Abstract
Posttranslational modification of cell cycle regulators with ubiquitin chains is essential for eukaryotic cell division. Such chains can be connected through seven lysine residues or the amino-terminus of ubiquitin, thereby allowing the assembly of eight homogenous and multiple mixed or branched conjugates. While functions of homogenous chain types have been described, physiological roles of branched structures are unknown. Here, we report that the anaphase-promoting complex (APC/C) efficiently synthesizes branched conjugates that contain multiple blocks of K11-linked chains. Compared to homogenous chains, the branched conjugates assembled by the APC/C strongly enhance substrate recognition by the proteasome, thereby driving the degradation of cell cycle regulators during early mitosis. Our work, therefore, identifies an enzyme and substrates for modification with branched ubiquitin chains and points to an important role of these conjugates in providing an improved signal for proteasomal degradation.
Keywords: ubiquitin, branched ubiquitin chain, K11-linkage, K48-linkage, proteasome
Introduction
Ubiquitylation controls critical signaling pathways in eukaryotes and is essential for cell proliferation, differentiation, and survival (Deshaies and Joazeiro, 2009; Schulman and Harper, 2009). The transfer of a single ubiquitin to a substrate, a reaction referred to as monoubiquitylation, typically alters interactions, localization, or activity of the modified protein (Dikic et al., 2009). Conversely, the attachment of multiple ubiquitin molecules results in polymeric chains that depending on their connectivity could have unique functions.
Ubiquitin chain formation can occur through seven lysine residues or the amino-terminus of ubiquitin, leading to the assembly of multiple chains with distinct topology (Komander and Rape, 2012). All linkages have been detected in cells, and their abundance changes during cell division or differentiation (Peng et al., 2003; Xu et al., 2009). The first chain types to be discovered, termed canonical ubiquitin chains, had distinct consequences for the modified protein: while chains connected through K48 of ubiquitin promoted proteasomal degradation, K63-linked chains regulated the assembly of oligomeric complexes (Chau et al., 1989; Johnson et al., 1995; Spence et al., 2000; Wang et al., 2001). Based on these observations, it was hypothesized that many ubiquitylation marks might trigger unique biological outcomes, reminiscent of a code.
Yet as roles of atypical conjugates are only beginning to emerge, the complexity of ubiquitin-dependent signaling remains poorly understood. In addition to the canonical conjugates, homogenous chains could also be formed by modification of M1, K6, K11, K27, K29, or K33 (Jin et al., 2008; Tokunaga et al., 2009). Several of these linkages can mediate proteasomal degradation, but the reason for this redundancy is unclear (Jin et al., 2008; Johnson et al., 1995; Koegl et al., 1999; Xu et al., 2009). Conjugates of more complex topology, such as mixed chains, are formed during endocytosis or immune signaling (Boname et al., 2010; Emmerich et al., 2013). Proteomic analyses also showed that a single ubiquitin molecule embedded within a chain can be modified at two or more sites, a process that leads to the assembly of branched conjugates (Kim et al., 2007; Peng et al., 2003). In vitro, branched linkages through K27, K29, or K33 of ubiquitin impede proteasomal recognition (Kim et al., 2007). However, as neither physiological enzymes nor substrates are known, it remains unclear whether branched conjugates play important roles in ubiquitin-dependent signaling.
The anaphase-promoting complex (APC/C) provides a powerful model to test for functions of atypical chains. While yeast APC/C modifies its substrates with canonical K48-linked chains (Rodrigo-Brenni and Morgan, 2007), the metazoan APC/C assembles atypical K11-linked conjugates to drive proteasomal degradation and mitotic exit (Jin et al., 2008; Matsumoto et al., 2010). In human cells, the APC/C initiates chain formation by using the E2 Ube2C (also known as UbcH10, UbcX, Vihar or E2C). Although Ube2C prefers to synthesize K11-linkages, it also connects ubiquitin molecules through K48 or K63 (Kirkpatrick et al., 2006; Williamson et al., 2011). Another APC/C-E2, Ube2S, recognizes substrate-attached ubiquitin to produce specific K11-linked chains (Wickliffe et al., 2011; Williamson et al., 2009; Wu et al., 2010). The abundance of K11-linkages rises dramatically during mitosis, when the APC/C is active, and this boost in K11-linked chain formation is dependent on Ube2S (Matsumoto et al., 2010; Wickliffe et al., 2011). However, as substrates that require Ube2S for degradation have not been reported, the physiological importance of K11-linked chains has not been fully addressed.
In this study, we have identified APC/C-substrates, including the kinase Nek2A, that require Ube2S for degradation. The reconstitution of Nek2A-ubiquitylation revealed that Ube2S does not simply extend a conjugate, but instead branches multiple K11-linked chains off the assemblies produced by Ube2C. Compared to homogenous chains, branched conjugates synthesized by the APC/C increase the efficiency of proteasomal substrate recognition, and accordingly, are required for the degradation of cell cycle regulators at times of limited APC/C-activity. Our work identifies the APC/C as an enzyme that synthesizes branched ubiquitin chains and ascribes an important role to these conjugates in providing an improved signal for proteasomal degradation.
Results
Prometaphase APC/C-substrates require Ube2S for degradation
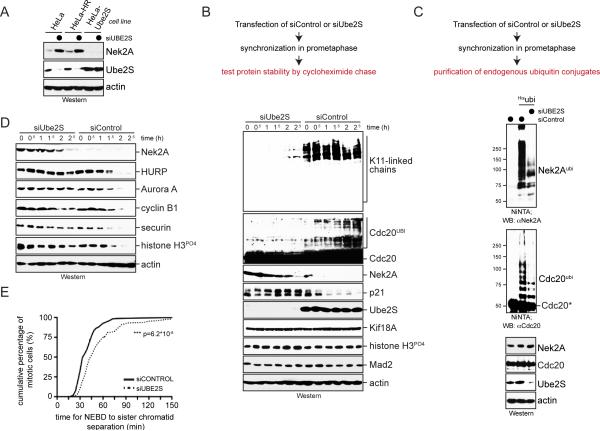
The metazoan APC/C cooperates with Ube2S to assemble K11-linked chains that drive the degradation of cell cycle regulators during mitosis and G1. However, as depletion of Ube2S showed few defects during mitosis, it is not fully understood whether K11-linked chains have essential roles for cell division (Garnett et al., 2009; Williamson et al., 2009; Wu et al., 2010). To address this issue, we systematically screened cells that expressed Ube2S at less than 2% of its normal abundance for changes in protein levels. Strikingly, depletion of Ube2S caused a strong increase in the abundance of Nek2A kinase during prometaphase, a cell cycle stage when the APC/C is partially inhibited by the spindle checkpoint (Figure 1A; Figure S1A). The accumulation of Nek2A was seen with multiple siRNAs against Ube2S and reversed by stable expression of siRNA-resistant Ube2S (Figure 1A; Figure S1B). Depletion of Ube2S neither changed the levels of Nek2A mRNA nor the mitotic stage of synchronized cells, suggesting that instead it affected the stability of the Nek2A protein (Figure S1C, D).
Figure 1. Ube2S is required for APC/C-substrate ubiquitylation in prometaphase.
A. Ube2S was depleted from prometaphase HeLa cells resistant to hygromycin (HeLa-HR) or stably expressing siRNA-resistant Ube2S, and analyzed for Nek2A-levels by Western. B. HeLa cells transfected with control or Ube2S-siRNAs were synchronized in prometaphase, treated with cycloheximide, and analyzed for protein abundance by Western. C. HeLa cells expressing Hisubiquitin and transfected with control- or Ube2S-siRNA were synchronized in prometaphase and treated with MG132. Purified ubiquitin conjugates were analyzed by αNek2A- or αCdc20-Western. D. Nocodazole-arrested HeLa cells treated with control- or Ube2S-siRNAs were released into drug-free medium and protein levels were analyzed by Western. E. Depletion of Ube2S delays anaphase initiation. HeLa cells stably expressing GFPtubulin and mCherryhistone H2B were transfected with control- or Ube2S-siRNA and monitored by live-cell imaging. The time from nuclear envelope breakdown to initiation of sister chromatid separation was determined for >130 cells per condition (n=3). See also Figure S1.
Based on these findings, we tested by cycloheximide chase experiments whether Ube2S was required for APC/C-substrate degradation during prometaphase. Indeed, the depletion of Ube2S resulted in a pronounced increase in the half-life of Nek2A as well as the CDK-inhibitor p21, another APC/C-substrate (Figure 1B). Accordingly, purification of ubiquitin conjugates from prometaphase cells revealed that Ube2S was required for the attachment of high molecular weight (MW) chains to endogenous Nek2A and p21 (Figure 1C; Figure S1E). Ube2S also mediated the ubiquitylation of Cdc20 (Figure 1B, C), a reaction that disassembles spindle checkpoint complexes (Foster and Morgan, 2012; Reddy et al., 2007; Uzunova et al., 2012; Williamson et al., 2009). By contrast, another prometaphase substrate, cyclin A, was not stabilized (Figure S1A), which might be due to incomplete Ube2S-depletion or redundant mechanisms governing degradation of this cell cycle regulator (Kikuchi et al., 2013). In agreement with our results, the loss of Ube2S had pronounced effects on early mitosis: it delayed sister chromatid separation, blocked release of cells from a spindle checkpoint induced arrest, and sensitized cells to low concentrations of nocodazole (Figure 1D, E; Figure S1F). These experiments, therefore, show that Ube2S is important during prometaphase, a cell cycle time when the spindle checkpoint reduces the overall APC/C-activity.
The APC/C synthesizes branched ubiquitin chains in vitro
To dissect the role of Ube2S during prometaphase, we reconstituted the ubiquitylation of Nek2A. Previous work had shown that Ube2S elongates K11-linked chains that were initiated by Ube2C or Ube2D (Garnett et al., 2009; Williamson et al., 2009; Wu et al., 2010). In agreement with this model, the APC/C and its E2s modified Nek2A with high-MW conjugates in the presence of wild-type, but not a K11R-mutant of ubiquitin (ubiR11; Figure 2A; Figure S2A). Chains derived with ubiR11, which cannot be used by Ube2S for chain formation, closely mirrored those seen in cells lacking Ube2S, showing that our reconstitution recapitulated the in vivo modification of Nek2A. Surprisingly, the MW of Nek2A-conjugates was strongly reduced in reactions that contained a ubiquitin mutant with K11 as the only site for chain formation (ubiK11; Figure 2A; Figure S2A). This was in contrast to substrates modified by Ube2S alone (Figure 2B) and suggested that formation of high-MW chains on Nek2A was dependent on multiple linkages.
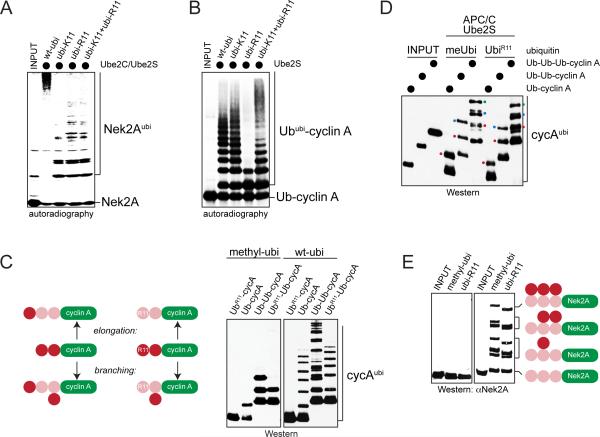
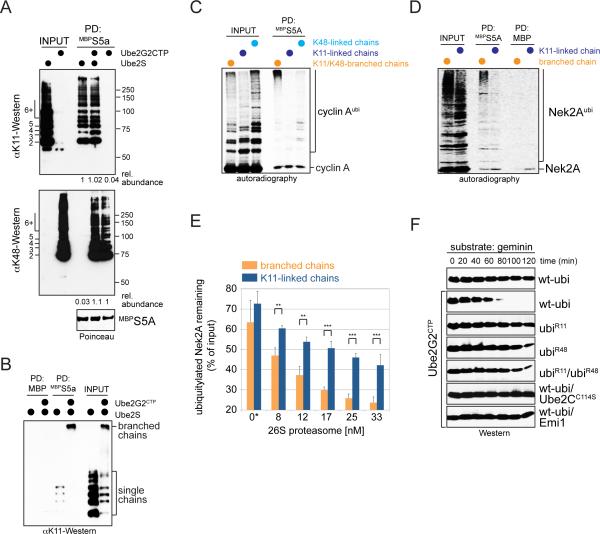
Figure 2. The APC/C assembles branched ubiquitin chains.
A. Nek2A is modified with branched chains in vitro. 35S-labeled Nek2A272-445 was incubated with APC/CCdc20, Ube2C, Ube2S, and ubiquitin mutants. Reactions were analyzed by autoradiography. B. 35S-labeled Ub-cyclin A was incubated with APC/CCdc20, Ube2S, and ubiquitin mutants. Reactions were analyzed by autoradiography. C. Ub-cyclin A, UbR11-cyclinA, Ub-Ubcyclin A and UbR11-Ub-cyclin A were incubated with APC/C, Ube2S, and methylubiquitin or ubiquitin. Left: reaction products if Ube2S would elongate or branch chains (pink: fused substrate-ubiquitin; red: ubiquitin added by Ube2S). Right: Reactions were analyzed by αHA-Western. D. Ub-cyclin AHA, Ub-Ub-cyclin AHA, or Ub-Ub-Ub-cyclin AHA were incubated with APC/CCdh1, Ube2S and methylubiquitin or ubiR11. Reaction products were analyzed by αHA-Western. Red dot: first added ubiquitin; blue dot: second added ubiquitin; green dot: third added ubiquitin. E. Ube2S efficiently modifies Nek2A with branched chains in vitro. Ub3-Nek2A was incubated with APC/C, Ube2S, and methylubiquitin or ubiR11, and analyzed by αNek2A-Western. See also Figure S2.
Conjugates with multiple linkages could be mixed chains, in which distinct connections are used within a single conjugate, or branched structures, in which at least one moiety received two or more ubiquitin molecules. To distinguish between these possibilities, we incubated Nek2A with mixtures of ubiR11 and ubiK11. If the APC/C assembled mixed chains, it could sequentially use ubiR11 and ubiK11 to build conjugates of similar MW as seen with ubiquitin. In contrast, branched conjugates that result from concurrent modification of K11 and another ubiquitin lysine could not be assembled; thus, if the APC/C were a branching enzyme, chains formed with ubiK11 and ubiR11 should be of lower MW than with wt-ubiquitin. Importantly, Nek2A was modified with low-MW conjugates in the presence of ubiR11 and ubiK11, suggesting that the APC/C decorated this substrate with branched chains (Figure 2A; Figure S2A).
As physiological enzymes for the assembly of branched chains were unknown, we wished to test the notion that the APC/C could produce such conjugates. To this end, we circumvented the initiation step and incubated APC/C and Ube2S with substrates already linked to linear chains of defined length. We investigated two prometaphase APC/C-substrates: Nek2A, which requires Ube2S for degradation, and cyclin A, which can be robustly ubiquitylated in vitro (Rape et al., 2006). We supplemented these reactions with methylubiquitin, which allows transfer of the first ubiquitin by Ube2S, but interferes with further chain extension. If Ube2S elongates preexisting chains, only the distal ubiquitin of our substrates should be modified, yet if it assembles branched conjugates, internal moieties could be ubiquitylated as well. Demonstrating the formation of branched chains, Ube2S added two methylubiquitin molecules to Ub-Ub-cyclin A, and it attached one ubiquitin to a substrate variant that lacked Lys11 in its distal moiety (UbR11-Ub-cyclin A; Figure 2C).
Ube2S synthesized the branched linkage on UbR11-Ub-cyclin A with the same specificity and efficiency as seen for elongation (Figure 2D; Figure S2B), and it branched ubiquitin molecules off substrates with longer chains, such as Ub3-cyclin A and Ub3-Nek2A (Figure 2D, E). If these assays were performed with wt-ubiquitin, Ube2S branched a block of ~6 K11-linked molecules off each ubiquitin containing a free K11, thereby effectively modifying Ub-Ub-cyclin A with chains that had twice the MW as those on Ub-cyclin A or UbR11-Ub-cyclin A (Figure 2C).
Ube2S acts after Ube2C, which decorates APC/C-substrates with short chains containing ~50% K11-, but also K48- and K63-linkages (Kirkpatrick et al., 2006). If Ube2C produced the latter two linkages, Ube2S could use an internal ubiquitin to branch off a K11-linked chain. To test this hypothesis, we incubated Ub-cyclin A with Ube2C at concentrations that led to formation of short chains (Ub3-cyclin A). We stopped the initiation step with an excess of Ube2CC114S, which does not affect Ube2S (Wickliffe et al., 2011), and then added methylubiquitin and Ube2S. Again demonstrating the assembly of branched conjugates, Ube2S attached up to three ubiquitin molecules to Ub3-cyclin A produced by Ube2C (Figure S2C). In addition, we performed single substrate-turnover reactions with ubiR48/63 or ubiK6/K11. These mutants support formation of K11-linked chains, but prevent the synthesis of K48- and K63-linkages by Ube2C that are required for branching by Ube2S. Importantly, Ube2S decorated APC/C-substrates with high-MW chains in the presence of Ube2C and WT-ubiquitin, but not if Ube2C was omitted or ubiR48/63 or ubiK6/K11 were used (Figure S2D, E). Thus, the APC/C modifies substrates with branched chains in vitro.
The APC/C synthesizes branched ubiquitin chains in cells
As seen by mass spectrometry, the APC/C decorated Nek2A with chains containing K11-, K48-, and K63-linkages (Figure S3A). Based on the linkage specificities of Ube2C and Ube2S (Kirkpatrick et al., 2006; Williamson et al., 2009), this finding implied that Ube2S assembles K11/K48- and K11/K63-branches, which - due to the multiple Lys and Arg residues separating the two attachment sites in the branched ubiquitin - cannot be detected by trypsin-based mass spectrometry. We, therefore, developed an alternative method to monitor formation of branched chains that allowed us to follow the synthesis of such conjugates in cells (Figure 3A): we identified two ubiquitin residues, Gly53 and Glu64, that permitted insertion of a TEV-cleavage site and FLAG-epitope and were still recognized by Ube2S and APC/C (Figure 3B, C; Figure S3B). We modified substrates with ubi53TEV and ubi64TEV/FLAG and then treated the reaction products with TEV-protease. This led to the collapse of long chains and the emergence of diagnostic FLAG-reactive peptides: while modification of a distal moiety produced a small FLAG-reactive cleavage product, branches could be seen as additional species of higher MW. We had to use both ubi53TEV and ubi64TEV/FLAG, as this mixture proved to be most efficient in competing with wt-ubiquitin in later in vivo experiments.
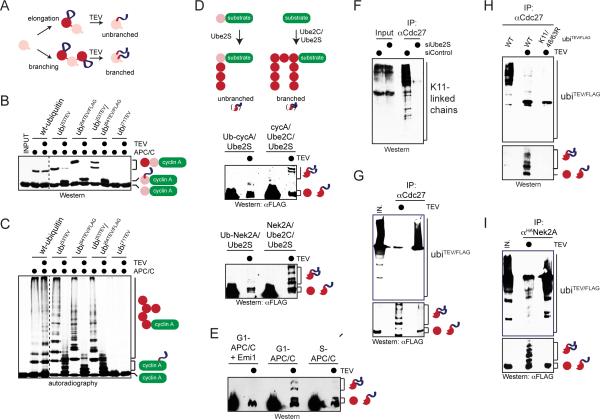
Figure 3. The APC/C assembles branched chains in cells.
A. Outline of a method to monitor synthesis of branched chains. A TEV-site and FLAG-epitope (ubiTEV/FLAG) were introduced into ubiquitin. Cleavage of ubiTEV/FLAG-conjugates reveals a ~2kDa stamp for elongation or multiple stamps for branching. B. ubiTEV/FLAG is recognized by Ube2S and APC/C and sensitive to TEV cleavage. Ub-cyclin AHA was incubated with APC/C, Ube2S, and wt-ubiquitin or ubiTEV mutants. Reactions were treated with TEV and analyzed by αHA-Western. C. 35S-cyclin A was incubated with APC/C, Ube2C, Ube2S, and ubiquitin, ubiTEV or ubiTEV/FLAG proteins. Reactions were treated with TEV and analyzed by autoradiography. D. Ub-cyclin A or Ub-Nek2A272-445 were incubated with APC/C, Ube2S, ubi53TEV and ubi64TEV/FLAG to add unbranched K11-linked chains; alternatively, cyclin A or Nek2A were modified by APC/C, Ube2C, Ube2S, ubi53TEV and ubi64TEV/FLAG with branched chains. Reactions were treated with TEV and analyzed by αFLAG-Western. E. Nek2A272-445 was incubated with APC/C, Ube2C, Ube2S, ubi53TEV and ubi64TEV/FLAG. APC/C was purified from G1 phase, when it is active, or S phase, when APC/C-activity is low. APC/C was also inhibited by Emi1. Reactions were treated with TEV and analyzed by αFLAG-Western. F. APC/C was purified from prometaphase HeLa cells transfected with control-or Ube2S-siRNA and analyzed for co-purifying K11-linked chains using αK11-Western. G. APC/C synthesizes branched chains in vivo. APC/C was purified from prometaphase cells expressing FLAGubi53TEV and Hisubi64TEV/FLAG, treated with TEV, and analyzed by αFLAG-Western. Upper panel: conjugates co-purifying with APC/C. Lower panel: MW-range for diagnostic peptides (longer exposure). Ubiquitin detected in reactions without TEV is produced by DUBs that co-precipitate with APC/C. H. APC/C was purified from cells WT- or Lys-mutant FLAGubi53TEV and HISubi64TEV/FLAG, treated with TEV, and analyzed for co-purifying conjugates (upper panel) or diagnostic peptides (lower panel). I. Nek2A is modified with branched chains in cells. HANek2A was purified from cells expressing FLAGubi53TEV and Hisubi64TEV/FLAG and treated with TEV. Reactions were analyzed by αFLAG-Western. See also Figure S3.
Importantly, diagnostic branched peptides were observed on Nek2A and cyclin A, further demonstrating that APC/C, Ube2C, and Ube2S modify substrates with branched chains (Figure 3D). By contrast, no branches were detected if we incubated Ub-cyclin A or Ub3-Nek2A with APC/C and Ube2S, which leads to attachment of unbranched K11-linked chains (Figure 3D). Moreover, branched conjugates were absent if Nek2A was incubated with ubiR11/48/63, a mutant that inhibits the formation of linkages used by Ube2C and Ube2S; if the APC/C was purified from S phase cells, when it is inactive; or if active APC/C was inhibited by Emi1 (Figure 3E; Figure S3F).
To determine whether the APC/C synthesizes branched chains in cells, we built on the observation that mitotic APC/C co-purifies with proteins modified with K11-linked chains. Formation of these conjugates was dependent on Ube2S (Figure 3F), and they were eliminated by treatment with the K11-specific DUB Cezanne (Figure S3C). The K11-linked chains could be washed off the APC/C with high-salt buffers (Figure S3D), indicating that they were attached to endogenous substrates that remained bound to the APC/C following their modification. Therefore, we expressed FLAGubi53TEV and ubi64TEV/FLAG under conditions that did not change ubiquitin levels (Figure S3E), purified the APC/C with co-precipitating substrates, and treated the samples with TEV. These experiments showed that endogenous APC/C-substrates were modified with branched chains, with the branching pattern reflecting that of our in vitro analyses (Figure 3G). The formation of branched linkages was severely reduced upon expression of ubiR11/48/63, a mutant that obliterated branching in vitro, or upon synchronization of cells in S phase, when APC/C is inactive (Figure 3H; Figure S3G). To test whether the APC/C modifies a specific substrate with branched chains, we purified ubiquitylated Nek2A from cells expressing ubiTEV-constructs. Treatment of Nek2A-purifications with TEV revealed a high fraction of branched chains, similar to the modification of Nek2A with such conjugates in vitro (Figure 3I). These findings were supported by the observation that expression of ubiK6/11, a mutant that allows formation of K11-linked chains, but prevents branched conjugates, strongly impaired the modification of APC/C-substrates in cells (Figure S3H). We conclude that the APC/C assembles branched chains in cells, with Nek2A being an efficient substrate for this modification.
Altering the linkage-specificity of metazoan APC/C
To determine specific functions of branched chains, we required an experimental system that allowed us to compare the fate of a substrate decorated with different homogenous chains or branched conjugates that resulted from a combination of the previous chain types, respectively. Such experiments are difficult to perform with Ube2C, as this E2 produces chains of mixed topology and length. To overcome this limitation, we engineered a K48-specific APC/C-E2 that together with the physiological E2s enabled us to modify substrates either with K11-, K48-linked, or K11/K48-branched chains.
To generate a K48-specific APC/C-E2, we focused on the C-terminal peptide (CTP) of Ube2S: whereas deletion of the CTP abrogated binding of Ube2S to the APC/C, its fusion was sufficient to deliver a heterologous protein to the APC/C (Figure S4A-E). If we appended the CTP to the K48-specific E2 Ube2G2, the resulting Ube2G2CTP bound the APC/C (Figure S4F, G) and ubiquitylated substrates with similar efficiency as Ube2S (Figure 4A; Figure S4H). The activity of Ube2G2CTP towards the APC/C was inhibited by mutation of its active site (Figure S4I, J), addition of pseudosubstrate or competitive APC/C-inhibitors (Figure 4B), or mutation of APC/C-degrons in substrates (Figure 4C). Importantly, assays with ubiquitin mutants or linkage-specific antibodies demonstrated that Ube2G2CTP modified APC/C-substrates with K48-linked chains (Figure 4D-F).
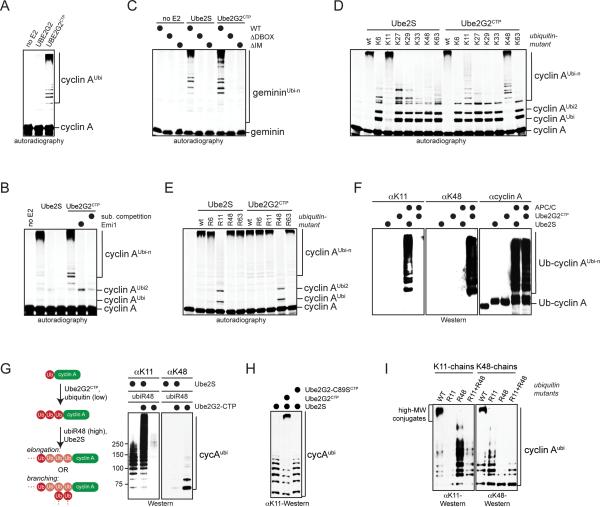
Figure 4. Synthesis of branched chains with defined composition.
A. 35S-cyclin A was incubated with APC/CCdh1 and no E2, Ube2G2, or Ube2G2CTP. Reactions were analyzed by autoradiography. B. 35S-labeled cyclin A was incubated with APC/CCdh1, low levels of Ube2C, and Ube2G2CTP or Ube2S. APC/C was inhibited by Emi1 or an excess of recombinant APC/C-substrate. Reactions were analyzed by autoradiography. C. 35S-geminin or mutants in its D-box or initiation motif (IM) were incubated with APC/C, low levels of Ube2C, and Ube2G2CTP or Ube2S. Reactions were analyzed by autoradiography. D. Ube2G2CTP catalyzes chain formation with ubiK48. 35S-cyclin A was incubated with APC/C, low levels of Ube2C, Ube2S or Ube2G2CTP, and ubiquitin mutants. Reactions were analyzed by autoradiography. E. Ube2G2CTP requires K48 of ubiquitin for chain formation. 35S-cyclin A was incubated with APC/C, low levels of Ube2C, Ube2S or Ube2G2CTP, and ubiquitin variants. Reactions were analyzed by autoradiography. F. Ube2G2CTP catalyzes K48-linked chain formation with APC/C. Ub-cyclin A was incubated with APC/C, either Ube2S or Ube2G2CTP, and ubiquitin. Reaction products were analyzed by Western. G. Ube2S and Ube2G2CTP synthesize high-MW conjugates. Ubcyclin A was mixed with Ube2G2CTP and low levels of ubiquitin, which leads to addition of two K48-linked ubiquitin molecules (Ub3-cyclin A). Next, an excess of ubiR48 and Ube2S were added. Ub-cyclin A was also directly incubated with Ube2S and ubiR48. Reactions were analyzed by Western. H. High-MW chains are seen in single substrate-turnover assays. An excess of Ub-cyclin A (1μM) over APC/CCdh1 (~10nM) was incubated with Ube2S, Ube2S/Ube2G2CTP, or Ube2S and Ube2G2CTP-C89S. Reactions were analyzed by Western. I. High-MW conjugates assembled by Ube2S and Ube2G2CTP are branched chains. Ub-cyclin A was incubated with APC/C, Ube2S, Ube2G2CTP, and ubiquitin mutants under single substrate-turnover conditions and analyzed by Western. Mixtures of ubiR11 and ubiR48 allow Ube2S and Ube2G2CTP to synthesize mixed chains, yet prevent introduction of branches. See also Figures S4 and S5.
To test whether we could use the engineered E2 to produce branched chains, we incubated Ub-cyclin A with APC/C, Ube2G2CTP, and ubiquitin under conditions that led to attachment of two K48-linked ubiquitin molecules (Ub3-cyclin A). We then inactivated Ube2G2CTP by an excess of ubiR48, before we added Ube2S. As expected for the assembly of branched chains, Ube2S decorated Ub3-cyclin A with conjugates of much higher MW than those formed on Ub-cyclin A, a substrate that can only be modified with a single K11-linked chain (Figure 4G). Similar high-MW conjugates were detected in single substrate-turnover assays that contained Ube2G2CTP and Ube2S at the same time, but were absent if only one E2 was used or the active site of Ube2G2CTP was mutated (Figure 4H). High-MW chains were also seen in extracts and cells, if both Ube2S and Ube2G2CTP were present (Figure S5A, B). Sequential purification of these conjugates or treatment with the K11-specific DUB Cezanne revealed that they contained K11- and K48-linkages (Figure S5C, D). Importantly, Ube2S and Ube2G2CTP did not produce high-MW chains if incubated with mixtures of ubiR11 and ubiR48 (Figure 4I). Under these conditions, mixed chains with K11- and K48-linkages can be synthesized, but – as no ubiquitin contains K11 and K48 at the same time – K11/K48-branched chains cannot be produced. Thus, these experiments show that Ube2S and Ube2G2CTP efficiently produce K11/K48-branched chains on APC/C-substrates.
Branched chains are stronger proteolytic signals
To determine the functional consequences of branched chains, we depleted Ube2S from cells to prevent formation of such conjugates under physiological conditions. However, as loss of Ube2S also interferes with chain elongation, we used the physiological and engineered APC/C-E2s as a complementary approach to modify substrates with distinct chain types in vitro or to increase branched chain formation in cells. The combination of both strategies should allow us to pinpoint important roles of branched ubiquitin chains in vitro and in cells.
We first tested how modification with distinct chains affects substrate recognition by effectors, such as the proteasome or p97. Intriguingly, whereas substrates modified with either K11- or K48-linked chains were similarly recognized by receptors of the proteasome or p97 (Figure 5A; Figure S6A-C), the same proteins decorated with K11/K48-branched chains associated with these effectors much more efficiently (Figure 5B, C; Figure S6D). The preferential recognition of branched chains was best seen in reactions that compared homogenous and branched conjugates at the same time (Figure 5B; Figure S6D), and it was not a mere function of the number of ubiquitin molecules, as branched conjugates were recognized more efficiently by the proteasomal receptor S5A than K48-linked chains of equal MW (Figure 5C). Similar effects were seen for physiological APC/C-E2s, which targeted Nek2A more efficiently to S5A by modifying it with branched, rather than homogenous chains (Figure 5D). As expected from these results, branched chains were more efficient than linear K11-linked conjugates in driving the degradation of Nek2A by purified proteasomes (Figure 5E). In contrast, the DUBs Cezanne or Usp37 similarly processed homogenous or branched linkages (Figure S6E-F), showing that branched chains do not improve substrate recognition by all effectors of ubiquitin-dependent signaling.
Figure 5. Branched chains enhance proteasomal degradation.
A. Ub-cyclin A was modified by the APC/C and Ube2S or Ube2G2CTP with either K11- or K48-linked chains. Reaction products were incubated with immobilized MBPS5a either alone or together, and analyzed by Western. B. Proteins decorated with K11/K48-linked branched chains bind more efficiently to S5a than those modified with single chains. Ub-cyclin A modified with K11-linked or K11/K48-branched chains were incubated with MBPS5a under stringent conditions, and binding was monitored by Western. C. Preferential binding of branched chains is independent of chain length. 35S-labeled cyclin A was modified by APC/C, low levels of Ube2C, Ube2S and/or Ube2G2CTP, and ubiquitin, ubiK11, or ubiK48 to assemble K11-, K48-linked, or K11/K48-branched chains, and incubated with S5a. D. Branched chains increase the affinity of Nek2A for the proteasomal receptor S5a. 35S-labeled Nek2A272-445 was modified by APC/C, Ube2C and Ube2S with homogenous or branched chains (using ubiK11 or wild-type ubiquitin) and incubated with MBPS5a. Reactions were analyzed by autoradiography. E. Branched chains improve the efficiency of proteasomal degradation in vitro. 35S-HANek2A272-445 was modified with branched or homogenous K11-linked chains and added to increasing concentrations of 26S proteasomes. Five independent titrations were monitored by autoradiography and quantified using ImageJ (n=5; data are average +/- SD; ** p<0.002 in paired t-test; *** p<0.001). The asterisk marks background degradation by proteasomes that co-purify with APC/C. F. Branched chains promote degradation more efficiently. Prometaphase extracts with endogenous Ube2S were treated with Ube2G2CTP in the presence of ubiquitin mutants. The stability of geminin was monitored by Western. Geminin was stable in extracts containing mixtures of ubiR11 and ubiR48, a condition that allows Ube2S and Ube2G2CTP to assemble mixed, but not branched chains. See also Figure S6.
We next compared the distinct chain types in extracts, a system that had allowed the discovery of K11-linked chains and Ube2S (Jin et al., 2008; Williamson et al., 2009). Extracts that either contained K11- or K48-specific APC/C-E2 supported the degradation of anaphase substrates with comparable efficiency (Figure S6G, H). However, neither E2 by itself was able to promote degradation of a substrate with low affinity for the APC/C or trigger substrate turnover under conditions of low APC/C-activity (Figure 5F; Figure S6I). In contrast, combination of K11- and K48-specific APC/C-E2s rapidly induced degradation even under challenging conditions (Figure 5F; Figure S6I). As this was not observed with mixtures of ubiR11 and ubiR48, the improved proteolytic capacity of combined APC/C-E2s required the formation of branched chains.
To analyze signaling by branched chains in vivo, we compared stable cell lines that expressed Ube2S to produce K11-linked chains; Ube2G2CTP and siRNAs against Ube2S to synthesize K48-linked chains; or both K11- and K48-specific APC/C-E2s to assemble K11/K48-branched chains. To exclude overexpression artifacts, we selected cells that expressed less Ube2G2CTP than endogenous Ube2S, and we generated cell lines that contained an extra copy of Ube2S as controls. Confirming our in vitro analyses, either K11- or K48-specific E2 promoted substrate degradation during anaphase, a time of high APC/C-activity (Figure S7A-C). Still, branched chains appeared to be stronger signals: in normal cells, ubiquitylated substrates remain temporarily bound to the APC/C. By contrast, in cells assembling K11/K48-branched chains in addition to the physiological branched conjugates, such proteins were only detected on the APC/C if the proteasome was inhibited, suggesting that an increased formation of branched chains facilitated substrate extraction from APC/C and degradation by the proteasome (Figure 6A). Accordingly, the levels of a model substrate were reduced upon co-expression of endogenous Ube2S and Ube2G2CTP (Figure S7D). For Nek2A, the same results were obtained with physiological E2s: under conditions that supported formation of branched chains, Nek2A was rapidly extracted from the APC/C in a proteasome-dependent manner (Figure 6B, C). By contrast, if Ube2S was depleted and formation of branched chains was inhibited, Nek2A remained bound to the APC/C (Figure 6B), coinciding with its stabilization and hyperphosphorylation of an APC/C-subunit (Figure 1B; Figure 6B-D).
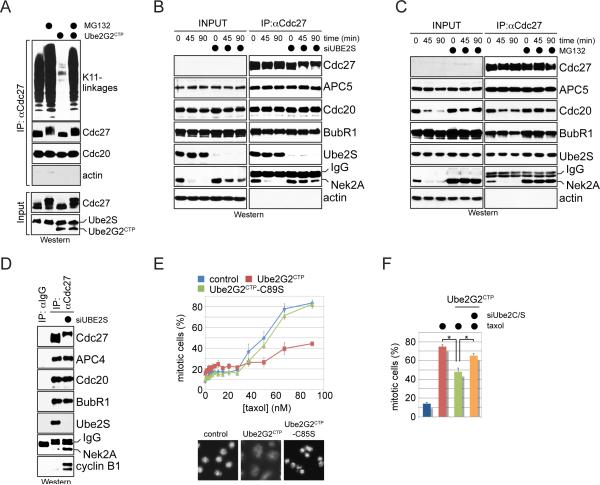
Figure 6. Branched chains promote proteasomal degradation in cells.
A. Branched chains facilitate extraction of ubiquitylated substrates from the APC/C. HeLa cells stably expressing Ube2G2CTP and endogenous Ube2S were released from nocodazole-induced mitotic arrest and treated with proteasome inhibitor MG132, as indicated. Co-purifying K11-conjugates were monitored by Western. B. Branched chains facilitate extraction of Nek2A from the APC/C. HeLa cells transfected with control- or Ube2S-siRNA were synchronized in prometaphase and treated with cycloheximide. APC/C was purified at indicated times after cycloheximide addition, and co-purifying proteins were analyzed by Western. C. The stability of Nek2A-binding to the APC/C was analyzed in prometaphase HeLa cells treated with cycloheximide and proteasome inhibitors. D. APC/C was purified from prometaphase HeLa cells treated with control- or Ube2S-siRNA, and levels of co-purifying proteins were determined by Western. E. Stable expression of Ube2G2CTP, but not Ube2G2CTP-C89S, causes spindle checkpoint bypass. Upper panel: HeLa cells stably expressing indicated proteins were treated with taxol. Mitotic arrest was scored by automated microscopy (n=3; >1000 cells/condition; data are average +/- SD; unpaired t-tests p<0.0005). Lower panel: Images of representative cells stained with Hoechst. F. Checkpoint bypass requires Ube2G2CTP and Ube2C/Ube2S. HeLa cells or stable cell lines expressing Ube2G2CTP were treated with siRNAs against Ube2C and Ube2S as well as taxol, and the mitotic index was determined by automated microscopy (n=3; 1000 cells/condition; data are average +SD; paired t-test p<0.02). See also Figure S7.
Consistent with these observations, constitutive formation of K11/K48-branched chains resulted in spindle checkpoint bypass and aneuploidy, known consequences of increased APC/C-activity (Figure 6E; Figure S7E). These phenotypes were observed most strongly at equimolar levels of Ube2G2CTP and Ube2S (Figure S7F), and hence at conditions of most efficient branching, but much less dramatically if either K11- or K48-specific E2 was absent (Figure 6F) or if an extra copy of Ube2S was expressed (Figure S7G). These findings corroborated our Ube2S-depletion studies: as mentioned before, loss of Ube2S and concomitant inhibition of branched chain formation impeded Cdc20-ubiquitylation, the reaction that drives checkpoint disassembly (Figure 1C); sensitized cells to nocodazole (Figure S1F); and blocked the release of cells from a spindle checkpoint-dependent arrest (Figure 1D). These experiments, therefore, support the notion that formation of branched chains increases the proteolytic capacity of the APC/C, allowing it to efficiently degrade its prometaphase substrates.
Discussion
In this study, we identified the APC/C as an enzyme that synthesizes branched ubiquitin chains and discovered a critical role for such conjugates in driving efficient proteasomal degradation. Branched conjugates are synthesized during mitosis when Ube2S adds multiple K11-linked chains to mixed assemblies produced by the initiating E2s of APC/C (Figure 7). Branched chains strongly improve proteasomal substrate recognition, and accordingly, they are required for degradation at times of limited APC/C-activity. We have observed similar branching activities for K48- and K63-specific enzymes (data not shown), suggesting that branched chains might frequently be employed in ubiquitin-dependent signaling.
Figure 7. Enhanced protein degradation by branched ubiquitin chains.
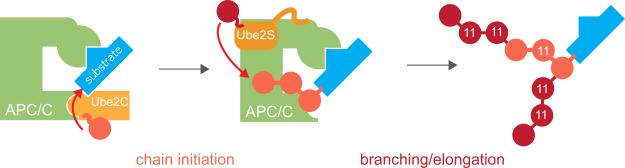
APC/C-substrates are initially modified by Ube2C with short chains containing K11-, K48-, and K63-linkages. Ube2S adds blocks of ~6 K11-linked ubiquitin molecules to each chain moiety with an unmodified K11. This process leads to the formation of branched ubiquitin chains that increase the proteolytic capacity of the initial ubiquitylation mark.
How are branched ubiquitin chains synthesized?
The assembly of branched chains depends on the cooperation of ubiquitylation enzymes with different linkage specificities, a condition fulfilled by the E2s of metazoan APC/C. Ube2C initiates chain formation by modifying substrates with short conjugates containing 50% K11-, but also K48- and K63-linkages (Kirkpatrick et al., 2006). At least in vitro, the APC/C is also able to nucleate chains with E2s of the Ube2D-family, which synthesize ubiquitin chains with little linkage preference (Kirkpatrick et al., 2006). If the initiating E2s use K48- or K63-linkages, K11 of an internal ubiquitin remains unmodified and provides a site for Ube2S to branch off a K11-linked chain. Ube2S is also able to modify ubiquitin molecules embedded in linear or completely K48-linked chains. This indicates that the presence of a free K11, but not overall chain linkage or structure, determines formation of a branch point. Consistent with this notion, Ube2S synthesized a branch with the same efficiency and specificity as it promoted chain elongation at the distal ubiquitin.
It is interesting to note that Ube2S synthesizes a branched linkage with such high efficiency. As documented with linkage-specific antibodies, the accumulation of K11-linked chains during mitosis depends entirely on Ube2S (Matsumoto et al., 2010; Wickliffe et al., 2011), indicating that its linkage specificity is different from most, if not all, mitotic ubiquitylation enzymes. Combined with its efficiency in synthesizing branched linkages, this observation implies that Ube2S could also branch K11-linked chains off conjugates initiated by other enzymes that interact with the APC/C. If this were the case, our findings raise the interesting possibility that the K11-specificity of metazoan APC/C did not arise due to a particular function of this topology, but because it allowed the formation of branched chains on as many initial conjugates as possible.
What is the signaling function of branched ubiquitin chains?
To decipher the information encoded by branched chains, we generated a toolkit of E2s that allowed us to modify substrates with K11-, K48-linked or K11/K48-branched chains. A key result of these studies, we could show for the same protein that K11- and K48-linked chains mediate substrate binding to the proteasome with similar efficiency, and accordingly, both chain topologies trigger degradation at times when the APC/C is fully active. These findings confirm that K11-linked chains are powerful degradation signals (Jin et al., 2008; Matsumoto et al., 2010). Moreover, they show that the K11-specificity of the APC/C is not essential for the turnover of anaphase substrates, while K48-linked chains are not a prerequisite for substrate targeting to the proteasome. Our observations thus support studies that found most linkages to accumulate in response to proteasome inhibition, indicative of a proteolytic role for the majority of chain topologies (Dammer et al., 2011; Peng et al., 2003; Xu et al., 2009).
Providing a potential explanation for this apparent redundancy, we propose that the combination of different chain types allows formation of branched conjugates with improved signaling capacity. Branched conjugates with multiple blocks of K11-linked chains, as generated by the APC/C, increased the efficiency of substrate recognition by the proteasome, and accordingly, promoted degradation in purified systems, extracts or cells even under conditions of low APC/C-activity. Thus, distinct from randomly branched chains that impair degradation (Kim et al., 2007), the branched conjugates assembled by the APC/C act as particularly powerful proteolytic signals.
Why branched chains with multiple blocks of K11-linkages facilitate proteasomal degradation requires further analysis. We favor the idea that the higher signal capacity of branched chains originates from increased levels of substrate-attached ubiquitin, which overcome the modest affinity of effectors towards ubiquitin. As longer ubiquitin chains do not provide a similar effect (Figure 5C; (Thrower et al., 2000)), it is likely that ubiquitin has to be concentrated close to the substrate, thus promoting multivalent recognition by effectors with several ubiquitin-binding domains in the same complex. Indeed, the proteasome and p97, both of which efficiently bind branched chains, contain multiple binding domains for ubiquitylated substrates (Finley, 2009; Lander et al., 2012; Richly et al., 2005). If this hypothesis were correct, we expect that multiple short chains attached to neighboring substrate lysines should also stimulate degradation. However, few substrates have clusters of Lys residues allowing the formation of such structures. Moreover, attachment of the first ubiquitin to a substrate Lys is often the rate-limiting step of chain formation (Pierce et al., 2009; Williamson, 2011), and cleavage of the proximal ubiquitin represents the rate-limiting step of degradation (Finley, 2009). The capacity of branched chains to increase the ubiquitin density without introducing more rate-limiting steps could provide an explanation for why these conjugates generate a particularly powerful proteolytic signal.
Evolutionary implications of branched chain formation
Our discovery that human APC/C assembles branched chains might provide insight into the changes in linkage specificity that occurred between yeast and metazoan APC/C. As yeast APC/C initiates chain formation with Ubc4/5, a set of E2s that add single ubiquitin molecules to substrates (Rodrigo-Brenni and Morgan, 2007), it is unlikely to support branching. Metazoan APC/C synthesizes branched chains with Ube2C and Ube2S, two E2s that are not present in yeast. In this study, we found that Ube2S plays an important role in prometaphase, a cell cycle stage that is different in yeast, which undergoes closed mitosis, and metazoans, where nuclear envelope breakdown dictates that spindle assembly occurs without the chromosomes being confined to a subcellular compartment. Possibly reflecting these differences, few substrates are degraded during prometaphase in yeast, whereas in metazoans several examples of APC/C-substrate degradation have been reported at this cell cycle time (Amador et al., 2007; Hayes et al., 2006; Sedgwick et al., 2013; Song and Rape, 2010; Wolthuis et al., 2008). In addition, prometaphase APC/C continuously disassembles spindle checkpoint complexes, resulting in a dynamic interplay between the APC/C and the checkpoint. Checkpoint disassembly is strongly promoted by a metazoan-specific protein, p31comet, which might point to an increased extent of checkpoint disassembly in these organisms (Foster and Morgan, 2012; Reddy et al., 2007; Uzunova et al., 2012; Varetti et al., 2011; Williamson et al., 2009). We therefore speculate that the increased proteolytic capacity of branched chains, formation of which required the emergence of Ube2C and Ube2S, allowed the metazoan APC/C to cope with higher demands on its activity during early stages of cell division.
Experimental Procesdures
an extended version can be found in the Supplemental Information.
Plasmids
Ube2G2CTP was generated by fusing human Ube2G2 to residues 186-222 of Ube2S. To produce Ub-Ub-cyclin A, ubiquitin1-74 was fused to Ub-cyclin A with a GSEDLYFQSG-linker. To generate Ub3-cyclin A or Ub3-Nek2A272-445, a 3x-ubiquitin-DNA template was synthesized and fused to the substrate. TEV-ubiquitin-derivatives were created by inserting a TEV site after G53 (SENLYFQGS); E64 (NLYFQG); or L71 (GSENLYFQGS). A FLAG-epitope was added after E64 to create TEV/FLAG. Mutagenesis to create K11R, K48R and K63R mutations was performed as described.
APC/C ubiquitylation
35S-labeled substrates were synthesized by IVT/T. APC/C was purified from G1-HeLa cells, and endogenous Ube2S was washed off by 500mM NaCl and 0.1% Triton. Beads were incubated for 30min with E1, 60μM ubiquitin, 3mM ATP, 22.5mM creatine phosphate, 1mM DTT, and substrate. Ube2S, Ube2G2, and Ube2G2CTP were at 10μM; Ube2C in combination with Ube2S or Ube2G2CTP at 100nM; Ube2C by itself at 10μM.
Branched chain detection
To detect branched linkages on Nek2A, 5μM HisNek2A272-445 was incubated with APC/C, 100nM E1, 3mM ATP, 22.5mM creatine phosphate, 1mM DTT, 10μM Ube2C, 10μM Ube2S, 60μM ubiTEV and 60μM ubiTEV/Flag for 30min at 37°C. TEV was added for 1h at 34°C. Reactions were resolved by 20% Tricine-SDS-PAGE. To detect branched linkages in vivo, Flagubi53TEV and Hisubi64TEV/Flag were expressed in 293T cells. After 48h, lysates were subjected to immunoprecipitation by αCdc27- or αHA-antibody. After washes, TEV was added for 1h at 34°C. Reactions were resolved by 20% Tricine-SDS-PAGE.
Purification of endogenous ubiquitin conjugates
To purify ubiquitylated proteins, eight 15cm-dishes HeLa cells were transfected with 5nM control- or Ube2S-siRNA (Lipofectamine RNAimax; Invitrogen). After 18h, cells were also transfected with Hisubiquitin. After 21h, cells were treated with 100ng/mL nocodazole for 16h and 15μM MG132 for the last 2h of synchronization. Cells were resuspended in 8M Urea and lysed by sonication. Hisubiquitin-conjugates were purified under denaturing conditions on NiNTA agarose and detected by Western.
Supplementary Material
Highlights.
The K11-specific Ube2S is required for APC/C-substrate degradation in prometaphase.
The APC/C and Ube2S assemble branched conjugates with blocks of K11-linked chains.
Branched chains improve proteasomal substrate recognition.
Branched chains drive substrate degradation under conditions of limiting APC/C-activity.
Acknowledgements
We dedicate this work to the memory of Gabriel Hayes. We thank Xianfang Donna Xia for early contributions and Rebecca Heald for fluorescently tagged HeLa cells. We are grateful to Julia Schaletzky and members of the Rape lab for comments on the manuscript and encouraging discussions. This work was funded by grants from the NIH to MR. MR is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Molecular cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Morgan DO. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Molecular cell. 2012;47:921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, Fry AM. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Ohata H, Ohoka N, Kawabata A, Naito M. Apollon Promotes Early Mitotic Cyclin A Degradation Independent of the Spindle Assembly Checkpoint. J Biol Chem. 2013 doi: 10.1074/jbc.M113.514430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick GG, Hayward DG, Di Fiore B, Pardo M, Yu L, Pines J, Nilsson J. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. The EMBO journal. 2013;32:303–314. doi: 10.1038/emboj.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nature structural & molecular biology. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Molecular cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit e2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A. Regulation of ubiquitin chain initiation to determine the timing of substrate degradation. Mol Cell. 2011;42:744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Molecular cell. 2011;42:744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.