Abstract
Membrane binding of proteins such as short chain dehydrogenases reductases or tail-anchored proteins relies on their N- and/or C-terminal hydrophobic transmembrane segment. In this review, we propose guidelines to characterize such hydrophobic peptide segments using spectroscopic and biophysical measurements. The secondary structure content of the C-terminal peptides of retinol dehydrogenase 8, RGS9-1 anchor protein, lecithin retinol acyl transferase, and of the N-terminal peptide of retinol dehydrogenase 11 has been deduced by prediction tools from their primary sequence as well as by using infrared or circular dichroism analyses. Depending on the solvent and the solubilization method, significant structural differences were observed, often involving α-helices. The helical structure of these peptides was found to be consistent with their presumed membrane binding. Langmuir monolayers have been used as membrane models to study lipid-peptide interactions. The values of maximum insertion pressure obtained for all peptides using a monolayer of 1,2-dioleoyl-sn-glycero-3-phospho-ethanolamine (DOPE) are larger than the estimated lateral pressure of membranes, thus suggesting that they bind membranes. Polarization modulation infrared reflection absorption spectroscopy has been used to determine the structure and orientation of these peptides in the absence and in the presence of a DOPE monolayer. This lipid induced an increase or a decrease in the organization of the peptide secondary structure. Further measurements are necessary using other lipids to better understand the membrane interactions of these peptides.
Keywords: Transmembrane hydrophobic peptide, lecithin retinol acyltransferase, retinol dehydrogenase, R9AP, monolayer, circular dichroism, infrared spectroscopy
1. Introduction
Biological processes involving different types of proteins take place at the cell membrane surface, such as signal transduction cascades. For example, the membrane-embedded G-protein coupled receptors have been shown to activate peripheral membrane G-proteins which, in turn, stimulate several additional membrane-associated proteins (for reviews, see [1–6]). In fact, biological membranes contain a large number of different integral membrane proteins, which collectively constitute ~30% of the cellular proteome [7, 8]. Proteins can bind membranes by use of one or several hydrophobic transmembrane segments [9–11], fatty acylation [12–14], or a small hydrophobic stretch located at the protein surface [15]. In particular, N- and/or C-terminal membrane-anchoring segments allow membrane binding of proteins of the short chain dehydrogenase/reductase (SDR) family [9, 10, 16, 17]. Moreover, tail-anchored (TA) proteins constitute an additional class of integral membrane proteins that are held in the phospholipid bilayer by a single C-terminal transmembrane stretch of hydrophobic amino acids (for reviews, see [18–27]). They constitute ~5% of eukaryotic membrane proteins and they are found in all cellular membranes [28]. In addition, they carry out fundamental cell biological phenomena such as membrane biogenesis, apoptosis, vesicular trafficking, protein degradation and many others [24–26]. The N- and/or C-terminal transmembrane segments, presumably helical, of SDR and TA proteins allow their localization at the membrane surface and dictate their membrane topology [27]. The transmembrane segments may vary considerably in length, from less than 15 to nearly 40 amino acid residues [29–32]. In the present review, the properties of the peptides corresponding to the transmembrane segment of SDR and TA proteins are compared to improve our understanding of their intricate interactions with membranes.
Retinol dehydrogenases 8 (RDH8) and 11 (RDH11) are members of the SDR family of proteins. Although some SDR enzymes share common substrates, they differ in regard to their subcellular localization, cofactor specificity, substrate affinity, and tissue distribution. It is commonly agreed that SDR enzymes preferring the NAD+ cofactor act as dehydrogenases, whereas those preferring NADPH function as reductases [33]. RDH8 and RDH11 are involved in the visual cycle of retinoids (for reviews, see [34–40]). During the visual cycle, the chromophore of rhodopsin, 11-cis retinal, is recycled from all-trans retinal by a series of enzymatic reactions taking place at the membrane of the photoreceptor outer segments as well as of the microsomes in the retinal pigment epithelium (RPE). RDH8 localizes exclusively to photoreceptor outer segments [41]. It has been shown to perform the reduction of all-trans retinal to all-trans retinol [42, 43]. The RDH8 gene encodes a protein with a molecular mass of ~34 kDa [41]. Moreover, Luo et al. [44] have demonstrated that membrane association of RDH8 was confered by its C-terminal segment. Their data also suggested that one or more of the three C-terminal cysteines is fatty acylated which could mediate membrane association of RDH8. RDH11 is a ~35 kDa enzyme that uses NADPH as a cofactor [45]. It is expressed in various tissues such as the prostate, brain, heart, liver and testis, but also in the RPE [46]. RDH11 is partly responsible for the production of 11-cis-retinal from 11-cis-retinol in vivo [47, 48]. However, it catalyzes the reduction of retinal ~50-fold more efficiently than it does the oxidation of retinol in vitro [49]. Protease protection assays allowed to suggest that RDH11 is anchored to the membrane by use of its N-terminal signal-anchor domain [50].
Lecithin retinol acyltransferase (LRAT) catalyzes the esterification of all-trans retinol to retinyl esters in the RPE as well as in other tissues including testis, liver, and intestine [51–54]. It has a molecular mass of ~25 kDa [55]. Its primary sequence is novel and it does not show any homology to enzymes that catalyze similar reactions nor to any protein of known function [55]. LRAT is thus the founder member of a new family of proteins [56]. The analysis of its amino acid sequence allowed to postulate that its N- and C-terminal segments could include hydrophobic transmembrane domains [55]. Only the C-terminal transmembrane domain has been recently proposed to be essential for membrane targeting [57]. However, both N- and C-terminal segments have been shown to behave similarly; indeed their binding, secondary structure content and orientation in lipid monolayers is almost identical [58]. It has thus been postulated that both N- and C-terminal hydrophobic α-helical peptides could serve to anchor LRAT to the membrane [58].
The regulator of G-protein signaling (RGS)-9-1 is a GTPase accelerating protein. It allows to accelerate the GTPase activity of the α subunit (Gα) of the visual G-protein called transducin when bound to phosphodiesterase [59]. RGS9-1 plays an essential role in the light response of vertebrate photoreceptors [60]. RGS9-1-anchor protein (R9AP) is a photoreceptor-specific phosphoprotein of ~25 kDa [61]. It was shown to interact with the N-terminal domain of RGS9-1. R9AP has been proposed to contain a transmembrane helix at its C-terminus on the basis of its hydropathy index, which would allow anchoring of RGS9-1 to photoreceptor membranes [61]. R9AP is also a member of the tail-anchored membrane proteins [28]. Membrane attachment of a protein complex (RGS9-1-Gα) by R9AP is also very important because it has been shown to allow protection of this complex from intracellular proteolysis in photoreceptors [62].
The retinoids are very hydrophobic molecules. Therefore, RDH8, RDH11 and LRAT must be closely associated to membranes to perform their enzymatic activities. In this regard, retinol dehydrogenases, devoided of their C-terminal tails, display greatly reduced enzymatic activity [63]. Moreover, R9AP stimulates the activity of RGS9-1-Gαβ complex ~4–30-fold by membrane targeting [64, 65]. Therefore, the postulated membrane anchoring of RDH8, RDH11, LRAT and R9AP by their N- and/or C-terminal transmembrane segments is very critical for their function. The present review will thus focus on recent results gathered by our group on the spectroscopic and monolayer-binding properties of the C-terminal peptide of bovine RDH8 (RDH8-Cter), the N-terminal peptide of human RDH11 (RDH11-Nter), the C-terminal peptide of human LRAT (LRAT-Cter) and the C-terminal peptide of human R9AP (R9AP-Cter) to improve our understanding of the properties of these peptides.
2. Primary structure of the hydrophobic membrane binding peptides and prediction of their secondary structure
The hydrophobic peptides postulated to anchor the proteins RDH8, RDH11, LRAT and R9AP to membranes are shown in Table 1. This table is showing the amino acid sequence of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides, which respectively include 32, 35, 30 and 29 amino acids. The analysis of the properties of the amino acids of these peptides allowed to determine that the R9AP-Cter peptide is the most hydrophobic one (76% hydrophobic residues) whereas the RDH8-Cter peptide is the least hydrophobic one (50% hydrophobic residues) (Table 1). The RDH11-Nter and LRAT-Cter peptides contain ~70% hydrophobic amino acids. The RDH8-Cter and RDH11-Nter peptides respectively contain in total 35 and 26% of both polar and charged amino acids. In contrast, the LRAT-Cter and R9AP-Cter peptides both include only 17% polar and charged amino acids. In addition, it should be stressed that there are only 3% polar amino acids in the R9AP-Cter peptide. Moreover, these hydrophobic peptides include a rather small amount of charged amino acids (between 9 and 16%) except for the LRAT-Cter peptide which entails no charged amino acids (Table 1). It is particularly interesting to analyze the tendency of the amino acids of these hydrophobic peptides to be found in transmembrane helices. It can thus be seen that 69, 80, 93 and 83% of the amino acids of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides, respectively, are typically found in transmembrane helices (TM helix, Table 1).
Table 1.
Analysis of the amino acid content of the selected peptides
| Peptide | Protein amino acids | Amino acid sequence | Proportion of amino acids† | |||
|---|---|---|---|---|---|---|
| Hydrophobic | Polar | Charged | TM helix* | |||
| RDH8-Cter | 281–312 |
|
50% 16/32 |
19% 6/32 |
16% 5/32 |
69% 22/32 |
| RDH11-Nter | 2–36 |
|
69% 24/35 |
17% 6/35 |
9% 3/35 |
80% 28/35 |
| LRAT-Cter | 201–230 |
|
70% 21/30 |
17% 5/30 |
0% 0/30 |
93% 28/30 |
| R9AP-Cter | 207–235 |
|
76% 22/29 |
3% 1/29 |
14% 4/29 |
83% 24/29 |
Hydrophobic amino acids are presented in red, polar amino acids in blue and charged amino acids in green. The amino acid tyrosine (Y) has been considered as a hydrophobic residue even though it presents a dual character (hydrophobic and hydroxylated). Glycine (G) and cysteine (C) are presented in black as “non classified amino acids” because their properties are changing depending of their environment (v.g. disulfide bond for cysteine).
These properties have been determined using textbooks of biochemistry (v.g. [15, 352]) as well as the paper of Dufourc et al. [353].
Amino acid residues that have tendency to be found in a transmembrane (TM) helix (underlined amino acids): I-L-F-V-C-M-A-T-Y-W-G-S (values ≤1 in [354] and reviewed in [355]). These data are in good agreement with those generated when a similar analysis is performed using the “Fraction of time amino acids are found buried in protein structure” [356] as well as using the “Amino acid frequency in transmembrane segments of tail-anchored proteins” [28].
The online tool « Heliquest » [66] is very often used to predict the secondary structure of amphiphilic peptides [67]. This algorithm allows to detect the existence of an uninterrupted hydrophobic face of at least five adjacent residues on a helical wheel. If such a face exists, it illustrates whether the residues in the opposite face are polar. The projection of the amino acid sequence of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter hydrophobic peptides in a helical wheel is presented in Fig. S1 together with peptides known as amphipathic helices. It can be seen that the amphiphilic helices Endophilin1 and GMAP-210 have a well defined hydrophobic face, which includes all hydrophobic amino acids, as well as a large hydrophilic face where charged and polar residues can be found. This is however not the case for the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides. Indeed, as can be seen in Fig. S1, hydrophobic amino acids are evenly distributed in the helical wheel. Therefore, because these transmembrane hydrophobic peptides are not amphipathic, this well known tool can not be used to appropriately predict their structure.
The Heliquest lipid-binding feature can however be combined with the Eisenberg plot approach [68–70] to generate a useful graphical representation of the mean hydrophobic moment as a function of the mean hydrophobicity [71]. It uses the Fauchere and Pliska scale [72] instead of the normalized scale of Eisenberg [69], thereby providing a user-friendly approach [71]. The mean hydrophobicity corresponds to a measure of the overall hydrophobicity of the amino acid sequence whereas the mean hydrophobic moment is a measure of the way polar and non-polar amino acids are distributed in the same amino acid sequence [71]. This plot provides interesting information on whether the peptide sequences correspond to globular, surface seeking or transmembrane segments [68–71]. The plot has been divided in three different regions (globular, surface seeking and transmembrane) on the basis of protein databases. As shown in Fig. S2, the RDH11-Nter, LRAT-Cter and R9AP-Cter peptides are located in the transmembrane region (TM) whereas the RDH8-Cter is a « surface seeking » peptide. This analysis is consistent with the content in hydrophobic amino acids of these peptides (Table 1).
The primary structure of the hydrophobic peptides has been analyzed using different online tools [73–78] to predict their secondary structure, which yielded very similar data (Fig. 1). The amino acid sequence of the R9AP-Cter peptide was thus predicted to form an α-helix (Fig. 1), which is consistent with its large content in hydrophobic residues (Table 1). Moreover, the RDH11-Nter peptide was predicted to include two α-helices separated by 2 amino acids (Fig. 1); the largest one is made of a large stretch of hydrophobic amino acids (LMFPLLLLLLPFLLYM, Fig. 1). In contrast, the RDH8-Cter and LRAT-Cter peptides are predicted to include an α-helix as well as one or two β-strands (Fig. 1); this is however inconsistent with the tendency of the amino acids of these peptides to be found in transmembrane α-helices (Table 1). Therefore, spectroscopic methods must be used to find out whether these analyses and predictions make sense.
Fig 1. Predicted secondary structure of the peptides obtained using the I-TASSER server.
Each sequence has been analyzed using two online tools: I-TASSER [73, 75] and Proteinprediction [76]. The two predictions resulted in similar data except for the LRAT-Cter peptide. In the case of this peptide, three additional online tools were used: Sspro [77], SSpro8 [77] and PSIPRED [78], resulting in consistent data with the I-TASSER prediction. The secondary structure presented is that obtained with the highest confidence score (Conf.Score). The confidence scores range from 0 to 9 which correspond respectively to the lowest and the highest confidence. H, helix; S, strand or β-sheet; C, coil.
3. Secondary structure of the selected hydrophobic peptides
Several methods can be used to estimate the secondary structure content of peptides. Infrared spectroscopy and circular dichroism (reviewed in [79–86]) are well established methods to determine the secondary structure content of proteins and peptides. Circular dichroism is very accurate for α-helix predictions whereas infrared spectroscopy is better for β-sheet estimation [87].
3.1 The challenge of solubilizing hydrophobic peptides
The first step before characterizing peptides consists in finding an appropriate solvent allowing their solubilization. This task is very challenging for hydrophobic peptides such as those reviewed in the present paper. However, solvents such as trifluoroethanol (TFE), hexafluoroisopropanol (HFIP), dimethyl sulfoxide (DMSO) or dimethyl formamide (DMF) very often allow to solubilize such hydrophobic peptides. Nevertheless, care must be taken when using TFE and HFIP because it is generally believed that they enhance the population of helical conformations of peptides and proteins. In particular, since the pioneering work of Tamburro et al. [88], fluorinated alcohols, such as TFE and HFIP, have been shown to promote the formation of α-helices for a large number of proteins and peptides (see, for example, references [89–103] and the references cited therein). It has been proposed that fluoroalcohol complexes would displace water molecules from the surface of the peptide; the acidity or hydrogen-bonding ability and hydrophobicity of the fluoroalcohols appear to play an important role [104]. It is thus very important to compare the spectroscopic properties of peptides in different solvents as this allows to assess the relationship between their α-helical content and the solvent used to solubilize the peptides. Solubilization assays of the peptides were thus performed in different solvents. DMF and DMSO have not been used because the former prevents a proper observation of the amide I band of the peptide in infrared spectroscopy whereas the latter is too little volatile. Because the peptides are injected into an aqueous subphase in the lipid monolayer binding experiments (sections 4 and 5), solvents miscible with water, preferably methanol, were first assayed. The RDH8-Cter peptide was soluble in methanol but not in HFIP, which is consistent with its lower content in hydrophobic residues (50%) than the other peptides (Table 1). When the peptide was not soluble in methanol, a procedure can be used where HFIP serves first to promote H bonding with the peptide. HFIP is then evaporated and the peptide is further solubilized in methanol (labeled H-MeOH; see Fig. 2 and section 3.2). This procedure has been particularly useful for the LRAT-Cter and R9AP-Cter peptides because they were not readily soluble in methanol. A similar procedure has previously been used to prepare a monomeric solution of amyloid beta peptide. After the evaporation of HFIP, this amyloid peptide was dissolved in water, which resulted in a random coil secondary structure [105].
Fig 2. Circular dichroism and infrared spectra of the RDH8-Cter (A-B), RDH11-Nter (C–D), LRAT-Cter (E–F) and R9AP-Cter (G–H) peptides in different solvents.
The RDH8-Cter, RDH11-Nter and R9AP-Cter peptides were purchased from Peptide 2.0 (Chantilly, VA) whereas the LRAT-Cter peptide was from Anaspec (Fremont, CA). The purity of the peptides was tested by mass spectroscopy and HPLC: 97% purity for RDH8-Cter, 93% for RDH11-Nter, >70% for LRAT-Cter and 92% for R9AP-Cter. Circular dichroic spectra were collected on a Jasco spectropolarimeter (Model J-815, Jasco, Easton, MD) at a peptide concentration of 150 μM. The spectra have been normalized to take into account the number of amino acids of each individual peptide. Indeed, the molar ellipticity is expressed in degree cm2 dmol−1 × 104. The concentration in dmol−1 is obtained as follows: protein concentration (g/mL) × number of amino acids/molar mass of each individual peptide. The spectra have been measured in different solvents depending on their solubility: Methanol (MeOH, black curves), HFIP (blue curves) and H-MeOH* (solubilized first in HFIP and evaporation of HFIP and then solubilized in MeOH; pink curves). The buffer contribution was subtracted and the corrected spectra were analyzed in the 190–260 nm range. Infrared spectra were recorded using a Nicolet Magna 850 Fourier transform infrared spectrometer from Thermo Scientific (Madison, WI) equipped with a liquid nitrogen cooled narrow-band mercury cadmium telluride detector and a Golden Gate ATR accessory. Infrared spectra were recorded using a peptide concentration of 150 μM for RDH8-Cter (B), RDH11-Nter (D) and LRAT-Cter (F) and R9AP-Cter (H) in the same solvents as those used in circular dichroism. Infrared spectra are the result of a subtraction of the spectrum of the appropriate solvent from that of the peptide in solution. The infrared spectrum of each peptide powder (orange curves) is also presented in this figure. The maximum of the amide I bands has been determined by performing a second derivative (or Fourier deconvolution) of the spectra with the software Omnic which is available with the spectrometer. A 3D structural model of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides obtained using the I-TASSER server is shown in inset of figures B, D, F and H, respectively. These models are built based on multiple-threading alignments by LOMETS and interactive TASSER simulations; function insights are then derived by matching the predicted models with protein function databases [73]. Several models were uploaded from the I-TASSER online server for the peptides, but a single 3D model has been selected for each peptide because of its consistency with the secondary structure prediction and the analysis of the infrared spectra.
3.2 Circular dichroism and infrared spectroscopy of the hydrophobic peptides
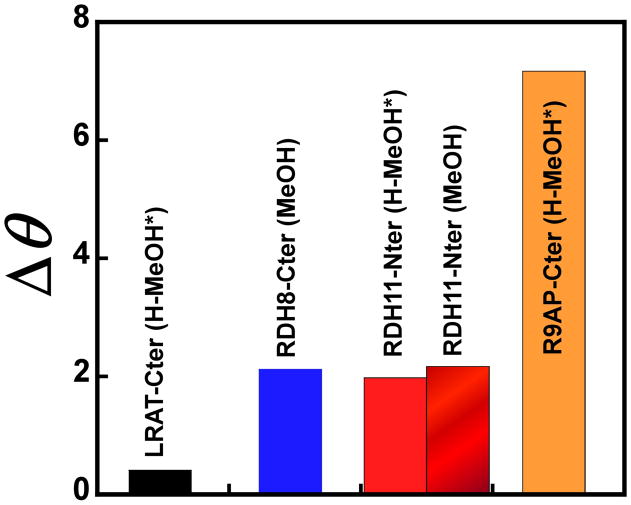
Fig. 2 is showing circular dichroism and infrared spectra of the hydrophobic peptides in different solvents. The shape of the circular dichroism spectra allows to qualitatively determine the secondary structure content of peptides; for example, minima at ~206 and ~220 nm are typical for α-helices [79]. The shape of the spectra shown in Figs. 2A, C, E and G suggests that all peptides contain an α-helical secondary structure component except for the LRAT-Cter peptide in H-MeOH. The quantitative analysis of the secondary structure content of these peptides in MeOH or H-MeOH has been performed with the CDSSTR online tool [106]. Table S1 is showing interesting features of the secondary structure of the peptides in MeOH or H-MeOH : 1) the RDH8-Cter peptide contains almost as much α-helical (40%) and random (32%) structural elements, 2) the RDH11-Nter peptide mainly comprehends α-helices (52–56%), 3) the LRAT-Cter peptide contains very little α-helices (4%) in this polar solvent, and 4) the R9AP-Cter peptide includes a very large amount of α-helices (63%).The α-helical secondary structure content of the peptides can be qualitatively analyzed using the ratio [94]) or the difference in molar ellipticity (Δθ) between the band at ~190–199 nm and that at ~204–208 nm (Δθ = [θ]max − [θ]min, (Fig. 3). It can be seen that the Δθ of the peptides in MeOH or H-MeOH increases as follows : LRAT-Cter < RDH8-Cter ≈ RDH11-Nter ⋘ R9AP-Cter (Fig. 3). These data are quite consistent with those reported in Table S1 except for the secondary structure analysis of the RDH8-Cter peptide. Indeed, the values of Δθ of the RDH8-Cter and RDH11-Nter peptides are very similar (Fig. 3) whereas a significantly larger content in α-helices is obtained for the RDH11-Nter peptide (52–56%) than for the RDH8-Cter peptide (40%) (Table S1). The data of Table S1 and Fig. 3 are consistent with regards to the large α-helical secondary structure content predicted in Fig. 1 for the R9AP-Cter peptide.
Fig. 3. Comparison between the difference in molar ellipticity of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides calculated from the circular dichroism spectra shown in Fig. 2.
The difference in molar ellipticity (Δθ) between the maximum at ~190–199 nm and the minima at ~204–208 nm (Δθ = [θ]max(190-199) − [θ]min(204-208)) is shown for each peptide. The peptides were solubilized as described in the legend of Fig. 2. The comparison is made with MeOH because the RDH8-Cter peptide is only soluble in this solvent.
The propensity of HFIP to favor the α-helical secondary structure is only clearly demonstrated with the LRAT-Cter peptide where α-helices can be obviously seen when solubilized in this solvent (Fig. 2E). However, the molar ellipticity of the RDH11-Nter peptide remained almost unchanged when solubilized in MeOH or HFIP (Fig. 2C). In contrast, the molar ellipticity of the R9AP-Cter peptide is much smaller in HFIP than in H-MeOH (Fig. 2G), which is not consistent with previous observations that this solvent favors the formation of α-helical structures (see section 3.1) [89–103].
The assignment of infrared bands to particular secondary structure components has been previously reviewed in details [80–82, 107–112]. Infrared spectroscopy also allowed to measure the spectrum of the peptide powders prior to solubilization, in addition to spectra in different solvents (Fig. 2). The amide I band of the RDH8-Cter peptide powder and after its solubilization in methanol is broad and includes several components (Fig. 2B). The maximum of the amide I band of the spectrum of the RDH8-Cter peptide powder is located at 1656 cm−1; a shoulder can also be seen at ~1628 cm−1 (Fig. 2B). These components of the amide I band can respectively be attributed to α-helices and β-sheets. When solubilized in methanol, a strong additional component appears in the amide I band at 1679 cm−1, which indicates the presence of turns. A model of the RDH8-Cter peptide produced by the online tool I-TASSER [73, 74] is also shown in Fig. 2B. It includes a small α-helix but the largest share of this model peptide structure is not ordered. It is quite consistent with the predicted secondary structure content of this peptide (Fig. 1) as well as the circular dichroism (Figs. 2A and 3) and infrared (Fig. 2B) spectra of the RDH8-Cter peptide in MeOH.
Very similar infrared spectra are obtained with the RDH11-Nter peptide powder or after its solubilization in different solvents (Fig. 2D). The amide I band is symmetrical and centered at 1653 or 1658 cm−1, which can be attributed to α-helices. This is consistent with the secondary structure prediction for this peptide (Fig. 1). It is noteworthy that the amide I band of the RDH11-Nter peptide powder and solubilized in HFIP overlap and is centered at 1653 cm−1 whereas that of the peptide solubilized in methanol and H-MeOH also overlap but are centered at 1658 cm−1. It has been shown that the position of the amide I band for α-helices depends on its length [81, 113]. Indeed, a maximum close to 1650 cm−1 indicates a long and rigid α-helical structure. This position can shift up to 1662 cm−1 when the α-helix progressively unwinds. This takes place in particular in polar solvents such as MeOH where the ends of the α-helix transiently unwind as a result of hydrogen bonding between the amide groups and the polar solvent. This phenomenon is less favorable in hydrophobic solvents because they have much less affinity for the polar amide groups. The shift from 1653 in HFIP to 1658 cm−1 in MeOH can thus be explained by a partial unwinding of the ends of the α-helix of the RDH11-Nter peptide in MeOH. A shift is also observed when the position of the positive band of the circular dichroism spectra of this peptide in MeOH and HFIP are compared (Fig. 2C). These data suggest that HFIP and MeOH respectively favor ordering and disordering of the α-helices of the RDH11-Nter peptide. As can be seen in Fig. S3A, the amide I band of the RDH11-Nter peptide in HFIP is much broader than that of the R9AP-Cter peptide in H-MeOH. The R9AP-Cter peptide was predicted to adopt a single α-helical structure (Fig. 1) and its Δθ is the largest one (Fig. 3). The larger width of the amide I band of the RDH11-Nter peptide thus suggests that its structure is less ordered than that of the R9AP-Cter peptide. The possible presence of two interconnected α-helices, such as those shown in Fig. 1 and in the model presented in Fig. 2D, could explain the increased disorder of the RDH11-Nter peptide compared to the R9AP-Cter peptide. In fact, such interconnected α-helices shall allow flexibility; band broadening has also been attributed to flexible structures (for a review, see [80]). This conclusion is also consistent with the smaller Δθ observed with the RDH11-Nter peptide compared to the R9AP-Cter peptide (Fig. 3). In addition, it is well known that vibrational spectroscopy provides a snapshot of the sample conformer population because of its short characteristic time scale (~10−13 s). The band position of a given vibration is typically slightly different for every conformer. This results in a heterogeneous band broadening. Therefore, flexible structures will give broader bands than rigid structures. The band width is thus a measure of conformational freedom. Accordingly, band broadening can also suggest the formation or the presence of different types of secondary structure elements. For example, the band position of disordered structures (1642–1657 cm−1) partly overlaps that of α-helices (1648–1657 cm−1) (for a review, see [80, 82, 109]). Therefore, amide I band broadening of the RDH11-Nter peptide in HFIP compared to the R9AP-Cter peptide in MeOH could also originate from the presence of disordered structures although the existence of flexible structures is likely a more appropriate explanation in view of the model presented in Fig. 2D.
The infrared spectra of the LRAT-Cter peptide powder and solubilized in two different solvents are shown in Fig. 2F. The peaks of these three spectra are very well separated from each other, which indicates that different structural components are present in these different conditions. It can be seen that the infrared spectrum of the powder is broad and not symmetrical. The maximum of its amide I band at 1632 cm−1 indicates that the LRAT-Cter peptide powder mainly contains β-sheets. In addition, the width and the shape of the amide I band of the LRAT-Cter peptide powder suggest that it also contains α-helices and turns. In contrast, the amide I band of this peptide solubilized in HFIP is symmetrical and centered at 1652 cm−1, indicating that this peptide adopts a long and rigid α-helical structure in this solvent. The amide I band of this peptide in H-MeOH is centered at 1678 cm−1, which corresponds to turns; shoulders can also be clearly seen at positions typical of α-helices and β-sheets. This observation is consistent with the shape of the circular dichroism spectrum in this solvent shown in Fig. 2E. The model of the LRAT-Cter peptide structure presented in Fig. 2F does not fit well with any of the infrared spectra of this peptide; indeed, the spectrum in HFIP indicates a long and rigid α-helix whereas that in H-MeOH and the powder respectively reveal a large content in turns and β-sheets. This raises also the issue that HFIP could favor the formation of α-helical structures in the case of this peptide [89–103].
The main difference between the infrared spectra of the R9AP-Cter peptide powder and solubilized in two different solvents is the width of its amide I band (Fig. 2H). The amide I band of this peptide is centered at ~1655 cm−1, indicating that it adopts an α-helical structure whatever it is in a powder or solubilized in a solvent. However, the width of the amide I band increases as follows : H-MeOH < powder < HFIP. The larger bandwidth in HFIP correlates well with the decrease in elipticity of this peptide in this solvent compared to that in H-MeOH (Figs. 2G and H) whereas the much smaller amide I bandwidth in H-MeOH (Fig. 2H) is also in very good agreement with its much larger Δθ (Figs. 2G and 3). In addition, the observation of a shoulder at 1625 cm−1 indicates the presence of β-sheets in the powder, whereas a shoulder typical of turns can be seen at 1679 cm−1 in H-MeOH (Fig. 2H). The structural prediction and model of this peptide presented in Figs. 1 and 2H, respectively, are coherent with the infrared spectrum of the R9AP-Cter peptide in H-MeOH, although no turn can be seen in the structure.
It is interesting to compare the data obtained by circular dichroism and infrared spectroscopy. As shown in Fig. S3B, the width of the amide I band of the LRAT-Cter, RDH11-Nter and R9AP-Cter peptides in HFIP is very similar. However, the molar ellipticity of the R9AP-Cter peptide is larger than that of the RDH11-Nter peptide in HFIP (Figs. 2G and C, respectively), which is much larger than that of the LRAT-Cter peptide (Fig. 2E). There is however a good agreement between the molar ellipticity of these peptides and the width of their amide I band in H-MeOH. Indeed, as shown in Fig. S3C, the smallest bandwidth of the amide I band of the R9AP-Cter peptide in H-MeOH is consistent with its largest Δθ (Fig. 3). Accordingly, when compared to the R9AP-Cter peptide in H-MeOH, the much larger bandwidth of the amide I band of the RDH11-Nter peptide (Fig. S3C) is consistent with its much smaller value of Δθ (Fig. 3), which is however incoherent with the analysis of its content in α-helices (52–56% for the RDH11-Nter peptide compared to 63% for the R9AP-Cter peptide, Table S1). Nevertheless, the large bandwidth of the amide I band and the shift of this band of the LRAT-Cter peptide to larger wavenumbers (Fig. S3C) in H-MeOH is consistent with its very small Δθ (Fig. 3) and content in α-helices (Table S1).
4. Binding of hydrophobic peptides to lipid monolayers
4.1 Monolayers as a model membrane to study hydrophobic transmembrane peptides
Phospholipid monolayers are very useful model membranes to study lipid-protein or lipid-peptide interactions (for a review, see [114–125]). Indeed, it allows to control several physical parameters such as the density of lipids and the surface pressure, the subphase content, the lipid composition, etc. Moreover, there is a direct thermodynamic relationship between bilayers and monolayers [126, 127]. In addition, a large number of spectroscopic (UV-visible absorption [128–134], fluorescence [128, 131–136], and vibrational [137–143], (see [120, 144–152] for a review)), microscopic (Brewster angle [153, 154] and fluorescence [155–157], (see [158–164] for a review)), and different additional physical methods [165–169] (see [117, 163, 170–173] for a review) have been developed to characterize monolayers. A large share of these methods has been reviewed by Dynarowicz-Latka et al. [174]. An additional major advantage of this model membrane is that it is the only one which can allow to independently determine the affinity of compounds for the inner or the outer leaflet of the membrane bilayer. Indeed, the lipids of cell membranes are known to be asymmetrically distributed in the membrane of most cell organelles including the plasma membrane (reviewed in [175]). Therefore, the monolayer model membrane can allow to independently assay binding of compounds to either the lipids found in the outer (phosphatidylcholine, sphingomyelin and glycosphingolipids) or the inner (phosphatidylethanolamine, phosphatidylinositol and phosphatidylserine) membrane leaflet. Monolayers have thus been extensively used to study the orientation and structure of different types of peptides upon lipid binding (see section 4.1 for an extensive review). More specifically, monolayers have additionally been used to characterize the structure and organization of hydrophobic transmembrane peptides such as a transmembrane segment of the nicotinic membrane receptor [176], of signal sequence peptides allowing translocation of proteins to membranes [177], of the gramicidin transmembrane ion channel [178–187], of a model peptide for an anesthetic-binding membrane protein [188, 189], of transmembrane α-helical model peptides [190, 191], of transmembrane peptides derived from the domain M of an enzyme [192], of the transmembrane segment of the Vpu protein [193–195], as well as of the synaptobrevin 1/VAMP1 and syntaxin 1 [196], and of the N- and C-terminal transmembrane peptides of LRAT [58].
Nevertheless, one can wonder whether lipid monolayers represent proper model membranes for transmembrane peptides, which span the entire thickness of a bilayer whereas monolayers correspond to one membrane leaflet. In fact, it has been shown that the orientation of transmembrane α-helical hydrophobic peptides is dependent on the hydrophobic mismatch in lipid bilayers [197–200]. For example, the tilt of a transmembrane peptide was shown to increase from 27 to 35 and 51° with respect to the normal of bilayers of phospholipids containing, respectively, the following decreasing fatty acyl chain lengths : myristoyl (C14:0), lauroyl (C12:0) and dodecanoyl (C10:0) fatty acyl chains [197]. Therefore, the shorter is the thickness of the bilayer, the larger is the tilt angle of the transmembrane α-helical hydrophobic peptide. Therefore, this change in tilt angle of a transmembrane helix is a compensation mechanism for hydrophobic mismatch [197], which is a driving force for α-helical transmembrane peptide orientation [201]. The same is true for lipid monolayers as discussed previously for the N- and C-terminal peptides of LRAT [58]. Indeed, lipid monolayers are not thick enough to accommodate transmembrane α-helical peptides that span the entire thickness of the membrane lipid bilayer. As a result, the transmembrane α-helical N- and C-terminal peptides of LRAT are tilted at angles of 40–45° with respect to the normal of monolayers of phospholipids containing palmitoyl (C16:0) fatty acyl chains [58]. This large tilt angle nonetheless resulted in the observation of the typical α-helical secondary structure of transmembrane peptides [58]. Interestingly, this value of 40–45° of tilt angle is similar to that observed for the Vpu transmembrane peptide in lipid bilayers with short fatty acyl chains [197]. Moreover, oblique oriented α-helical peptides can be found in membranes and are now recognized as a class of peptides playing specific membrane functions (for reviews, see [202–204]). Altogether, these data suggest that the monolayer model membrane is appropriate to characterize transmembrane hydrophobic peptides.
4.2 Preparation of lipid monolayers to study hydrophobic transmembrane peptides
Lipid monolayers are spread from a known volume of solvent at the surface of a buffer poured in a Langmuir trough (Fig. 4). The lipid solution must be spread until the desired initial surface pressure (Πi) is reached (Fig. 4B). Alternatively, the lipid monolayer is compressed to the same Πi. A peptide solution is then injected into the subphase (Fig. 4B). Monolayer binding of the peptide can then be measured at either a constant surface area or a constant surface pressure. Unfortunately, these two approaches have never been systematically compared although they could potentially provide complementary information. The observation of an increase in surface pressure is restricted by the maximum insertion pressure (MIP) of the peptide (see section 4.5; for a review, see [116]). The MIP corresponds to the maximum surface pressure of the monolayer up to which the peptide can insert in the monolayer and beyond which no insertion takes place. Therefore, beyond this value of MIP, no increase in surface pressure should be observed at a constant area. The determination of the MIP can provide information on the affinity of a peptide for a given type of lipid [205] as well as on its extent of binding to membranes. Indeed, given that the membrane lateral pressure has been estimated in the range of 30–35 mN/m [206–213], one can postulate that a MIP larger than this value will suggest that the peptide could bind membranes. The synergy is obtained by adding 1 to the slope of the plot of the surface pressure increase (ΔΠ) as a function of the initial surface pressure (Πi) [116]. We have previously shown that a positive synergy corresponds to a favorable binding of proteins. The MIP in this case corresponds to an insertion surface pressure [205]. In contrast, when the synergy is close to zero, the binding of the protein is neither favored nor disfavored by the lipid monolayer because the equilibrium surface pressure (Πe) remains almost unchanged with increasing values of Πi. It is preferable to use very small troughs to determine the MIP and synergy of peptides because measurements must be performed at several initial surface pressures. Significant amounts of peptides will be spent unless if microtroughs can be used. Therefore, in order to save peptide, commercially available microtroughs are used to perform these measurements, such as that from Kibron (DeltaPi4) (Fig. 4A). Moreover, four measurements of peptide monolayer binding can be measured at the same time with this apparatus using microtroughs containing a buffer subphase of 500 μL (Fig. 4A). Prior to describing how to perform MIP measurements, one must figure out whether the peptide should preferably be injected into the subphase or spread as a lipid-peptide mixture at the surface of the buffer subphase. It should however be stressed that MIP values can only be obtained if the peptide is injected into the subphase.
Fig. 4. Instrument and procedure to prepare lipid monolayers and to study peptide binding using the Langmuir model membrane system.

A) Picture of the microtroughs sold by the company Kibron (Helsinki, Finland) which have been used for the measurement of peptide binding to lipid monolayers. B) Schematic diagram of the adsorption of peptides at the lipid/water interface. All surface pressure measurements in monolayer were performed with the Delta Pi4 instrument from Kibron.
4.3 Spreading the peptide-lipid mixture at the surface or injecting the peptide into the subphase underneath the lipid monolayer : which approach is most appropriate?
The monolayer methodology can be used to determine the affinity of a peptide for a given type of lipid as well as its organization and structure in the presence and absence of lipids. Two approaches can be used to perform such measurements : 1) spreading a lipid at the surface of the buffer and then injecting the peptide into this monolayer subphase or 2) preparing a lipid-peptide mixture in an organic solvent and then spreading a known volume of this solution at the surface of the buffer. One can wonder whether the same results can be obtained with these two approaches and which one is most appropriate. There is approximately twice as many papers reporting measurements where peptides are injected into the subphase [105, 125, 176, 192, 214–294] as compared to papers reporting data after spreading lipid-peptide mixtures at the air-water interface [176–178, 180–185, 190, 191, 193, 194, 218, 220, 229, 289, 295–326]. It is most relevant to find out whether the same peptide structure is obtained when using these two approaches. In situ infrared spectroscopy of monolayers has thus been used to clarify this issue (see section 5 for more details on this method).
Fig. 5 is showing the comparison between the spectra of the R9AP-Cter peptide when spread at the surface or injected into the subphase in the absence (Fig. 5A) or in the presence (Fig. 5B) of DOPE (1,2-dioleoyl-sn-glycero-3-phospho-ethanolamine) monolayer. It can be seen that very similar spectra are obtained when the individual peptide is adsorbed at the surface after its injection into the subphase or when it is spread at the surface (Fig. 5A). The amide I band is centered at 1656 cm−1, which indicates that the peptide structure is α-helical. However, shoulders are appearing at 1625 and 1695 cm−1 when the spread peptide is compressed to a surface pressure close to that obtained upon injecting the peptide into the subphase (Fig. 5A). The observation of these shoulders indicates that compression results in the formation of antiparallel β-sheets. Contrasting data have been obtained in the presence of a DOPE monolayer. The spreading and compression of the DOPE-peptide mixture results in a large amide I bandwidth centered at 1651 cm−1 as well as shoulders at 1634 and 1693 cm−1, which are respectively typical of α-helices and antiparallel β-sheets (Fig. 5B). However, the spectrum of the peptide injected into the subphase is showing an amide I band with a maximum typical of an α-helical structure (1657 cm−1) as well as a small shoulder at 1634 cm−1 for β-sheets. The amide I band of the spread peptide is much broader than that of the injected peptide, which strongly suggests that the spreading procedure leads to the formation of disordered structures. Previous reports have shown a slight or no effect of monolayer compression with some lipid-peptide mixtures [295, 311] whereas a large modification of the peptide structure was found to take place for other lipid-peptide mixtures [177, 313, 325]. Although measurements with additional peptides should be performed to definitively clarify this issue, the present data suggest that it is preferable to perform measurements by injecting peptides into the lipid monolayer subphase. However, the spreading of a lipid-peptide mixture is better suited to study their thermodynamics of mixing in as much as the peptide structure remains unaltered by this procedure. Nevertheless, peptides must be injected into the subphase to observe their extent of lipid monolayer binding and to determine their MIP. It is then necessary to evaluate which peptide concentration should be used to perform MIP measurements.
Fig 5. PM-IRRAS spectra of the R9AP-Cter peptide measured either after spreading the peptide (or the DOPE-peptide mixture) at the air-water interface or after injecting the peptide into the subphase of the DOPE monolayer.

The trough and PM-IRRAS have been described previously [357]. The subphase buffer contains 50 mM Tris HCl, 150 mM NaCl and 5 mM β-mercaptoethanol (pH 7.4). The final concentration of the injected peptide is 5 μM. A ratio of 20/1 (3.2/0.16 nmoles) has been used for the spread DOPE-peptide mixture. The spectra have been normalized to facilitate their comparison. A) Comparison of the PM-IRRAS spectra of the individual peptide either injected into the subphase (Πe = 16.8 mN/m, black curve) or spread at the air-water interface (Πe = 7.8 mN/m, red curve) and then compressed to 12.9 mN/m (blue curve) (in the absence of a lipid monolayer). B) Comparison between the PM-IRRAS spectra of the peptide injected underneath a DOPE monolayer (Πe = 21 mN/m, orange curve) and of a DOPE-peptide mixture spread at the air-water interface and compressed to a Πe=13 mN/m (green curve).
4.4 Determination of the peptide concentration to perform monolayer measurements
MIP measurements must be performed at a peptide concentration where surface saturation has been reached. The first step when performing measurements of peptide monolayer binding thus consists in determining the dependence of the surface pressure increase on peptide concentration. Therefore, measurements of surface pressure as a function of time must be performed with several peptide concentrations either in the presence or in the absence of a lipid monolayer, usually without much difference. A typical adsorption isotherm of a peptide is presented in the inset of Fig. 6 which shows that surface pressure increases until Πe is reached. Then, the values of Πe are plotted as a function of protein concentration (Fig. 6). An exponential plot is typically obtained and the optimal protein concentration is that where no increase in surface pressure is observed when protein concentration is further increased, which indicates that surface saturation has been reached. For example, in Fig. 6, a saturating protein concentration has been obtained at a peptide concentration ranging between 3 and 5 μM.
Fig. 6. Determination of the saturating surface concentration of peptides for the monolayer measurements.
Typical example of the extent of peptide adsorption using the R9AP-Cter peptide. The final subphase concentrations are 0.25, 0.3, 0.8, 2, 5, 12 μM. The R9AP-Cter peptide solubilized in H-MeOH has been injected into the subphase of a microtrough of 500 μL (Kibron) in the absence of a lipid monolayer. The surface pressure at equilibrium (Πe) obtained after ~0.5 hour of adsorption is plotted as a function of the concentration of the R9AP-Cter peptide. The optimal concentration of R9AP-Cter peptide is lying between 3 and 5 μM. Inset: Typical adsorption kinetics of the R9AP-Cter peptide at the surface of a buffer containing 50 mM Tris HCl, 150 mM NaCl, and 5 mM β-mercaptoethanol (pH 7.4) at a final peptide concentration of 2 μM (only one kinetics of adsorption has been shown for clarity).
4.5 Determination of the maximum insertion pressure and synergy of the hydrophobic peptides in the presence of a lipid monolayer
The buffer is poured in the 500 μL microtroughs shown in Fig. 4A. Lipids are then spread at the surface of this buffer (Fig. 4B) until the desired Πi is reached (inset of Fig. 7A). A period of time is then allowed for the spreading solvent to evaporate and for the film to reach equilibrium. This waiting time varies with the type of lipid, the spreading volume, the initial surface pressure and the lipid concentration. A saturating quantity of peptide is then injected into the subphase underneath the lipid monolayer (section 4.4; Figs. 4B and 6). As a consequence, depending on the surface activity of the peptide and its affinity for the lipid monolayer, surface pressure will increase until Πe is reached (inset of Fig. 7A). This allows calculation of the surface pressure increase (ΔΠ = Πe − Πi) (inset of Fig. 7A). The magnitude of this surface pressure change will depend on the initial surface pressure and can be used to compare the extent of protein- or peptide-lipid interactions (see [116] for a review). Injection of the peptide is performed at different Πi of the lipid monolayer (Fig. 7A). Then, the plot of ΔΠ as a function of Πi allows the determination of the MIP by extrapolating the regression of the plot to the x axis, as shown in Fig. 7A. It is meaningful to calculate the uncertainty of the MIP and synergy data, which allows to make proper conclusions. This experimental error must absolutely be calculated, as previously described [116, 205, 327, 328]. The data are otherwise very difficult to properly interpret. We have recently created a freely available webpage where this experimental error can be readily calculated (http://www.crchudequebec.ulaval.ca/BindingParametersCalculator/), which should facilitate this operation.
Fig. 7. Determination of the maximum insertion pressure and the synergy of peptides in the presence of a DOPE monolayer.
A) The MIP of the peptides has been determined by extrapolating the plot of the surface pressure increase (ΔΠ) as a function of the initial surface pressure (Πi) where the curve reaches a value of 0 on the x-axis. The surface pressure increase corresponds to ΔΠ = Πe − Πi. The curves have been obtained by measuring the binding of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides onto a DOPE monolayer at different Πi as a function of time (shown in the inset). Inset: Typical adsorption kinetics after the injection of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides at a final subphase concentration of 2.5, 1.25, 1.25 and 5 μM, respectively, underneath a DOPE monolayer at a Πi of ~10 mN/m. The subphase buffer comprises 50 mM Tris, 150 mM NaCl, and 5 mM β-mercaptoethanol (pH 7.4) for all peptides. Histograms of the MIP (B) and of the synergy (C) values of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides in the presence of a DOPE monolayer. The synergy is obtained by adding 1 to the slope of the curves shown in (A).
It is very important to properly select the type of lipid that will be used to study the binding of the peptide. One should thus select lipids that can be found in the membranes where the peptide is located [175]. In the case of the present hydrophobic peptides which have been postulated to anchor proteins to membranes, one should look for the lipid composition of these membranes. It is also interesting to compare the behavior of the peptide in the presence of unsaturated, polyunsaturated as well as saturated phospholipids. Indeed, the different physical states of these phospholipids could drive specificity of peptide binding such as previously observed for different proteins [205, 327]. The specific binding of a peptide to a saturated phospholipid could also suggest that this peptide is located in rafts since these structures contain a large percentage of this type of fatty acyl chain (for a review, see [329–332]). One should also be careful when using polyunsaturated phospholipids because they are very sensitive to oxidation [328]. In our experiments, typical MIP measurements were performed with the same unsaturated phospholipid (DOPE) in order to compare the behavior of the different peptides with the same lipid as well as because this lipid is a major component of the membranes where RDH11 and LRAT are located (retinal pigment epithelium, [333, 334]) and a minor component of photoreceptor membranes [335–337] where RDH8 and R9AP can be found.
As can be seen in the inset of Fig. 7A, a similar value of Πe has been obtained when the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides are injected underneath a DOPE monolayer at the same initial surface pressure. Also, similar intercepts with the x axis and slopes of the plot of ΔΠ as a function of Πi have been obtained for these peptides except for the R9AP-Cter peptide which is quite different (Fig. 7A). The MIP and synergy values are easier to compare using histograms (Figs. 7B and 7C). Fig. 7B is showing that there is no significant difference between the values of MIP obtained for the RDH8-Cter (46.7 ± 3.0 mN/m), RDH11-Nter (44.7 ± 4.8 mN/m) and LRAT-Cter (43.2 ± 3.4 mN/m), which are very different from that of the R9AP-Cter peptide (33.5 ± 3.5 mN/m). Accordingly, no significant difference can be observed between the synergy values of the RDH8-Cter (0.34 ± 0.03), LRAT-Cter (0.32 ± 0.05) and RDH11-Nter (0.38 ± 0.05) peptides whereas a much smaller synergy has been obtained for the R9AP-Cter peptide (0.17 ± 0.06). Nevertheless, the values of MIP reported in Fig. 7B are all larger than the lateral pressure of membranes, thereby suggesting that the RDH8-Cter, LRAT-Cter, RDH11-Nter and R9AP-Cter peptides bind membranes. This observation strongly supports the postulated role of these transmembrane peptide segments in anchoring proteins to membranes.
5. Determination of peptide structure and orientation in monolayer by infrared spectroscopy
Two approaches have been developed to determine the structure and orientation of peptides in monolayers at the air-water interface by infrared spectroscopy : infrared reflection absorption spectroscopy (IRRAS) [141–143] and polarization modulation infrared reflection absorption spectroscopy (PM-IRRAS, Fig. 8) [137]. In order to get rid of the water absorption bands, in particular that in the amide region, IRRAS and PM-IRRAS are using different strategies : 1) the IRRAS spectra are resulting from the ratio between spectra measured in the presence and absence of a monolayer. This is achieved by alternatively moving a Langmuir trough from regions where the monolayer or the bare substrate are located (see [144, 146] for a review). 2) The PM-IRRAS spectra are resulting from the fast polarization modulation of the incident beam between parallel (p) and perpendicular (s) polarizations using a photoelastic modulator (Fig. 8) [137, 152]. The two-channel processing of the signal allows to obtain the differential reflectivity spectrum:
| (1) |
where Rp and Rs are the reflectivity of the interface for polarized light, J0 and J2 are the zero and second-order Bessel functions, whereas Φ0 is the maximum dephasing given by the photoelastic modulator. Moreover, to remove the isotropic contributions from bulk water and water vapor, experimental drifts and to get rid of the dependence of the signal on the Bessel function, the spectrum of the phospholipid monolayer in the presence of the adsorbed peptide is divided by that of the subphase (alternatively, the spectrum of the lipid-peptide monolayer can be divided or subtracted by that of the lipid monolayer, see section 5.1) to produce the resulting normalized PM-IRRAS spectrum:
| (2) |
Fig. 8. Schematic representation of the instrument used to perform polarization modulation infrared reflection absorption spectroscopy (PM-IRRAS) measurements.

The PM-IRRAS has been developed to determine the structure and orientation of peptides in monolayers at the air-water interface [137, 357].
The signal-to-noise ratio of the technique is therefore enhanced because water vapor molecules do not contribute to the PM-IRRAS signal. However, oriented and bound water molecules can not be removed from the spectra for both IRRAS and PM-IRRAS.
The main advantages of IRRAS is that the absolute infrared signal of individual bands can be quantitatively analyzed and that drifts of the signal are minimized by ratioing in real time the spectrum of the sample with that of the bare monolayer area. However, the disadvantage of this method is that time-consuming measurements at several angles of incidence must be performed to determine the orientation of molecules. In contrast to what was argued in a recent review [146], there is no need with PM-IRRAS to run spectra at different angles of incidence to determine the orientation of compounds. Indeed, at an incidence angle of 75°, transition moments preferentially oriented in the plane of the interface give intense and upward oriented bands, while perpendicular transition moments give weaker and downward oriented bands. The orientation of molecules can thus potentially be determined from a single spectrum. For example, changes in the orientation of an α-helical peptide can be observed from the variation of the ratio between the amide I and amide II bands at a single angle of incidence. Simulations of the experimental spectra are necessary to get absolute values of orientation of compounds such as α-helical peptides as described previously [338]. Then, the theoretical normalized PM-IRRAS signal must be calculated for an anisotropic peptide monolayer, using a general software program [339]. Values of the optical anisotropic indices of the film are thus generated by taking into account the relative infrared absorptions and dichroism reported for the amide I and amide II absorptions of an α-helical polypeptide [340, 341]. The results of the simulations at different angles of orientation of an α-helical polypeptide relative to the water surface are shown in Fig. S4. On the basis of the PM-IRRAS selection rules on a dielectric surface [342], a strong positive amide I band and a weak positive amide II band can be seen when the helix axis is parallel to the interface (90°, Fig. S4). Conversely, a strong negative amide I band and a strong positive amide II band are obtained when the α-helix is perpendicular to the monolayer surface (0°, Fig. S4). Similar simulations have been performed for other secondary structures of peptides [343]. The PM-IRRAS spectra of the hydrophobic peptides (section 5.2) have thus been analyzed using this procedure. This dependence of the PM-IRRAS signal on the orientation of compounds, however, renders difficult a quantitative analysis of the absolute infrared signal of individual bands, in contrast to IRRAS.
5.1 Treatment of infrared spectra
As mentioned above, the PM-IRRAS spectrum of the phospholipid-peptide monolayer can be either divided by that of the subphase or, alternatively, by that of the lipid monolayer. Subtracting the spectrum of the subphase, instead of dividing, can also be assayed with typically better results. It can happen that no amide I band can be observed by subtracting the spectrum of the subphase from that of the phospholipid-peptide monolayer as can be seen in Fig. 9. Indeed, in this case, the C=O ester band of the phospholipid monolayer can be observed at 1737 cm−1 as well as the typical broad negative band in the 1700–1640 cm−1 region. The latter band is due to an abrupt variation of the refractive index of the aqueous subphase in this range of frequency [344]. Indeed, the water absorption band is centered at approximately 1640 cm−1 where the PM-IRRAS spectrum changes abruptly showing a positive band followed immediately by a negative band. For unknown reasons, this sometimes results in difficulties to observe amide I and amide II bands, as shown in Fig. 9. Moreover, poorly defined positive bands are also seen in the amide region which prevent a proper monitoring of peptide secondary structure components. Information from the amide I can thus hardly be extracted from such spectra. Alternatively, amide I and II bands can very often be clearly observed when one subtracts the PM-IRRAS spectrum of the phospholipid monolayer from that of the phospholipid-peptide monolayer. A typical spectrum resulting from this treatment allows to observe well defined amide I and II bands (Fig. 9). When this alternative procedure is not successful, one must perform measurements on deuterated water which will most likely allow to solve the issue.
Fig. 9. Comparison between two different ways to treat PM-IRRAS spectra of a peptide bound to a lipid monolayer.
The spectrum « Subtraction of buffer » is resulting from the subtraction of the spectrum of the subphase from that of the peptide-phospholipid monolayer. The spectrum « Subtraction of lipid » is resulting from the subtraction of the spectrum of the phospholipid monolayer from that of the peptide-phospholipid monolayer. The RDH11-Nter peptide dissolved in HFIP has been injected into the subphase (50 mM Tris HCl, 150 mM NaCl, and 5 mM β-mercaptoethanol, pH 7.4) of a DOPE monolayer at an initial surface pressure of 21 mN/m. The final concentration of the peptide was 1.25 μM.
5.2 Infrared spectra of the peptides in the absence and in the presence of a lipid monolayer
The PM-IRRAS spectra of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides in the presence and in the absence of a DOPE monolayer are shown in Fig. 10. The amide I band of the RDH8-Cter peptide (Fig. 10A) is broader in the absence than in the presence of the DOPE monolayer. This indicates that the binding of the peptide to the lipid monolayer improves the order of this peptide. In the presence of DOPE, the amide I band is centered at ~1655 cm−1 which suggests that the RDH8-Cter peptide adopts mainly an α-helical secondary structure. However, as mentioned in section 3.2, the amide I band of disordered structures largely overlaps that of α-helices. Given that the RDH8-Cter peptide has the largest amide I bandwidth among the 4 hydrophobic peptides (Fig. S5A), it likely includes both α-helical and disordered structures in the presence of a DOPE monolayer. These spectroscopic data could thus hardly be used to estimate the orientation of the RDH8-Cter peptide because the simulations presented in Fig. S4 have been made to determine the orientation of pure α-helical peptides. The infrared spectrum of the RDH8-Cter peptide in the presence of a DOPE monolayer has been compared to that of this peptide dissolved in methanol in Fig. S6A. Although the amide I band is rather broad, binding of the peptide to the DOPE monolayer significantly improved its order. Indeed, the shoulders attributed to β-sheets and turns originally observed in methanol can not be readily seen in the spectrum of the peptide in the presence of the DOPE monolayer (Fig. S6A).
Fig. 10. PM-IRRAS spectra of the peptides in monolayers.
RDH8-Cter (A), RDH11-Nter (B), LRAT-Cter (C) and R9AP-Cter (D) peptides in the absence and in the presence of a DOPE monolayer. The spectra have been normalized to facilitate their comparison. The subphase buffer is composed of 50 mM Tris HCl, 150 mM NaCl, and 5 mM β-mercaptoethanol (pH 7.4) for all peptides. The final concentration of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides is 2.5, 1.25, 1.25 and 5 μM, respectively.
The spectra of the RDH11-Nter peptide are presented in Fig. 10B. In the absence of a DOPE monolayer, the amide I band of the RDH11-Nter peptide is symmetrical and centered at 1655 cm−1. This peptide thus mainly adopts an α-helical structure. However, the presence of the DOPE monolayer leads to the appearance of a β-sheet component at ~1633 cm−1 in the RDH11-Nter peptide and thus to a small modification of the secondary structure of this peptide. This is contrasting with the RDH8-Cter peptide where the DOPE monolayer improved its order (compare Figs. 10A and 10B). The orientation of RDH11-Nter peptide can not be appropriately estimated in the presence of the DOPE monolayer because a β-sheet component was observed in this spectrum (Fig. 10B). However, the RDH11-Nter peptide orientation can be evaluated in the absence of a DOPE monolayer. An AI/AII ratio of 1.7 has been calculated in these conditions. The orientation of the RDH11-Nter peptide is thus very close to 50° (AI/AII ratio of 2.1, Fig. S4) with respect to the normal. The infrared spectrum of the RDH11-Nter peptide in HFIP is compared to that in the presence of a DOPE monolayer in Fig. S6B. It can be seen that the amide I bands are very similar except that the one in HFIP is more symmetrical but a little broader and devoided of a shoulder when compared to that in the presence of a DOPE monolayer (Fig. S6B). As mentioned in section 3.2, this band broadening could be attributed to a larger flexibility of the structure of the RDH11-Nter peptide in HFIP.
Fig. 10C is showing the spectra of the LRAT-Cter peptide in the presence and in the absence of a DOPE monolayer. The amide I band of the peptide in the presence of the DOPE monolayer is centered at 1652 cm−1 which shows that this peptide adopts an α-helical structure. A shoulder at 1630 cm−1, which is almost as intense as the α-helical component, can additionally be clearly seen in the spectrum of the peptide in the absence of the DOPE monolayer, thereby showing that a large β-sheet structural component is formed in these conditions in addition to α-helices. The presence of the DOPE monolayer thus highly favored the formation of an α-helical structure of the LRAT-Cter peptide at the expense of β-sheets. The orientation of the LRAT-Cter peptide in the presence of a DOPE monolayer (AI/AII ratio of 1.9) is very close to 50° (AI/AII ratio of 2.1, Fig. S4) with respect to the normal. The infrared spectra of the LRAT-Cter peptide solubilized in HFIP and in the presence of a DOPE monolayer are compared in Fig. S6C. The spectra are very similar and almost perfectly overlap. However, it must be stressed that, differently from all other peptides, which were injected into the monolayer subphase in a methanolic solution, this spectrum was measured after the injection of the LRAT-Cter peptide solubilized in HFIP. This spectrum of the LRAT-Cter peptide bound to a DOPE monolayer is almost the same as that of the peptide dissolved in HFIP (Fig. S6C). It is however very different from that of the peptide solubilized in H-MeOH (Fig. S6C). It thus raises the issue of whether the type of solvent could dictate the final structure of this peptide upon binding lipid monolayers. More measurements are necessary to clarify this issue.
The spectra of the R9AP-Cter peptide in the presence and in the absence of a DOPE monolayer are presented in Fig. 10D. These spectra are very similar. Indeed, the amide I band of both spectra is very thin and centered at 1657 cm−1 (α-helices) and their AI/AII ratio is almost the same. In the same manner as for the RDH11-Nter peptide (Fig. 10B), a β-sheet component at ~1634 cm−1 appears in the presence of the DOPE monolayer, thus resulting in a small modification of the secondary structure of the R9AP-Cter peptide compared to when it is injected into the subphase in the absence of a lipid monolayer (Fig. 10D). The orientation of the R9AP-Cter peptide in the absence of a DOPE monolayer (AI/AII ratio of 1.8) is very close to 50° (AI/AII ratio of 2.1, Fig. S4) with respect to the normal. The infrared spectrum of the R9AP-Cter peptide in H-MeOH is compared to that in the presence of a DOPE monolayer in Fig. S6D. It can be seen that the amide I band of both spectra is very thin and centered almost at the same position. The presence of the DOPE monolayer resulted in the disappearance of the turns observed in H-MeOH but to the appearance of a β-sheet structural component. Therefore, the injection of the R9AP peptide in this solvent resulted in a slightly different structure of this peptide (Fig. S6D).
The PM-IRRAS spectra of the RDH8-Cter, RDH11-Nter, LRAT-Cter and R9AP-Cter peptides in the presence of a DOPE monolayer have been compared in Fig. S5A. It can be seen that the width of the amide I band of these peptides decreases as following : RDH8-Cter > LRAT-Cter > RDH11-Nter > R9AP-Cter. One can thus conclude that the R9AP-Cter peptide is most ordered whereas the RDH8-Cter peptide is most disordered among these 4 peptides in the presence of a DOPE monolayer.
6. Concluding remarks
In this review, the spectroscopic properties of hydrophobic peptides have been characterized in different solvents as well as their binding, structure and orientation in the presence and in the absence of a lipid monolayer. Several issues have been raised concerning 1) the usefulness of the available online tools to provide structural information for transmembrane peptides, 2) the role of the solvent used to solubilize the peptides on their structure, and 3) the effect of the lipid monolayer on the peptide structure and its relationship with the maximum insertion pressure.
6.1 Usefulness of the available online tools to provide structural information for transmembrane peptides
It is very difficult to provide reliable information on the secondary structure and properties of peptides on the basis of their amino acid sequence. The challenge is even greater with hydrophobic transmembrane peptides because of the particular membrane environment where they are located, which can influence their structure. Indeed, the hydrocarbon core of the membrane bilayer is not a favorable environment for charged amino acids [345]. Conversely, it can be expected that hydrophobic amino acids will be more confortable when surrounded by the fatty acyl chains of the hydrocarbon core than in the lipid head group region. Fortunately, the exponentially growing number of high-resolution membrane protein structures provided a deeper understanding of the topology and structure of transmembrane helices [346]. Statistical free energies of membrane insertion of amino acids have thus been calculated from these high-resolution structures [347]. This allowed to draw distinct distribution profiles of the amino acids along the transmembrane helices. For example, hydrophobic residues (A, I, V, L) as well as the amino acids F, G and M prefer to be located in the center of the hydrophobic core of the membrane bilayer [348]. Moreover, the aromatic residues H, W and Y are mainly found in the polar head region at the membrane interface whereas charged and polar residues are largely absent from the membrane interior. Therefore, the analysis of the primary structure of peptides should be useful and can not be ignored. In this regard, it is interesting to compare the data shown in Table 1 for the hydrophobic peptides reviewed in this paper with those resulting from the analysis of the amino acid content of the 7 transmembrane α-helical segments of a typical membrane protein, the GPCR rhodopsin (Table S2A), on the basis of its high-resolution structure [349]. These data have then been compared in Table S2B. It can be seen that the percentage of amino acids of the transmembrane α-helical segments (TMS) of rhodopsin and of the hydrophobic peptides (HP) found in “TM helix” (Table S2B) are quite close to each other when removing the data of the RDH8-Cter peptide : hydrophobic peptides = 80–93% compared to 78–84% for rhodopsin. Among the four transmembrane peptides studied in this review, the RDH8-Cter peptide is the least hydrophobic (Table 1, Fig. S2). It is also the only one that is solely soluble in methanol which is consistent with its small content in α-helices predicted in Figs. 1 and 2B. Indeed, pure α-helices, such as that of R9AP-Cter, are typically not soluble in polar solvents (unless if it is first solubilized in HFIP). These prediction models have also been found to be quite coherent with the assignment of the amide I band of the infrared spectrum of this R9AP-Cter peptide in the same solvent. In fact, the structural models presented in Fig. 1 as well as in Figs. 2B, D, F and H all fit quite well with the experimental data except for the LRAT-Cter peptide. The latter peptide is however the one for which it has been necessary to use several online tools to get more consistent data. One should stress that these tools were optimized for proteins and they might be less efficient for peptides. Nevertheless, one can argue that altogether the available online tools can provide useful structural information. However, experimental data must be obtained to make sure that these predictions are reliable.
6.2 Role of the solvent on the peptide structure
Different solvents have been used to solubilize the four transmembrane peptides reviewed in this paper, except for the RDH8-Cter peptide, which was only soluble in methanol, as mentioned above. The solvent has a minor effect on the secondary structure of the RDH11-Nter peptide, a slightly larger effect on the R9AP-Cter peptide but a strong effect on the LRAT-Cter peptide (Figs. 2D, H and F, respectively). HFIP did not favor formation of an α-helical secondary structure for the RDH11-Nter and R9AP-Cter peptides (Figs. 2D and H, respectively). Interestingly, solubilization of the RDH11-Nter, LRAT-Cter, R9AP-Cter peptides in HFIP resulted in a very similar amide I band (Fig. S3B), suggesting a similar structural effect of this solvent on these peptides. In fact, the thinnest amide I band with measured with the R9AP-Cter peptide has been obtained when solubilized in H-MeOH which is rather surprising in view of its properties to progressively unwind α-helical structures (Fig. 2H). In contrast, the solubilization of the LRAT-Cter peptide in H-MeOH resulted in a large shift of the amide I band to higher wavenumbers, compared to that in HFIP, and thus to the formation of a rather disordered peptide in this polar solvent (Fig. 2F). As shown in Fig. S6C, a very similar spectrum is obtained for the LRAT-Cter peptide solubilized in HFIP and bound to a DOPE monolayer after its injection into the subphase using a HFIP solution. Would similar results be obtained if this peptide were injected into the subphase in a H-MeOH solution? Would one then get the same spectrum as that in H-MeOH (Fig. S6C)? In this regard, as can be seen in Figs. S6A, B and D, quite similar spectra have been obtained for the RDH8-Cter, RDH11-Nter and R9AP-Cter peptides when comparing the solution spectrum with that in monolayer (using the same solvent to inject the peptide into the subphase). However, as previously stated, the presence of the lipid monolayer resulted in a significant ordering of the RDH8-Cter peptide (Fig. S6A). Finally, one can wonder whether a similar spectrum for the peptides can be obtained in solution and as a pure monolayer (in the absence of a lipid monolayer). As shown in Fig. S7, all peptides are more ordered in monolayer than in solution except for the LRAT-Cter peptide. All of these peptides contain at least one Cys. One could then expect that peptide aggregation could be observed when pure peptides are sitting as a pure monolayer at the air-water interface. This is clearly not the case for the RDH11-Nter, LRAT-Cter and R9AP-Cter peptides. However, the amide I band of the RDH8-Cter peptide is wide enough to include some disordered structures even though β-mercaptoethanol was present in the subphase. The solvent thus plays a moderate to critical role in modulating the secondary structure of transmembrane hydrophobic peptides but measurements with additional peptides would allow to draw more general conclusions.
6.3 Effect of the lipid monolayer on peptide structure and its relationship with the maximum insertion pressure
The RDH8-Cter and LRAT-Cter peptides are the most disordered ones in the absence of a lipid monolayer. However, their order was significantly improved in the presence of the DOPE monolayer (Figs. 10A and C, respectively). In contrast, the highly ordered RDH11-Nter and R9AP-Cter peptides in the absence of a lipid monolayer (the amide I bandwidth of the RDH11-Nter and R9AP-Cter peptides is very similar and very small, Fig. S5B) become less ordered in the presence of a DOPE monolayer. Indeed, lipid monolayer binding by these peptides resulted in the formation of a β-sheet structural component in addition to α-helices (Figs. 10B and D, respectively). Only one type of phospholipid has been assayed within the scope of this review. DOPE forms a fluid phospholipid monolayer in the liquid-expanded state. Among the four peptides assayed, only the RDH11-Nter and R9AP-Cter peptides contain a long α-helical segment. One could thus postulate that peptides containing such α-helical segments could suffer from binding lipids in a fluid physical state as for the DOPE monolayer. Measurements of PM-IRRAS spectra in the presence of phospholipids in the solid-condensed state, such as DSPC (1,2-distearoyl-sn-glycero-3-phospho-choline), could allow to determine if this physical state could permit to retain the secondary structure of these peptides. In addition, in order to get a clear idea of the importance of lipids on the anchoring of proteins bearing a transmembrane segment to membranes, measurements must be performed using a wide range of different lipids found in the membrane where the protein is located. Also, measurements with charged lipids could allow to find out the peptide preference for the inner leaflet of the membrane, whereas measurements with mixtures of saturated phospholipids, sphingomyelin and cholesterol could allow to figure out whether it is located in rafts. Additional measurements using the protein with its transmembrane hydrophobic segment would then be necessary to confirm the conclusions drawn with the peptides. Moreover, we have previously shown that MIP measurements could allow to determine the preference of proteins for saturated or unsaturated phospholipids [205]. However, it is most useful to find out whether a relationship can be found between the values of MIP and the peptide secondary structure observed in monolayers. Similar values of MIP have been obtained with the RDH8-Cter, RDH11-Nter and LRAT-Cter peptides whereas a significantly lower value of MIP has been obtained with the R9AP-Cter peptide (Fig. 7B). In contrast, the R9AP-Cter peptide is the most highly ordered one with the thinnest amide I bandwidth in the presence of a DOPE monolayer. Nevertheless, the MIP of this peptide is larger than the postulated lateral pressure of membranes [206–213]. Measurements with additional lipids in different physical states could allow to improve our understanding of the relationship between MIP values and peptide secondary structure. An orientation close to 50° with respect to the normal of the monolayer has been estimated for the RDH11-Nter and R9AP-Cter peptides (in the absence of a DOPE monolayer) as well of the LRAT-Cter peptide (in the presence of a DOPE monolayer) (section 5.2). It has been previously argued that the tilt of a single transmembrane α-helix depends on the hydrophobic length of the peptide: a greater length of peptide would result in a greater angle of the helix axis with respect to the bilayer normal [350]. Similarly, the tilt of a transmembrane peptide with respect to the normal of the bilayer was shown to decrease with the length of the fatty acyl chains of phospholipids [197]. The thinner is the bilayer, the larger is the tilt angle of the transmembrane α-helical hydrophobic peptide. For example, a tilt angle of 51° has been observed for a bilayer made of phospholipids with very short fatty acyl chains (C10:0) [197], which is very close to the value of 50° observed for the RDH11-Nter, LRAT-Cter and R9AP-Cter peptides in monolayers. A survey of 45 transmembrane helices has shown that their helices are tilted with respect to the bilayer normal up to 35° [351]. Likewise, the longer transmembrane α-helices of rhodopsin are tilted up to 33° with respect to the normal of the bilayer plane presumably because of the hydrophobic mismatch between their length and the thickness of the bilayer [349]. The orientation of transmembrane α-helical hydrophobic peptides has been shown to depend on hydrophobic mismatch in lipid bilayers [197–200]. As previously postulated [58] and discussed in section 4.1, the orientation of the RDH11-Nter, LRAT-Cter and R9AP-Cter peptides could result from differences between the thickness of the lipid monolayer and the length of these peptides.
6.4 Peptides showing properties typical of hydrophobic transmembrane segments
It is useful to summarize the data to figure out which peptides reviewed in this paper most likely allow membrane anchoring of proteins. Transmembrane hydrophobic peptides typically have an α-helical structure [346]. The RDH8-Cter peptide contains the smallest percentage of hydrophobic amino acids and of residues having the tendency to be found in a transmembrane helix (Table 1). It has been classified in the surface seeking category with the Eisenberg plot (Fig. S2) and its structure prediction and model included only a small α-helical segment (Figs. 1 and 2B). Its structure in solution includes several different components (Fig. 2B). In addition, the bandwidth of its amide I band in monolayer is by far the largest compared to the other peptides (Figs. 10A and S5A), thereby suggesting that it includes a fair amount of disordered structures together with α-helical structures. Therefore, it is unlikely that this peptide could serve by itself to anchor the RDH8 protein. It could however allow membrane anchoring of RDH8 through acylation of one or more of the three C-terminal cysteines of this peptide, as previously postulated [44]. The RDH11-Nter, LRAT-Cter and R9AP-Cter peptides contain a similar percentage of hydrophobic amino acids (Table 1) and they are all found in the transmembrane category with the Eisenberg plot (Fig. S2). The RDH11-Nter peptide was predicted to contain mainly α-helical structural elements (Figs. 1 and 2D). This was quite consistent with the infrared and circular dichroism spectra in solution (Figs. 2C and D), although the length of these α-helices is most likely reduced in methanol (see section 3.2). This α-helical structure is maintained in monolayers but a β-sheet component is appearing in the presence of a fluid phospholipid monolayer (Fig. 10B). This peptide thus behaves as expected for a transmembrane peptide and could therefore allow membrane anchoring of the RDH11 protein. The LRAT-Cter peptide was predicted to contain a partial α-helical structure (Figs. 1 and 2F). However, it contains the largest number of residues having the tendency to be found in a transmembrane helix (93%, Table 1). Among the four studied peptides, the secondary structure of the LRAT-Cter peptide is the only one which is highly solvent- (Fig. 2F) and lipid-dependent (Fig. 10C). It is also the only one that might be influenced by the propensity of HFIP to favor the formation of α-helical structure. Nevertheless, a large β-sheet component is observed in monolayers when injected into the subphase using HFIP whereas only an α-helical structure is observed in the presence of a phospholipid monolayer (Fig. 10C). Altogether, these data suggest, as previously proposed [58], that this peptide is showing properties typical of transmembrane peptides and that it could allow anchoring of LRAT to the membrane. Finally, the R9AP-Cter peptide contains the largest number of hydrophobic residues (Table 1). It is also the only one which is predicted to include a long α-helix (Figs. 1 and 2H). This is consistent with the observation of the largest value of ellipticity and the smallest amide I bandwidth for this peptide both in solution and in monolayers compared to the other ones (Figs. 2G-H, 3, and S5A). Therefore, this peptide also behaves as a transmembrane peptide and it could thus be involved in the membrane anchoring of R9AP.
Supplementary Material
Acknowledgments
The work described here was supported by the Canadian Institutes of Health Research (CIHR). S. Bernier and S. Bussières are the recipients of the Canada Frederick Banting and Charles Best Graduate Scholarship from the CIHR. S. Bussières also held a travel fellowship from the Réseau FRQS de recherche en santé de la vision (RRSV) and S. Bernier was awarded scholarships from the FRQS, PROTEO and the RRSV. M. Lhor was holding a graduate scholarship from PROTEO. The authors thank Mario Méthot for his help in the measurement of the infrared spectrum of the powder of RDH11-Nter peptide.
Abbreviations
- DMF
dimethyl formamide
- DMSO
dimethyl sulfoxide
- DOPE
1,2-dioleoyl-sn-glycero-3-phospho-ethanolamine
- DSPC
1,2-distearoyl-sn-glycero-3-phospho-choline
- Gα
α subunit of transducin
- GMAP-210
Golgi microtubule associated-protein 210
- HFIP
hexafluoroisopropanol
- H-MeOH
peptide in methanol after solubilisation and evaporation of HFIP
- LRAT
Lecithin retinol acyltransferase
- MeOH
methanol
- MIP
maximum insertion pressure
- PM-IRRAS
polarization modulation infrared reflection absorption spectroscopy
- RDH
retinol dehydrogenase
- RGS
regulator G-protein signaling
- RPE
retinal pigment epithelium
- R9AP
RGS9-1-anchor protein
- SDR
short chain dehydrogenase/reductase
- TA
tail-anchored
- TFE
trifluoroethanol
- TM helix
transmembrane helix
References
- 1.Yeagle PL, Albert AD. Biochim Biophys Acta. 2007;1768:808. doi: 10.1016/j.bbamem.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Chini B, Parenti M, Poyner DR, Wheatley M. Biochem Soc Trans. 2013;41:135. doi: 10.1042/BST20120344. [DOI] [PubMed] [Google Scholar]
- 3.Katritch V, Cherezov V, Stevens RC. Annu Rev Pharmacol Toxicol. 2013;53:531. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audet M, Bouvier M. Cell. 2012;151:14. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, et al. Trends Biochem Sci. 2009;34:540. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Prog Retin Eye Res. 2013;36:24. doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallin E, von Heijne G. Protein Sci. 1998;7:1029. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagerberg L, Jonasson K, von Heijne G, Uhlen M, Berglund L. Proteomics. 2010;10:1141. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 9.Liden M, Eriksson U. J Biol Chem. 2006;281:13001. doi: 10.1074/jbc.R500027200. [DOI] [PubMed] [Google Scholar]
- 10.Liden M, Tryggvason K, Eriksson U. Mol Aspects Med. 2003;24:403. doi: 10.1016/s0098-2997(03)00036-0. [DOI] [PubMed] [Google Scholar]
- 11.Orgel JP. Curr Protein Pept Sci. 2006;7:553. doi: 10.2174/138920306779025666. [DOI] [PubMed] [Google Scholar]
- 12.Magee T, Seabra MC. Curr Opin Cell Biol. 2005;17:190. doi: 10.1016/j.ceb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Resh MD. Biochim Biophys Acta. 1999;1451:1. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 14.Resh MD. Subcell Biochem. 2004;37:217. doi: 10.1007/978-1-4757-5806-1_6. [DOI] [PubMed] [Google Scholar]
- 15.Devlin TM. Textbook of biochemistry with clinical correlations. 3. New York: Wiley-Liss; 1992. [Google Scholar]
- 16.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, et al. Biochemistry. 1995;34:6003. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 17.Belyaeva OV, Kedishvili NY. Genomics. 2006;88:820. doi: 10.1016/j.ygeno.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Borgese N, Fasana E. Biochim Biophys Acta. 2011;1808:937. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Borgese N, Brambillasca S, Colombo S. Curr Opin Cell Biol. 2007;19:368. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Borgese N, Brambillasca S, Soffientini P, Yabal M, Makarow M. Biochem Soc Trans. 2003;31:1238. doi: 10.1042/bst0311238. [DOI] [PubMed] [Google Scholar]
- 21.Borgese N, Colombo S, Pedrazzini E. J Cell Biol. 2003;161:1013. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutay U, Hartmann E, Rapoport TA. Trends Cell Biol. 1993;3:72. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 23.High S, Abell BM. Biochem Soc Trans. 2004;32:659. doi: 10.1042/BST0320659. [DOI] [PubMed] [Google Scholar]
- 24.Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, et al. EMBO J. 2005;24:2533. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegde RS, Keenan RJ. Nat Rev Mol Cell Biol. 2011;12:787. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ. Nature. 2011;477:61. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wattenberg B, Lithgow T. Traffic. 2001;2:66. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- 28.Kalbfleisch T, Cambon A, Wattenberg BW. Traffic. 2007;8:1687. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- 29.Monne M, von Heijne G. FEBS Lett. 2001;496:96. doi: 10.1016/s0014-5793(01)02415-2. [DOI] [PubMed] [Google Scholar]
- 30.Krishnakumar SS, London E. J Mol Biol. 2007;374:671. doi: 10.1016/j.jmb.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.London E, Shahidullah K. Curr Opin Struct Biol. 2009;19:464. doi: 10.1016/j.sbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Senes A, Gerstein M, Engelman DM. J Mol Biol. 2000;296:921. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 33.Labrie F, Luu-The V, Lin SX, Labrie C, Simard J, Breton R, et al. Steroids. 1997;62:148. doi: 10.1016/s0039-128x(96)00174-2. [DOI] [PubMed] [Google Scholar]
- 34.Kiser PD, Golczak M, Maeda A, Palczewski K. Biochim Biophys Acta. 2012;1821:137. doi: 10.1016/j.bbalip.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saari JC. Invest Ophthalmol Vis Sci. 2000;41:337. [PubMed] [Google Scholar]
- 36.Saari JC. Annu Rev Nutr. 2012;32:125. doi: 10.1146/annurev-nutr-071811-150748. [DOI] [PubMed] [Google Scholar]
- 37.Lamb TD, Pugh EN., Jr Prog Retin Eye Res. 2004;23:307. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 38.von Lintig J, Kiser PD, Golczak M, Palczewski K. Trends Biochem Sci. 2010;35:400. doi: 10.1016/j.tibs.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Prog Retin Eye Res. 2001;20:469. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 40.Parker RO, Crouch RK. Exp Eye Res. 2010;91:788. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rattner A, Smallwood PM, Nathans J. J Biol Chem. 2000;275:11034. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- 42.Maeda A, Maeda T, Imanishi Y, Kuksa V, Alekseev A, Bronson JD, et al. J Biol Chem. 2005;280:18822. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Thompson DA, Koutalos Y. J Biol Chem. 2012;287:24662. doi: 10.1074/jbc.M112.354514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo W, Marsh-Armstrong N, Rattner A, Nathans J. J Neurosci. 2004;24:2623. doi: 10.1523/JNEUROSCI.5302-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin B, White JT, Ferguson C, Wang S, Vessella R, Bumgarner R, et al. Cancer Res. 2001;61:1611. [PubMed] [Google Scholar]
- 46.Kasus-Jacobi A, Ou J, Birch DG, Locke KG, Shelton JM, Richardson JA, et al. J Biol Chem. 2005;280:20413. doi: 10.1074/jbc.M413789200. [DOI] [PubMed] [Google Scholar]
- 47.Haeseleer F, Jang GF, Imanishi Y, Driessen CA, Matsumura M, Nelson PS, et al. J Biol Chem. 2002;277:45537. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim TS, Maeda A, Maeda T, Heinlein C, Kedishvili N, Palczewski K, et al. J Biol Chem. 2005;280:8694. doi: 10.1074/jbc.M413172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kedishvili NY, Chumakova OV, Chetyrkin SV, Belyaeva OV, Lapshina EA, Lin DW, et al. J Biol Chem. 2002;277:28909. doi: 10.1074/jbc.M202588200. [DOI] [PubMed] [Google Scholar]
- 50.Belyaeva OV, Stetsenko AV, Nelson P, Kedishvili NY. Biochemistry. 2003;42:14838. doi: 10.1021/bi035288u. [DOI] [PubMed] [Google Scholar]
- 51.MacDonald PN, Ong DE. J Biol Chem. 1988;263:12478. [PubMed] [Google Scholar]
- 52.Saari JC, Bredberg DL. J Biol Chem. 1989;264:8636. [PubMed] [Google Scholar]
- 53.Schmitt MC, Ong DE. Biol Reprod. 1993;49:972. doi: 10.1095/biolreprod49.5.972. [DOI] [PubMed] [Google Scholar]
- 54.Barry RJ, Canada FJ, Rando RR. J Biol Chem. 1989;264:9231. [PubMed] [Google Scholar]
- 55.Ruiz A, Winston A, Lim YH, Gilbert BA, Rando RR, Bok D. J Biol Chem. 1999;274:3834. doi: 10.1074/jbc.274.6.3834. [DOI] [PubMed] [Google Scholar]
- 56.Bok D, Ruiz A, Yaron O, Jahng WJ, Ray A, Xue L, et al. Biochemistry. 2003;42:6090. doi: 10.1021/bi0342416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moise AR, Golczak M, Imanishi Y, Palczewski K. J Biol Chem. 2007;282:2081. doi: 10.1074/jbc.M608315200. [DOI] [PubMed] [Google Scholar]
- 58.Bussieres S, Cantin L, Desbat B, Salesse C. Langmuir. 2012;28:3516. doi: 10.1021/la203896n. [DOI] [PubMed] [Google Scholar]
- 59.He W, Cowan CW, Wensel TG. Neuron. 1998;20:95. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 60.Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. Nature. 2000;403:557. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 61.Hu G, Wensel TG. Proc Natl Acad Sci USA. 2002;99:9755. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gospe SM, 3rd, Baker SA, Kessler C, Brucato MF, Winter JR, Burns ME, et al. J Neurosci. 2011;31:14660. doi: 10.1523/JNEUROSCI.3516-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tryggvason K, Romert A, Eriksson U. J Biol Chem. 2001;276:19253. doi: 10.1074/jbc.M100215200. [DOI] [PubMed] [Google Scholar]
- 64.Hu G, Zhang Z, Wensel TG. J Biol Chem. 2003;278:14550. doi: 10.1074/jbc.M212046200. [DOI] [PubMed] [Google Scholar]
- 65.Martemyanov KA, Lishko PV, Calero N, Keresztes G, Sokolov M, Strissel KJ, et al. J Neurosci. 2003;23:10175. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gautier R, Douguet D, Antonny B, Drin G. Bioinformatics. 2008;24:2101. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 67.Drin G, Antonny B. FEBS Lett. 2010;584:1840. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 68.Eisenberg D, Weiss RM, Terwilliger TC. Proc Natl Acad Sci USA. 1984;81:140. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 70.Eisenberg D, Weiss RM, Terwilliger TC. Nature. 1982;299:371. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 71.Keller RC. Int J Mol Sci. 2011;12:5577. doi: 10.3390/ijms12095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fauchere JL, Pliska V. Eur J Med Chem. 1983;18:369. [Google Scholar]
- 73.Roy A, Kucukural A, Zhang Y. Nat Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y. Proteins. 2009;77(Suppl 9):100. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.http://zhanglab.ccmb.med.umich.edu/I-TASSER/.
- 76.https://http://www.predictprotein.org.
- 77.http://scratch.proteomics.ics.uci.edu.
- 78.http://bioinf.cs.ucl.ac.uk/psipred/.
- 79.Whitmore L, Wallace BA. Biopolymers. 2008;89:392. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 80.Barth A. Biochim Biophys Acta. 2007;1767:1073. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Goormaghtigh E, Cabiaux V, Ruysschaert JM. Subcell Biochem. 1994;23:329. doi: 10.1007/978-1-4615-1863-1_8. [DOI] [PubMed] [Google Scholar]
- 82.Goormaghtigh E, Cabiaux V, Ruysschaert JM. Subcell Biochem. 1994;23:405. doi: 10.1007/978-1-4615-1863-1_10. [DOI] [PubMed] [Google Scholar]
- 83.Clarke DT. Methods Mol Biol. 2011;752:59. doi: 10.1007/978-1-60327-223-0_5. [DOI] [PubMed] [Google Scholar]
- 84.Ranjbar B, Gill P. Chem Biol Drug Des. 2009;74:101. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 85.McPhie P. Anal Biochem. 2008;375:379. doi: 10.1016/j.ab.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelly SM, Jess TJ, Price NC. Biochim Biophys Acta. 2005;1751:119. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 87.Pancoska P, Bitto E, Janota V, Urbanova M, Gupta VP, Keiderling TA. Protein Sci. 1995;4:1384. doi: 10.1002/pro.5560040713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamburro AM, Scatturin A, Rocchi R, Marchiori F, Borin G, Scoffone E. FEBS Lett. 1968;1:298. doi: 10.1016/0014-5793(68)80137-1. [DOI] [PubMed] [Google Scholar]
- 89.Buck M, Radford SE, Dobson CM. Biochemistry. 1993;32:669. doi: 10.1021/bi00053a036. [DOI] [PubMed] [Google Scholar]
- 90.Chiti F, Taddei N, Webster P, Hamada D, Fiaschi T, Ramponi G, et al. Nat Struct Biol. 1999;6:380. doi: 10.1038/7616. [DOI] [PubMed] [Google Scholar]
- 91.Dong-Pyo Hong MH, Kuboi Ryoichi, Goto Yuji. J Amer Chem Soc. 1999;121:8427. [Google Scholar]
- 92.Dyson HJ, Rance M, Houghten RA, Wright PE, Lerner RA. J Mol Biol. 1988;201:201. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 93.Gast K, Zirwer D, Muller-Frohne M, Damaschun G. Protein Sci. 1999;8:625. doi: 10.1110/ps.8.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ha SW, Asakura T, Kishore R. Biomacromolecules. 2006;7:18. doi: 10.1021/bm050783m. [DOI] [PubMed] [Google Scholar]
- 95.Lehrman SR, Tuls JL, Lund M. Biochemistry. 1990;29:5590. doi: 10.1021/bi00475a025. [DOI] [PubMed] [Google Scholar]
- 96.Luo P, Baldwin RL. Biochemistry. 1997;36:8413. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 97.Nelson JW, Kallenbach NR. Proteins. 1986;1:211. doi: 10.1002/prot.340010303. [DOI] [PubMed] [Google Scholar]
- 98.Reiersen H, Rees AR. Protein Eng. 2000;13:739. doi: 10.1093/protein/13.11.739. [DOI] [PubMed] [Google Scholar]
- 99.Segawa S, Fukuno T, Fujiwara K, Noda Y. Biopolymers. 1991;31:497. doi: 10.1002/bip.360310505. [DOI] [PubMed] [Google Scholar]
- 100.Shiraki K, Nishikawa K, Goto Y. J Mol Biol. 1995;245:180. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- 101.Sonnichsen FD, Van Eyk JE, Hodges RS, Sykes BD. Biochemistry. 1992;31:8790. doi: 10.1021/bi00152a015. [DOI] [PubMed] [Google Scholar]
- 102.Storrs RW, Truckses D, Wemmer DE. Biopolymers. 1992;32:1695. doi: 10.1002/bip.360321211. [DOI] [PubMed] [Google Scholar]
- 103.Xue R, Wang S, Wang C, Zhu T, Li F, Sun H. Biopolymers. 2006;84:329. doi: 10.1002/bip.20478. [DOI] [PubMed] [Google Scholar]
- 104.Chatterjee C, Gerig JT. Biopolymers. 2007;87:115. doi: 10.1002/bip.20796. [DOI] [PubMed] [Google Scholar]
- 105.Maltseva E, Brezesinski G. Chemphyschem. 2004;5:1185. doi: 10.1002/cphc.200400045. [DOI] [PubMed] [Google Scholar]
- 106.http://lamar.colostate.edu/~sreeram/CDPro/CDPro.htm.
- 107.Goormaghtigh E, Cabiaux V, Ruysschaert JM. Subcell Biochem. 1994;23:363. doi: 10.1007/978-1-4615-1863-1_9. [DOI] [PubMed] [Google Scholar]
- 108.Vigano C, Manciu L, Buyse F, Goormaghtigh E, Ruysschaert JM. Biopolymers. 2000;55:373. doi: 10.1002/1097-0282(2000)55:5<373::AID-BIP1011>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 109.Goormaghtigh E, Gasper R, Benard A, Goldsztein A, Raussens V. Biochim Biophys Acta. 2009;1794:1332. doi: 10.1016/j.bbapap.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 110.Haris PI, Chapman D. Biopolymers. 1995;37:251. doi: 10.1002/bip.360370404. [DOI] [PubMed] [Google Scholar]
- 111.Surewicz WK, Mantsch HH, Chapman D. Biochemistry. 1993;32:389. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- 112.Jackson M, Mantsch HH. Crit Rev Biochem Mol Biol. 1995;30:95. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 113.Nevskaya NA, Chirgadze YN. Biopolymers. 1976;15:637. doi: 10.1002/bip.1976.360150404. [DOI] [PubMed] [Google Scholar]
- 114.Möhwald H. In: Handbook of Biological Physics. 1. Lipowsky R, Sackmann E, editors. North-Holland; 1995. [Google Scholar]
- 115.Kaganer VM, Mohwald H, Dutta P. Rev Mod Phys. 1999;71 [Google Scholar]
- 116.Calvez P, Bussieres S, Demers E, Salesse C. Biochimie. 2009;91:718. doi: 10.1016/j.biochi.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 117.Giner-Casares JJ, Brezesinski G, Möhwald H. Current Opinion in Colloid & Interface Science. 2013 In press. [Google Scholar]
- 118.Moghaddam B, Ali MH, Wilkhu J, Kirby DJ, Mohammed AR, Zheng Q, et al. Int J Pharm. 2011;417:235. doi: 10.1016/j.ijpharm.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 119.Vollhardt D, Fainerman VB. Adv Colloid Interface Sci. 2000;86:103. doi: 10.1016/s0001-8686(00)00034-8. [DOI] [PubMed] [Google Scholar]
- 120.Boucher J, Trudel E, Methot M, Desmeules P, Salesse C. Colloids Surf B: Biointerfaces. 2007;58:73. doi: 10.1016/j.colsurfb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 121.Brezesinski G, Möhwald H. Adv Colloid Interface Sci. 2003;100–102:563. doi: 10.1016/s0001-8686(02)00071-4. [DOI] [PubMed] [Google Scholar]
- 122.Brockman H. Curr Opin Struct Biol. 1999;9:438. doi: 10.1016/S0959-440X(99)80061-X. [DOI] [PubMed] [Google Scholar]
- 123.Maget-Dana R. Biochim Biophys Acta. 1999;1462:109. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]
- 124.Bussières SBJ, Desmeules D, Grandbois M, Desbat B, Salesse C. In: Structure & Dynamics of Membranous Interfaces. Nag K, editor. Chapter 7 Hoboken: John Wiley & Sons; 2008. [Google Scholar]
- 125.Thakur G, Micic M, Leblanc RM. Colloids Surf B: Biointerfaces. 2009;74:436. doi: 10.1016/j.colsurfb.2009.07.043. [DOI] [PubMed] [Google Scholar]
- 126.Feng SS. Langmuir. 1999;15:998. [Google Scholar]
- 127.MacDonald RC, Simon SA. Proc Natl Acad Sci U S A. 1987;84:4089. doi: 10.1073/pnas.84.12.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gallant J, Lavoie H, Tessier A, Munger G, Leblanc RM, Salesse C. Langmuir. 1998;14:3954. [Google Scholar]
- 129.Sergeeva TI, Zaitsev SY, Tsarkova MS, Gromov SP, Vedernikov AI, Kapichnikova MS, et al. J Colloid Interface Sci. 2003;265:77. doi: 10.1016/s0021-9797(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 130.Wang S, Vidon S, Leblanc RM. J Colloid Interface Sci. 1998;207:303. doi: 10.1006/jcis.1998.5803. [DOI] [PubMed] [Google Scholar]
- 131.Dutta AK, Lavoie H, Ohta K, Salesse C. Langmuir. 1997;13:801. [Google Scholar]
- 132.Dutta AK, Salesse C. Langmuir. 1997;13:5401. [Google Scholar]
- 133.Tsukanova V, Grainger DW, Salesse C. Langmuir. 2002;18:5539. [Google Scholar]
- 134.Tsukanova V, Lavoie H, Harata A, Ogawa T, Salesse C. J Phys Chem B. 2002;106:4203. [Google Scholar]
- 135.Orbulescu J, Mello SV, Huo Q, Sui GD, Kele P, Leblanc RM. Langmuir. 2001;17:1525. [Google Scholar]
- 136.Itoh T, Tsujii Y, Fukuda T, Miyamoto T, Ito S, Asada T, et al. Langmuir. 1991;7:2803. [Google Scholar]
- 137.Blaudez D, Buffeteau T, Cornut JC, Desbat B, Escafre N, Pezolet M, et al. Thin Solid Films. 1994;242:146. [Google Scholar]
- 138.Gericke A, Michailov AV, Hühnerfuss H. Vib Spectrosc. 1993;4:335. [Google Scholar]
- 139.Hunt RD, Mitchell ML, Dluhy RA. J Mol Struct. 1989;214:93. [Google Scholar]
- 140.Flach CR, Brauner JW, Mendelsohn R. Biophys J. 1993;65:1994. doi: 10.1016/S0006-3495(93)81276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dluhy RA, Reilly KE, Hunt RD, Mitchell ML, Mautone AJ, Mendelsohn R. Biophys J. 1989;56:1173. doi: 10.1016/S0006-3495(89)82764-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dluhy RA, Wright NA, Griffiths PR. Appl Spectrosc. 1988;42:138. [Google Scholar]
- 143.Dluhy RA, Cornell DG. J Phys Chem. 1985;89:3195. [Google Scholar]
- 144.Mendelsohn R, Mao G, Flach CR. Biochim Biophys Acta. 2010;1798:788. doi: 10.1016/j.bbamem.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blaudez D, Buffeteau T, Desbat B, Turlet JM. Curr Opin Colloid Interface Sci. 1999;4:265. [Google Scholar]
- 146.Blume A, Kerth A. Biochim Biophys Acta. 2013;1828:2294. doi: 10.1016/j.bbamem.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 147.Lavoie H, Gallant J, Grandbois M, Blaudez D, Desbat B, Boucher F, et al. Mater Sci Eng C. 1999;10:147. [Google Scholar]
- 148.Sung W, Kim D, Shen YR. Curr Appl Phys. 2013;13:619. [Google Scholar]
- 149.Mendelsohn RaCRF. In: Handbook of Vibrational Spectroscopy. 2. Griffiths JMCaPR., editor. John Wiley & Sons; 2002. [Google Scholar]
- 150.Dluhy RA, Mendelsohn R. Analytical chemistry. 1988;60:269A. doi: 10.1021/ac00155a001. [DOI] [PubMed] [Google Scholar]
- 151.Blaudez D, Turlet J-M, Dufourcq J, Bard D, Buffeteau T, Desbat B. J Chem Soc Faraday Trans. 1996;92:525. [Google Scholar]
- 152.Blaudez DCS, Desbat B. In: Biointerface characterization by advanced IR spectroscopy. Pradier CMCY, editor. Chapter 2 Amsterdam: Academic Press; 2011. [Google Scholar]
- 153.Hénon S, Meunier J. Rev Sci Instrum. 1991;62:936. [Google Scholar]
- 154.Hönig D, Möbius D. J Phys Chem. 1991;95:4590. [Google Scholar]
- 155.Peters R, Beck K. Proc Natl Acad Sci USA. 1983;80:7183. doi: 10.1073/pnas.80.23.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lösche MSE, Möhwald H. Ber Busenges Phys Chem. 1983;87:848. [Google Scholar]
- 157.McConnell HM, Tamm LK, Weis RM. Proc Natl Acad Sci USA. 1984;81:3249. doi: 10.1073/pnas.81.10.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weis RM. Chem Phys Lipids. 1991;57:227. doi: 10.1016/0009-3084(91)90078-p. [DOI] [PubMed] [Google Scholar]
- 159.Stottrup BL, Nguyen AH, Tuzel E. Biochim Biophys Acta. 2010;1798:1289. doi: 10.1016/j.bbamem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 160.Fanani ML, Hartel S, Maggio B, De Tullio L, Jara J, Olmos F, et al. Biochim Biophys Acta. 2010;1798:1309. doi: 10.1016/j.bbamem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 161.Knobler CM. Science. 1990;249:870. doi: 10.1126/science.249.4971.870. [DOI] [PubMed] [Google Scholar]
- 162.Vollhardt D, Fainerman VB. Adv Colloid Interface Sci. 2010;154:1. doi: 10.1016/j.cis.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 163.Vollhardt D, Fainerman VB. Adv Colloid Interface Sci. 2006;127:83. doi: 10.1016/j.cis.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 164.Roldan-Carmona C, Giner-Casares JJ, Perez-Morales M, Martin-Romero MT, Camacho L. Adv Colloid Interface Sci. 2012;173:12. doi: 10.1016/j.cis.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 165.Vaknin D, Kjaer K, Als-Nielsen J, Lösche M. Biophys J. 1991;59:1325. doi: 10.1016/S0006-3495(91)82347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Möhwald H. Thin Solid Films. 1988;159:1. [Google Scholar]
- 167.Grundy MJ, Richardson RM, Roser SJ, Penfold J, Ward RC. Thin Solid Films. 1988;159:43. [Google Scholar]
- 168.Heinz TF, Chen CK, Ricard D, Shen YR. Chem Phys Lett. 1981;83:180. [Google Scholar]
- 169.Ducharme D, Salesse C, Leblanc RM. Thin Solid Films. 1985;132:83. [Google Scholar]
- 170.Langevin D, Monroy F. Curr Opin Colloid Interface Sci. 2010;15:283. [Google Scholar]
- 171.Vollhardt D. Mater Sci Eng C. 2002;22:121. [Google Scholar]
- 172.Sriram I, Schwartz DK. Surf Sci Rep. 2012;67:143. [Google Scholar]
- 173.Sanyal MK. In: Encyclopedia of Condensed Matter Physics. Franco B, Gerald LL, Peter W, editors. Oxford: Elsevier; 2005. [Google Scholar]
- 174.Dynarowicz-Latka P, Dhanabalan A, Oliveira ON., Jr Adv Colloid Interface Sci. 2001;91:221. doi: 10.1016/s0001-8686(99)00034-2. [DOI] [PubMed] [Google Scholar]
- 175.Zhao H, Lappalainen P. Mol Biol Cell. 2012;23:2823. doi: 10.1091/mbc.E11-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ambroggio EE, Villarreal MA, Montich GG, Rijkers DT, De Planque MR, Separovic F, et al. Biophys Chem. 2006;121:171. doi: 10.1016/j.bpc.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 177.Ambroggio EE, Fidelio GD. Biochim Biophys Acta. 2013;1828:708. doi: 10.1016/j.bbamem.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 178.Broniatowski M, Obidowicz K, Vila Romeu N, Broniatowska E, Dynarowicz-Latka P. J Colloid Interface Sci. 2007;313:600. doi: 10.1016/j.jcis.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 179.Broniatowski M, Suarez MN, Romeu NV, Dynarowicz-Latka P. J Phys Chem B. 2006;110:19450. doi: 10.1021/jp0623138. [DOI] [PubMed] [Google Scholar]
- 180.Diociaiuti M, Bordi F, Motta A, Carosi A, Molinari A, Arancia G, et al. Biophys J. 2002;82:3198. doi: 10.1016/S0006-3495(02)75662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lewis RN, Prenner EJ, Kondejewski LH, Flach CR, Mendelsohn R, Hodges RS, et al. Biochemistry. 1999;38:15193. doi: 10.1021/bi9912342. [DOI] [PubMed] [Google Scholar]
- 182.Tournois H, Gieles P, Demel R, de Gier J, de Kruijff B. Biophys J. 1989;55:557. doi: 10.1016/S0006-3495(89)82849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ulrich WP, Vogel H. Biophys J. 1999;76:1639. doi: 10.1016/S0006-3495(99)77323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Van Mau N, Trudelle Y, Daumas P, Heitz F. Biophys J. 1988;54:563. doi: 10.1016/S0006-3495(88)82990-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Weis M, Vanco M, Vitovic P, Hianik T, Cirak J. J Phys Chem B. 2006;110:26272. doi: 10.1021/jp064555d. [DOI] [PubMed] [Google Scholar]
- 186.Ducharme D, Vaknin D, Paudler M, Salesse C, Riegler H, Möhwald H. Thin Solid Films. 1996;284–285:90. [Google Scholar]
- 187.Lavoie H, Blaudez D, Vaknin D, Desbat B, Ocko BM, Salesse C. Biophys J. 2002;83:3558. doi: 10.1016/s0006-3495(02)75356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ye S, Strzalka J, Churbanova IY, Zheng S, Johansson JS, Blasie JK. Biophys J. 2004;87:4065. doi: 10.1529/biophysj.104.051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Churbanova IY, Tronin A, Strzalka J, Gog T, Kuzmenko I, Johansson JS, et al. Biophys J. 2006;90:3255. doi: 10.1529/biophysj.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gustafsson M, Vandenbussche G, Curstedt T, Ruysschaert JM, Johansson J. FEBS Lett. 1996;384:185. doi: 10.1016/0014-5793(96)00290-6. [DOI] [PubMed] [Google Scholar]
- 191.Nilsson G, Gustafsson M, Vandenbussche G, Veldhuizen E, Griffiths WJ, Sjovall J, et al. Eur J Biochem. 1998;255:116. doi: 10.1046/j.1432-1327.1998.2550116.x. [DOI] [PubMed] [Google Scholar]
- 192.Johnson JE, Rao NM, Hui SW, Cornell RB. Biochemistry. 1998;37:9509. doi: 10.1021/bi980340l. [DOI] [PubMed] [Google Scholar]
- 193.Zheng S, Strzalka J, Jones DH, Opella SJ, Blasie JK. Biophys J. 2003;84:2393. doi: 10.1016/S0006-3495(03)75045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zheng S, Strzalka J, Ma C, Opella SJ, Ocko BM, Blasie JK. Biophys J. 2001;80:1837. doi: 10.1016/S0006-3495(01)76154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sun F. J Mol Model. 2003;9:114. doi: 10.1007/s00894-003-0123-3. [DOI] [PubMed] [Google Scholar]
- 196.Yassine W, Taib N, Federman S, Milochau A, Castano S, Sbi W, et al. Biochim Biophys Acta. 2009;1788:1722. doi: 10.1016/j.bbamem.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 197.Park SH, Opella SJ. J Mol Biol. 2005;350:310. doi: 10.1016/j.jmb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 198.Kandasamy SK, Larson RG. Biophys J. 2006;90:2326. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Esteban-Martin S, Strandberg E, Fuertes G, Ulrich AS, Salgado J. Biophys J. 2009;96:3233. doi: 10.1016/j.bpj.2008.12.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Strandberg E, Esteban-Martin S, Salgado J, Ulrich AS. Biophys J. 2009;96:3223. doi: 10.1016/j.bpj.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Holt A, Killian JA. European biophysics journal : EBJ. 2010;39:609. doi: 10.1007/s00249-009-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Thomas A, Brasseur R. Curr Protein Pept Sci. 2006;7:523. doi: 10.2174/138920306779025594. [DOI] [PubMed] [Google Scholar]
- 203.Lins L, Decaffmeyer M, Thomas A, Brasseur R. Biochim Biophys Acta. 2008;1778:1537. doi: 10.1016/j.bbamem.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 204.Harris F, Daman A, Wallace J, Dennison SR, Phoenix DA. Curr Protein Pept Sci. 2006;7:529. doi: 10.2174/138920306779025602. [DOI] [PubMed] [Google Scholar]
- 205.Calvez P, Demers E, Boisselier E, Salesse C. Langmuir. 2011;27:1373. doi: 10.1021/la104097n. [DOI] [PubMed] [Google Scholar]
- 206.Demel RA, Geurts van Kessel WS, Zwaal RF, Roelofsen B, van Deenen LL. Biochim Biophys Acta. 1975;406:97. doi: 10.1016/0005-2736(75)90045-0. [DOI] [PubMed] [Google Scholar]
- 207.Blume A, Eibl H. Biochim Biophys Acta. 1979;558:13. doi: 10.1016/0005-2736(79)90311-0. [DOI] [PubMed] [Google Scholar]
- 208.Moreau H, Pieroni G, Jolivet-Reynaud C, Alouf JE, Verger R. Biochemistry. 1988;27:2319. doi: 10.1021/bi00407a012. [DOI] [PubMed] [Google Scholar]
- 209.Seelig A. Biochim Biophys Acta. 1987;899:196. doi: 10.1016/0005-2736(87)90400-7. [DOI] [PubMed] [Google Scholar]
- 210.Silvius JR. Biochim Biophys Acta. 2003;1610:174. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 211.Boguslavsky V, Rebecchi M, Morris AJ, Jhon DY, Rhee SG, McLaughlin S. Biochemistry. 1994;33:3032. doi: 10.1021/bi00176a036. [DOI] [PubMed] [Google Scholar]
- 212.Marsh D. Biochim Biophys Acta. 1996;1286:183. doi: 10.1016/s0304-4157(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 213.Salesse C, Ducharme D, Leblanc RM, Boucher F. Biochemistry. 1990;29:4567. doi: 10.1021/bi00471a010. [DOI] [PubMed] [Google Scholar]
- 214.Nasir MN, Besson Fβ. J Colloid Interface Sci. 2012;387:187. doi: 10.1016/j.jcis.2012.07.091. [DOI] [PubMed] [Google Scholar]
- 215.Brehmer T, Kerth A, Graubner W, Malesevic M, Hou B, Bruser T, et al. Langmuir. 2012;28:3534. doi: 10.1021/la204473t. [DOI] [PubMed] [Google Scholar]
- 216.Dietrich U, Kruger P, Kas JA. Chem Phys Lipids. 2011;164:266. doi: 10.1016/j.chemphyslip.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 217.Haro I, Gomara MJ, Galatola R, Domenech O, Prat J, Girona V, et al. Biochim Biophys Acta. 2011;1808:1567. doi: 10.1016/j.bbamem.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 218.Larios C, Minones J, Jr, Haro I, Alsina MA, Busquets MA, Trillo JM. J Phys Chem B. 2006;110:23292. doi: 10.1021/jp0628582. [DOI] [PubMed] [Google Scholar]
- 219.Neville F, Cahuzac M, Konovalov O, Ishitsuka Y, Lee KY, Kuzmenko I, et al. Biophys J. 2006;90:1275. doi: 10.1529/biophysj.105.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sanchez-Martin MJ, Haro I, Alsina MA, Busquets MA, Pujol M. J Phys Chem B. 2010;114:448. doi: 10.1021/jp906900k. [DOI] [PubMed] [Google Scholar]
- 221.Travkova OG, Brezesinski G. Chem Phys Lipids. 2013;167–168:43. doi: 10.1016/j.chemphyslip.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 222.Varas M, Sanchez-Borzone M, Sanchez JM, Barioglio SR, Perillo MA. J Phys Chem B. 2008;112:7330. doi: 10.1021/jp7111236. [DOI] [PubMed] [Google Scholar]
- 223.Ishitsuka Y, Pham DS, Waring AJ, Lehrer RI, Lee KY. Biochim Biophys Acta. 2006;1758:1450. doi: 10.1016/j.bbamem.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 224.Gidalevitz D, Ishitsuka Y, Muresan AS, Konovalov O, Waring AJ, Lehrer RI, et al. Proc Natl Acad Sci USA. 2003;100:6302. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Olak C, Muenter A, Andra J, Brezesinski G. J Pept Sci. 2008;14:510. doi: 10.1002/psc.954. [DOI] [PubMed] [Google Scholar]
- 226.Rocha S, Loureiro JA, Brezesinski G, Pereira MdC. Biochem Biophys Res Commun. 2012;420:136. doi: 10.1016/j.bbrc.2012.02.129. [DOI] [PubMed] [Google Scholar]
- 227.Protopapa E, Maude S, Aggeli A, Nelson A. Langmuir. 2009;25:3289. doi: 10.1021/la803368r. [DOI] [PubMed] [Google Scholar]
- 228.Lahdo R, Coillet-Matillon S, Chauvet JP, de La Fourniere-Bessueille L. Eur J Biochem. 2002;269:2238. doi: 10.1046/j.1432-1033.2002.02882.x. [DOI] [PubMed] [Google Scholar]
- 229.Lahdo R, De La Fourniere-Bessueille L. Biochem J. 2004;382:987. doi: 10.1042/BJ20040777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ji SR, Wu Y, Sui SF. Biochemistry (Mosc) 2002;67:1283. doi: 10.1023/a:1021361607611. [DOI] [PubMed] [Google Scholar]
- 231.Bringezu F, Majerowicz M, Maltseva E, Wen S, Brezesinski G, Waring AJ. Chembiochem. 2007;8:1038. doi: 10.1002/cbic.200600503. [DOI] [PubMed] [Google Scholar]
- 232.Engel MF, Yigittop H, Elgersma RC, Rijkers DT, Liskamp RM, de Kruijff B, et al. J Mol Biol. 2006;356:783. doi: 10.1016/j.jmb.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 233.Dyck M, Losche M. J Phys Chem B. 2006;110:22143. doi: 10.1021/jp056697y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sood R, Domanov Y, Pietiainen M, Kontinen VP, Kinnunen PK. Biochim Biophys Acta. 2008;1778:983. doi: 10.1016/j.bbamem.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 235.Ege C, Lee KY. Biophys J. 2004;87:1732. doi: 10.1529/biophysj.104.043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Eeman M, Berquand A, Dufrene YF, Paquot M, Dufour S, Deleu M. Langmuir. 2006;22:11337. doi: 10.1021/la061969p. [DOI] [PubMed] [Google Scholar]
- 237.Volkov V, Bonn M. J Phys Chem B. 2013 doi: 10.1021/jp405852r. [DOI] [PubMed] [Google Scholar]
- 238.Dennison SR, Harris F, Morton LH, Phoenix DA. FEMS Microbiol Lett. 2013 doi: 10.1111/1574-6968.12212. [DOI] [PubMed] [Google Scholar]
- 239.Fantini J, Yahi N, Garmy N. Front Physiol. 2013;4:120. doi: 10.3389/fphys.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Keszthelyi T, Hill K, Kiss E. J Phys Chem B. 2013;117:6969. doi: 10.1021/jp401533c. [DOI] [PubMed] [Google Scholar]
- 241.Arouri A, Kerth A, Dathe M, Blume A. Langmuir. 2011;14:14. doi: 10.1021/la104887s. [DOI] [PubMed] [Google Scholar]
- 242.Bellet-Amalric E, Blaudez D, Desbat B, Graner F, Gauthier F, Renault A. Biochim Biophys Acta. 2000;1467:131. doi: 10.1016/s0005-2736(00)00218-2. [DOI] [PubMed] [Google Scholar]
- 243.Castano S, Desbat B, Delfour A, Dumas JM, da Silva A, Dufourcq J. Biochim Biophys Acta. 2005;1668:87. doi: 10.1016/j.bbamem.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 244.Castano S, Desbat B, Laguerre M, Dufourcq J. Biochim Biophys Acta. 1999;1416:176. doi: 10.1016/s0005-2736(98)00220-x. [DOI] [PubMed] [Google Scholar]
- 245.Dennison SR, Phoenix DA. Biochemistry. 2011;50:10898. doi: 10.1021/bi201267v. [DOI] [PubMed] [Google Scholar]
- 246.Dennison SR, Phoenix DA. Biochemistry. 2011;50:1514. doi: 10.1021/bi101687t. [DOI] [PubMed] [Google Scholar]
- 247.Dietrich U, Kruger P, Gutberlet T, Kas JA. Biochim Biophys Acta. 2009;1788:1474. doi: 10.1016/j.bbamem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 248.Erbe A, Kerth A, Dathe M, Blume A. Chembiochem. 2009;10:2884. doi: 10.1002/cbic.200900444. [DOI] [PubMed] [Google Scholar]
- 249.Hill K, Penzes CB, Schnoller D, Horvati K, Bosze S, Hudecz F, et al. Physical chemistry chemical physics : PCCP. 2010;12:11498. doi: 10.1039/c002737e. [DOI] [PubMed] [Google Scholar]
- 250.Hinz A, Galla HJ. European biophysics journal : EBJ. 2005;34:285. doi: 10.1007/s00249-004-0450-z. [DOI] [PubMed] [Google Scholar]
- 251.Ionov R, El-Abed A, Angelova A, Goldmann M, Peretti P. Biophys J. 2000;78:3026. doi: 10.1016/S0006-3495(00)76841-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Levy M, Garmy N, Gazit E, Fantini J. The FEBS journal. 2006;273:5724. doi: 10.1111/j.1742-4658.2006.05562.x. [DOI] [PubMed] [Google Scholar]
- 253.Lin MS, Chen XB, Wang SS, Chang Y, Chen WY. Colloids Surf B: Biointerfaces. 2009;74:59. doi: 10.1016/j.colsurfb.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 254.Lourenzoni MR, Namba AM, Caseli L, Degreve L, Zaniquelli ME. J Phys Chem B. 2007;111:11318. doi: 10.1021/jp067127g. [DOI] [PubMed] [Google Scholar]
- 255.Neville F, Ishitsuka Y, Hodges CS, Konovalov O, Waring AJ, Lehrer R, et al. Soft matter. 2008;4:1665. doi: 10.1039/b718295c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Perez S, Minones J, Jr, Espina M, Alsina MA, Haro I, Mestres C. J Phys Chem B. 2005;109:19970. doi: 10.1021/jp0516240. [DOI] [PubMed] [Google Scholar]
- 257.Roes S, Seydel U, Gutsmann T. Langmuir. 2005;21:6970. doi: 10.1021/la048218c. [DOI] [PubMed] [Google Scholar]
- 258.Rzeznicka, Sovago M, Backus EH, Bonn M, Yamada T, Kobayashi T, et al. Langmuir. 2010;26:16055. doi: 10.1021/la1028965. [DOI] [PubMed] [Google Scholar]
- 259.Tsanova A, Jordanova A, Georgiev GA, Pajpanova T, Golovinsky E, Lalchev Z. Amino acids. 2012;42:253. doi: 10.1007/s00726-010-0803-0. [DOI] [PubMed] [Google Scholar]
- 260.Salay LC, Nobre TM, Colhone MC, Zaniquelli ME, Ciancaglini P, Stabeli RG, et al. J Pept Sci. 2011;17:700. doi: 10.1002/psc.1392. [DOI] [PubMed] [Google Scholar]
- 261.Volinsky R, Kolusheva S, Berman A, Jelinek R. Langmuir. 2004;20:11084. doi: 10.1021/la0477486. [DOI] [PubMed] [Google Scholar]
- 262.Zhang L, Rozek A, Hancock REW. J Biol Chem. 2001;276:35714. doi: 10.1074/jbc.M104925200. [DOI] [PubMed] [Google Scholar]
- 263.Zhang L, Scott MG, Yan H, Mayer LD, Hancock RE. Biochemistry. 2000;39:14504. doi: 10.1021/bi0011173. [DOI] [PubMed] [Google Scholar]
- 264.Sevcsik E, Pabst G, Richter W, Danner S, Amenitsch H, Lohner K. Biophys J. 2008;94:4688. doi: 10.1529/biophysj.107.123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wagner K, Van Mau N, Boichot S, Kajava AV, Krauss U, Le Grimellec C, et al. Biophys J. 2004;87:386. doi: 10.1529/biophysj.103.036921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Palgunachari MN, Mishra VK, Lund-Katz S, Phillips MC, Adeyeye SO, Alluri S, et al. Arteriosclerosis thrombosis and vascular biology. 1996;16:328. doi: 10.1161/01.atv.16.2.328. [DOI] [PubMed] [Google Scholar]
- 267.Nasir MN, Besson F. Biochim Biophys Acta. 2012;1818:1302. doi: 10.1016/j.bbamem.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 268.Dwyer MD, He L, James M, Nelson A, Middelberg AP. Journal of the Royal Society Interface / the Royal Society. 2013;10:20120987. doi: 10.1098/rsif.2012.0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hendrickson HS, Fan PC, Kaufman DK, Kleiner DE. Arch Biochem Biophys. 1983;227:242. doi: 10.1016/0003-9861(83)90367-3. [DOI] [PubMed] [Google Scholar]
- 270.Kerth A, Brehmer T, Meister A, Hanner P, Jakob M, Klosgen RB, et al. Chembiochem. 2012;13:231. doi: 10.1002/cbic.201100458. [DOI] [PubMed] [Google Scholar]
- 271.Mendez MA, Nazemi Z, Uyanik I, Lu Y, Girault HH. Langmuir. 2011;27:13918. doi: 10.1021/la202970g. [DOI] [PubMed] [Google Scholar]
- 272.Al-Kurdi R, Gulino-Debrac D, Martel L, Legrand JF, Renault A, Hewat E, et al. J Mol Biol. 2004;337:881. doi: 10.1016/j.jmb.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 273.Bucki R, Pastore JJ, Randhawa P, Vegners R, Weiner DJ, Janmey PA. Antimicrobial agents and chemotherapy. 2004;48:1526. doi: 10.1128/AAC.48.5.1526-1533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Datta G, Epand RF, Epand RM, Chaddha M, Kirksey MA, Garber DW, et al. J Biol Chem. 2004;279:26509. doi: 10.1074/jbc.M314276200. [DOI] [PubMed] [Google Scholar]
- 275.Isarankura Na Ayudhya C, Prachayasittikul V, Galla HJ. European biophysics journal : EBJ. 2004;33:522. doi: 10.1007/s00249-004-0393-4. [DOI] [PubMed] [Google Scholar]
- 276.Burger KN, Wharton SA, Demel RA, Verkleij AJ. Biochim Biophys Acta. 1991;1065:121. doi: 10.1016/0005-2736(91)90221-s. [DOI] [PubMed] [Google Scholar]
- 277.Demel RA, Peelen T, Siezen RJ, De Kruijff B, Kuipers OP. Eur J Biochem. 1996;235:267. doi: 10.1111/j.1432-1033.1996.00267.x. [DOI] [PubMed] [Google Scholar]
- 278.Tamm LK, Tomich JM, Saier MH., Jr J Biol Chem. 1989;264:2587. [PubMed] [Google Scholar]
- 279.Flach CR, Prendergast FG, Mendelsohn R. Biophys J. 1996;70:539. doi: 10.1016/S0006-3495(96)79600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Barzyk W, Campagna S, Wieclaw K, Korchowiec B, Rogalska E. Colloids Surf A: Physicochem Eng Aspects. 2009;343:104. [Google Scholar]
- 281.Brandenburg K, Harris F, Dennison S, Seydel U, Phoenix D. Eur J Biochem. 2002;269:5414. doi: 10.1046/j.1432-1033.2002.03225.x. [DOI] [PubMed] [Google Scholar]
- 282.Dennison SR, Dante S, Hauss T, Brandenburg K, Harris F, Phoenix DA. Biophys J. 2005;88:3008. doi: 10.1529/biophysj.104.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Dennison SR, Morton LH, Harris F, Phoenix DA. Chem Phys Lipids. 2008;151:92. doi: 10.1016/j.chemphyslip.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 284.Gutsmann T, Fix M, Larrick JW, Wiese A. The Journal of membrane biology. 2000;176:223. doi: 10.1007/s00232001092. [DOI] [PubMed] [Google Scholar]
- 285.Gutsmann T, Hagge SO, Larrick JW, Seydel U, Wiese A. Biophys J. 2001;80:2935. doi: 10.1016/S0006-3495(01)76259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Mura M, Dennison SR, Zvelindovsky AV, Phoenix DA. Biochim Biophys Acta. 2013;1828:586. doi: 10.1016/j.bbamem.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 287.Matar G, Besson F. Biochim Biophys Acta. 2011;1808:2534. doi: 10.1016/j.bbamem.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 288.Matar G, Nasir MN, Besson F. J Colloid Interface Sci. 2010;352:520. doi: 10.1016/j.jcis.2010.08.075. [DOI] [PubMed] [Google Scholar]
- 289.Sanchez-Martin MJ, Cruz A, Busquets MA, Haro I, Alsina MA, Pujol M. Int J Pharm. 2012;436:593. doi: 10.1016/j.ijpharm.2012.07.051. [DOI] [PubMed] [Google Scholar]
- 290.Seelig A, Alt T, Lotz S, Holzemann G. Biochemistry. 1996;35:4365. doi: 10.1021/bi952434q. [DOI] [PubMed] [Google Scholar]
- 291.Wang X, Han X, Yang F. Mol Cell Biochem. 2004;262:61. doi: 10.1023/b:mcbi.0000038216.13105.08. [DOI] [PubMed] [Google Scholar]
- 292.Lecorche P, Walrant A, Burlina F, Dutot L, Sagan S, Mallet JM, et al. Biochim Biophys Acta. 2012;1818:448. doi: 10.1016/j.bbamem.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 293.Ta HP, Berthelot K, Coulary-Salin B, Desbat B, Gean J, Servant L, et al. Langmuir. 2011;27:4797. doi: 10.1021/la103788r. [DOI] [PubMed] [Google Scholar]
- 294.Walrant A, Vogel A, Correia I, Lequin O, Olausson BE, Desbat B, et al. Biochim Biophys Acta. 2012;1818:1755. doi: 10.1016/j.bbamem.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 295.Bi X, Flach CR, Perez-Gil J, Plasencia I, Andreu D, Oliveira E, et al. Biochemistry. 2002;41:8385. doi: 10.1021/bi020129g. [DOI] [PubMed] [Google Scholar]
- 296.Nakahara H, Lee S, Sugihara G, Shibata O. Langmuir. 2006;22:5792. doi: 10.1021/la060194h. [DOI] [PubMed] [Google Scholar]
- 297.Issaurat B, Teissie J. Biochim Biophys Acta. 1988;944:516. doi: 10.1016/0005-2736(88)90523-8. [DOI] [PubMed] [Google Scholar]
- 298.Flach CR, Gericke A, Keough KM, Mendelsohn R. Biochim Biophys Acta. 1999;1416:11. doi: 10.1016/s0005-2736(98)00205-3. [DOI] [PubMed] [Google Scholar]
- 299.Ambroggio EE, Separovic F, Bowie J, Fidelio GD. Biochim Biophys Acta. 2004;1664:31. doi: 10.1016/j.bbamem.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 300.Konovalov O, Myagkov I, Struth B, Lohner K. European biophysics journal : EBJ. 2002;31:428. doi: 10.1007/s00249-002-0233-3. [DOI] [PubMed] [Google Scholar]
- 301.Sospedra P, Alsina MA, Espina M, Reig F, Haro II, Mestres C. J Colloid Interface Sci. 2000;221:230. doi: 10.1006/jcis.1999.6585. [DOI] [PubMed] [Google Scholar]
- 302.Deleu M, Paquot M, Jacques P, Thonart P, Adriaensen Y, Dufrene YF. Biophys J. 1999;77:2304. doi: 10.1016/S0006-3495(99)77069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kruger P, Schalke M, Wang Z, Notter RH, Dluhy RA, Losche M. Biophys J. 1999;77:903. doi: 10.1016/S0006-3495(99)76941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schwarz G, Taylor SE. Biophys J. 1999;76:3167. doi: 10.1016/S0006-3495(99)77468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Van Mau N, Vie V, Chaloin L, Lesniewska E, Heitz F, Le Grimellec C. The Journal of membrane biology. 1999;167:241. doi: 10.1007/s002329900488. [DOI] [PubMed] [Google Scholar]
- 306.Trommeshauser D, Galla HJ. Chem Phys Lipids. 1998;94:81. doi: 10.1016/s0009-3084(98)00047-4. [DOI] [PubMed] [Google Scholar]
- 307.Ma J, Koppenol S, Yu H, Zografi G. Biophys J. 1998;74:1899. doi: 10.1016/S0006-3495(98)77899-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Nakahara H, Lee S, Shibata O. Biochim Biophys Acta. 2010;1798:1263. doi: 10.1016/j.bbamem.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 309.Nakahara H, Dudek A, Nakamura Y, Lee S, Chang CH, Shibata O. Colloids Surf B: Biointerfaces. 2009;68:61. doi: 10.1016/j.colsurfb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 310.Na Nakorn P, Meyer MC, Flach CR, Mendelsohn R, Galla HJ. European biophysics journal : EBJ. 2007;36:477. doi: 10.1007/s00249-006-0102-6. [DOI] [PubMed] [Google Scholar]
- 311.Agirre A, Flach C, Goni FM, Mendelsohn R, Valpuesta JM, Wu F, et al. Biochim Biophys Acta. 2000;1467:153. doi: 10.1016/s0005-2736(00)00214-5. [DOI] [PubMed] [Google Scholar]
- 312.Ambroggio EE, Austen B, Fidelio GD. J Pept Sci. 2007;13:245. doi: 10.1002/psc.838. [DOI] [PubMed] [Google Scholar]
- 313.Castano S, Desbat B. Biochim Biophys Acta. 2005;1715:81. doi: 10.1016/j.bbamem.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 314.Deleu M, Paquot M, Nylander T. J Colloid Interface Sci. 2005;283:358. doi: 10.1016/j.jcis.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 315.Dieudonné D, Gericke A, Flach CR, Jiang X, Farid RS, Mendelsohn R. J Amer Chem Soc. 1998;120:792. [Google Scholar]
- 316.Eeman M, Deleu M, Paquot M, Thonart P, Dufrene YF. Langmuir. 2005;21:2505. doi: 10.1021/la0475775. [DOI] [PubMed] [Google Scholar]
- 317.Eeman M, Pegado L, Dufrene YF, Paquot M, Deleu M. J Colloid Interface Sci. 2009;329:253. doi: 10.1016/j.jcis.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 318.Fidelio GD, Maggio B, Cumar FA. Biochim Biophys Acta. 1986;862:49. [Google Scholar]
- 319.Ma G, Allen HC. Photochemistry and photobiology. 2006;82:1517. doi: 10.1562/2006-06-30-IR-958. [DOI] [PubMed] [Google Scholar]
- 320.Nakahara H, Lee S, Shibata O. Biophys J. 2009;96:1415. doi: 10.1016/j.bpj.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Nakahara H, Lee S, Shibata O. Biochim Biophys Acta. 2013;1828:1205. doi: 10.1016/j.bbamem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 322.Nakahara H, Lee S, Sugihara G, Chang CH, Shibata O. Langmuir. 2008;24:3370. doi: 10.1021/la703180x. [DOI] [PubMed] [Google Scholar]
- 323.Shanmukh S, Biswas N, Waring AJ, Walther FJ, Wang Z, Chang Y, et al. Biophys Chem. 2005;113:233. doi: 10.1016/j.bpc.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 324.Krishnaswamy R, Rathee V, Sood AK. Langmuir. 2008;24:11770. doi: 10.1021/la8019765. [DOI] [PubMed] [Google Scholar]
- 325.Agopian A, Castano S. Biochim Biophys Acta. 2013;1838:117. doi: 10.1016/j.bbamem.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 326.Taylor SE, Desbat B, Blaudez D, Jacobi S, Chi LF, Fuchs H, et al. Biophys Chem. 2000;87:63. doi: 10.1016/s0301-4622(00)00182-4. [DOI] [PubMed] [Google Scholar]
- 327.Boisselier E, Calvez P, Demers E, Cantin L, Salesse C. Langmuir. 2012;28:9680. doi: 10.1021/la301135z. [DOI] [PubMed] [Google Scholar]
- 328.Boisselier E, Calvez P, Demers E, Cantin L, Salesse C. Colloids Surf B: Biointerfaces. 2013;109:109. doi: 10.1016/j.colsurfb.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 329.Coskun U, Simons K. FEBS Lett. 2010;584:1685. doi: 10.1016/j.febslet.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 330.Fan J, Sammalkorpi M, Haataja M. FEBS Lett. 2010;584:1678. doi: 10.1016/j.febslet.2009.10.051. [DOI] [PubMed] [Google Scholar]
- 331.George KS, Wu S. Toxicol Appl Pharmacol. 2012;259:311. doi: 10.1016/j.taap.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Heberle FA, Feigenson GW. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Chen H, Anderson RE. Curr Eye Res. 1992;11:793. doi: 10.3109/02713689209000752. [DOI] [PubMed] [Google Scholar]
- 334.Gulcan HG, Alvarez RA, Maude MB, Anderson RE. Invest Ophthalmol Vis Sci. 1993;34:3187. [PubMed] [Google Scholar]
- 335.Salesse C, Boucher F, Leblanc RM. Anal Biochem. 1984;142:258. doi: 10.1016/0003-2697(84)90462-7. [DOI] [PubMed] [Google Scholar]
- 336.N’Soukpoe-Kossi CN, Salesse C, Leblanc RM, Boucher F. Anal Biochem. 1985;151:409. doi: 10.1016/0003-2697(85)90196-4. [DOI] [PubMed] [Google Scholar]
- 337.Fliesler SJ, Anderson RE. Prog Lipid Res. 1983;22:79. doi: 10.1016/0163-7827(83)90004-8. [DOI] [PubMed] [Google Scholar]
- 338.Cornut I, Desbat B, Turlet JM, Dufourcq J. Biophys J. 1996;70:305. doi: 10.1016/S0006-3495(96)79571-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Buffeteau T, Desbat B. Appl Spectrosc. 1989;43:1027. doi: 10.1366/000370203769699135. [DOI] [PubMed] [Google Scholar]
- 340.Ishida KP, Griffiths PR. Appl Spectrosc. 1993;47:584. [Google Scholar]
- 341.Tsuboi M, Mitsui Y, Wada A, Miyazawa T, Nagashima N. Biopolymers. 1963;1:297. [Google Scholar]
- 342.Blaudez D, Buffeteau T, Cornut JC, Desbat B, Escafre N, Pezolet M, et al. Appl Spectrosc. 1993;47:869. [Google Scholar]
- 343.Banc A, Desbat B, Renard D, Popineau Y, Mangavel C, Navailles L. Langmuir. 2007;23:13066. doi: 10.1021/la702037k. [DOI] [PubMed] [Google Scholar]
- 344.Grandbois M, Desbat B, Salesse C. Biophys Chem. 2000;88:127. doi: 10.1016/s0301-4622(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 345.von Heijne G. J Intern Med. 2007;261:543. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 346.von Heijne G. Annu Rev Biochem. 2011;80:157. doi: 10.1146/annurev-biochem-111910-091345. [DOI] [PubMed] [Google Scholar]
- 347.Ulmschneider MB, Sansom MS, Di Nola A. Proteins. 2005;59:252. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]
- 348.von Heijne G. Nat Rev Mol Cell Biol. 2006;7:909. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 349.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Biochemistry. 2001;40:7761. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. J Mol Biol. 2000;295:117. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 351.Bowie JU. J Mol Biol. 1997;272:780. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 352.Voet D, Voet JG. Biochemistry. 4. Wiley; 2011. [Google Scholar]
- 353.Dufourc EJ, Buchoux S, Toupe J, Sani MA, Jean-Francois F, Khemtemourian L, et al. Curr Protein Pept Sci. 2012;13:620. doi: 10.2174/138920312804142138. [DOI] [PubMed] [Google Scholar]
- 354.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, et al. Nature. 2005;433:377. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 355.Wolfenden R. J Gen Physiol. 2007;129:357. doi: 10.1085/jgp.200709743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Chothia C. J Mol Biol. 1976;105:1. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 357.Bourque H, Laurin I, Pezolet M, Klass JM, Lennox RB, Brown GR. Langmuir. 2001;17:5842. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.