G, Bondeson ML. Noonan and Cardio-facio-cutaneous syndromes: Two Clinically and Genetically Overlapping Disorders. J Med Genet. 2008;45:500–506. doi: 10.1136/jmg.2008.057653. [DOI] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, Lopez Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- Rauen KA. Distinguishing Costello versus cardio-facio-cutaneous syndrome: BRAF mutations in patients with a Costello phenotype. Am J Med Genet A. 2006;140:1681–1683. doi: 10.1002/ajmg.a.31315. [DOI] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, Fujiwara Y, Matsushima M, Mizuno K, Tokuyama M, Hirota H, Muneuchi J, Higashinakagawa T, Matsuoka R. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- Roberts A, Allanson J, Jadico SK, Kavamura MI, Noonan J, Opitz JM, Young T, Neri G. The cardiofaciocutaneous syndrome. J Med Genet. 2006;43:833–842. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- Sarkozy A, Digilio MC, Zampino G, Dallapiccola B, Tartaglia M, Gelb BD. LEOPARD syndrome: clinical aspects and molecular pathogenesis. In: Zenker M, editor. Monographs in Human Genetics - Vol. 17. Noonan syndrome and related disorders: A matter of deregulated RAS signaling. Basel: Karger Press; 2009. pp. 55–65. [Google Scholar]
- Schubbert S, Lieuw K, Rowe SL, Lee CM, Li X, Loh ML, Clapp DW, Shannon KM. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–317. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Bollag G, Lyubynska N, Nguyen H, Kratz CP, Zenker M, Niemeyer CM, Molven A, Shannon K. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27:7765–7770.a. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308.b. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Schulz AL, Albrecht B, Arici C, van der Burgt I, Buske A, Gillessen-Kaesbach G, Heller R, Horn D, Hubner CA, Korenke GC, Konig R, Kress W, Kruger G, Meinecke P, Mucke J, Plecko B, Rossier E, Schinzel A, Schulze A, Seemanova E, Seidel H, Spranger S, Tuysuz B, Uhrig S, Wieczorek D, Kutsche K, Zenker M. Mutation and phenotypic spectrum in patients with Cardio-Facio-Cutaneous and Costello syndrome. Clin Genet. 2008;73:62–70. doi: 10.1111/j.1399-0004.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- Shchelochkov OA, Patel A, Weissenberger GM, Chinault AC, Wiszniewska J, Fernandes PH, Eng C, Kukolich MK, Sutton VR. Duplication of chromosome band 12q24.11q24.23 results in apparent Noonan syndrome. Am J Med Genet A. 2008;146:1042–1048. doi: 10.1002/ajmg.a.32215. [DOI] [PubMed] [Google Scholar]
- Stumpf DA, Alksne JF, Annegers JF, Brown SS, Conneally PM, Housman D, Leppert MF, Miller JP, Moss ML, Pileggi AJ, Rapin I, Strohman RC, Swanson LW, Zimmerman A. Neurofibromatosis: conference statement. Arch Neurol. 1988;45:575–578. [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, Zampino G, Burgt I, Palleschi A, Petrucci TC, Sorcini M, Schoch C, Foa R, Emanuel PD, Gelb BD. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, Ustaszewska A, Martin J, Bristow J, Carta C, Lepri F, Neri C, Vasta I, Gibson K, Curry CJ, Siguero JP, Digilio MC, Zampino G, Dallapiccola B, Bar-Sagi D, Gelb BD. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Molecular genetics of Noonan syndrome. In: Zenker M, editor. Monographs in Human Genetics - Vol. 17. Noonan syndrome and related disorders: A matter of deregulated RAS signaling. Basel: Karger Press; 2009. pp. 20–39. [Google Scholar]
- Van der Burgt I, Berends E, Lommen E, van Beersum S, Hamel B, Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet. 1994;53:187–191. doi: 10.1002/ajmg.1320530213. [DOI] [PubMed] [Google Scholar]
- Voron DA, Hatfield HH, Kalkhoff RK. Multiple lentigines syndrome. Case report and review of the literature. Am J Med. 1976;60:447–456. doi: 10.1016/0002-9343(76)90764-6. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Zampino G, Pantaloni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, De Feo E, Delogu A, Sarkozy A, Atzeri F, Selicorni A, Rauen KA, Weksberg R, Dallapiccola B, Ballabio A, Gelb BD, Neri G, Tartaglia M. Diversity, parental germline origin and phenotypic spectrum of de novo HRAS misssense changes in Costello syndrome. Hum Mutat. 2007;28:265–272. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
- Zenker M, Lehmann K, Schulz AL, Barth H, Hansmann D, Koenig R, Korinthenberg R, Kreiss-Nachtsheim M, Meinecke P, Morlot S, Mundlos S, Quante AS, Raskin S, Schnabel D, Wehner LE, Kratz CP, Horn D, Kutsche K. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.