Abstract
Specific chemicals can prime the plant immune system for augmented defence. β-aminobutyric acid (BABA) is a priming agent that provides broad-spectrum disease protection. However, BABA also suppresses plant growth when applied in high doses, which has hampered its application as a crop defence activator. Here we describe a mutant of Arabidopsis thaliana that is impaired in BABA-induced disease immunity (ibi1) but hypersensitive to BABA-induced growth repression. IBI encodes an aspartyl-tRNA synthetase. Enantiomer-specific binding of R-BABA to IBI1 primed the protein for non-canonical defence signalling in the cytoplasm after pathogen attack. This priming was associated with aspartic acid accumulation and tRNA-induced phosphorylation of translation initiation factor eIF2α. However, mutation of eIF2α-phosphorylating GCN2 kinase did not affect BABA-induced immunity, but relieved BABA-induced growth repression. Hence, BABA-activated IBI1 controls plant immunity and growth via separate pathways. Our results open new opportunities to separate broad-spectrum disease resistance from the associated costs on plant growth.
INTRODUCTION
Plants have evolved sophisticated strategies to defend themselves against pathogens. Apart from pre-existing defence barriers, plants can mobilise structural and chemical defence barriers that become active after pathogen attack. These inducible defences are controlled by the plant’s innate immune system, which provides protection against the majority of potentially harmful micro-organisms. Regulation of plant immunity involves small-molecule hormones, such as salicylic acid (SA) and jasmonic acid (JA)1. To optimally adapt to hostile environments, plants can sensitize their basal immune system in response to specific alarm signals2,3. This so-called ‘priming of defence’ mediates a faster and stronger defence reaction to future pathogen attacks. A classic example comes from systemic acquired resistance (SAR), which is associated with priming of SA-inducible defences in distal plant parts after localised pathogen attack4,5. Although priming of defence rarely provides complete protection against disease and is associated with fitness costs6,7, it has the benefit that it boosts multi-genic basal resistance that relies on perception of multiple microbe- and damage-associated molecular patterns (MAMPs and DAMPS, respectively)8,9. Accordingly, priming of defence offers broad-spectrum disease protection that is difficult to break by pathogens. Furthermore, priming typically provides long-lasting disease protection and can even be transmitted epigenetically to following generations10,11. Taken together, these characteristics make priming attractive for integration in sustainable crop protection8,12.
The search for resistance-inducing chemicals in plants has yielded various compounds. These molecules are often (derivatives of) MAMPs, or plant-endogenous signalling molecules, such as (methyl)SA or azeleic acid13. When applied in relatively moderate quantities, these chemicals prime SA-inducible defences. There are also plant-xenobiotic chemicals that can prime plant defence2,14. A well-known example is β-aminobutyric acid (BABA). This non-protein amino acid protects plants against an exceptionally broad spectrum of stresses, including crop diseases that are difficult to control by conventional strategies of disease management15. The broad-spectrum effectiveness of BABA-induced resistance (BABA-IR) is based on simultaneous priming of SA-dependent and SA-independent defence mechanisms16,17. The SA-dependent component of BABA-IR leads to augmented activation of SA-inducible defence genes and requires a functional NPR1 protein16. Previously, we identified three Arabidopsis mutations that differentially affect SA-dependent and SA-independent BABA-IR17. Recently, two additional Arabidopsis mutants in SA-dependent BABA-IR have been identified: the lerc mutant that is affected in a receptor kinase gene18, and the ald1 mutant that is impaired in pipecolic acid production19. SA-independent BABA-IR is associated with priming of pathogen-induced callose, and requires intact biosynthesis and perception of the plant hormone abscisic acid (ABA)17,20. This priming of cell wall defence has also been linked to induced systemic resistance (ISR) after root colonization by Pseudomonas fluorescens WCS417r21.
A major disadvantage of chemical priming agents is that over-stimulation can lead to stress that affects plant growth6. These stress reactions have been attributed to the costs of direct induction of defence mechanisms, which often develops upon treatment with relatively high doses of the priming agent7. However, the BABA-induced stress response appears to be more specific, and has been related to perturbations in amino acid homeostasis22. This undesirable side-effect of BABA has hindered exploitation of the agent as a crop defence activator, despite its extraordinary wide range of effectiveness15.
In this study, we have identified the IMPAIRED IN BABA-INDUCED IMMUNITY 1 (IBI1) gene as a new master regulator of BABA-IR. IBI1 encodes an aspartyl-tRNA synthetase (AspRS). We provide evidence that the active R-enantiomer of BABA binds to the IBI1 protein and interferes with canonical AspRS activity. This interaction primes IBI1 for non-canonical defence activity against pathogen attack and simultaneously activates a separate stress pathway that results in plant growth repression. Our study shows that the benefits of broad-spectrum disease resistance can be separated from the associated costs, providing new opportunities to improve chemical induction of broad-spectrum disease resistance without affecting crop growth.
RESULTS
The impaired in BABA-induced immunity1 (ibi1) mutant
BABA-treated Arabidopsis develops full immunity against the biotrophic downy mildew pathogen H. arabidopsidis, which acts independently from the SA/NPR1-dependent pathway16,17. To search for novel signalling components in this SA-independent immune response, we screened an EMS-mutagenized M2 population of SA non-accumulating Col-0 NahG23 for mutants impaired in BABA-induced immunity (ibi) against H. arabidopsidis. Evaluation of 34,200 plants from 57 M2 pools yielded two putative ibi mutants from different pools, whose phenotype could be confirmed in the M3 generation. No complementation of the ibi phenotype was observed in F2 progeny from a cross between both mutants (n > 50), while F2 progeny from a backcross between NahG ibi1-1 with NahG displayed monogenic 1: 3 segregation of the mutant phenotype (χ2 = 0.144; p=0.704). These results indicate that both mutations, termed ibi1-1 and ibi1-2, are recessive and allelic.
Microscopic analysis of trypan-blue stained leaves confirmed that leaves of NahG ibi1-1, unlike NahG, failed to arrest H. arabidopsidis colonization after treatment of the roots with 150 μM BABA (Fig. 1a, Supplementary Results, Supplementary Fig. 1a). Epifluoresence microscopy of aniline blue/calcofluor-stained leaves revealed that BABA-treated NahG ibi1-1 did not deposit augmented levels of pathogen-arresting callose at 2 days post inoculation (dpi) with H. arabidopsidis conidiospores (Supplementary Fig. 1b). Hence, IBI1 controls SA-independent priming of cell wall defence. The NahG ibi1-1 mutant was also impaired in SA-independent BABA-IR against the necrotrophic fungus Plectosphaerella cucumerina (Supplementary Fig. 1c), demonstrating that IBI1 controls BABA-IR against biotrophic and necrotrophic pathogens. BABA-induced priming of cell wall defence against necrotrophic fungi requires intact responsiveness to the plant hormone abscisic acid (ABA)20,24. However, both NahG and NahG ibi1-1 developed equal levels of ABA-induced resistance against P. cucumerina (Supplementary Fig. 1c). It can thus be concluded that the ibi1-1 mutation does not affect ABA-induced defence, which suggests that IBI1 acts upstream of ABA in the BABA-IR pathway.
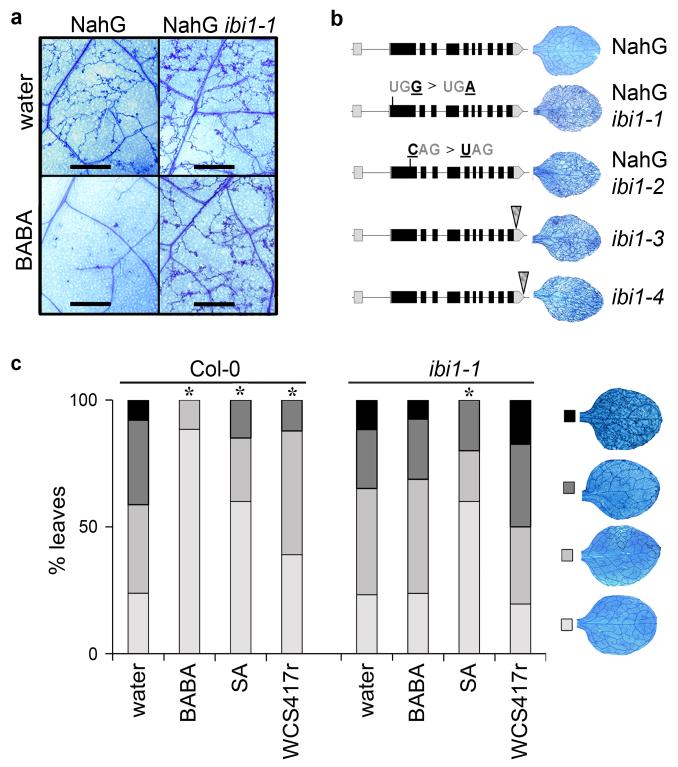
Figure 1. Identification and characterization of the Arabidopsis impaired in BABA-induced immunity1 (ibi1) mutant.
(a) The NahG ibi1-1 mutant does not express induced resistance against H. arabidopsidis WACO9 after root treatment with BABA (150 μM). Photographs of trypan-blue stained leaves show representative differences in pathogen colonization at 7 days after inoculation. Scale bar = 1 mm. (b) Genomic structure of IBI1 (At4g31180) and locations of ibi1-1 (EMS), ibi1-2 (EMS), ibi1-3 (T-DNA, SALK), and ibi1-4 (T-DNA, SAIL) mutations. Insets illustrate representative levels of H. arabidopsidis colonization in leaves of BABA-treated plants. (c) Levels of induced resistance against H. arabidopsidis WACO9 in Col-0 and ibi1-1 after root treatment with BABA (150 μM), Pseudomonas fluorescens WCS417r (5×107 cells/g. soil), or shoot treatment with SA (0.5 mM). Insets show different classes of pathogen colonization; asterisks indicate statistically significant differences in class distribution relative to water-treated plants (Fisher’s exact test, p<0.01, n=50-100).
IBI1 encodes a class II aspartyl-tRNA synthetase
Mapping of the ibi1-1 mutation was performed in a F2 population from a cross between NahG ibi1-1 and the Landsberg erecta (Ler) rpp5 mutant25, which is susceptible to H. arabidopsidis WACO9. Apart from the ibi1-1 allele and the NahG transgene, other genetic determinants from Col-0 and Ler parents were segregating in this mapping population, causing a gradient of susceptibility phenotypes to H. arabidopsidis. Approximately 14,000 F2 plants were screened for loss of BABA-IR against H. arabidopsidis, of which only 1,204 individuals displayed sporulation by 7 dpi. Using sequence length polymorphism markers and (derived) cleaved amplified polymorphic sequence markers26, the ibi1-1 mutation was mapped to a region on the lower arm of chromosome IV, encompassing 64 candidate genes. Sanger sequencing of cDNA-derived PCR products identified a stop mutation in the 1st translated exon of At4g31180 (Fig. 1b), which encoded a class II aspartyl-tRNA synthetase (AspRS). Subsequent sequencing of At4g31180 from NahG ibi1-2 revealed a different stop mutation in the same exon of the gene (Fig. 1b).
IBI1 controls priming of multiple defence responses
To confirm that mutations in At4g31180 are responsible for the ibi1 phenotype, we evaluated phenotypes of two additional T-DNA insertion mutants, ibi1-3 and ibi1-4 (Fig. 1b). Despite the fact that these mutations are in the genetic background of the SA-producing Col-0 accession, both mutants failed to express BABA-IR against H. arabidopsidis (Supplementary Fig. 2a). Similarly, the ibi1-1 mutation crossed into the Col-0 background blocked BABA-IR against H. arabidopsidis (Fig. 1c, Supplementary Fig. 2a). The absence of BABA-IR in ibi1-1 was associated with lack of augmented cell wall defence (Supplementary Fig. 2b). Hence, mutations in At4g31180 cause the ibi1 phenotype and can block BABA-IR in the background of SA-producing Col-0. Considering that SA-dependent resistance is highly effective against H. arabidopsidis27, this suggests that IBI1 controls both SA-dependent and SA-independent BABA-IR. However, application of 0.5 mM SA to shoots of ibi1-1 induced wild-type levels of resistance against H. arabidopsidis (Fig. 1c), demonstrating that IBI1 does not regulate SA-induced defence directly. Since SA-dependent BABA-IR is based on priming of SA-dependent genes, rather than direct induction of SA-dependent defence16,17,21, we tested BABA-induced priming of SA-induced PR1 gene expression. BABA-treated ibi1-1 failed to show augmented PR1 gene induction after SA application to the leaves, whereas BABA-treated Col-0 showed augmented PR1 induction compared to water-treated plants at 4 hours after SA application (Supplementary Fig. 2c). BABA-treated ibi1-1 also failed to display augmented PR1 gene induction after inoculation with H. arabidopsidis (Supplementary Fig. 2d). Thus, the ibi1-1 mutation blocks BABA-induced priming of both SA-dependent and SA-independent defence.
Induced systemic resistance (ISR) against H. arabidopsidis after root colonization by Pseudomonas fluorescens WCS417r is associated with SA-independent priming of cell wall defence, and shares signal transduction components with BABA-IR21. To examine possible involvement of IBI1 in ISR, wild-type and ibi1-1 plants were grown on P. fluorescens WCS417r-containing soil and evaluated for resistance against H. arabidopsidis. As is shown in Fig. 1c, P. fluorescens WCS417r elicited ISR in the wild-type, but not in ibi1-1. Hence, IBI1 is not only critical for BABA-IR, it also controls rhizobacteria-mediated ISR.
IBI1 responds to pathogen attack
The IBI1 gene encodes a class II aspartyl-tRNA synthetase (AspRS), which catalyses biosynthesis of aspartyl-tRNA through esterification of L-aspartic acid (L-Asp) to cognate tRNAsAsp 28. The Arabidopsis genome contains two close AspRS gene homologues: IBI1 and At4g26870. The corresponding proteins show 74% identify and 84% similarity in amino acid sequence (Supplementary Fig. 3a). RT-qPCR analysis revealed that At4g26980 is expressed to only 6% of that of IBI1 (Supplementary Fig. 3b), which is in agreement with publically available transcriptome data. A homozygous T-DNA mutant in At4g26870 (SALK_030485) was fully capable of expressing BABA-IR (Supplementary Fig. 3c), demonstrating that the AspRS encoded by At4g26870 has no alternative defence activity. Interestingly, IBI1 gene transcription was not influenced by BABA (Supplementary Fig. 4). This indicates that BABA-IR by IBI is regulated by post-transcriptional processes. In contrast, IBI1, unlike At4g26870, showed increased transcription in response to H. arabidopsidis, (Fig. 2a), suggesting defence regulatory activity independently of BABA.
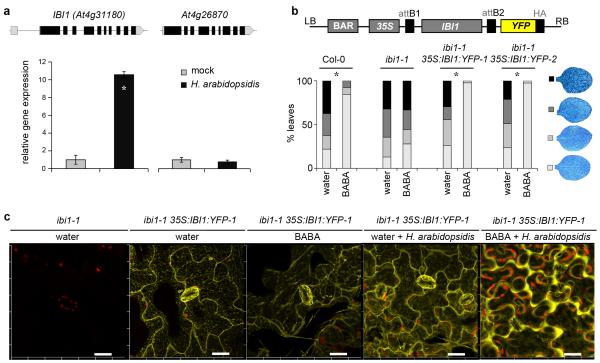
Figure 2. Characterization of the defence function of IBI1.
(a) Transcript accumulation of IBI1 and its close homologue At4g26870 in Col-0 at 5 dpi with H. arabidopsidis WACO9. Data represent average gene expression values (± SEM), relative to mock-inoculated plants. Asterisks indicates a statistically significant difference between treatments (Student’s t-test, p<0.05, n=3). (b) Transformation of ibi1-1 with 35S:IBI1:YFP restores induced resistance against H. arabidopsidis WACO9 after root treatment with BABA (150 μM). Insets show different classes of pathogen colonization; asterisks indicate statistically significant differences in class distribution relative to water-treated plants (Fisher’s exact test, p<0.01, n=50-100). (c) Confocal laser-scanning microscopy analysis of subcellular IBI1:YFP in 35S:IBI1:YFP-1 plants at 2 days after root treatment with water or BABA (150 μM), and 5 days after subsequent H. arabidopsidis WACO9 inoculation. Yellow indicates fluorescence by IBI1:YFP; red indicates chlorophyll fluorescence from chloroplasts. Left panel shows lack of YFP fluorescence in un-transformed ibi1-1 plants. Scale bar = 20 μm.
To study the subcellular localisation of IBI1, we created transgenic lines in the ibi1-1 background, expressing recombinant IBI1:YFP under constitutive control by the 35S cauliflower mosaic virus promoter. Two independent lines displayed a 31- and 32-fold increase in IBI1 transcript levels compared to un-infected Col-0 plants, respectively (Supplementary Fig. 5a). This level of IBI1 expression was sufficient to fully restore BABA-IR against H. arabidopsidis (Fig. 2b), demonstrating cellular functionality of recombinant IBI1-YFP protein. Both over-expression lines also displayed increased basal resistance to H. arabidopsidis, which was particularly pronounced at 5 dpi and associated with augmented expression of pathogen-induced cell wall defence and SA-induced PR1 transcription (Supplementary Figs. 5c-d). Considering that IBI1 transcription was enhanced during H. arabidopsidis infection (Fig. 2a), these results demonstrate that elevated IBI1 transcription contributes to basal resistance in the absence of BABA. Confocal laser scanning microscopy revealed that IBI1-YFP is predominantly localised at the endoplasmic reticulum (ER) and cytoplasmic strands surrounding the ER (Fig. 2c). This subcellular localisation was confirmed by co-localization analysis of IBI1-YPF with CFP- and GFP-tagged ER markers29 in double transgenic F1 progenies from a cross between ibi1-1 35S:IBI1:YFP-1 and ER-marker lines (Supplementary Fig. 6). BABA treatment alone did not alter the subcellular localization of IBI1-YFP (Fig. 2c). However, inoculation of un-primed plants with H. arabidopsidis caused a minor shift of IBI1-YFP to the cytoplasmic periphery of the cell (Fig. 2c). Interestingly, this subcellular response to H. arabidopsidis was dramatically increased in BABA-treated plants, where IBI1-YFP was almost exclusively translocated to the peripheral cytoplasm (Fig. 2c, Supplementary Fig. 7). Since plant aminoacyl-tRNA proteins are targeted to either plastids, mitochondria or cytoplasm30, we conclude that BABA primes pathogen-inducible translocation of IBI1 within the cytoplasmic domain, from the cytoplasmic strands surrounding the ER to the cytoplasmic periphery of the cell.
BABA binds to IBI1 in an enantiomer-specific manner
The chemical structure of L-Asp is strikingly similar to that of the R-enantiomer of BABA (R-BABA; Fig. 3a). Previous studies have shown that R-BABA is the active enantiomer for induced resistance in tobacco, cauliflower and lettuce31-33. The similarity between L-Asp and R-BABA prompted us to investigate whether BABA-IR in Arabidopsis is also determined by its R-enantiomer. Of the two enantiomers, only R-BABA induced resistance against H. arabidopsidis (Fig. 3a). A similar enantiomer-specific effect was found for BABA-IR in tomato against Botrytis cinerea (Supplementary Fig. 8). Hence, BABA is perceived in a mechanistically similar manner across taxonomically unrelated plant species. Co-application of 150 μM BABA with L-Asp to Arabidopsis mildly reduced BABA-IR to H. arabidopsidis (Supplementary Fig. 9a), suggesting competition between BABA and L-Asp. Treatment with 150μM L-Asp alone failed to induce disease resistance (Fig. 3a), although a 10-fold higher concentration of L-Asp (1.5 mM) resulted in a relatively weak increase in resistance (Supplementary Fig. 9b). However, the latter resistance response was also present in ibi1-1 plants, suggesting an IBI1-independent stress response to application of high levels of L-Asp.
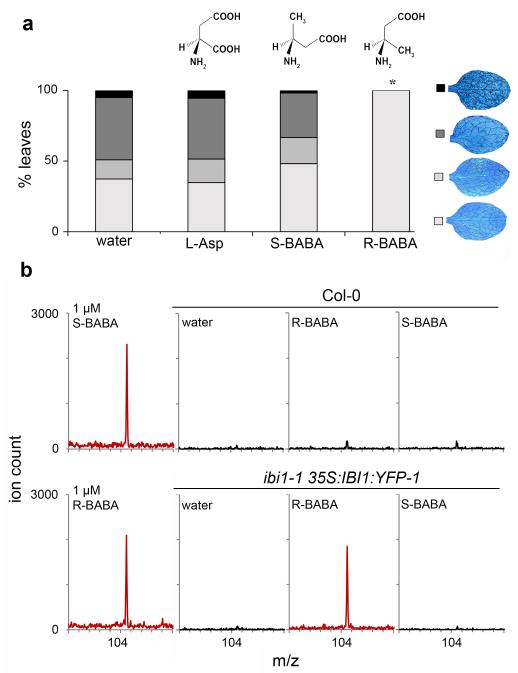
Figure 3. Enantiomer-specific activity of BABA to IBI1.
(a) Levels of induced resistance against H. arabidopsidis WACO9 in Arabidopsis (Col-0) after root treatment with L-Asp (150μM), R-BABA (150μM), or S-BABA (150μM). Insets show different classes of pathogen colonization; asterisk indicates a statistically significant difference in class distribution relative to water-treated plants (Fisher’s exact test, p<0.01, n=50-100). (b) Mass spectrometry analysis of BABA from immunoprecipitated IBI1:YFP of ibi1-1 35S:IBI1:YFP-1 or un-transformed Col-0 (negative control), using MALDI-qTOF ([M+H]; m/z=104,025). Left panels show standards (R-BABA and S-BABA; 1 μM). Immunoprecipitation (IP) was performed with protein extracts from leaves after two successive root treatments with water (control), R-BABA (1.2 mM), or S-BABA (1.2 mM). MALDI-qTOF analysis of IP extracts from ibi1-1 35S:IBI1:YFP-1 plants after R- or S-BABA applications was repeated twice from material of independent experiments, yielding similar results.
To explore the possibility that R-BABA binds to the L-Asp-interacting site of IBI1, we modelled protein-amino acid interactions, using information from crystallised AspRS proteins of Pichia pastoris and Pyrococcus kodakaraensis. Previous site-directed mutagenesis studies with P. pastoris have revealed the L-Asp-, ATP- and tRNA-interacting protein domains of AspRS34, which are conserved between Arabidopsis IBI1, P. pastoris AspRS, and P. kodakaraensis AspRS (Supplementary Fig. 10). Due to the availability of a crystallised co-structure of P. kodakaraensis AspRS and L-Asp at relatively high resolution (1.9 Å)35, our computational docking models of BABA to the L-Asp-binding domain were based on P. kodakaraensis AspRS (PDB: 3NEL). Binding of L-Asp to AspRS produced the lowest energy complex in comparison to R-BABA and S-BABA (Supplementary Fig. 11). The difference in binding energy between L-Asp and R-BABA was 7.25 kcal.mol−1, which can be attributed to the loss of two weak hydrogen-bonding interactions between the side-chain carboxylic acid group of L-Asp and the AspRS binding residues SER487 and ASP331. However, the stronger backbone interactions between ASP348 and SER307 (via a bridging water molecule) were retained for R-BABA. Furthermore, Van der Waals/hydrophobic interactions between the proximal methyl group of BABA and the binding site would partially compensate for the loss of the carboxylic acid group in comparison of L-Asp. The energy difference between L-Asp and S-BABA binding was 11.74 kcal.mol−1. This relatively weak binding affinity between S-BABA and IBI1 is due to the loss of another H-bond between the bridging water molecule and the amino group of S-BABA. Consequently, S-BABA orientated in a non-favourable direction for H-bonding and structural fitting into the protein’s binding cavity (Supplementary Fig. 11). Our models suggest that R-BABA binds with a greater affinity than S-BABA to the Asp-binding pocket of AspRS.
To experimentally confirm enantiomer-specific binding of BABA to IBI1, wild-type (Col-0) and functionally complemented ibi1-1 35S:IBI1:YFP-1 were treated with water, R-BABA, or S-BABA. Subsequently, IBI1:YFP in plant protein extracts was immunoprecipitated and analysed by mass spectrometry for the presence of BABA. Using both MALDI-qTOF and ESI-qTOF analysis, we detected significant quantities of BABA in the immunoprecipitated IBI1:YFP fraction from R-BABA-treated ibi1-1 35S:IBI1:YFP-1 plants (Fig. 3b, Supplementary Fig. 12). Conversely, we detected only traces of BABA in the immunoprecipitated IBI1:YFP fraction of S-BABA-treated ibi1-1 35S:IBI1:YFP-1 plants, which were comparable to levels from R-BABA-treated Col-0 lacking the YFP epitope for immunoprecipitation (Fig. 3b, Supplementary Fig. 12). Thus, BABA binds in an enantiomer-specific manner to IBI1 in planta.
BABA blocks canonical AspRS activity of IBI1
To explore the impact of BABA binding to IBI1, we examined the effects of BABA on AspRS activity. Since IBI1:YFP functionally restores BABA-IR in the ibi1-1 mutant (Fig. 2b), we used immunoprecipitated IBI1:YFP to assay in vitro AspRS activity. However, despite various experimental variations of the assay (Supplementary Table 1)36-39, we failed to detect in vitro ATP-pyrophosphate (PPi) exchange activity by IBI1:YFP. Subsequent experiments revealed that IBI1:YFP loses enantiomer-specific binding activity to BABA during protein purification (Supplementary Fig. 13), indicating that IBI1 functionality is affected when the protein is extracted from its cellular context, probably due to lack of interacting co-factors under in vitro conditions. We therefore continued with an alternative in planta approach to study the impact of BABA on AspRS activity.
In both yeast and Arabidopsis, the protein kinase GCN2 phosphorylates eukaryotic translation initiation factor eIF2α in response to hyper-accumulation of uncharged tRNA40,41. Accordingly, we used eIF2α phosphorylation to test whether BABA inhibits AspRS activity, thereby increasing accumulation of uncharged tRNA substrate. Western blot analysis confirmed that BABA induces eIF2α phosphorylation in Col-0, ibi1-1 and Ler plants, but not in the gcn2-1 mutant42 (Fig. 4a, Supplementary Fig. 14). Hence, BABA activates GCN2, presumably by hindering in planta aminoacylation activity of tRNA synthetases. Interestingly, ibi1-1 plants displayed faint eIF2α phosphorylation in the absence of BABA and increased eIF2α phosphorylation after BABA treatment (Fig. 4a; Supplementary Fig. 14). This suggests that AspRS activity from the relatively lowly expressed At4g26870 gene is also sensitive to perturbation by BABA. It is plausible that the limited AspRS activity of the ibi1-1 mutant increases its sensitivity to BABA-induced AspRS inhibition, leading to augmented accumulation of uncharged tRNA and GCN2 activity.
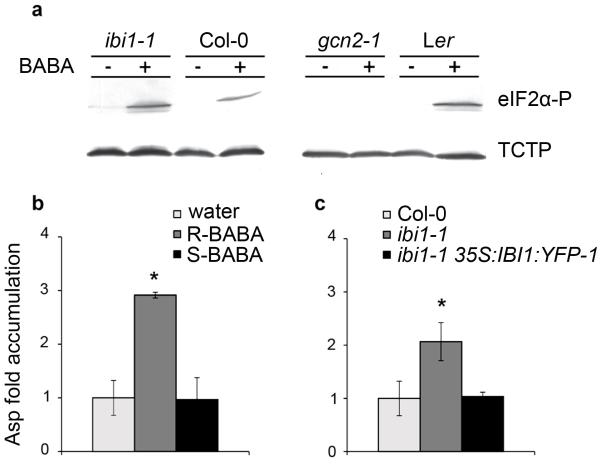
Figure 4. BABA interferes with canonical AspRS activity.
(a) Western blot analysis of eIF2α phosphorylation (Ser51) and TCTP (loading control) in ibi1-1, Col-0, gcn2-1 and Ler at 2 days after spraying the leaves with BABA (250 μM). (b) MALDI-qTOF quantification of Asp levels in leaves of Col-0 at 2 days after root treatment with water, R-BABA (250 μM), or S-BABA (250 μM). (c) MALDI-qTOF quantification of Asp levels in leaves of Col-0, ibi1-1 and ibi1-1 35S:IBI1:YFP-1. Data in (b) and (c) represent average fold-change values in comparison to basal Asp levels in water-treated Col-0. Asterisks indicate statistically significant differences in comparison to water-treated Col-0 (Student’s t-test, p<0.05, n=3-5).
To confirm the impact of BABA on AspRS activity, we employed MALDI-qTOF analysis to measure Asp levels in leaf extracts of wild-type plants following root treatment with either R-BABA, or S-BABA. Treatment with active R-BABA increased in planta Asp levels by 3-fold, whereas the inactive S-enantiomer had no effect (Fig. 4b). Since Asp is a substrate of AspRS, these results strengthen our conclusion that BABA obstructs canonical AspRS activity.
Finally, we compared Asp levels in leaf tissues between wild-type, ibi1-1 and ibi1-1 35S:IBI1:YFP-1 plants to verify in planta AspRS activity by IBI1 (Fig. 4c). As can be expected for a genotype affected in AspRS activity, ibi1-1 displayed enhanced Asp accumulation, which was reverted to wild-type levels in ibi1-135S:IBI1:YFP-1 plants. Together with the observation that ibi1-1 displays augmented activity of tRNA-induced GCN2 (Fig. 4a), these results confirm that IBI1 functions as an AspRS enzyme.
IBI1 controls resistance and growth via separate pathways
Apart from inducing broad-spectrum resistance, BABA is also known to suppress plant growth6,22. To test the role of IBI1 in this stress response, growth rates of wild-type and ibi1-1 seeds were assayed on BABA-containing agar plates (400 μM). Surprisingly, ibi1-1 seedlings on BABA-containing agar stopped growing immediately after seed germination, or failed to germinate all together, while wild-type seeds managed to germinate and grow under these conditions (Supplementary Fig. 15a). This hypersensitivity to BABA was only apparent upon treatment with the R-enantiomer of BABA, and functions independently from SA, since NahG ibi1-1 displayed similar stress sensitivity as ibi1-1 (Supplementary Fig. 15a-b). The difference in BABA-induced stress between wild-type and ibi1-1 plants was also apparent at later developmental stages: one week after BABA application to roots of three-week-old plants, ibi1-1 displayed 50% lower shoot fresh weight values than control-treated ibi1-1, whereas BABA-treated wild-type only exhibited a 26% reduction in shoot fresh weight (Fig. 5a). In a separate experiment, the SALK_030485 mutant in the IBI1 homologue At4g26870 displayed wild-type levels of BABA-induced growth repression (Supplementary Fig. 16a). Considering that At4g26870 is expressed to only 6% of IBI1 (Supplementary Fig. 3b), we conclude that the relatively small reduction in total AspRS of the SALK_030485 mutant is not sufficient to cause detectable levels of hypersensitivity to BABA-induced stress. By contrast, over-expression of IBI1 in ibi1-1 35S:IBI1:YFP-1 provided enhanced tolerance to BABA-induced growth repression (Supplementary Fig. 16b), as can be expected from a genotype with increased BABA-binding capacity.
Figure 5. Genetic separation of BABA-IR and growth repression.
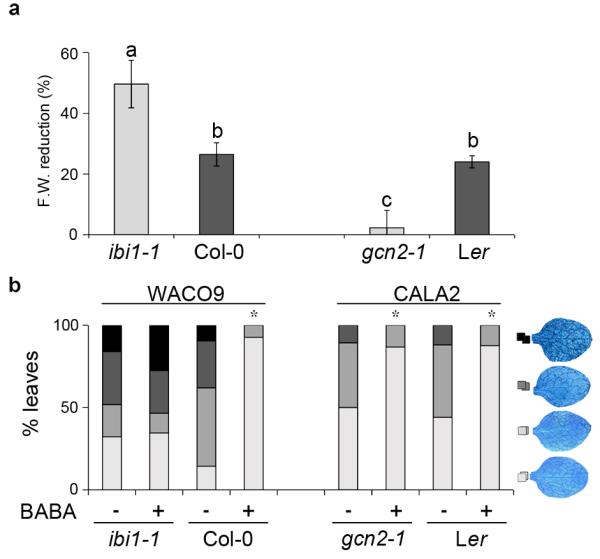
(a) BABA-induced growth suppression in ibi1-1, Col-0, gcn2-1 and Ler at 1 week after root treatment with BABA (400 μM). Shown are mean percentages (± SEM) of fresh weight (F.W.) reduction in BABA-treated plants relative to water-treated plants. Different letters indicate statistically significant differences between genotypes (Fisher’s least significant differences test; p<0.05). (b) Levels of induced resistance against H. arabidopsidis WACO9 (Col-0 and ibi1-1) and H. arabidopsidis CALA2 (Ler and gcn2-1) after root treatment with BABA (150 μM). Insets show different classes of pathogen colonization; asterisks indicate statistically significant differences in class distribution relative to water-treated plants (Fisher’s exact test, p<0.01, n=50-100). (Fisher’s exact test, p<0.01, n=50-100).
BABA-induced plant stress is associated with perturbations in amino acid homeostasis in Arabidopsis22. Considering that eIF2α phosphorylation by GCN2 impacts amino acid metabolism in yeast, we investigated the possibility that GCN2 controls BABA-induced growth repression. Comparison of the gcn2-1 mutant to its corresponding wild-type (Ler) revealed that the gcn2-1 mutation is more tolerant to BABA-induced growth repression (Fig. 5a), confirming that GCN2 indeed regulates BABA-induced stress. By contrast, gcn2-1 seedlings were not impaired in BABA-IR against H. arabidopsidis CALA2, which is virulent on Ler genotypes43 (Fig. 5b). Hence, GCN2 does not regulate BABA-IR. Together, these results show that BABA-IR and BABA-induced growth reduction are controlled by separate signalling pathways.
DISCUSSION
Our study has shown that BABA-induced disease protection requires an intact IBI1 gene (Figs. 1, 2b, 3a, 4b, Supplementary Figs. 1, 2, 3c). In combination with our demonstration that only the active R-enantiomer of BABA binds to IBI1 in planta (Fig. 3b; Supplementary Fig. 12), our results provide plausible evidence that IBI1 is the plant receptor of BABA. Computational docking studies indicated that R-BABA binds to the conserved L-Asp binding site of AspRS in a similar spatial orientation as L-Asp (Supplementary Figs. 10, 11). The canonical L-Asp substrate of the enzyme is constantly processed and released as charged Asp-tRNAasp. Conversely, R-BABA cannot be aminoacylated and would eventually replace L-Asp from the binding pocket, thereby ‘jamming’ the protein’s canonical AspRS activity. In support of this, BABA triggered enhanced Asp accumulation in an enantiomer-specific manner (Fig. 4b) and stimulated GCN2-dependent eIF2α phosphorylation (Fig. 4a), which marks cellular accumulation of uncharged tRNA40-42.
Our study raises one important question: why have plants evolved a receptor of a xenobiotic compound that is not produced by the plant? One possible explanation comes from our finding that P. fluorescens WCS417r bacteria failed to elicit ISR in ibi1-1 plants (Fig. 1c). Considering the critical role of IBI1 in BABA perception, it is plausible that ISR-eliciting rhizobacteria produce BABA, or a functional analogue thereof. Indeed, P. fluorescens WCS417r-ISR and BABA-IR against H. arabidopsidis share similar signal transduction components and are both associated with priming of callose deposition21. It is also plausible that IBI1 contributes to basal disease resistance. Of the two Arabidopsis genes encoding non-plastid AspRS proteins, only IBI1 showed increased transcription in response to H. arabidopsidis inoculation (Fig. 2a). Moreover, transgenic over-expression of IBI1 increased basal resistance in the absence of BABA (Fig. 2b, Supplementary Fig. 5). In combination with selective uptake of Asp by the pathogen, increased IBI1 expression during pathogen infection would lead to more IBI1 protein that is deprived of AspRS activity. We propose that this denial of canonical activity, together with pathogen-induced translocation to the cytoplasm (Fig. 2c, Supplementary Fig. 7), triggers non-canonical defence activity by IBI1. R-BABA partially mimics this process by blocking the Asp-binding pocket of IBI1, thereby depriving it from AspRS activity and priming its non-canonical defence response to pathogen attack.
Aminoacyl-tRNA synthetases are known for their ability to express non-canonical functions beyond translation, which have been attributed to their ability to interact with other proteins44. Interaction between IBI1 and (proteinaceous) co-factors could explain why purified IBI1:YFP lacks in vitro activity (Supplementary Table 1; Supplementary Fig. 13). Specific IBI1-protein interactions could also be responsible for the non-canonical defence activity of IBI1 in the cytoplasm (Fig. 2c, Supplementary Fig. 7). Human glutaminyl-tRNA synthetase exerts anti-apoptotic activity through inhibitory interaction with the mitogen-activated protein triple kinase (MAPKKK) ASK145. Therefore, it is tempting to speculate that IBI1 inhibits cytoplasm-localized plant MAPKKKs with repressive activity on plant immunity, such as MEKK1 and EDR146,47. Interestingly, amino acid alignments between IBI1 and the AspRS homologue At4g26870 revealed relatively high dissimilarity at the N-terminus (Supplementary Fig. 3a). Since At4g26870 does not regulate BABA-IR (Supplementary Fig. 3c), we propose that the non-canonical defence activity of IBI1 is determined by its unique N-terminal sequence., This hypothesis is in agreement with previous findings that non-canonical functions of aminoacyl-tRNA synthetases are determined by variable N- or C-terminal domains44,48.
We conclude that plant perception of BABA is mediated by IBI1. The enzyme can switch from canonical AspRS activity to non-canonical defence activity upon pathogen infection. This response is strongly augmented by R-BABA, which blocks the L-Asp binding site of the enzyme, thereby depriving the protein from AspRS activity and priming it for defence activity (Fig. 6). BABA-induced inhibition of AspRS activity also increases cellular accumulation of uncharged tRNAAsp, which triggers GCN2-dependent eIF2α phosphorylation and stress-related growth repression, independently from BABA-IR (Fig. 6). Hence, broad-spectrum disease protection by BABA can be uncoupled from the concurring plant stress response. Since BABA is perceived in a mechanistically similar manner by taxonomically unrelated plant species31-33 (Supplementary Fig. 8), our discovery of the plant receptor of BABA opens new possibilities to protect crops against economically important crop diseases.
Figure 6. The BABA receptor IBI1 controls BABA-IR and plant growth suppression via separate pathways.
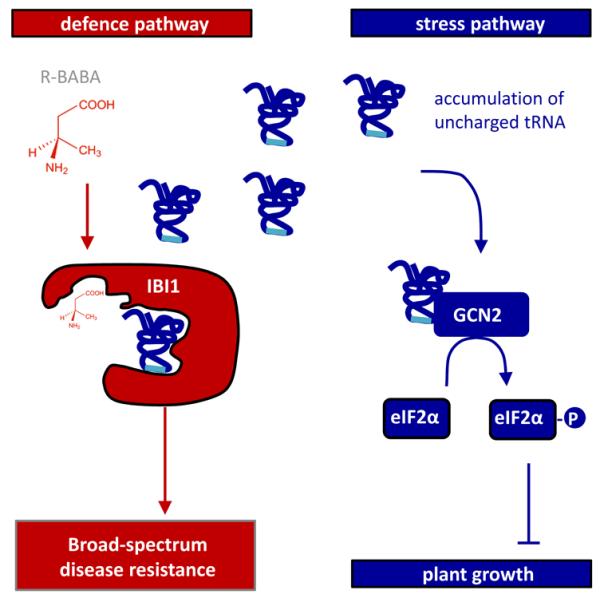
Binding of the R-enantiomer of BABA to the L-Asp-binding site of IBI1 blocks canonical aspartyl-tRNA synthetase (AspRS) activity, which primes the protein for pathogen-inducible defence activity in the cytoplasm (red). Concurrently, the inhibitory activity of R-BABA on AspRS activity leads to accumulation of uncharged tRNAs, which triggers GCN2-dependent phosphorylation of the translation initiation factor eIF2α and suppression of plant growth (blue).
ONLINE METHODS
Plant material and growth conditions
Genotypes of the Arabidopsis lines used are presented in Supplementary table 2. Unless stated otherwise, seeds for plants in bioassays and gene expression experiments were planted in 60 mL pots containing peat/sand mixture (2:1), stratified at 4° C in darkness for 2 days, and cultivated at 8.5h light (150 μmol.m−2.s−1; 20°C) and 15.5h darkness (18°C). For experiments with plants older than 3 weeks, 10-day old seedlings were transplanted individually to 60 mL pots. To minimise environmental variation, plants for immunoblot blot analysis of eIF2α phosphoryation (Westerns) were cultivated on ½-strenth MS agar (0.8%). Solanum lycopersicum (cv. Micro-Tom) seeds were germinated in water for 5 days (darkness; 28°C), transplanted to 100 mL-pots containing Scotts Levington M3 soil, and cultivated at 16h light (150 μmol.m−2.s−1; 26°C) and 8h darkness (21°C).
Chemical treatments
Induction treatments with S/R-β-aminobutyruc acid (BABA; Sigma-Aldrich; A4420-7), R-BABA (Astatech 62363), S-BABA (Astatech; 62359), (±)-Abscisic acid (ABA; Sigma-Aldrich; A1049), or L-Asp (Sigma-Aldrich; A9256) were performed by injecting 10× concentrated 6-mL quantities into 60-mL pots once. Final soil concentrations are indicated in figure legends. Sodium-SA (Sigma-Aldrich; S3007) was applied by spraying a 0.5 mM solution onto 3-week-old plants. Unless stated otherwise, secondary elicitation treatments were performed 2 days after primary induction treatment. For in planta analysis of BABA-IBI1:YFP interaction, 5-week-old Col-0 wild-type and ibi1-1 35S:IBI1:YFP-1 plants were treated twice with water, 1.2 mM R-BABA or 1.2 mM S-BABA at 3 days apart. Five days after the second treatment, leaf material was harvested for protein extraction, immunoprecipitation and mass spectrometry analysis. Agar-grown seedlings for Western blot analysis (2-week-old) were sprayed once with 250 μM S/R-BABA to ensure synchronised activation of BABA-induced stress.
EMS mutagenesis, mutant screening and mapping
Approximately 10,000 Col-0 NahG seeds were EMS mutagenized as described49. Seeds from 25±5 M1 plants/pot were pooled in M2 families for screening. Of each M2 family, 600 seedlings (~10-days old) were transplanted into 200-well trays (Teku; JP3050/230). Three-week-old plants were soil-drenched to a final concentration of 150 μM BABA. Two days later, plants were spray-inoculated with H. arabidopsidis (5×104 sporangiospores/mL) and maintained at 100% R.H. At 5-7 days post inoculation (dpi), plants were evaluated for their phenotype and sporulating plants were transplanted and rescued with 0.1 mg/mL ridomil Gold® (Syngenta). Loss of BABA-IR was confirmed in the M3 generation, as described below. The screen identified 2 impaired in BABA-induced immunity (ibi) mutants (NahG ibi1-1 and NahG ibi1-2), which were confirmed to be recessive and allelic on the basis of segregation analysis of F2 progeny from a backcross with NahG cross and a complementation cross, respectively. NahG ibi1-1 was crossed with the Ler rpp5 mutant25 to produce a F2 mapping population, which was screened for ibi phenotypes. Initial mapping of ibi1-1 was based on 30 susceptible F2 plants, using 22 SSLP markers26. Fine-mapping was based on a selection of 621 of the most susceptible individuals from a total of 1204 sporulating F2 plants isolated from the screen, using additional SSLP, CAPS and dCAPS markers, which were designed from polymorphisms between Col-0 and Ler genomic sequences, using the TAIR Polymorphism/Allele search tool (www.arabidopsis.org). Candidate genes in the fine-mapped region (At4g31560-At4g32105) were amplified from mutant cDNA by PCR using Phusion® DNA polymerase (Finnzymes F-530S), sequenced, and aligned to wild-type DNA using Vector NTI® Advance 11 computer software (Invitrogen).
Selection for ibi1 mutant genotypes in the Col-0 background
F2 progeny from a cross between NahG ibi1-1 and Col-0 were analysed by PCR, using primers against NahG, wild-type IBI1, and the mutant ibi1-1 allele (Supplementary table 2). Screening of 95 F2 plants revealed one heterozygous ibi1-1/IBI1 recombinant without NahG, suggesting that NahG in line B15 is genetically linked to IBI1. PCR-based selection of selfed progeny provided the homozygous Col-0 ibi1-1 mutant. Mutants ibi1-3 and ibi1-4 are derived from SALK (SALK-103893) and SAIL (SAIL-228-H03), respectively, and were selected for homozygous insertions using one T-DNA specific primer and two gene-specific primers flanking the T-DNA insertions (Supplementary table 2).
Resistance assays, stress assays and statistics
Hyaloperonospora arabidopsidis strain WACO9 (compatible with Ws and Col genotypes), or CALA-2 (compatible with Ler), were maintained on Ws NahG or Ler plants, respectively. H. arabidopsidis assays were performed as described21. Plants (2- to 3-week old) were inoculated with H. arabidopsidis (5×104 conidiospores/mL) at 2 days after chemical treatment. Unless stated otherwise, H. arabidopsidis colonisation was determined microscopically at 7 dpi in trypan-blue stained leaves, as described10. Differences in class distribution between treatments were analysed for statistically significant differences by Fisher’s exact tests (α=0.05; SPSS, v19.0).
Callose depositions were examined at 2 dpi in analine-blue/calcofluor-stained leaves, using UV-epifluorescence microscopy as described17. Effectiveness of callose was quantified by percetnage of conidiospores, from which the proximal end of the emerging germ tube was encapsulated in callose. Statistical analyses were performed with average percentages per collected leaf (Student’s t-tests; α=0.05; SPSS, v19.0). Additionally, [total number of callose-arrested spores]/[total number of non-arrested spores] ratios were tested for statistical significance against corresponding ratios from water-treated wild-type plants (binomial test; α=0.05; SPSS, v19.0).
Assays to assess priming of PR1 transcription were performed with 3-week-old plants. SA (0.5 mM) or H. arabidopsidis (5×104 conidiospores/mL) was sprayed onto shoots at 2 days after root treatment with BABA (150 μM) and shoot samples were collected at different time-points after SA application, or 2 dpi with H. arabidopsidis. Statistical analysis was performed with normalised gene expression values (Student’s t-test; α=0.05; SPSS, v19.0).
Plectosphaerella cucumerina bioassays were performed as described20. Five-week old plants were soil-drenched with BABA or ABA and 2 days later inoculated (2.5×106 spores/mL). Disease and colonisation were evaluated microscopically at 7 dpi by assigning trypan-blue stained leaves to different classes: I, no cell death or fungal colonization; II, moderate necrosis at inoculation site but no pathogen colonization; III, full necrosis at the site of inoculation with restricted growth of fungal hyphae IV, spreading necrosis and hyphal colonization beyond the sites of inoculation. Differences in class distribution between treatments were analysed for statistically significant differences by Fisher’s exact tests (α=0.05; SPSS, v19.0).
Induced resistance assays with tomato and Botrytis cinerea were performed largely as described50. Disease and colonisation by B. cinerea were examined microscopically at 3 dpi in trypan-blue stained leaves and assigned to different classes: I, contained lesion and no hyphal growth outside the necrotic lesion; II, contained lesion with minimal hyphal growth beyond the lesion; III, spreading lesion with extensive hyphal growth beyond the lesion. Differences in class distribution between treatments were analysed for statistically significant differences by χ2 or Fisher’s exact tests (α=0.05; SPSS, v19.0).
BABA-induced stress in young seedlings was recorded by digital photography at 10 days or 3 weeks after planting surface-sterilised seeds on control- and BABA-supplemented agar (500 μM BABA, ½-MS; 0.8% agar). BABA-induced growth inhibition was determined in 2- to 3-week-old plants over a 7-day time interval after soil-drench treatment with BABA, and expressed as percentage fresh weight reduction compared to control-treated plants. Shapiro-Wilk tests for normality and a Levene’s test for homogeneity of variances confirmed that these values meet the criteria for ANOVA. Post-hoc analysis was performed using Fisher’s least significant differences test (α=0.05; SPSS, v19.0).
Generation of transgenic plants
Gateway® cloning was used to produce an overexpression construct for a C-terminal IBI1:YFP fusion protein (Fig. 2b). The IBI1 gene (1681bp; without stop) was amplified by PCR from Col-0 cDNA (Fw: 5′-CACCATGTCGTCGGAATCTGAAAT Rv: 5′-GGGTGAAAGCCTTTGAGGGTCA), and cloned into pENTR (Invitrogen; 11791). A PCR-amplified fragment (m13/m13rev) from pENTR containing IBI1 cDNA with boarding attL1 and attL2 sequences was used for recombination into pEarleyGate-10151. The binary plasmid was extracted (QIAprep Spin Miniprep Kit Cat. 27104) and sequenced (FW: CAATATGTGGTGACTTGCGA; RV1 TCTCCATCTCCACATCAAGA; RV2: AAAGGATTCGCGACTCAGCT) to confirm the correct insert. Agrobacterium tumefaciens GV3101 and Arabidopsis ibi1-1 (Col-0) were transformed as described previously52,53. Selection Arabidopsis transformants was performed by multiple spray applications of 120 mg/L glufosinate-ammonium PESTANAL® (BASTA; Sigma; 45520). Selected T1 and T2 progenies were verified by PCR, using BAR-specific primers (Fw: 5′-GTCTGCACCATCGTCAAC; Rv: 5′-GAAGTCCAGCTGCCAGAA). Single insertion lines were selected on the basis of monogenic segregation of BASTA resistance in T2 progeny (3R:1S). Two independent homozygous over-expression lines were obtained and verified for enhanced IBI1 transcription (Supplementary fig. 4a), using RT-Q-PCR analysis with IBI1-specific primers (FW: 5′-GAGCGAGTGGTCATGCTTTTC; RV: 5′-CGAGGGAAGAGGGATGTTTTC).
Confocal laser scanning microscopy
YFP protein localisation was analysed in leaves of ibi1-1 35S:IBI1-1 plants, using a Zeiss 780 Laser Scanning Microscope and Zen 2010 software. Leaves were placed on a slide with the upper epidermis facing the cover-slip. YFP was excited with the 514 nm laser and emission was collected from 525-600 nm. Chloroplast autofluorescence was excited with the 633 nm laser and the emission was collected from 650-750 nm. Gain and offset were maintained at the same settings. Co-localization analysis of IBI1-YFP and ER marker protein was performed with an Inverted Zeiss LSM510-NLO microscope and a 40×/1.2 water immersion lens. Plants (ibi1-1 35S:IBI1:YFP-1) were crossed with ER-marker lines ER-ck or ER-gk29 to obtain double transgenic F1 progenies: ibi1-1 35:IBI1:YFP-1/ER-ck and ibi1-1 35S:IBI1:YFP-1/ER-gk. Fluorescence spectra of CFP, GFP and YFP were collected from cotyledons of ER-ck, ER-gk and ibi1-1 35S:IBI1:YFP single transgenic lines, respectively. Fluorescent proteins (FPs) were excited with a Ar 30 mW laser (458, 488 and 514 nm) or with a Coherent Chameleon multi-photon laser. Emission spectra were collected from 478-500 nm for CFP, 480-520 nm for GFP, and 525-600 nm for YFP. Epidermal peels from ibi1-1 35S:IBI1:YFP-1/ER-gk were obtained by removing the abaxial cell layer from fully expanded leafs. Fluorescence spectra of CFP, GFP and YFP in double transgenic F1 progenies were isolated, using previously recorded spectra from each FP-expressing single line with the “linear un-mixing” tool in LSM software (v4.2).
Protein binding assays, immunoprecipitation and immunoblotting
For in planta analysis of BABA-IBI1:YFP binding, proteins were extracted from 1 g pooled leaf material of water- and BABA-treated Col-0 and ibi1-1 35S:IBI1:YFP-1, using a standard protein extraction buffer (50 mM TRIS pH 7.5, 150 mM NaCl, 10% Glycerol, 10 mM MgCl2, mM EDTA, 5 mM DTT, 0.6 mM PMSF, protease inhibitor cocktail; Sigma P9599). Extracts were centrifuged twice for 15 min. at 4°C and 14,000g. Protein concentrations in the final supernatant (1 ml) were quantified by Bradford reagent (Sigma, B6916), and standardized to equal levels (2 mg/ml) before immunoprecipitation (IP). IP was performed using an anti-YFP antibody cross-linked to sepharose beads (Abcam ab69314), following manufacturer’s specifications, with slight modifications: prior to IP, sepharose beads were washed with 0.1% bovine serum albumine (BSA)-containing phosphate-buffered saline (PBS). After overnight IP on a rocking platform at 4°C, beads were pelleted by centrifugation (3000g, 1 min). Equal amounts of IBI1:YFP in IP pellets from ibi1-1 35S:IBI1:YFP-1 plants were verified by YFP fluorescence. Proteins in pelleted sepharose beads were denatured by incubation for 5 min. at 100°C in 50 μl elution buffer (8 mM Tris, 4% SDS and 6μl/ml of β-mercaptoethanol), after which the beads were pelleted by centrifugation (14000g, 1 min) and the eluted supernatant was analysed for BABA by mass spectromtry, as detailed below.
For in vitro analysis of BABA-IBI1:YFP binding, BABA enantiomers were applied at three different stages of IBI1:YFP protein purification from 35S:IBI1:YFP-1 plants: crude protein extracts, sephadex-purified protein extracts (PD-10 columns; Amersham Pharmacia Biotech, 281710), and immunoprecipitated IBI1:YFP protein. IBI1:YFP concentrations in all extracts were standardized on the basis of YFP fluorescence (Ex.: A-485, Em.: A-520; FLUOstar OPTIMA, BMG-ABTECH). Following application of 100 μM R-BABA or S-BABA, protein extracts were incubated at RT for 30 min. on a rocking platform. IBI1:YFP in crude and sephadex-purified extracts were immunoprecipitated, as described above. Before protein denaturation in elution buffer, pellets were washed twice in protein extraction buffer (see above for buffer details). Quantification of BABA in the eluted samples was performed by mass spectrometry, as detailed below.
Protein extraction and immunoblotting for phosphorylation analysis of eIf2α was performed as described54, using anti-Phospho-eIF2α (Ser51) antibody (Cell Signalling Technology, USA, 9721S), Anti-TCTP antibody54, and anti-rabbit IgG secondary antibody (Promega, S3731) in 1:1,000, 1:10,000 and 1:5,000 dilutions, respectively. Chemi-luminescent detection was performed using the Western-Star Immunodetection System (Applied Biosystems, Australia) and a VersaDoc MP 4000 CCD detector system with Quantity One software (Bio-Rad).
In vitro AspRS activity assays
Purified IBI1:YFP protein was immunoprecipitated from protein extracts of over-expression plants (ibi1-1 35S:IBI1:YFP-1), using YFP-specific antibody (Abcam ab69314). Activity assays were performed with immunoprecipitated IBI1:YFP in suspension with different reaction buffer components and conditions, as summarized in supplementary table 1. Asp (A9256), ATP (02055) and crude tRNA from wheat germ (R7876) were purchased from Sigma. tRNAAsp was synthesized through in vitro MEGAscript T7 transcription (Ambion, AM1333) of PCR products from Arabidopsis tRNAAsp genes (FW: TAATACGACTCACTATAGGGTCGTTGTAGTATAGTGGTAAG, RV: TGGCGCCGTTGCCGGGGATCG). tRNAAsp structure was given by incubating the tRNAAsp for 10 mins. at 37° C and subsequently maintained by the addition of MgCl2 to a final concentration of 2 mM, as previously described39. AspRS activity was analysed by a fluorometric pyrophosphate assay kit to detect the production of inorganic pyrophosphate, following manufacture’s specifications (Abcam, ab112155) and using a plate reader (FLUOstar OPTIMA, BMG-ABTECH; Ex.: A-340; Em.: A-460). Pyrophosphate fluorescence in IBI1:YFP samples was compared to background fluorescence of immunoprecipitated samples from un-transformed Col-0 lacking IBI1:YFP protein.
Mass spectrometry detection of Asp and BABA
Solvents used for mass spectrometry (MS) analyses were HPLC and/or MS grade: methanol (MeOH; Fluka; 34966), formic acid (HCOOH; Fluka; 06450), trifluoroacetic acid (TFA; Sigma-Aldrich; 302031), water (H2O; Waters; 232141B1). Amino acid extraction for Asp quantification was performed from biologically replicated plant material (n=3) as reported previously55, with slight modifications: the lyophilised residues were re-suspended into MeOH:H2O:HCOOH (50:49.9:0.1) and vortexed for 2 min. prior to mass spectrometry analysis. Mass spectra of Asp were obtained by Matrix-Assisted Laser Desorption/Ionization coupled to quadruple Time Of Flight Mass Spectrometry (MALDI-qTOF-MS) analysis, using a Synapt G2 mass spectrometer (Waters; Manchester; UK) in positive ionization mode ([M+H]; m/z = 133.03) and in ‘target enhancement mass’ function. MALDI was powered by a solid state laser emitting at 355 nm with a repetition rate of 2.5 kHz. The lock mass for MALDI-qTOF-MS was sulfadoxine methoxide (Fluka; S1950000) and was acquired for 120s, while samples were acquired for 180s, using a spiral pattern with the laser firing conditions optimised at a plate speed of 50Hz and attenuation of 140. Ion acquisition was performed at 10kV with 11kV hexapole bias and 7.0 mm. aperture. Mass ranges of scans (1s) were defined from 50 to 1200 Da. Matrix solution (α-Cyano-4-hydroxycinnamic acid; CHCA; Sigma-Aldrich; C2020) was prepared at 5mg.mL−1 in MeOH:TFA (99.9:0.1). Each standard/sample was mixed with an equal volume of matrix solution (1:1) and spotted (0.2μL) in triplicate onto 96-well stainless steel MALDI target plates (Waters). In parallel, sulfadoxine methoxide (Fluka; S1950000) was prepared at 10μg.mL−1 in MeOH:H2O (50:50; v/v) and spotted as lock mass for MALDI/TOF-MS analyses ([M+H]; m/z=311.08). Droplets were allowed to dry, using a dry block-heater (Dri-block DB3; Tecam; Italy) at 50°C. Raw data were processed by Masslynx software (v4.1; Waters) and Asp levels were quantified by extrapolating peak areas from a calibration curve, and corrected for fresh weight. Stock solutions of Asp (Sigma-Aldrich; A93100) were prepared in MeOH:H2O:HCOOH (50:49.9:0.1; v/v) and used for calibration curves.
BABA was detected by both MALDI-qTOF-MS and ESI-qTOF-MS (Zspray™ with NanoLockSpray™; Waters), using a Synapt G2 mass spectrometer in positive ionization mode ([M+H] = 104.025) and in ‘target enhancement mass’ function. Samples from the IBI1:YFP pull-down assay (10μL) were mixed with 10μL of MeOH:H2O:HCOOH (50:49.9:0.1; v/v). For MALDI-qTOF-MS, the diluted extract was mixed with 20μL CHCA matrix (5mg.mL−1) and analyses were carried out at described above for Asp. For ESI-qTOF-MS, source voltage at the injection capillary was set at 4kV, the sampling cone was set at 20kV and the extraction cone was set at 4kV. Source and desolvation temperatures were set at 120°C and 150°C, respectively. Gas flows were adjusted at 20L/h (cone gas), 0.5Bar (nano-flow gas) and 600L/h (purge and desolvation gas). Each sample was run out at 100nL/min using a syringe pump (SP100iZ; WPI; UK) and data were acquired for 30 min. In parallel, leucine enkephalin ([M+H]; m/z=556.08; Sigma-Aldrich; L9133) at 2μg/mL in MeOH:H2O (50:50; v/v;) was infused as lock mass. Standards of R- and S-BABA (Astatech) were prepared in protein extraction buffer (see above) and diluted to appropriate concentrations with MeOH:H2O:HCOOH (50:49.9:0.1; v/v) before analysis.
RNA extraction, reverse transcription and quantitative real-time PCR analysis
RNA extraction, cDNA synthesis and quantitative PCR reactions and data analysis was performed as described before10,21. After PCR amplification, melting curve analysis was performed to verify amplification of single PCR products. Relative transcript quantities were calculated according to (1+E)ΔCt, where ΔCt = Ct(sample) − Ct(calibrator sample), and normalised to (1+E)ΔCt values of two reference genes, At1g13440 and At5g2576056.
Modelling of interactions between BABA enantiomers and Asp-RS proteins
BLAST of Arabidopsis IBI1 (UniProt: Q9M084) against the protein databank (PDB) identified protein crystal structures with a high sequence similarity. Two proteins from P. pastoris and P. kodakaraensis (PDB codes: 1EOV and 3NEL, respectively) were selected on the basis of sequence similarity and availability of crystal structures. Sequence alignment was performed using the Protein Modelling protocol in Discovery Studio 3.5 (DS)57. Crystal structures were prepared for docking studies using the ‘Prepare_Protein’ tools, and binding sites were defined using the ‘Define_and_Edit_Binding_Site’ tool in DS. A 15 angstrom binding sphere was creased around previously identified binding residues of 1EOV40, 41, and the co-crystallised asp coordinates in 3NEL. Ligands (L-Asp, R-BABA, and S-BABA) were prepared using the ‘Prepare_Ligands’ tool in DS for generating 3D coordinates and pH-based ionisation forms. Docking was performed using DS CDOCKER, which is a grid-based molecular docking method employing CHARMm58. Random ligand conformations were generated from the initial ligand structure, using high temperature molecular dynamics at 1000 K, followed by random rotations. Ligand confirmations were optimised using grid-based simulated annealing, followed by a final full force field minimization. Each docked ligand pose was assigned a score, which includes the internal force-field energy of the ligand, conformational entropy loss of the ligand, receptor-ligand hydrogen-bond interaction, solvation electrostatic energy change, hydrogen-bond donor/acceptor desolvation, and hydrophobic energy. Comparisons were made between top-ranked ligand scores, on the basis of generated 3D interaction maps.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Saskia van Wees, Nigel Halford, John Lucas, John Pickett, Ivo Feussner, Xuebin Zhang, Wouter Kegge, Dusica Acoska, Mike Roberts, Victoria Pastor, Jordi Gamir, Brigitte Mauch-Mani for fruitful discussions, practical assistance, and/or helpful feedback. Arabidopsis NahG B15 and Ler rpp5 seeds were kindly provided by John Ryals (Research Triangle Park, USA) and Jane Parker (MPI, Cologne), respectively. The research was supported by a VENI grant to J.T. (no. 863.04.019) from the Netherlands Organisation of Scientific Research (NWO), a BBSRC Institute Career Path Fellowship (no. BB/E023959/1) to J.T., a consolidator grant from the European Research Council (ERC; no. 309944-Prime-A-Plant) to J.T., a Research Leadership Award from the Leverhulme Trust (no. RL-2012-042) to J.T., European Union 7th Framework Programme (FP7/2007-2013; n°265865-PURE) to J.T., a grant from the Felix Thornley Cobbold Agricultural Trust to J.T. and E.L. and a VICI grant (no. 865.04.002) to C.M.J.P. from NWO.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal Modulation of Plant Immunity. Annual Review of Cell and Developmental Biology. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 2.Conrath U, et al. Priming: getting ready for battle. Molecular Plant Microbe Interact. 2006;19:1062–71. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- 3.Pastor V, Luna E, Mauch-Mani B, Ton J, Flors V. Primed plants do not forget. Environmental and Experimental Botany. 2013;94:46–56. [Google Scholar]
- 4.Kohler A, Schwindling S, Conrath U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiology. 2002;128:1046–1056. doi: 10.1104/pp.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- 6.Van Hulten M, Pelser M, van Loon L, Pieterse CMJ, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proceedings of the National Academy of Sciences. 2006;103:5602–7. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters D, Heil M. Costs and trade-offs associated with induced resistance. Physiological and Molecular Plant Pathology. 2007;71:3–17. [Google Scholar]
- 8.Ahmad S, Gordon-Weeks R, Pickett J, Ton J. Natural variation in priming of basal resistance: from evolutionary origin to agricultural exploitation. Molecular Plant Pathology. 2010;11:817–827. doi: 10.1111/j.1364-3703.2010.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrath U. Molecular aspects of defence priming. Trends In Plant Science. 2011;16:524–531. doi: 10.1016/j.tplants.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. Next-Generation Systemic Acquired Resistance. Plant Physiology. 2012;158:844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slaughter A, et al. Descendants of Primed Arabidopsis Plants Exhibit Resistance to Biotic Stress. Plant Physiology. 2012;158:835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckers GJM, Conrath U. Priming for stress resistance: from the lab to the field. Current Opinion in Plant Biology. 2007;10:425–431. doi: 10.1016/j.pbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Gao QM, Kachroo A, Kachroo P. Chemical inducers of systemic immunity in plants. J Exp Bot. 2014 doi: 10.1093/jxb/eru010. [DOI] [PubMed] [Google Scholar]
- 14.Walters DR, Ratsep J, Havis ND. Controlling crop diseases using induced resistance: challenges for the future. Journal of Experimental Botany. 2013 doi: 10.1093/jxb/ert026. [DOI] [PubMed] [Google Scholar]
- 15.Cohen YR. β-Aminobutyric Acid-Induced Resistance Against Plant Pathogens. Plant Disease. 2002;86:448–457. doi: 10.1094/PDIS.2002.86.5.448. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerli L, Jakab G, Métraux J-P, Mauch-Mani B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proceedings of the National Academy of Sciences. 2000;97:12920–12925. doi: 10.1073/pnas.230416897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ton J, et al. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. The Plant Cell. 2005;17:987–99. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh P, et al. The Lectin Receptor Kinase-VI.2 Is Required for Priming and Positively Regulates Arabidopsis Pattern-Triggered Immunity. The Plant Cell Online. 2012;24:1256–1270. doi: 10.1105/tpc.112.095778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Návarová H, Bernsdorff F, Döring A-C, Zeier J. Pipecolic Acid, an Endogenous Mediator of Defense Amplification and Priming, Is a Critical Regulator of Inducible Plant Immunity. The Plant Cell Online. 2012 doi: 10.1105/tpc.112.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ton J, Mauch-Mani B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. The Plant Journal. 2004;38:119–30. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- 21.Van der Ent S, et al. Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytologist. 2009;183:419–431. doi: 10.1111/j.1469-8137.2009.02851.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu C-C, Singh P, Chen M-C, Zimmerli L. L-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. Journal of Experimental Botany. 2010;61:995–1002. doi: 10.1093/jxb/erp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaney TP, et al. A Central Role of Salicylic Acid in Plant Disease Resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 24.Flors V, et al. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. The Plant Journal. 2008;54:81–92. doi: 10.1111/j.1365-313X.2007.03397.x. [DOI] [PubMed] [Google Scholar]
- 25.Parker JE, et al. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. The Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell CJ, Ecker JR. Assignment of 30 Microsatellite Loci to the Linkage Map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 27.Mauch-Mani B, Slusarenko AJ. Production of Salicylic Acid Precursors Is a Major Function of Phenylalanine Ammonia-Lyase in the Resistance of Arabidopsis to Peronospora parasitica. The Plant Cell Online. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nature Reviews Molecular Cell Biology. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–36. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 30.Duchene AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen Y. 3-Aminobutyric acid induces systemic resistance against peronospore tabacina. Physiological and Molecular Plant Pathology. 1994;44:273–288. [Google Scholar]
- 32.Silue D, Pajot E, Cohen Y. Induction of resistance to downy mildew (Peronospora parasitica) in cauliflower by DL-beta-amino-n-butanoic acid (BABA) Plant Pathology. 2002;51:97–102. [Google Scholar]
- 33.Cohen Y, Rubin AE, Kilfin G. Mechanisms of induced resistance in lettuce against Bremia lactucae by DL-beta-amino-butyric acid (BABA) European Journal of Plant Pathology. 2010;126:553–573. [Google Scholar]
- 34.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13:327–37. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt E, et al. Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. Embo Journal. 1998;17:5227–5237. doi: 10.1093/emboj/17.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrift A, Bechard D, Harcup C. Utilization of Selenocysteine by a Cysteinyl-tRNA Synthetase from Phaseolus aureus. Plant Physiol. 1976;58:248–52. doi: 10.1104/pp.58.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulmasov B, Topin A, Chen Z, He SH, Folk WR. Identity elements and aminoacylation of plant tRNATrp. Nucleic Acids Research. 1998;26:5139–5141. doi: 10.1093/nar/26.22.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd AJ, Thomann HU, Ibba M, Soll D. A broadly applicable continuous spectrophotometric assay for measuring aminoacyl-tRNA synthetase activity. Nucleic Acids Res. 1995;23:2886–92. doi: 10.1093/nar/23.15.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cestari I, Stuart K. A Spectrophotometric Assay for Quantitative Measurement of Aminoacyl-tRNA Synthetase Activity. Journal of Biomolecular Screening. 2013;18:490–497. doi: 10.1177/1087057112465980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dever TE, Hinnebusch AG. GCN2 whets the appetite for amino acids. Molecular Cell. 2005;18:141–142. doi: 10.1016/j.molcel.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Li MW, AuYeung WK, Lam HM. The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biology. 2013;15:13–18. doi: 10.1111/j.1438-8677.2012.00606.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J Exp Bot. 2008;59:3131–41. doi: 10.1093/jxb/ern169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holub EB, Beynon LJ, Crute IR. Phenotypic and Genotypic Characterization of Interactions between Isolates of Peronospora-Parasitica and Accessions of Arabidopsis-Thaliana. Molecular Plant-Microbe Interactions. 1994;7:223–239. [Google Scholar]
- 44.Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol. 2013;9:145–53. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko Y-G, et al. Glutamine-dependent Antiapoptotic Interaction of Human Glutaminyl-tRNA Synthetase with Apoptosis Signal-regulating Kinase 1. Journal of Biological Chemistry. 2001;276:6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 46.Frye CA, Tang D, Innes R.r.W. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proceedings of the National Academy of Sciences. 2001;98:373–378. doi: 10.1073/pnas.98.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong Q, et al. The MEKK1-MKK1/MKK2-MPK4 Kinase Cascade Negatively Regulates Immunity Mediated by a Mitogen-Activated Protein Kinase Kinase Kinase in Arabidopsis. The Plant Cell Online. 2012;24:2225–2236. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–74. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y, Schumaker K, Zhu J-K. EMS Mutagenesis of Arabidopsis. In: Salinas J, Sanchez-Serrano J, editors. Arabidopsis Protocols. Vol. 323. Humana Press; 2006. pp. 101–103. [DOI] [PubMed] [Google Scholar]
- 50.Audenaert K, De Meyer GB, Hofte MM. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiology. 2002;128:491–501. doi: 10.1104/pp.010605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 52.Lin JJ. Optimization of the Transformation Efficiency of Agrobacterium-Tumefaciens Cells Using Electroporation. Plant Science. 1994;101:11–15. [Google Scholar]
- 53.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 54.Berkowitz O, Jost R, Pollmann S, Masle J. Characterization of TCTP, the Translationally Controlled Tumor Protein, from Arabidopsis thaliana. Plant Cell. 2008;20:3430–3447. doi: 10.1105/tpc.108.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pétriacq P, et al. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. The Plant Journal. 2012;70:650–665. doi: 10.1111/j.1365-313X.2012.04920.x. [DOI] [PubMed] [Google Scholar]
- 56.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Accelrys SI. Discovery Studio Modeling Environment. Release 3.5. San Diego: 2012. [Google Scholar]
- 58.Wu GS, Robertson DH, Brooks CL, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER - A CHARMm-based MD docking algorithm. Journal of Computational Chemistry. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.