SUMMARY
Pluripotent stem cells have distinct metabolic requirements, and reprogramming cells to pluripotency requires a shift from oxidative to glycolytic metabolism. Here, we show that this shift occurs early during reprogramming of human cells and requires Hypoxia Inducible Factors in a stage-specific manner. HIF1α and HIF2α are both necessary to initiate this metabolic switch and for acquisition of pluripotency, and stabilization of either protein during early phases of reprogramming is sufficient to induce the switch to glycolytic metabolism. In contrast, stabilization of HIF2α during later stages represses reprogramming, due at least in part to up-regulation of TNF-related apoptosis-inducing ligand (TRAIL). TRAIL inhibits iPSC generation by repressing apoptotic caspase 3 activity specifically in cells undergoing reprogramming, but not hESCs, and inhibiting TRAIL activity enhances hiPSC generation. These results shed light on the mechanisms underlying the metabolic shifts associated with acquisition of a pluripotent identity during reprogramming.
INTRODUCTION
In contrast to differentiated cells, human embryonic stem cells (hESC) rely mainly on glycolysis for their source of energy, regardless of oxygen availability (Folmes et al., 2011; Panopoulos et al., 2012; Prigione and Adjaye, 2010; Varum et al., 2011; Zhang et al., 2011; Zhou et al., 2012). Pluripotent cells share this metabolic particularity with cancer cells (Warburg effect, Cairns et al., 2011). In both cell types glycolytic genes are up-regulated, mitochondrial activity is reduced and lactate production is significantly increased (Panopoulos et al., 2012; Prigione et al., 2010; Varum et al., 2011; Yanes et al., 2010). Further, it has been proposed recently that the metabolic properties of stem cells and cancer cells are important for their identity (Greer et al., 2012; Rafalski et al., 2012). However, it is not yet clear how stem cells gain this metabolic signature and how they again activate mitochondrial oxidative phosphorylation pathways during differentiation.
The bioenergetics of pluripotent cells can vary depending on their developmental stage. For example, mouse epiblasts stem cells, that are believed to be at the same primed stage than hESC, are also highly glycolytic while more naïve mouse ESC are bivalent in their energy production, switching from glycolysis to mitochondrial respiration on demand (Zhou et al., 2012). Human induced pluripotent stem cells (iPSC) are usually reprogrammed from somatic cells to a primed stage and are very similar metabolically to hESC (Panopoulos et al., 2012; Suhr et al., 2010; Varum et al., 2011). Therefore, a metabolic switch from oxidative to highly glycolytic needs to take place during iPSC formation. Supporting this idea, inhibition of glycolysis reduces the reprogramming efficiency while stimulation of glycolytic activity enhances iPSC generation (Folmes et al., 2011; Panopoulos et al., 2012; Zhu et al., 2010). How iPSCs establish a Warburg-like metabolic phenotype during the reprogramming process is largely unknown.
The dependency of stem cells on glycolysis to produce ATP could be an adaptation to low oxygen tensions in vivo since hypoxia has appeared as a key feature of the stem cell niche (Mohyeldin et al., 2010; Suda et al., 2011). Further, low oxygen levels are beneficial for embryonic stem cells (hESC), adult stem cells (Danet et al., 2003; Ezashi et al., 2005; Morrison et al., 2000; Simsek et al., 2010; Studer et al., 2000) and cancer cells (Axelson et al., 2005; Cabarcas et al., 2011; Mathieu et al., 2011; Takubo and Suda, 2012). Cellular adaptation to hypoxic conditions is mainly mediated through the activation of the oxygen-sensitive transcription factors, Hypoxia-Inducible Factors (HIFs). In normoxia, HIF1α and HIF2α undergo prolyl-hydroxylation that leads to specific binding to the ubiquitin E3 ligase VHL, poly-ubiquitination and proteasomal degradation. However HIF1α and HIF2α are stabilized in low oxygen, dimerize with HIF1β and control the transcription of multiple target genes, including genes involved in glucose metabolism (Pouyssegur et al., 2006; Semenza, 2003). HIF1α is expressed ubiquitously, while HIF2α expression is more tissue-restricted and both factors have essential roles during development (Compernolle et al., 2002; Iyer et al., 1998; Ryan et al., 1998). Increasing evidence suggests that HIFs can activate factors involved in pluripotency and regulate the stem cell phenotype, both in normal and cancer cells (Ezashi et al, 2005, Takubo & Suda, 2012, Covello et al., 2006; Mathieu et al., 2011, Mathieu et al, 2013). In addition, hypoxia enhances the generation of iPSC (Yoshida et al., 2009). However the mode of function of HIFs in the process is not fully understood. Because HIF2α has been shown to activate Oct4, and HIF2α deficient embryos have severely reduced numbers of primordial germ cells (Covello et al., 2006), it is believed to be the HIF family member that regulates stem cells (Das et al., 2012; Franovic et al., 2009; Heddleston et al., 2009; Li et al., 2009; Mohyeldin et al., 2010). However, recent data indicate that HIF1α can also regulate stem cell properties (Takubo et al., 2010; Wang et al., 2011). Therefore, it is important to dissect whether HIF1α and HIF2α, are involved in the acquisition of stem cell fate and in particular in the mechanism underlying the hypoxia effect in reprogramming, and whether HIFs are responsible for the metabolic shift during reprogramming.
We now show that both HIF2α and HIF1α are essential for the metabolic changes required early for iPSC generation in human. We further show that HIF2α is required at early stages but is detrimental at later stages of reprogramming. HIF1α and HIF2α are sufficient to induce the stem cell specific metabolic switch. However, prolonged HIF2α stabilization represses reprogramming due to up-regulation of TRAIL. These data reveal similarity between normal reprogramming and cancer progression; both require early metabolic switch, induced by HIF1α and HIF2α and both are sensitive to the presence of TRAIL ligand.
RESULTS
A metabolic switch occurs early during the reprogramming process
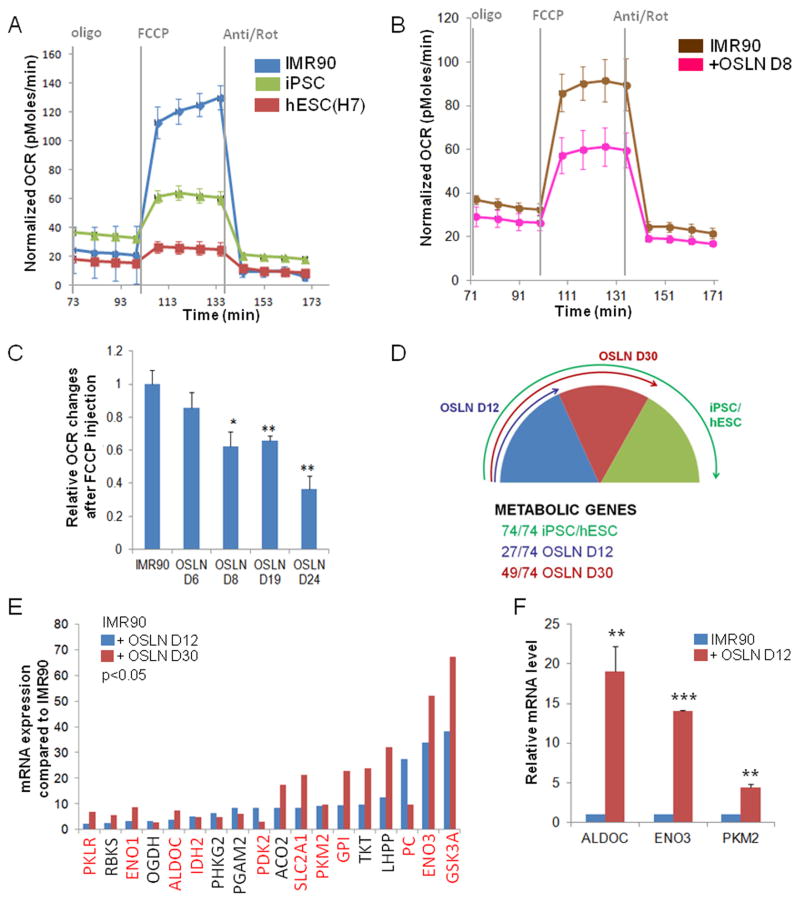
The metabolism of primed pluripotent stem cells differs from the one observed in naïve pluripotent stem cells or many differentiated cells (Varum et al., 2011; Zhou et al., 2012). Primed stem cells show reduced mitochondrial activity and rely on glycolysis, while differentiated cells produce the majority of their ATP by mitochondrial oxidative phosphorylation. We used the reprogramming assay (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007) to analyze the requirements for acquisition of the metabolic state of primed pluripotent stem cells. Human fibroblasts were reprogrammed into induced pluripotent stem cell (iPSC) state using the four reprogramming factors Oct4, Sox2, Lin28 and Nanog (OSLN). To characterize the metabolic profiles of the cells we measured a metabolic parameter that mainly defines the mitochondrial respiration levels, oxygen consumption rate (OCR) under various conditions and treatments using Seahorse Extracellular Flux analyzer (Zhou et al., 2012). We treated the cells with mitochondrial ATP synthase inhibitor, oligomycin, followed by a proton gradient discharger, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) to measure the maximal turnover of the electron transport chain uncoupled from ATP synthesis. This analysis reveals the maximal mitochondrial reserve in the presence of glucose (Goldsby and Heytler, 1963; Heytler, 1963). Fibroblasts showed significantly higher level of maximal respiratory capacity than hESCs and iPSCs (Fig. 1A; Fig. S1A). These results, in line with previous findings (Folmes et al., 2011; Prigione and Adjaye, 2010; Hansson et al., 2012), together suggest that a switch from oxidative to glycolytic metabolism occurs during iPSC reprogramming process. We further examined the kinetics of the metabolic switch by analyzing the OCR and ECAR of the cells at different time points of the reprogramming process and found that the FCCP response following oligomycin treatment was already significantly reduced at day 8 in reprogramming fibroblasts compared to control fibroblasts (Fig. 1B–C, Fig. S1B–D). These data indicate that the metabolic changes initiate early in the reprogramming process and support the previous gene expression and proteomics analysis in mouse and human reprogramming (Folmes et al., 2011; Prigione and Adjaye, 2010; Hansson et al., 2012).
Figure 1.
Metabolic switch occurs early in reprogramming process. (A) Metabolic profile comparing hESC H7 and iPSC to fibroblasts (IMR90). A representative trace of OCR changes is shown under mitochondrial stress protocol. (B) IMR90 OSLN reprogramming cells have reduced OCR change at day 8 in response to FCCP following oligomycin treatment compared to the IMR90 fibroblasts. (C) Kinetics of changes in oxidative metabolism in the entire IMR90 fibroblast population subjected to OSLN factors over the time-course of reprogramming process is shown. Significant change is first observed in D8 (day 8) reprogramming cells as shown by relative OCR changes after FCCP injection. (D) Metabolic gene (Table S1) expression pattern display dynamic changes in early (D12) and late (D30, p=0.001) reprogramming cells compared to IMR90 fibroblasts. 74 metabolic genes show higher expression level in iPSC/hESC compared to IMR90 fibroblasts, and among those 74 genes, 27 genes are up-regulated in early (D12) reprogramming cells while additional 22 genes (total of 49) are up-regulated in late (D30) reprogramming cells. Arrows indicate the proportion of the 74 up-regulated genes in each category. (E) Microarray expression data are shown for metabolic genes that are up-regulated more than 2 fold in early (OSLN D12) and late (OSLN D30) IMR90 reprogramming cells compared to the IMR90 fibroblasts. Hypoxia responsive genes are highlighted in red. (F) Quantitative RT-PCR validates the up-regulation of some of these hypoxia responsive genes in early reprogramming fibroblasts compared to control IMR90 fibroblasts. P values were calculated using Student’s t-test. *, P< 0.05; **, P< 0.01 and ***, P<0.001. Bars show SEM for at least 3 separate experiments.
See also Figure S1.
Reprogramming process has hypoxic expression signature
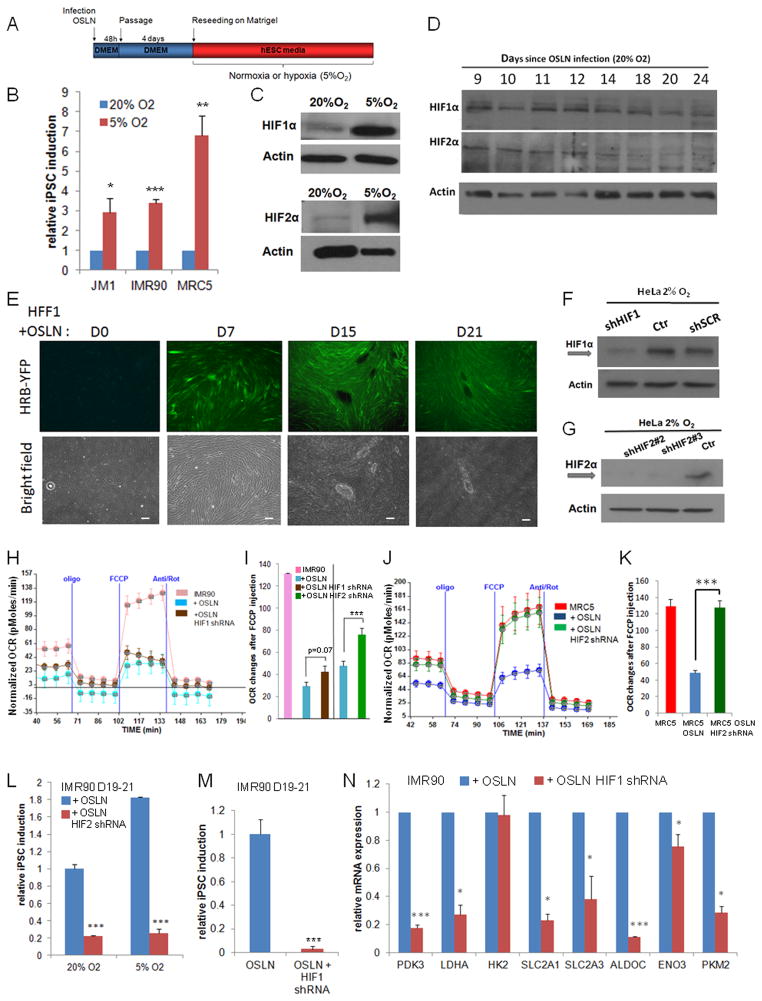
To reveal the key metabolic genes involved in the transition, we analyzed the gene expression profiles of reprogramming cells and observed an increased expression of metabolic genes in early (day 12) and further, in late (day 30) time points of the entire fibroblast population exposed to reprogramming factors (Fig. 1D, Fig. S1E–F, Table S1). Majority of the increased metabolic genes during early reprogramming are hypoxia responsive genes (Fig. 1E). We validated the early up-regulation in reprogramming process for ALDOC, ENO3 and PKM2 by qPCR (Fig. 1F). Since the reprogramming experiments were performed in normoxia, we tested whether performing the process in hypoxic conditions would be beneficial (Fig. 2A). Indeed, the reprogramming process is significantly more efficient when performed in hypoxic conditions. Compared to 20% O2, both 2% and 5% O2 promote iPSC colony formation in various cell lines, including IMR90, MRC5 and JM1 (Fig. 2A–B and Fig. S2A–C), confirming that hypoxia is beneficial for iPSC induction (Yoshida et al., 2009). We showed by Western blot analysis that the two main hypoxia responsive factors, HIF1α and HIF2α, were stabilized in hypoxic reprogramming process (Fig. 2C and Fig. S2D–E).
Figure 2.
HIFs are required for the metabolic switch in early reprogramming. (A–B) Hypoxia (5%O2) enhances iPSC formation in three cell lines, JM1, IMR90 and MRC5. Colonies were counted at day 21 after OSLN infection. (C) Comparison of HIF1α and HIF2α protein levels in the reprogramming cells under normoxia (20% O2, at day 9) and under hypoxia (5% O2, day 7). (D) HIF1α and HIF2α protein stabilization occurs in reprogramming fibroblasts (MRC5) under normoxia as shown by western blots. (E) HFF1 cells harboring a YFP hypoxia reporter show an increase of HIF activity during the course of reprogramming. Bars represent 200μm. Western blots validate the knock-down of HIF1α and HIF2α using shRNA against HIF1α (F) and HIF2α (G), respectively in HeLa cells. (H–I) The changes in oxidative metabolism are shown for IMR90, IMR90 infected with OSLN and IMR90 with OSLN+HIF1α shRNA or OSLN+HIF2α shRNA at day 8 of the reprogramming process, with a representative trace of OCR under mitochondrial stress protocol (H) and relative OCR changes after FCCP injection (I). (J–K) The changes in oxidative metabolism are shown for MRC5, MRC5 infected with OSLN and MRC5 with OSLN+HIF2α shRNA at day 8 of the reprogramming process, with a representative trace of OCR under mitochondrial stress protocol (J) and relative OCR changes after FCCP injection (K). OCR change in MRC5 with OSLN+HIF2α shRNA is significantly different from that in MRC5 infected with OSLN. (L) shRNA against HIF2α decreases iPSC colony formation in both 20% and 5% O2 as compared to the control. (M) shRNA against HIF1α reduces the number of colonies observed at day 19–21. (N) Expression of metabolic genes are reduced at day 23 in IMR90 with OSLN+HIF1α shRNA as compared to the control. P values were calculated using Student’s t-test. *, P< 0.05; **, P< 0.01 and ***, P<0.001. Bars show SEM for at least 3 separate experiments.
See also Figures S2 and S3.
To test whether HIF1α and HIF2α are stabilized in the normoxic (20% O2) reprogramming process, we analyzed multiple timepoints in normal reprogramming (Fig. 2D, Fig. S2F). Both HIF1α and HIF2α proteins were transiently stabilized during normoxic reprogramming (Fig. 2C–D, Fig. S2F). To test whether HIFs are also transcriptionally active during the reprogramming in normoxic oxygen environment, we introduced a HIF-reporter that contains six HIF minimal binding sites in front of eYFP (Zhou et al., 2011) into the fibroblasts used for reprogramming assay (HFF1). This sensor has shown to react to hypoxia and both HIF1α and HIF2α activity (Zhou et al., 2011, Fig. S2G). By day 7 of reprogramming the fluorescent signal had increased significantly, suggesting that HIF1α and/or HIF2α are activated during the reprogramming process (Fig. 2E). However, when iPSC colonies were formed, the signal was highly reduced, suggesting that the viral construct was inactivated in the stem cell stage as has been observed before (Takahashi et al., 2007; Xia et al., 2007; Yu et al., 2007).
HIF2α is required for the metabolic switch during early reprogramming
To test whether HIF1α and/or HIF2α are required for the metabolic switch observed during reprogramming, we used HIF1α and HIF2α shRNA constructs that resulted in a significant reduction of HIF1α and HIF2α protein levels, respectively (Li et al., 2007; Fig. 2F–G; Fig. S3A–B). Importantly, when either HIF1α or HIF2α were significantly reduced starting from the initiation of the reprogramming process, at 8–10 day time point the total OCR increase after FCCP addition was abnormal. While HIF1α shRNA effect in the metabolic switch was not significant, HIF2α knock-down (KD) significantly blocked the process (Fig. 2H–K). At day 8 the reprogramming cells with HIF2α KD showed a high OCR increase after FCCP treatment, comparable to fibroblasts. This is with a stark contrast to the highly reduced OCR detected in the control reprogramming samples (Fig. 2I–K, Fig. S3C). Furthermore, at the end of the reprogramming process, the number of iPSC colonies was also significantly reduced when HIF2α was knocked-down during the process both in normoxia and in hypoxia (Fig. 2L, Fig. S3D). The iPSC colony formation was also highly reduced when HIF1α was knocked-down during the process (Fig. 2M). The reduction observed in iPSC induction when HIF1α or HIF2α were knocked-down was not due to significant reduced proliferation rates (Fig. S3E). We further showed a reduction in the key metabolic, hypoxia responsive genes in the reprogramming cells in which HIF1α or HIF2α was knocked-down (Fig. 2N, Fig. S3F). Later in the reprogramming process, an increase of small iPSC-like colonies was observed in some of the HIF1α KD plates (Fig. S3G–J). Altogether, these data suggest that both HIF2α and HIF1α are required for the metabolic switch that initiates early during the reprogramming process and that this step is necessary for iPSC colony formation.
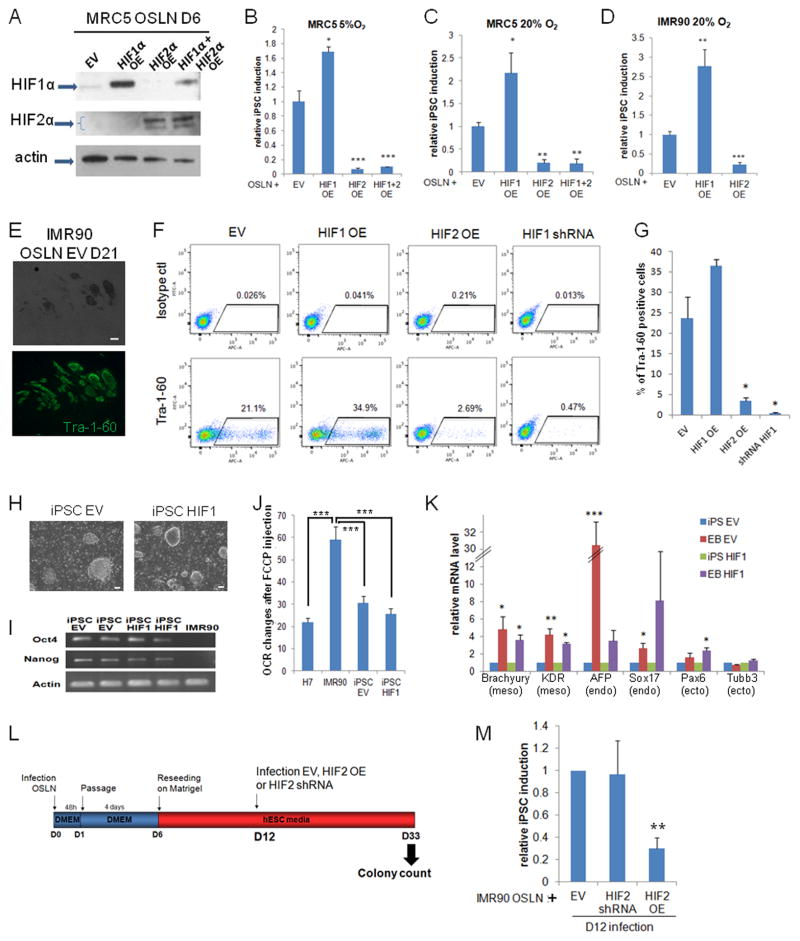
Reprogramming process requires controlled HIF2α activity
Since hypoxia is beneficial for reprogramming and HIF2α and HIF1α are required for the process, we tested whether constitutive stabilization of HIF1α and/or HIF2α would enhance iPSC generation by over-expressing non-degradable forms of HIF1α and HIF2α during reprogramming (Pro402, 564/Ala mutations and Pro405,531/Ala mutations, respectively; Yan et al., 2007; Fig. 3A, Fig. S4A). HIF1α over-expression (OE) during reprogramming significantly increased the efficiency of iPSC colony formation in norrmoxia as well as in hypoxia in IMR90 and MRC5 fibroblast cell lines (Fig. 3B–D). However, surprisingly prolonged stabilization of HIF2α significantly repressed the reprogramming process in these conditions (Fig. 3B–D), even in combination with HIF1α (Fig. 3B–C). To confirm that the morphologically- and AP staining-identified colonies consist of true pluripotent cells, we stained all cells late in reprogramming process with TRA-160 and quantified the positive cells by FACS analysis (Fig. 3E–G, Fig. S4B–F). Importantly, similar to the morphological analysis, TRA-160 positive cell number was increased when HIF1α was stabilized but significantly reduced with HIF2α stabilization (Fig. 3F–G). As expected, TRA-160 FACS analysis also validated the loss-of function data (Fig. 3F–G; Fig. 2M). This difference in HIF1α OE and HIF2α OE effect on reprogramming was not caused by differences in cell proliferation rates (Fig. S3E). The iPSC colonies derived from HIF1α expressing or control samples displayed hESC-like morphology, self-renewal capacity, AP activity and endogenous NANOG and OCT4 mRNA expression (Fig. 3H–I, Fig. S4G). In addition, their metabolic profile resembled the hESC metabolic profile, as evident in the diminished mitochondrial functional reserves induced by FCCP following oligomycin treatment (Fig. 3J). Cells in these iPSC colonies were pluripotent since they had the capacity to differentiate into all three germ layers, as indicated by up-regulation of mesoderm, endoderm and ectoderm markers in Embryoid Body (EB) assays (Fig. 3K, Yu et al., 2007). Despite several attempts, we were not able to maintain the few colonies derived from the HIF2α over-expressing cells. Together, these data show that while continuous and prolonged OE of HIF1α is beneficial for reprogramming process, prolonged HIF2α activation represses iPSC formation.
Figure 3.
HIF1α and HIF2α over-expression have opposite effects on reprogramming. (A) Over-expression (OE) of non-degradable HIF1α and HIF2α was confirmed by western blot analysis in MRC5 6 days after OSLN infection. (B–D) HIF1α OE promotes iPSC colony formation, while HIF2α OE inhibits colony formation in 5% O2 (B) and 20% O2 (C) in MRC5 as well as in 20% O2 in IMR90 (D, n=7 independent experiments). Colonies were counted at day 21 of reprogramming. (E) Immunofluorescence microscopy shows that colonies counted at 21 post-infection express the stem cell marker Tra-1-60. Scale bar represents 250μm. (F) Flow cytometry analysis of Tra-1-60 at 21 days post-OSLN infection in IMR90. (G) Quantification of Tra-1-60 positive cells detected by flow cytometry at day 21 of reprogramming. Bars show SEM for triplicate. Colony count and flow cytometry analysis of those triplicates are presented in Fig. S4F and Fig. S4D, respectively. (H) iPSC colonies reprogrammed from HIF1α OE IMR90 fibroblasts can self-renew, (I) express endogenous Oct4 and Nanog, and (J) display a glycolytic profile similar to hESCs (H7) that is different from the parental fibroblasts (IMR90). Bars represent 200μm. (K) The differentiation abilities of both EV iPSC and HIF1α iPSC derived EBs are demonstrated by RT-qPCR analysis of genes representative of three germ layers. Brachyury and KDR are markers for mesoderm, AFP and Sox17 for endoderm, and Pax6 and Tubb3 for ectoderm. (L) Experimental scheme is shown to test the requirement and specificity of HIF2α at the later stage of iPSC reprogramming process. (M) When infected at day 12(D12), HIF2α OE reduces colony formation while HIF2α knock-down by shRNA shows no significant change in colony formation as compared to the EV infected cells. P values were calculated using Student’s t-test. *, P< 0.05; **, P< 0.01 and ***, P<0.001. Bars show SEM for at least 3 separate experiments.
See also Figure S4.
HIF2α function at different stages of reprogramming
We showed that loss of HIF2α is detrimental for reprogramming (Fig. 2L, Fig. S3D). Surprisingly, while continuous HIF1α activation is beneficial, continuous activation of HIF2α represses iPSC induction, suggesting that the reprogramming process requires tightly controlled HIF2α activity. To understand the timing of HIF2α function in the process, instead of reducing HIF2α levels from the beginning of reprogramming, we infected the cells with shRNA against HIF2α in the middle of the reprogramming process (Day 12) and measured late in reprogramming process (Day 33) the total number of iPSC colonies formed (Fig. 3L–M). Interestingly, while HIF2α shRNA reduced iPSC colony number significantly when introduced early (D0, Fig. 2L), no obvious defect was observed when HIF2α was knocked-down in the middle (D12) of the reprogramming process (Fig. 3L–M). However, when the non-degradable form of HIF2α was over-expressed in the middle of the reprogramming process, the iPSC colony number was significantly reduced (Fig. 3L–M). These data suggest that HIF2α is required early, but not late in iPSC induction. Further, HIF2α needs to be down-regulated at the latter part of the reprogramming process since constitutive activation of HIF2α late in the process causes a significant repression of reprogramming.
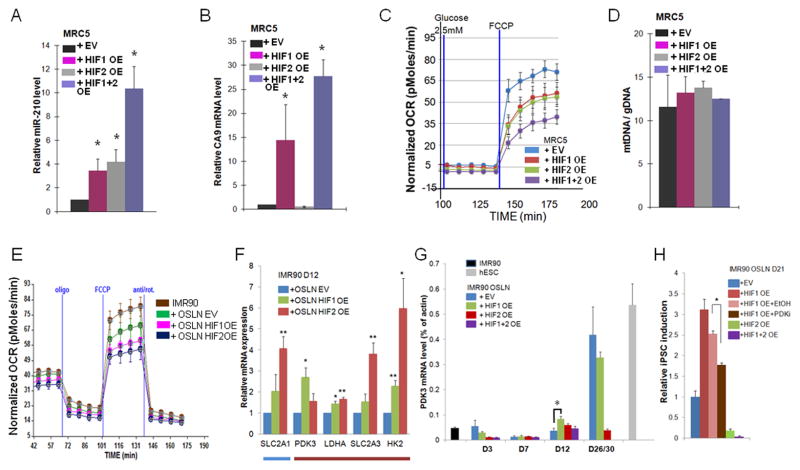
HIF1α and HIF2α over-expression is sufficient to induce metabolic changes
To analyze the mechanism of HIF1α and HIF2α action in somatic cells, we expressed constitutively active HIF1α and HIF2α in fibroblasts without the reprogramming factors. We first validated the HIF1α and HIF2α activity by showing that in MRC5 fibroblasts HIF1α and/or HIF2α OE results in up-regulation of HIF target genes, Carbonic Anhydrase IX (CA9) and miR-210 (Fig. 4A–B). To test whether over-activation of HIF1α or HIF2α could affect the metabolism of the fibroblasts without the addition of the reprogramming factors, we analyzed the maximum OCR change after FCCP addition in these conditions. Importantly, OE of HIF1α or HIF2α reduced the oxidative metabolism, and this metabolic change was more prominent when both transcription factors were activated simultaneously (Fig. 4C). We further showed that the observed reduction on mitochondrial activity was not due to the reduction of mitochondrial number by utilizing mitochondrial DNA copy number assay (Fig. 4D). Since no significant change in mitochondrial copy number was found in HIF1α and HIF2α OE cells (Fig. 4D), mitochondrial activity regulated by gene expression rather than mitochondrial number is critical for the metabolic switch observed in fibroblasts.
Figure 4.
HIF1α and HIF2α over-expression are sufficient to induce a metabolic change. (A–D) In MRC5 fibroblasts infected with HIF1α OE alone, HIF2α OE alone or HIF1α OE and HIF2α OE together, (A) Carbonic anhydrase 9 (CA9) expression was induced only in HIF1α over-expressing cells, and (B) miR-210 expression was induced both in HIF1α and HIF2α over-expressing cells, confirming the functionality of the over-expressed HIF proteins. (C) Both HIF1α and HIF2α over-expression in MRC5 fibroblasts without OSLN factors induced glycolytic metabolism similar to hESC H1. Shown are representative SeaHorse traces. (D) Mitochondrial DNA copy number was not changed in HIF1α or HIF2α over-expressing MRC5 fibroblasts. (E) Both HIF1α and HIF2α over-expression in reprogramming cells (IMR90+OSLN) at day 8 timepoint resulted in a further decrease in the OCR. (F) qPCR validation of the expression level of metabolic genes that were up-regulated by HIF1α or HIF2α OE in D12 sample compared to the control (blue and red bars indicate the comparative expression in control as illustrated in Fig 1D). (G) Kinetics of PDK3 mRNA level in IMR90 reprogramming cells is shown as the percentage of actin. (H) Treatment of PDK3 inhibitor (PDKi) on HIF1α OE cells reduces colony formation as compared to HIF1α OE cells in the vehicle control (EtOH). P values were calculated using Student’s t-test. *, P< 0.05, and **, P< 0.01. Bars show SEM for at least 3 separate experiments.
See also Figure S5.
Similarly, OCR reduction was accelerated when HIF1α or HIF2α were over-expressed with OSLN during the reprogramming assay (Fig. 4E). We therefore tested whether HIF1α and/or HIF2α OE would accelerate the increase in expression patterns of metabolic genes observed during reprogramming (Fig. 1D). In order to examine the change in gene expression that occurs at various stages of the reprogramming process when HIF2α is OE during OSLN-induced reprogramming, we performed a microarray analysis (Fig. S5A). We first validated the microarray data by showing that stem cell markers were highly enriched in reprogramming cells and hypoxia target genes were up-regulated in HIF2α OE reprogramming cells (Fig. S5B–C). Our microarray analysis revealed that hypoxia target genes were enriched in HIF2α OE samples compared to fibroblasts (Fig. S5D–E). Furthermore, HIF2α OE and HIF1α OE accelerated the metabolic gene expression during the reprogramming process (Fig. S5F). qPCR validation revealed that many of the metabolic genes normally up-regulated late in reprogramming process were already up-regulated at 12 day time point due to HIF2α and HIF1α overexpression (Fig. 4F), suggesting hastened kinetics of the metabolic switch in the reprogramming cells.
Among the metabolic genes up-regulated late (D30) during normal reprogramming that were expressed earlier (D12) in HIF1α and HIF2α OE cells was Pyruvate Dehydrogenase Kinase 3 (PDK3; Fig. 4F, Fig. S5F–H). PDK3, an enzyme that blocks the conversion of pyruvate to acetyl-CoA is significantly enriched in hESCs and hypoxic tumor cells (Mathieu et al., 2011; Stadler et al., 2010). Unlike other PDKs, PDK3 is not inhibited by excess pyruvate and therefore is considered a key regulator for a switch from oxidative to glycolytic metabolism. We studied the kinetics of PDK3 up-regulation when HIF1α and/or HIF2α are over-expressed during reprogramming by quantitative RT-PCR. While in control an eight fold increase in PDK3 expression was observed at day 30 of reprogramming process, HIF1α and HIF2α accelerated this process since up-regulation of PDK3 was observed at day 7 or day 12 in reprogrammed MRC5 or IMR90 cells, (Fig. S5H; Fig. 4F–G). We further tested whether inhibition of PDK3 could suppress the HIF1α induced increase in iPSC formation. Using the chemical inhibitor Radicicol (Kato et al., 2007) we showed that reduction of PDK3 activity reduced the number of iPSC colonies in HIF1α over-expressing reprogramming cells as well as in normal reprogramming cells (Fig. 4H; Suppl. Fig. 5I), suggesting an important functional role for PDK3 in HIF1α induced increase in iPSC induction. However, since Radicicol did not fully eliminate iPSC induction, additional HIF targets probably are involved in the process.
These data show that HIF1α and HIF2α together are sufficient to induce the metabolic switch in fibroblasts. Similarly, HIF1α and HIF2α stabilization in hypoxic cells is shown to be invariably sequential (Keith et al., 2012; Zhou et al., 2011). These data support the hypothesis that HIF1α and HIF2α have a combinatorial, perhaps sequential role in the metabolic switch observed in early iPSC induction. However, while HIF1α stabilization is beneficial for the process, surprisingly HIF2α stabilization late in the process is detrimental for iPSC induction.
HIF2α inhibits reprogramming through TRAIL
In accordance with the observation that HIF2α is required early, but not late in reprogramming, we also observed that both MRC5 and IMR90 fibroblasts with HIF2α OE give rise to approximately equal or larger number of pre-colonies in early time points compared to the controls (before Day 14, Fig. 5A, Fig. S6A–B), suggesting that HIF2α over-expression is beneficial during the early phase of reprogramming. However, a significant repression in colony formation and PDK3 expression was observed in HIF2α over-expressing samples at later time points (Fig. 4G–H, Fig. 5A, Fig. S6A). Similarly, HIF2α OE initiated in the middle of reprogramming process (day 12-14) dramatically reduced the iPSC colony formation (Fig. 3M), suggesting that HIF2α OE late during reprogramming is detrimental for iPSC formation. We therefore examined the gene expression profiles of HIF2α over-expressing and control cells at day 30 of the reprogramming process to identify the key HIF2α target that can repress iPSC formation.
Figure 5.
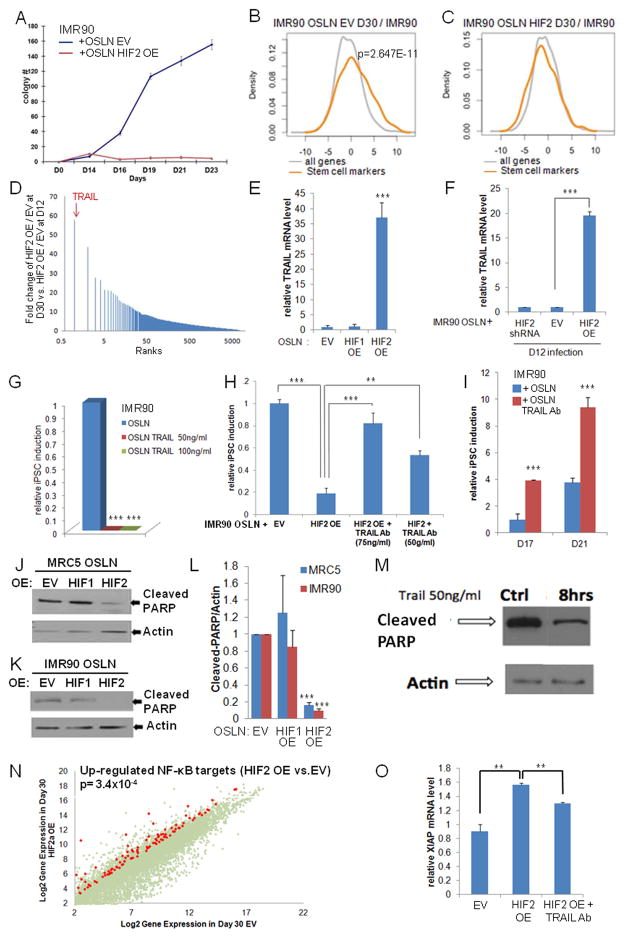
HIF2α inhibits iPSC formation through TRAIL activation. (A) Kinetics of iPSC colony formation in HIF2α over-expressing (OE) IMR90 fibroblasts (OSLN+HIF2α), showing an increase in early (before Day 14) but dramatic reduction of iPSC colony formation in late time points compared to the control (OSLN+EV). (B) Stem cell markers are significantly up-regulated in the control, but not in (C) HIF2α OE cells during reprogramming. (D) Microarray analysis reveals that TRAIL is the most up-regulated gene at D30 in HIF2α OE reprogramming cells compared to normal IMR90 reprogramming cells. (E) qPCR validation shows the high TRAIL mRNA expression in HIF2α, but not in HIF1α, OE reprogramming cells in late timepoints (D30). (F) TRAIL mRNA expression increased at late timepoints (D30) in HIF2α OE cells infected at day 12 of the reprogramming process compared to OSLN control in cells. (G) Administration of TRAIL during IMR90 reprogramming process (from Day6/7) represses iPSC formation, but not fibroblast growth. (H) Administration of TRAIL antibody on HIF2α over-expressing cells rescues iPSC formation in IMR90. Colonies were counted at day 21. (I) Administration of TRAIL antibody (75ng/ml) in the normal reprogramming process (in IMR90) promotes colony formation. In both MRC5 (J) and IMR90 (K) reprogramming cells, HIF2α OE represses PARP cleavage 5 fold (L) as compared to empty vector control and HIF1α OE cells (three independent experiments). (M) Western blots confirm that PARP cleavage is reduced in reprogramming cells treated with 50ng/ml TRAIL for 8 hours as compared to the control. (N) NF-κB target genes are visualized on a scatter plot comparing HIF2α OE at day 30 to EV for those that are 4 fold higher expressed in HIF2α OE cells. (O) Quantification of XIAP expression by real-time RT-PCR assay shows an increased XIAP expression in HIF2α OE reprogramming cells as compared to the EV at day 27. Importantly, such an increase is reduced in HIF2α OE reprogramming cells treated with TRAIL neutralizing antibody. P values were calculated using Student’s t-test. **, P< 0.01 and ***, P<0.001. Bars show SEM for at least 3 separate experiments.
See also Figure S6.
As expected, the expression of stem cell markers was only enriched in the control samples, not in HIF2α OE cells, validating the lack of iPSC colonies (Fig. 5B–C, Fig. S6C–D). Importantly, we identified TNF-related apoptosis-inducing ligand (TRAIL) as the most up-regulated gene with stabilized HIF2α at late stage of reprogramming compared to controls (Fig. 5D, Fig. S6E). This up-regulation of TRAIL was not observed early in reprogramming (Fig. S6F) and was specific for HIF2α OE since no effect in TRAIL expression was observed with stabilized HIF1α (Fig. 5E). Furthermore, the significant up-regulation of TRAIL was also observed when HIF2α OE was induced late in reprogramming (D12, Fig. 5F) and a trend of TRAIL down-regulation was observed in HIF2 KD late reprogramming samples (Fig. S6G).
To analyze whether TRAIL expression was sufficient to reproduce the repressive function of HIF2α, we administered TRAIL recombinant protein into the cell culture media during reprogramming process. Importantly, when 50ng/ml or 100ng/ml of TRAIL was administered to the reprogramming process, fibroblasts continued dividing but no iPSC colonies were produced (Fig. 5G, Fig. S6G–K), showing that TRAIL mimics HIF2α OE phenotype by repressing the iPSC induction but not fibroblast viability. We further showed that addition of TRAIL neutralizing antibody on HIF2α over-expressing cells is able to counteract HIF2α repressive function and rescue the iPSC formation (Fig. 5H, Fig. S6H–K). These data show that HIF2α exerts its dominant repression at late stage of reprogramming through TRAIL.
TRAIL effect in reprogramming
To test whether TRAIL is normally expressed and has a repressive effect during normal reprogramming, we added TRAIL neutralizing antibody (TRAIL Ab) during normal reprogramming (from day 7 on) and counted the iPSC colony number at day 17 and day 21 in the process. Importantly, the colony number was significantly increased when endogenous TRAIL was sequestered by TRAIL antibody (Fig. 5I). The colonies formed were able to undergo self-renewing division and expressed endogenous Oct4 (Fig. S6L). These data show that indeed normal reprogramming generates low levels of endogenous TRAIL that has a repressive property on the reprogramming process, hence we proceeded to reveal the mechanism of TRAIL action in the process.
TRAIL, by activating a specific set of receptors can either induce apoptosis through the caspase pathway or display an anti-apoptotic effect mostly through NF-κB signaling (Merino et al., 2007; Walczak et al., 1999). We tested whether HIF2α OE activates the caspase pathway through TRAIL in the reprogramming cells by analyzing the level of cleaved PARP, the cleavage target of active caspase 3. Instead of caspase activation, we observed repression of caspase activity indicated by significant reduction of cleaved PARP in HIF2α, but not HIF1α over-expressing reprogramming cells (Fig. 5J–L). We further tested whether exogenously added TRAIL protein in the reprogramming system can affect PARP cleavage, and found that 8 hours after TRAIL addition at day 17 of iPSC reprogramming, a reduction of PARP was also detected (Fig. 5M).
Since reprogramming cells require active caspases (Li et al., 2010), a reduction of caspase activity could be causal for the lack of iPSC formation in cells constitutively over-expressing HIF2α. Gene expression profiling data further revealed that expression of NF-κB targets, and in particular cFLIP and XIAP, inhibitors of caspase activation, are higher in HIF2α over-expressing cells than the control (Fig. 5N–O, Table S2, Fig. S6M), suggesting that TRAIL could induce anti-apoptotic activity instead of apoptotic signaling in reprogramming cells. Importantly, TRAIL neutralizing antibody significantly reduced the up-regulation of XIAP observed in HIF2α expressing reprogramming cells (Fig. 5O).
hESCs are not responsive to TRAIL
Since reprogramming cells are sensitive to TRAIL, we tested TRAIL effect on pluripotent hESCs and iPSCs and found that surprisingly these pluripotent cells can self-renew in the presence of TRAIL (Fig. 6A–B; Fig. S7A–D). We further found that this difference in TRAIL responsiveness correlated with differential expression of TRAIL receptors. While DR4 expression is 10 fold lower in reprogramming cells compared to hESCs, decoy receptor 2 (DcR2) expression is significantly higher in fibroblasts (IMR90: 8% of actin, Fig. S7E) and reprogramming cells (1.3% of actin) than in hESCs or iPSCs (0.05% of actin) (Fig. 6C; Fig. S7D). Previous data have shown that DcR2, upon binding to TRAIL, can induce NF-κB signaling, which imposes an inhibitory effect on caspase activation (Ehrhardt et al., 2003; Kim et al., 2002). A lower level of DcR2 in hESCs might protect these cells from TRAIL induced anti-caspase signals. Accordingly, in pluripotent cells cultured in the presence of TRAIL for prolonged periods, we found that PARP cleavage is not reduced compared to the controls (Fig. 6D; Fig. S7A–C). These results show that TRAIL does not inhibit caspase activity in hESCs or iPSCs.
Figure 6.
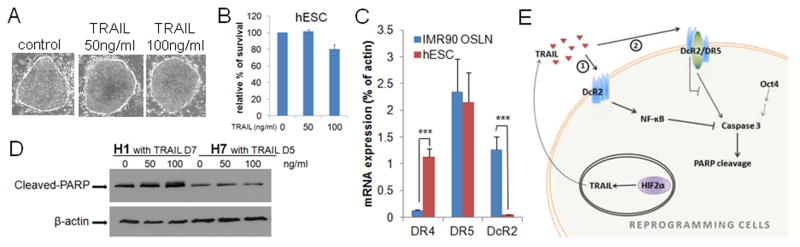
hESC bypass TRAIL effect due to low level of DcR2: (A–B) H1 hESC can self-renew in the presence of TRAIL in culture. Morphologies (A) and percentage of survival (B) are shown for H1 cells with 50ng/ml or 100ng/ml TRAIL in culture for 7 days compared to control H1 cells. Bars represent 200μm. (C) DcR2 mRNA expression is low in hESC compared to reprogramming cells (IMR90 OSLN). (D) TRAIL administration does not reduce PARP cleavage in H1 or H7. (E) Models for the repressive action of prolonged HIF2α OE on reprogramming: TRAIL inhibits caspase activity by binding DcR2 and activating NF-κB (1), or binding heteromeric complex of DcR2/DR5 (2). P values were calculated using Student’s t-test. ***, P<0.001. Bars show SEM for at least 3 separate experiments.
See also Figure S7.
DISCUSSION
This study shows that reprogramming has distinct stages. While both HIF1α and HIF2α are required, HIF2α has a stage specific function in the process. HIF2 is essential in early, but not late in reprogramming. When stabilized, HIF2 is beneficial in early reprogramming, but prolonged stabilization of HIF2α, in contrast to HIF1α dramatically blocks iPSC formation. We identified the mechanism for prolonged HIF2α repressive action in reprogramming. HIF2α, through TRAIL inhibits caspase 3 activity, thereby repressing iPSC formation. Unlike reprogramming cells, hESCs can self-renew in the presence of TRAIL in culture. This correlates with the low expression of DcR2 in hESCs.
Human embryonic stem cells are glycolytic (Panopoulos et al., 2012; Prigione et al., 2010; Varum et al., 2011; Zhou et al., 2012) and the reprogramming process undergoes and requires a metabolic switch from oxidative to glycolytic (Armstrong et al., 2010; Panopoulos et al., 2011; Panopoulos et al., 2012; Prigione et al., 2010; Prigione et al., 2013; Varum et al., 2011). This metabolic switch precedes the stem cell fate marker expression (Folmes et al., 2011; Zhou et al., 2012). Here we show by analyzing the metabolic flux that the metabolic switch begins early in the reprogramming process. These data are in accordance with recent expression analysis of mouse reprogramming cells and human reprogramming using cMyc as one of the reprogramming factors (Polo et al., 2012; Prigione et al., 2013). Interestingly, both HIF1α and HIF2α enhance, and are required for this switch, possibly through separate transcriptional targets (Keith et al., 2012; Loboda et al., 2010). An interesting question is whether the metabolic switch alone is sufficient to initiate the reprogramming process. We showed that prolonged stabilization of HIF1α and HIF2α in fibroblasts is sufficient to induce the metabolic switch from oxidative to highly glycolytic observed in stem cells, however, it was not sufficient to induce pluripotency. This could be due to the repressive effect of HIF2α in iPSC induction through TRAIL. It will be important for future research to test whether switching cell metabolism with more tightly controlled HIFα expression, alone and/or in combination with other factors, could be another revenue to drive somatic reprogramming.
Our findings show that HIF2α is required during the early iPSC reprogramming process to promote the metabolic switch. However, prolonged stabilization of HIF2α represses iPSC formation through TRAIL induced inhibition of caspase 3 signaling. Previously it was shown that activation of apoptotic caspases occurs during the reprogramming process and their inhibition prevents iPSC generation (Li et al., 2010). We now show that HIF2α OE inhibits the caspase 3 activity induced by the reprogramming factors. We also show that HIF2α inhibits iPSC formation through TRAIL. The binding of TRAIL to its cognate receptors DR4 and DR5 activates both apoptotic and non-apoptotic signaling while binding to its decoy receptor 2 (DcR2) only activates anti-apoptotic pathways (Degli-Esposti et al., 1997; Sanlioglu et al., 2005). Interestingly, reprogramming cells have a low level of DR4 but a high level of DcR2. In these cells TRAIL could activate DcR2 and thereby NF-κB pathway that can repress caspase activity (Ehrhardt et al., 2003; Kim et al., 2002). Further, since reprogramming cells require an active caspase pathway (Li et al., 2010), TRAIL dependent caspase inhibition could block iPSC formation (Fig. 6E). Our finding that NF-κB targets, and in particular the anti-apoptotic gene XIAP, are up-regulated in HIF2α OE reprogramming cells compared to controls (Fig. 5N-O) support this hypothesis. Alternatively, DcR2 could form a heteromeric complex with DR5 and inhibit caspase activation through steric hindrance (Merino et al., 2006; Fig. 6E). We showed that both fibroblasts and reprogramming cells have higher expression of DcR2 than hESCs, which may be due to low p53 expression in hESCs (Liu et al., 2005). Fibroblasts survive with TRAIL-induced inhibition of the caspase pathway while fibroblasts in the process of becoming iPSCs are sensitive to TRAIL, indicating that similar DcR2 levels in two cell types can lead to different outcomes due to cell-dependent responses to the downstream pathway.
This study presents an example of a significant difference between HIF1α and HIF2α function in the same cellular process. Our results suggest that HIF1α and HIF2α may play non-overlapping roles in iPSC reprogramming process: HIF1α and HIF2α are essential for the metabolic switch and induction of pluripotency. Additionally, prolonged stabilization of HIF1α increases iPSC induction, however surprisingly prolonged stabilization of HIF2α significantly represses reprogramming through TRAIL activation (Fig. 7). A fine balance exists between the differential roles of the two HIFα factors in early embryonic development, a period when physiological hypoxia is critical for the formation and early differentiation of both germ line and somatic stem cells (Dunwoodie, 2009). Our findings in this study provide further evidence on functional difference between HIF1α and HIF2α (Hu et al., 2007; Husa et al., 2010). Reprogramming assays can be used in the future to reveal how structural differences between the two HIFα factors lead to their diverse functional outcomes.
Figure 7.
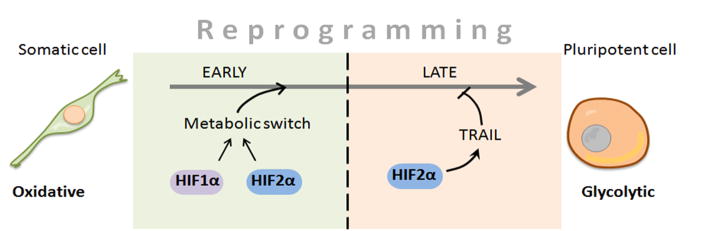
Model of HIF2α stage-dependent role during the reprogramming process. When oxidative fibroblasts are reprogrammed into glycolytic pluripotent cells, metabolic switch from oxidative to glycolytic occurs. Our study shows that such a metabolic switch takes place in early stage of the reprogramming process, and HIF1α and HIF2α are essential for this metabolic change. In contrast, constitutive HIF2α stabilization is detrimental for the reprogramming process at the later stage, as it promotes TRAIL expression, which prevents reprogramming.
This study and previous work show that TRAIL does not induce death in hESCs, cancer stem cells or adult stem cells (Kruyt and Schuringa, 2010). However, TRAIL is detrimental for cancer cells (Merino et al., 2007; Walczak et al., 1999). Our study now reveals that TRAIL also represses the iPSC reprogramming process. These data suggest a similar mechanism in response to TRAIL in cancer cells and reprogramming cells. In general, the results suggest that cells undergoing reprogramming process may have similar characteristics to the cells undergoing progression towards aggressive tumor cell allowing us to propose that cancer progression is a slow reprogramming process. First, cancer cells and cells under reprogramming (iPSC induction) both change their metabolism from oxidative to highly glycolytic early during the process. Second, HIFs regulate the switch in both cell types. Third, cancer cells and reprogramming cells are sensitive to TRAIL, while cancer stem cells and pluripotent stem cells are resistant to TRAIL. These similarities between cancer cells and reprogramming cells may allow us in the future to utilize somatic cell reprogramming process as a novel model to understand mechanisms and events involved in the cancer progression, and to learn from our current knowledge in cancer progression to facilitate understanding of the acquisition of pluripotency.
EXPERIMENTAL PROCEDURES
Cell culture and reprogramming
hESCs and isolated iPSCs were maintained as previously described (Ware et al., 2006). Reprogramming of human fibroblasts (MRC5, IMR90, HFF1) was carried out with Oct4, Sox2, Lin28 and Nanog lentiviruses (OSLN, generated from Addgene constructs #21162 and 21163, Yu et al 2009). To examine hypoxia effect on iPSC formation, from 7 days post-infection cells were cultured under 2% or 5% of O2. The number of iPSC colonies was defined as the number of Alkaline Phosphatase (AP) positive, Oct4-GFP positive and Tra-1-60 positive colonies. See details in Supplemental Experimental Procedures.
Over-expression and inhibition of HIF1α or HIF2α during reprogramming
Retroviruses expressing non-degradable HIF1α or HIF2α (Addgene constructs19005 and 19006, Yan et al., 2007) were infected together with OSLN on day 0. An empty vector construct was used as a control. To examine the role of HIF2α in the later stage of the reprogramming process, OSLN reprogramming cells were infected with HIF2α retrovirus at day 12. HIF1α and HIF2α knockdowns (KD) were obtained using shRNA constructs (shHIF1α compared to shRNA scramble control shSCR, Nemetski and Gardner, 2007; Addgene plasmids #22131/shHIF2#2 and #22132/shHIF2#3, Li et al., 2007). HIF1α and HIF2α OE and KD were validated by Western blot analysis.
iPSC formation with recombinant TRAIL, TRAIL antibody, or PDK3 inhibitor administration
During the iPSC reprogramming process, human TRAIL recombinant protein (R&D Systems; 50ng/ml or 100ng/ml), human TRAIL polyclonal antibody (R&D Systems; 75ng/ml or 50ng/ml) or PDK3 inhibitor Radicicol (Sigma, Kato et al., 2007; 36.5ng/ml) were added to OSLN-infected fibroblasts on day 7.
Western blot analysis, quantitative RT-PCR analysis and mitochondrial DNA copy number measurement
Standard protocols were used. See details in Supplemental Experimental Procedures. Primers used in our study are listed in Table S3.
Whole genome-wide microarray analysis
Total RNA isolated from IMR90 and MRC5 reprogramming cells infected with HIF2α OE virus or empty vector control at day 12 and day 30 were used in the microarray analysis. RNA qualification, Agilent microarray labeling, hybridization and scanning were performed in microarray facility at Institute for Systems Biology. Any intensity-dependent biases were removed in the data using the normalize.qspline function in the affy Bioconductor package.
OCR measurement using SeaHorse Cellular Flux assays
MitoStress and GlucoseStress assays were performed on fibroblasts and reprogramming fibroblasts (infected by OSLN) using the SeaHorse XF96 extracellular flux analyzer. The OCR values were further normalized to the number of cells, quantified by the Hoechst staining (HO33342; Sigma-Aldrich). See Supplemental Experimental Procedures for details.
In-vivo detection of HIF activity using eYFP reporting system (HBR-6U)
To test the level of HIF transcriptional activity during reprogramming, fibroblasts were first infected with HIF reporter HBR-6U lentivirus (Zhou et al., 2011) before undergoing normal reprogramming assay.
Statistical analysis
Throughout the paper, P values were calculated using Student’s t-test. *, P< 0.05; **, P< 0.01 and ***, P<0.001. Bars show Standard Error of the Mean (SEM) for at least 3 separate experiments.
Supplementary Material
Highlights.
HIF1α and HIF2α are required during early stages of reprogramming
HIF2α inhibits reprogramming when expressed during later stages of the process
HIF2α regulates expression of TRAIL, which inhibits reprogramming
TRAIL represses caspase 3 activity specifically in cells undergoing reprogramming
Acknowledgments
We thank Drs. Ware, Hockenbery, Horwitz and members of the Ruohola-Baker laboratory for helpful discussions throughout this work. We thank Dr. Stadler for work early in the project. We thank Michael Choi and Timothy Dosey for technical help. We thank Pamela Troisch from Institute for Systems Biology for the microarray gene expression service and Dr. Mecham from Sage bionetworks for some of the statistical analysis on microarray data.. We thank Dr. Gardner for providing the scramble shRNA and shRNA against HIF1α. We also thank members of Tom & Sue Ellison Stem Cell Core for help on iPSC reprogramming procedures and cell cultures. This work was supported by fellowships from the American Heart Association to JM, WZ and YX, Tietze award for KTK and grants from the National Institutes of Health R01GM097372, R01GM083867, R01GM083867-02S2 for HRB, U01HL100395 for ZA and RTM, and P01GM081619 for RTM and HRB.
Footnotes
Supplemental information for this article includes 7 figures, 3 tables and additional supplemental experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- Axelson H, Fredlund E, Ovenberger M, Landberg G, Pahlman S. Hypoxia-induced dedifferentiation of tumor cells--a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. 2005;16:554–563. doi: 10.1016/j.semcdb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bai RK, Perng CL, Hsu CH, Wong LJ. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann N Y Acad Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- Cabarcas SM, Mathews LA, Farrar WL. The cancer stem cell niche--there goes the neighborhood? Int J Cancer. 2011;129:2315–2327. doi: 10.1002/ijc.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nature medicine. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danet GH, Pan Y, Luongo JL, Bonnet DA, Simon MC. Expansion of human SCID-repopulating cells under hypoxic conditions. J Clin Invest. 2003;112:126–135. doi: 10.1172/JCI17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Bayat-Mokhtari R, Tsui M, Lotfi S, Tsuchida R, Felsher DW, Yeger H. HIF-2alpha suppresses p53 to enhance the stemness and regenerative potential of human embryonic stem cells. Stem Cells. 2012;30:1685–1695. doi: 10.1002/stem.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity. 1997;7:813–820. doi: 10.1016/s1074-7613(00)80399-4. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Developmental cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Ehrhardt H, Fulda S, Schmid I, Hiscott J, Debatin KM, Jeremias I. TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene. 2003;22:3842–3852. doi: 10.1038/sj.onc.1206520. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci U S A. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2alpha oncogenic axis. Proc Natl Acad Sci U S A. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby RA, Heytler PG. Uncoupling of Oxidative Phosphorylation by Carbonyl Cyanide Phenylhydrazones. Ii. Effects of Carbonyl Cyanide M-Chlorophenylhydrazone on Mitochondrial Respiration. Biochemistry. 1963;2:1142–1147. doi: 10.1021/bi00905a041. [DOI] [PubMed] [Google Scholar]
- Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. The EMBO journal. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency (2012) Cell Rep. 2(6):1579–92. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heytler PG. uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-Cl-CCP action on mitochondria and chloroplasts. Biochemistry. 1963;2:357–361. doi: 10.1021/bi00902a031. [DOI] [PubMed] [Google Scholar]
- Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Molecular biology of the cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husa M, Liu-Bryan R, Terkeltaub R. Shifting HIFs in osteoarthritis. Nature medicine. 2010;16:641–644. doi: 10.1038/nm0610-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Li J, Chuang JL, Chuang DT. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Schwabe RF, Qian T, Lemasters JJ, Brenner DA. TRAIL-mediated apoptosis requires NF-kappaB inhibition and the mitochondrial permeability transition in human hepatoma cells. Hepatology. 2002;36:1498–1508. doi: 10.1053/jhep.2002.36942. [DOI] [PubMed] [Google Scholar]
- Kruyt FA, Schuringa JJ. Apoptosis and cancer stem cells: Implications for apoptosis targeted therapy. Biochemical pharmacology. 2010;80:423–430. doi: 10.1016/j.bcp.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li F, He Z, Shen J, Huang Q, Li W, Liu X, He Y, Wolf F, Li CY. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell stem cell. 2010;7:508–520. doi: 10.1016/j.stem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang L, Zhang X, Yan Q, Minamishima YA, Olumi AF, Mao M, Bartz S, Kaelin WG., Jr Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Molecular and cellular biology. 2007;27:5381–5392. doi: 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue P, Khuri FR, Sun SY. Decoy receptor 2 (DcR2) is a p53 target gene and regulates chemosensitivity. Cancer Res. 2005;65:9169–9175. doi: 10.1158/0008-5472.CAN-05-0939. [DOI] [PubMed] [Google Scholar]
- Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Molecules and cells. 2010;29:435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhang Z, Nelson A, Lamba DA, Reh TA, Ware C, Ruohola-Baker H. Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells. 2013;31(9):1737–48. doi: 10.1002/stem.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D, Lalaoui N, Morizot A, Schneider P, Solary E, Micheau O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Molecular and cellular biology. 2006;26:7046–7055. doi: 10.1128/MCB.00520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D, Lalaoui N, Morizot A, Solary E, Micheau O. TRAIL in cancer therapy: present and future challenges. Expert Opin Ther Targets. 2007;11:1299–1314. doi: 10.1517/14728222.11.10.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell stem cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Csete M, Groves AK, Melega W, Wold B, Anderson DJ. Culture in reduced levels of oxygen promotes clonogenic sympathoadrenal differentiation by isolated neural crest stem cells. J Neurosci. 2000;20:7370–7376. doi: 10.1523/JNEUROSCI.20-19-07370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemetski SM, Gardner LB. Hypoxic regulation of Id-1 and activation of the unfolded protein response are aberrant in neuroblastoma. The Journal of biological chemistry. 2007;282:240–248. doi: 10.1074/jbc.M607275200. [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Ruiz S, Yi F, Herrerias A, Batchelder EM, Izpisua Belmonte JC. Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PloS one. 2011;6:e19743. doi: 10.1371/journal.pone.0019743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- Prigione A, Adjaye J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int J Dev Biol. 2010;54:1729–1741. doi: 10.1387/ijdb.103198ap. [DOI] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Prigione A, Rohwer N, Hoffman S, Mlody B, Drews K, Bukowiecki R, Blümlein K, Wanker EE, Ralser M, Cramer T, et al. HIF1α modulates reprogramming through early glycolytic shift and up-regulation of PDK1-3 and PKM2. Stem Cells. 2013 doi: 10.1002/stem.1552. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. Journal of cell science. 2012;125:5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. The EMBO journal. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlioglu AD, Dirice E, Aydin C, Erin N, Koksoy S, Sanlioglu S. Surface TRAIL decoy receptor-4 expression is correlated with TRAIL resistance in MCF7 breast cancer cells. BMC cancer. 2005;5:54. doi: 10.1186/1471-2407-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nature reviews Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell stem cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby C, Blau CA, Ware C, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem cells and development. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L, Csete M, Lee SH, Kabbani N, Walikonis J, Wold B, McKay R. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci. 2000;20:7377–7383. doi: 10.1523/JNEUROSCI.20-19-07377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell stem cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Tjong J, Alcasid N, Perkins GA, Goissis MD, Ellisman MH, Perez GI, Cibelli JB. Mitochondrial rejuvenation after induced pluripotency. PloS one. 2010;5:e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell stem cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Takubo K, Suda T. Roles of the hypoxia response system in hematopoietic and leukemic stem cells. Int J Hematol. 2012;95:478–483. doi: 10.1007/s12185-012-1071-4. [DOI] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CAt, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PloS one. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nature medicine. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Malek SN, Zheng P. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell stem cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem cells and development. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Bartz S, Mao M, Li L, Kaelin WG., Jr The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Molecular and cellular biology. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell stem cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. The EMBO journal. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. The EMBO journal. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Dosey TL, Biechele T, Moon RT, Horwitz MS, Ruohola-Baker H. Assessment of hypoxia inducible factor levels in cancer cell lines upon hypoxic induction using a novel reporter construct. PloS one. 2011;6:e27460. doi: 10.1371/journal.pone.0027460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell stem cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.