Abstract
From an evolutionary perspective, the major function of bone is to provide stable sites for muscle attachment and affording protection of vital organs, especially the heart and lungs (ribs) and spinal cord (vertebrae and intervertebral discs). However, bone has a considerable number of other functions: serving as a store for mineral ions, providing a site for blood cell synthesis and participating in a complex system-wide endocrine system. Not surprisingly, bone and cartilage cell homeostasis is tightly controlled, as is the maintenance of tissue structure and mass. While a great deal of new information is accruing concerning skeletal cell homeostasis, one relatively new observation is that the cells of bone (osteoclasts osteoblasts and osteocytes) and cartilage (chondrocytes) exhibit autophagy. The focus of this review is to examine the significance of this process in terms of the functional demands of the skeleton in health and during growth and to provide evidence that dysregulation of the autophagic response is involved in the pathogenesis of diseases of bone (Paget disease of bone) and cartilage (osteoarthritis and the mucopolysaccharidoses). Delineation of molecular changes in the autophagic process is uncovering new approaches for the treatment of diseases that affect the axial and appendicular skeleton.
Keywords: bone, cartilage, growth plate, autophagy, remodeling, osteoarthritis, Paget disease of bone, osteoclasts, chondrocytes, stem cells, mucopolysaccharidosis
Introduction to Skeletal Tissues
The fossil records and phylogenetic systematics indicate that mineralized tissues have existed in a myriad of chemical forms and in numerous configurations for millions of years. Close examination of fossilized long bones and vertebrae indicates that both osseous and cartilage-like tissues appeared early in evolution; indeed, it was thought that cartilage, a poorly organized tissue, may have predated bone. However, closer examination indicates that while a cartilaginous skeleton is an optimal structure for organisms living in aqueous environments, it was likely preceded by a denser and mechanically stronger tissue, mineralized bone. Romer noted in his paper to the New York Academy of Sciences that:
“the early vertebrates had a considerable degree of ossification which was followed in a majority of cases by a slump toward a cartilaginous condition. Bone is an ancient, rather than a relatively new, skeletal material in the history of vertebrates.”1
We are just now beginning to understand the complex interplay between all of these mineralized tissues and other tissues in the body. For example, Karsenty’s laboratory has shown that regulation of bone mass is coordinated with both energy metabolism and even fertility.2
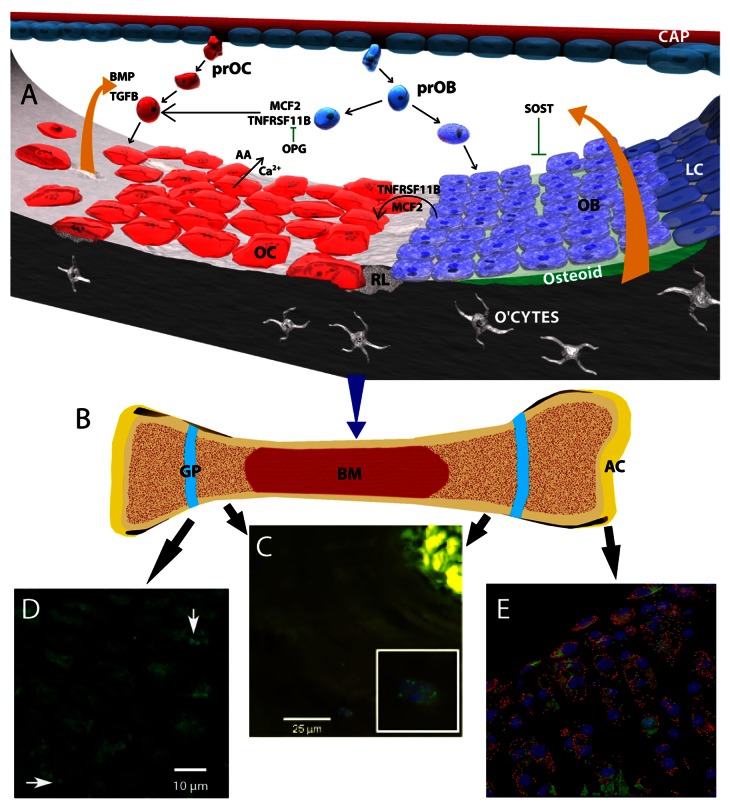
Not surprisingly, bone mass, structure, and function is tightly regulated. In the adult, this is achieved by two active and coupled processes, bone resorption and bone formation. Prime regulators of these processes include hormones, especially estrogen, PTH (parathyroid hormone) and PTHLH (parathyroid hormone-like hormone) and vitamin D metabolites. Bone marrow-derived osteoblasts are responsible for new bone formation while macrophage-like osteoclasts mediate the resorptive process. The activities of these cell types are linked through a bone-remodeling cycle (Fig. 1). Osteoclasts activate the cycle by inducing the resorption of old bone; this activity is followed by a formative phase during which osteoblasts synthesize new bone to replace the resorbed tissue. Those osteoblasts that become engulfed in the mineralized bone, osteocytes, play a pivotal role in functionally adapting the tissue to applied mechanical forces. As will be highlighted in this review, autophagy is relevant for the survival and function of each of these cell types within their highly specialized matrices.
Figure 1. The remodeling process in bone, and autophagy in bone, the growth plate and articular cartilage. (A) Schematic showing molecular control of bone remodeling. A basic multicellular unit consists of osteoblasts (OB) osteoclasts (OC) and bone lining cells (LC). Pro-osteoclasts (PrOC) and pro-osteoblasts (prOB) enter the unit through capillaries (CAP) and home to the bone surface where they undergo differentiation into OB and osteocytes (O’CTES) and OC cells, respectively. Under the influence of local factors including those secreted by osteoblasts (MCF2, TNFSF11), osteoclasts differentiate and resorb bone liberating Ca2+ and amino acids (AA) from the bone matrix. At these sites, a resorption lacuna is formed (RL). Another secreted factor, TNFRSF11B/osteoprotegrin (tumor necrosis factor receptor superfamily, member 11b), inhibits osteoclast-mediated bone resorption by serving as a physiological inhibitor of TNFSF11. A number of factors are released from the resorbing bone that include BMP and TGFB. These proteins promote osteoblast function and maturation. SOST, an inhibitor of osteoblast activity, is also released from the bone matrix. (B) Schematic of a long bone showing articular cartilage (AC), growth plates (GP) and bone marrow (BM). (C) Section through a demineralized osteon showing the presence of autophagic osteocytes (inset, stained with an anti-MAP1LC3 antibody). From Zahm et al., Cells Tissues Organs 2011; 194(2–4):274–8D, and with permission from S Karger AG. (D) Section through the rat growth plate showing the presence of autophagic pre-hypertrophic chondrocytes (arrows) stained with an antibody to MAP1LC3. From Srinivas et al., Cells Tissues Organs. 2009;189(1–4):88–92 and with kind permission from S Karger AG. (E) Chondrocyte autophagy in knee cartilage of GFP-MAP1LC3 transgenic mice. Confocal microscopy-rendered reconstruction using 3D IMARIS (Bitplane Inc.) indicating that the highest levels of GFP-MAP1LC3 signal are observed in chondrocytes in the superficial and upper middle zone of the articular cartilage. In contrast, only a few cells in the deep zone contain detectable levels of GFP-MAP1LC3 signal. Mag × 63
When the equilibrium between these two processes is disturbed, and there is excessive bone formation, this can lead to over-mineralization of bone, or osteopetrosis. More frequently, there is increased bone loss leading to a porotic state, i.e., osteopenia or osteoporosis. Osteoporosis, or more commonly postmenopausal osteoporosis, is a condition which increases bone fracture risk, thereby challenging the quality of life and longevity of elderly women and men.3 The relationship between the autophagic pathway and osteoporosis, was highlighted in a genome-wide association study of wrist bone mineral density; the pathway-based analysis showed significant associations with regulation-of-autophagy genes including ATG (autophagy-related) 5, ATG7, ATG12 PIK3C3 (phosphatidylinositol 3-kinase, catalytic subunit type 3), PRKAA2 (protein kinase, AMP-activated, α 2 catalytic subunit), GABARAPL1 [GABA(A) receptor–associated protein like 1], BECN1 (beclin 1, autophagy related), and IFNA13 (interferon, α 13).4 The authors related variants in these genes to the pathogenesis of osteoporosis in terms of modulating factors required for bone formation and/or remodeling.
While a great deal of new information is accruing concerning skeletal cell homeostasis, there is growing awareness that autophagy enables osteoblasts, osteoclasts, and chondrocytes to survive within a hypoxic, and even hypertonic environment. In this way, cells can overcome stressor challenges and nutrient deficiencies and, for osteocytes and articular chondrocytes, fulfill their fates as exceedingly long-lived terminally differentiated cells. The focus of this review is to examine the significance of the autophagic process in terms of the functional demands of the skeleton in growth and homeostasis, and to provide evidence that dysregulation of autophagy is involved in the pathogenesis of diseases of bone (Paget disease of bone, PDB) and cartilage (osteoarthritis, OA, and the mucopolysaccharidoses, MPS; see Table 1).
Table 1. Autophagic changes in relationship to diseases of bone and cartilage.
| Disease name | Target cells | Phenotype | Clinical changes | Autophagy gene/protein involvement | Autophagy change |
|---|---|---|---|---|---|
| Paget disease of bone | Osteoclasts and progenitors. Possibly osteoblasts | Increased and disorganized bone turnover; abnormal osteoclast function | Focal areas of bone abnormality with pain, fracture and deformity. Bone pain. | SQSTM1, OPTN VCP |
Dysregulation of autophagy and enhanced autophagosome formation |
| Osteoporosis | Osteoclasts and to a lesser extent osteoblasts | Decreased bone mineral density, disorganized bone | Increased fragility with increased fracture susceptibility | *PIK3C3, ATG12, PRKAA2, ATG5, GABARAPL1, BECN1, IFNA13, ATG7 | Increased autophagy |
| Osteoarthritis | Chondrocytes (possibly osteoblasts) | Loss of articular cartilage | Decreased joint junction | MAP1LC3, ULK1, BECN1 | Decreased autophagy and autophagic flux |
| Glucocorticoid- induced osteoporosis | Osteoblasts and to a lesser extent osteoclasts | Reduced osteoblast number, premature osteoblast apoptosis | Bone loss, bone fractures; osteonecrosis | MAP1LC3 | Increased osteocyte autophagic flux |
| Osteogenesis imperfecta | Osteoblast progenitors | Mutations in COL1A1 and COL1A2 | Fragile bones | ATG7, BECN1 MAP1LC3-II, CTSK | Increased autophagy |
| Multiple sulfatase deficiency | Chondrocytes, osteoblasts, fibroblasts | Mutations in SUMF1 gene. (required for removal of sulfate groups from GAGs) | Severe shortening of the axial and appendicular skeleton, dysostosis and neurodegeneration | MAP1LC3, GFP-MAP1LC3 | Marked increased in autophagosomes |
| Inclusion body myopathy with Pagets disease and frontotemporal dementia | Muscle, brain and bone cells | Muscle weakness, retardation | Multi-organ degeneration affecting muscle brain and bone | Mutation in VCP | Reduced autophagic flux |
Expression level changes relevant only to the wrist bone density.4
Osteoblast and Osteocyte Function and Autophagy
Signaling pathways regulating osteoblast activity and autophagy
Osteoblasts secrete the organic matrix of bone (osteoid) and participate in its mineralization. During its encasement in bone, the osteoblast becomes fully differentiated and assumes the morphology and function of an osteocyte (see later and Fig. 1). It is likely that the physiological stimulus for new bone formation is linked to intermittent shifts in PTH levels and the resorptive activities of osteoclasts which release growth factors such as TGFB1 (transforming growth factor, β 1), insulin-like growth factors and bone morphogenetic proteins (BMPs), from the extracellular matrix of the bone. In addition, a neuronal involvement cannot be excluded since bone mass is responsive to the sympathetic tone of the nervous system, through modulation of ADRB2 (adrenoreceptor β 2, surface/cAMP signaling system).5 During resorption, release of these buried agents from the resorbing tissue promotes osteoblast migration, activation, and even new bone formation.
The BMPs are clinically powerful bone-forming agents that bind to a common cognate receptor on osteoblasts, and transduce signals through the SMAD signaling system. The functional activity of the BMP ligands are regulated by extracellular protein antagonists that include NOG (noggin), CHRD (chordin), and SOST (sclerostin).6 That BMPs may regulate autophagy is suggested by the studies of Cao et al.7 who showed that NOG reduces MAP1LC3 (microtubule-associated protein 1 light chain 3)-II levels, albeit in acute pancreatitis cells; this was overcome by administration of BMP2, which increases the levels of both BECN1 and LAMP2 (lysosomal-associated membrane protein 2). Indeed, a mutation in the ACVRL1 (activin A receptor type II-like 1) kinase causes a rare and very disabling disease, fibrodysplasia ossificans progressiva. Whether disease progression is related to expression of an autophagic phenotype awaits further study.
Activities of proteins of the WNT-CTNNB1/β-catenin pathway are major regulators of chondrocyte and osteoblast function. WNT signaling plays a major role in regulating stem cell commitment to the osteoblast lineage and osteoblast differentiation. In tumor and hepatic cells, WNT signaling has been negatively linked to autophagy. This is mediated through the interaction of DVL2 (dishevelled segment polarity protein 2) which binds to the autophagy receptor SQSTM1 (sequestosome 1) which in turn facilitates MAP1LC3-mediated autophagosome recruitment, ubiquitination, and degradation.8 Likewise, activation of the WNT-CTNNB1 pathway is central to the pathogenesis of both rheumatoid arthritis and OA. While it is tempting to speculate that activation of this pathway suppresses autophagy and enhances osteoblast or chondrocyte death, this relationship has not as yet been fully established.
The functional role of autophagy in the osteoblast is still largely unexplored, particularly in vivo. However, several autophagy-related proteins have a profound impact on osteoblast biology. The autophagy receptor NBR1 (neighbor of BRCA1 gene 1) is involved with targeting ubiquitinated cargos to the autophagosome.9 It fulfills this function by interacting with MAP1LC3 protein family members through its LC3-interacting region (LIR) and with target proteins through the ubiquitin-like modifier activating enzyme (UBA) domain. A knock-in mouse model with deletion of both LIR and UBA domains in the NBR1 locus results in increased osteoblast differentiation and activity.10 This aberrant osteoblastic activity appears to be dependent on AHSA1 MAPK1 hyperactivation. The moderate increase in the autophagy receptor SQSTM1 in osteoblasts isolated from NBR1 knock-in mice can impair proteasomal function and activate the stress responsive transcription factor NFE2L2/NRF2, which results in the induction of many cytoprotective genes.
At least 2 families of transcription factors with known roles in autophagy control osteoblast survival and function. Of these, the family of FOXO (forkhead box O) transcription factors serves key roles in cell growth, cell proliferation, DNA repair, cell cycle arrest, reactive oxygen species (ROS) generation, energy homeostasis, and glucose metabolism.11 FOXO1, 3, 4, and 6 are downstream of early signaling events in the insulin pathway and negatively regulate AKT (v-akt murine thymoma viral oncogene homolog) signaling.12 FOXO activation potently induces autophagy by directly binding to the promoter regions of target genes. Genetic deletion of FOXOs in osteoblasts induces oxidative stress and increased apoptosis, mimicking the aging process. Conversely, FOXO3 overexpression prevents bone loss associated with aging.13 Since autophagy has an important cytoprotective role against oxidative stress and other aging-related phenotypes, and with glucosamine promotes autophagic flux in chondrocytes,14 it is tempting to consider that the role of FOXO in maintaining bone homeostasis is at least, in part, mediated by the induction of autophagy.
ATF4 (activating transcription factor 4), a member of the CREB (cAMP responsive element binding protein) family of B ZIP proteins, is required to maintain osteoblast function and promote terminal differentiation; it also protects cells from amino acid starvation and enhances amino acid import into cells. Changes in ATF4 activity have been linked to skeletal manifestations of 2 human genetic diseases, Coffin-Lowry syndrome and neurofibromatosis type I. This transcription factor induces osteoblast-specific gene expression in fibroblasts together with osteocalcin synthesis, thus predisposing these cells to aberrant mineral deposition.15 Interestingly, ATF4 is thought to promote cell survival through the transcription of several autophagy genes including MAP1LC3B and ATG5. Elefteriou et al.16 noted that the increased bone-mass phenotype resulting from NF1 (neurofibromin 1) deficiency can be rescued through nutritional restriction of protein intake, a finding that would strongly suggest that a link exists between expression of this protein and autophagic flux.
Autophagy and the osteoblast–osteocyte transition
In contrast to the short half-lives (days/weeks) of osteoblasts, osteocytes are very long-lived cells that exhibit an architecture closer to neurons than cartilage or bone cells. The basic feature of the osteocyte is a cell subsumed into mineralized bone lacunae with a large number of long dendritic processes contained in bone canals (canaliculi), many of which interconnect with other osteocytes to form a syncytium as well as interacting with osteoblasts and bone-lining cells (Fig. 1). It is likely that their primary role is mechanotransduction, i.e., converting mechanical forces on the bone into biological signals that serve to promote the remodeling process. While supporting little biological activity, there is evidence that like osteoblasts, these cells can express TNFSF11 [tumor necrosis factor (ligand) superfamily, member 11; see below] and thereby influence osteoclastogenesis and resorption. Osteocytes can also secrete SOST, which inhibits signaling and possibly enhances osteocyte apoptosis through the WNT signalng pathway. Not surprisingly, studies by Zahm et al.17 clearly showed that in situ a considerable number of osteonal osteocytes display a punctate distribution of MAPILC3A protein indicative of a basal level of autophagy (Fig. 1).
In addition, in culture, using pre-osteocyte-like murine (MLO-A5) cells, autophagy is upregulated following nutrient deprivation and hypoxic culture, stress conditions that osteocytes encounter in vivo (Fig. 1; Table 1).17 Furthermore, in response to calcium stress, HIF1A [hypoxia inducible factor 1, α subunit (basic helix-loop-helix transcription factor] regulates MLO-A5 autophagy, indicating that low oxygen pressure (pO2) may serve as a positive regulator of autophagy in this cell type. Last, it should be mentioned that low-dose glucocorticoid therapy profoundly influences osteocyte function and increases autophagic activity and antioxidative responsiveness 30-fold. High doses reduce both the expression of genes encoding anti-oxidant proteins and the number of autophagic osteocytes.18 Bone formation, measured by serum osteocalcin and surface-based histomorphometry, was greatly reduced by chronic or high dose glucocorticoid treatment. Xia et al.19 proposed that modification of the oxidative and autophagic pathways may provide promising new targets for maintaining bone formation in the presence of glucocorticoids while preserving bone mass.
Osteoclastogenesis, Osteoclast Function, and Autophagy
Regulation of osteoclastogenesis
At remodeling sites, hematopoietic mononuclear myeloid stem cells, mostly resident in the bone marrow, commit to the osteoclast phenotype and migrate to the tissue surface. When activated, the terminally differentiated osteoclast becomes tightly adherent to the bone surface. Attachment is mediated through one or more specialized structures termed podosomes. The podosome contains bands of actin filaments, as well as F-actin and actin monomers. Contained within the podosome, a ruffled border is formed by fusion of secretory lysosomes with the plasma membrane, and functions as a site of secretion and externalization of hydrochloric acid and proteases; the acid dissolves the mineral phase of bone, hydroxyapatite, while key proteases such as MMP9 [matrix metalloproteinase 9 (gelatinase B, 92 kDa type IV collagenase] and MMP13 [matrix metalloproteinase 13 (collagenase 3)] and CTSK (cathepsin K), hydrolyze the collagen-rich organic bone matrix.20
Differentiation of the adherent cells into an active osteoclast is dependent on CSF1 [colony stimulating factor 1 (macrophage)] and TNFSF11/RANKL [tumor necrosis factor (ligand) superfamily, member 11].21 Following fusion with other mononuclear precursors of the macrophage–monocyte lineage, they form multinucleated giant cells.22 Recruitment is chemokine-dependent, especially CXCL12 [chemokine (C-X-C motif) ligand 12] which regulates cell migration. Recently, a lysophospholipid-derived from sphingomyelin, sphingosine 1-phosphate (S1P), has been shown to be an osteoclast chemoattractant.23 Osteoclasts express S1PR (sphingosine-1-phosphate receptor), and by modulating the activity of mechanistic target of rapamycin (MTOR), S1P counteracts autophagy and promotes apoptosis.24 Lee et al.25 have shown that S1P levels are higher in postmenopausal women, and possibly due to increased bone resorption its action is associated with low bone mineral density.
While differentiating osteoclasts populate the surface and interior of bone trabeculae, the local environment is probably hypoxic. Arnett et al.26 showed that a low pO2 promoted increased expression of BNIP3 (BCL2/adenovirus E1B 19 kDa interacting protein 3). BNIP3, increased autophagic flux and MAP1LC3 recruitment to autophagosomes, and osteoclast differentiation. These observations fueled the speculation that a HIF1A-BNIP3 signaling pathway promotes osteoclast differentiation.27
Although environmental factors may play an important role in the differentiation process, the active osteoclast needs to both regulate the secretion of hydrolytic enzymes as well as promote the intracellular digestion of peptide residues of matrix proteins. DeSelm and colleagues showed that ATG5, ATG7, ATG4B, and MAP1LC3 are required for both osteoclastogenesis and activation of bone resorption. ATG5 and ATG7 promote bone resorptive activities both in vivo and in vitro, and serve to target lysosomes to the actin ring of the functioning osteoclast. However, neither protein influences osteoclastogenesis, the numbers of nuclei in the osteoclasts, the presence of secretory lysosomes or even the expression of actin ring proteins. ATG4B modulation of MAPILC3A, blocks both resorptive activity and expression of CTSK, an enzyme required for dissolution of matrix proteins. These findings lend support to the notion that lysosomal targeting is regulated by ATG5, ATG7, MAPILC3A, and ATG4B, whereas control of bone resorption is independent of autophagic activity.28 This concept was further developed by Chung et al.who knocked down MAPILC3A and confirmed that it does not influence multinucleation, although it inhibits actin ring formation and resorption activity, including CTSK release.29 Using the ATG5 knockdown, these workers showed that the lowered amount of protein levels caused a marked loss in CDC42 activity and actin ring disruption. Based on these findings, the possibility exists that MAP1LC3 can regulate bone-resorbing activity via CDC42-dependent actin ring formation and ruffled border organization.29 Overall, these observations confirm an important, noncanonical role for autophagy in the regulation of osteoclastogenesis.
Osteoclast function and autophagy
There is a considerable amount of new evidence that points to a role for the autophagic process in the clearance of cytotoxic protein aggregates, which accumulate in disease states due to impairment of the ubiquitin-proteasome system. Aberrant, misfolded proteins, along with chaperones, are commonly found in SQSTM1- and ubiquitin-positive aggregates (sequestosomes), which are precursors to the inclusion bodies seen in many age-related neurodegenerative and liver diseases.30 Several proteins that have been linked to autophagic regulation of protein aggregates include WDFY3/ALFY (WD repeat and FYVE domain containing 3), SQSTM1, NBR1, CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2), OPTN (optineurin) and VCP (valosin-containing protein). Currently, it is not clear if their involvement is limited to selective autophagy or bulk autophagy in response to starvation. However, as noted previously some of these proteins act as autophagy cargo receptors (e.g., NBR1 and SQSTM1), while others are scaffolds (e.g., WDFY3) that facilitate autophagosome membrane formation around the cargo to be degraded.31 In osteoclasts, WDFY3 interacts directly with SQSTM1 (via its PH-BEACH domain), ATG5 (via its WD40 repeat domain) and phosphatidylinositol-3-phosphate (PtdIns3P) (via its FYVE domains) and forms large cytoplasmic aggregates (see section 4).32
Using a WDFY3 siRNA knockdown Filimonenco et al.33 showed that while there is no direct effect on starvation-induced autophagic clearance of HTT poly Q protein, WDFY3 nuclear localization and shuttling may be the rate-limiting step for aggregate clearance in HeLa and N2a cells. When compared with mononuclear precursors, amino acid starvation of mature multinucleated cells results in WDFY3 rapidly relocating to the cytoplasm and interacting and colocalizing with SQSTM1 in cytoplasmic aggregates. Importantly, WDFY3 together with the proteins mentioned above can be linked to skeletal homeostasis; SQSTM1 and VCP mutations as well as OPTN genetic variants are linked to human disorders associated with a skeletal phenotype (see section 4), whereas NBR1 regulates osteoblast function (see section 2).10,34 The important role of autophagy in mature osteoclast function and survival is probably best illustrated by consideration of the osteoclast phenotype of SQSTM1 mutations observed in patients with Paget disease of bone.
Autophagy and the Pathogenesis of Paget Disease of Bone
Pathogenesis of PDB
PDB is a common age-dependent skeletal disorder characterized by focal areas of increased and disorganized bone turnover. Clinically, if left unchecked, PDB can cause bone deformity and fracture.35 PDB is principally a disorder of the osteoclast, characterized by an increase in their number, size, and activity within the bone lesion. PDB osteoclasts exhibit increased sensitivity to TNFSF11 in vitro and appear to be more resistant to apoptosis.36 The underlying cause(s) of the abnormal osteoclast activity and function in PDB is unclear, although evidence for a secondary involvement of osteoblasts in lesion development, is now emerging.35
The etiology of PDB involves a complex interplay between genetic and environmental factors37-39 with some studies implicating paramyxovirus infection.40 Interestingly, many viruses exert their effects by subverting autophagy, although to date no studies have directly determined the impact of viral infection on osteoclast autophagy. As many as 40% of patients have a positive family history of disease and PDB is 7–10 times more common in first-degree relatives of affected individuals.41 Current thinking is that PDB is caused by a combination of rare, high-penetrance variants in small number of genes, together with common variants in other genes which together increase the risk of developing the disorder.
The only gene to date identified as being causally related to PDB is SQSTM1, which encodes the autophagy receptor SQSTM1 protein.42 However, SQSTM1 mutations are found only in 5–20% of PDB patients.35 The functional domains of SQSTM1 include a UBA domain, an N-terminal PB1 domain and two internal regions representing an LIR and KIR [KEAP1 (kelch-like ECH-associated protein 1)-interacting region]. It colocalizes with MAP1LC3 in cellular “protein bodies,” including those containing aggregated mutant proteins, a process dependent on both the UBA and Phox and Bem1p (PB1) domains.30 The direct interaction (via the LIR) of SQSTM1 with MAP1LC3 facilitates the autophagic degradation of ubiquitin-modified cytosolic protein aggregates and organelles.43 However, the precise complement of autophagic substrates of SQSTM1 in vivo remains to be fully clarified.
Consistent with its role as a cargo receptor for the autophagic degradation of ubiquitin-modified targets, SQSTM1 is upregulated by various stressors including starvation, proteasome inhibition, and NFKB and NFE2L2 activation. Functionally, it cooperates with the autophagy receptor NBR1via PB1 domain-mediated interactions, and (as was noted above) in osteoclast-like cells with WDFY3 to facilitate degradation of misfolded proteins.32,44 Furthermore, interaction of SQSTM1 via its KIR region with KEAP1 regulates the levels of this scaffold protein.45,46 KEAP1 in turn controls levels and activity of the NFE2L2 transcription factor, which regulates the expression of cytoprotective genes, thus contributing to the cell’s capacity to defend itself against chemical and oxidative stress, and controls proliferation and differentiation of osteoblasts.47 Interestingly, KEAP1 is also reported to downregulate TNF/TNFα-induced NFKB activation through autophagic degradation of IKBKB/IKKβ.48 Recent work shows a direct and functional interaction between SQSTM1 and components of MTORC1, establishing it as a key regulator of nutrient sensing.49 In terms of regulation of SQSTM1-mediated autophagic function at the molecular level, phosphorylation of the UBA domain at Ser403 serves as a signal to promote autophagy.50 Ser403 phosphorylation appears to regulate the ubiquitin-binding ability of SQSTM1 and a phosphomimetic mutant (S403E) promotes the formation of “sequestosomes,” presumed to be precursors of autophagosomes.50 Genetic inactivation of Sqstm1 in mice results in impaired osteoclastogenesis in vitro and in vivo.51 and importantly, in a mouse carrying a P394L-SQSTM1 missense mutation (equivalent to the most common PDB-associated P392L human mutation), a PDB-like disorder is seen with altered osteoclast autophagy (see below).
Disease mechanisms
At the protein level, most of the disease-associated mutations affect the UBA domain of SQSTM1 and cause a loss of ubiquitin-binding activity.52,53 While the PDB-mutant SQSTM1 causes an increase in osteoclast activity involving NFKB signaling,54,55 little is known of the impact of the mutation on autophagy. As there is crosstalk between the 2 systems,55 changes in NFKB activity and autophagic function could be expected. For example, induction of autophagy in macrophages in response to TLR4 (toll-like receptor 4) is associated with TRAF6 (TNF receptor-associated factor 6, E3 ubiquitin protein ligase)-mediated ubiquitination of BECN1,56 and TRAF6 may be degraded by a form of autophagy that is dependent on CALCOCO2 (although not associated with the canonical conversion of MAP1LC3-I to MAP1LC3-II).57 Furthermore, the IKK complex, which is an essential mediator of the TNFRSF11A/RANK-NFKB pathway, contributes to the induction of autophagy and is activated by multiple autophagy inducers, without affecting NFKB nuclear translocation.58 Conversely, levels of NFKBIA/IκB, the inhibitor of NFKB, appear to be regulated by autophagy.59 Further emphasizing common pathways, under resting conditions BECN1 forms a complex with the MAP3K7-binding proteins TAB2 (TGF-β activated kinase 1/MAP3K7 binding protein 2) and TAB3; during autophagy, BECN1 activates IKBKB and induces autophagy,60,61 NFKB activation can also limit autophagy activators (BNIP3, MAPK8/JNK1, and ROS) and increase expression of autophagy inhibitors (TNFAIP3/A20, BCL2, and BCL2L1/BCLXL62).
PDB-associated mutations of SQSTM1 and autophagy
Indirect evidence supports the notion that alterations in autophagy are linked to the pathogenesis of PDB: disease-causing mutations increase osteoclast activity; autophagy positively regulates osteoclast activity,28 and SQSTM1, which is commonly mutated in PDB, is an autophagy receptor. Furthermore, PDB-associated mutations map to regions of SQSTM1, which are relevant for its autophagy-dependent function: principally the UBA domain,42,63 but also the LIR (D335E)64 and the KIR (S349T) domains.65
Earlier observations that SQSTM1 is overexpressed in PDB patient samples, regardless of SQSTM1 mutation status, were the first indication that autophagy may be altered in the pagetic state (Table 1).66 However, perhaps the best evidence of alterations in autophagic function comes from studies of the P394L-SQSTM1 mouse, which, as noted above, develops a PDB-like bone disorder with focal bone lesions.67 Osteoclast precursors from the mutant animals, exhibit increased expression of SQSTM1, ATG5, and MAP1LC3 along with increased MAP1LC3-II protein levels in the presence of bafilomycin A1 suggesting a possible increase in autophagic flux (and consistent with the known relationship between autophagic and osteoclastic activity).28 In a cell model of PDB with SQSTM1 mutations, a preliminary report describes alterations in autophagic flux.68
Other related disorders and autophagy
Curiously, SQSTM1 mutations, including some UBA domain mutations that are associated with PDB, have also been reported in patients with ALS (amyotrophic lateral sclerosis) and frontotemporal lobar degeneration.69,70 Interestingly, several of the recently identified new genes for ALS represent autophagy receptors including OPTN (also linked to PDB), UBQLN2, and VCP.70 The latter is perhaps the most relevant, given that VCP mutations are a cause of the multisystem disorder inclusion body myopathy associated with PDB and frontotemporal dementia. Although its precise role in autophagy is unclear, in muscle cells the mutant VCP appears to be linked to alterations in autophagy.71 Indeed, its expression is associated with accumulation of nondegradative autophagosomes and a failure to degrade aggregated proteins.72 In knockout animals with mutant VCP there are increased levels of MAP1LC3B-II in muscle cells, osteoclast precursors exhibit increased sensitivity to TNFSF11, and there are focal bone lesions.73 Finally, OPTN encodes an autophagy receptor, which been implicated in PDB74 with preliminary studies suggesting that it negatively regulates osteoclast activity.75
Autophagy in the Growth Cartilage
Function of the growth plate
The epiphyseal growth plate is a transient form of cartilage located at sites of long-bone growth and composed of chondrocytes embedded in a matrix containing COL2A1 (collagen, type II, α 1) and PRG/proteoglycan. In comparison with articular cartilage (see below), chondrocytes in the growth plate have a very short half-life (days, not years).76 During growth, a number of well-defined zones can be delineated; most obvious is a columnar cell zone that contains proliferating cells. As these cells mature, they become terminally differentiated hypertrophic chondrocytes. Mineralization of the mature cartilage begins in the deep hypertrophic zone in cell-derived particles, matrix vesicles. Enzymes in the matrix vesicle remove local inhibitors of mineralization, enhance calcium transport and localization, and promote mineral deposition.77 During the growth period, there is evidence of chondrocyte autophagy, and indeed treatment of rats with the autophagy activator rapamycin impairs longitudinal growth.78
Autophagy and chondrocyte function
Regulation of the cellular changes described above is complex and involves an interplay between agents generated by chondrocytes, (PTHLH and IHH/indian hedgehog), cytokines, and systemic factors including hormones of the hypothalamus-pituitary axis.79,80 Once mineralization has begun, there are autophagic changes in hypertrophic chondrocytes: reorganization of MAP1LC3-II and BECN1 proteins into punctate granules. In addition, transmission electron microscopy studies indicate the presence of double-membrane vacuoles. Not surprisingly, suppression of MTOR causes a marked increase in chondrocyte autophagy.81 Following autophagy, chondrocytes are deleted from the plate by the initiation of programmed cell death.82 These terminal changes and the control of growth through the WNT signaling pathway would suggest that autophagy serves not just to regulate the final stages of the chondrocyte life cycle, but also the rate at which chondrocytes enter the maturation process.
Metabolic control of chondrocyte autophagy
Chondrocytes generate metabolic energy through anaerobic glycolysis, an environmental adaptation that permits them to survive in the restricted vascular supply of cartilage.83 HIF1A is expressed at high levels in hypertrophic chondrocytes. Conditional inactivation of the HIF1A gene causes a reduction in the number of maturing hypertrophic cells, an elevation in numbers of apoptotic cells and a disorganized layer of subchondral (metaphyseal) bone.84 To further explore this activity and relate it to autophagy, a chondrocyte line was developed (N1511) that mimics many of the phenotypic changes expressed by epiphyseal chondrocytes. When serum-stressed, these cells robustly expressed punctate MAP1LC3 protein and BECN1.84
Not surprisingly, at a low pO2, the cultured chondrocytes are refractory to an apoptotic challenge. It was noted that HIF1A suppresses BECN1, leading to enhanced cell death. The observation that BECN1 suppression results in increased BID (BH3 interacting domain death agonist), cleavage, and CASP8/caspase 8 activation clearly links apoptosis with autophagy. Thus, BECN1 serves to maintain chondrocyte survival activity, possibly by regulating the activities of pro-apoptotic genes.84 Another HIF1A target is PRKAA2 (protein kinase, AMP-activated, α 2 catalytic subunit), a protein that responds to the energy status of the cell, and is activated in a HIF1A-dependent manner. As might be expected this protein is robustly expressed in the glycolytic growth plate.85 A number of studies have shown a connection between PRKAA2 and MTOR. Once activated, PRKAA2 phosphorylates TSC2 (tuberous sclerosis 2), which then suppresses MTOR and hence promotes autophagy. By downregulating the phosphorylation of targets RPS6KB1 and EIF4EBP1, MTOR integrates multiple signals including those from nutrients, as well as metabolic signals from glycolysis and ATP.86
From a physiological viewpoint, in the growth plate, autophagy is controlled by 2 environmental sensors: PRKAA2 and MTOR and probably EGLN1 (egl-9 hypoxia-inducible factor 1) via EGLN1 oxygen sensors.86 The hypoxic plate would foster the expression of HIF1A and the high glycolytic activity would elevate AMP levels and suppress MTOR. Once autophagy is activated, it would serve to maintain the life span of the hypertrophic cell, thus allowing the cells to reach their final maturation stage. Eventually, extended autophagic activity would lead to sensitization of terminally differentiated chondrocytes to local and intrinsic signals, resulting in apoptosis, deletion of cells from the growth plate and bone growth.87
Autophagy and the Pathogenesis of the Mucopolysaccharidoses (MPS)
Lysosomal storage disorders (LSDs)
LSDs are recognized as a cohort of nearly 60 different inherited disorders, each with a genetic defect that renders the lysosomal system dysfunctional and unable to degrade specific molecules. As a consequence, many tissues and organ systems are affected, including bone and cartilage.88 The MPSs comprise a group of LSDs caused by deficiency in the enzymes catalyzing the degradation of glycosaminoglycans (GAGs), which are long, repeating chains of complex sugar molecules, normally degraded in the lysosome.89 Multiple sulfatase deficiency is a very severe form of MPS due to mutations in the SUMF1 (sulfatase modifying factor 1) gene.90 The SUMF1 protein is responsible for an essential posttranslational modification of the sulfatase enzymes, a class of hydrolases that remove sulfate groups from different molecules including GAGs. Many sulfatases are lysosomal, and thus one of the major consequences of the lack of sulfatase activity is the accumulation of multiple sulfated substrates in the lysosomes.91 The sumf1−/− mouse recapitulates most of the features of the human multiple sulfatase deficiency disease and in particular displays a remarkable skeletal dysplasia, characterized by severe shortening of the axial and appendicular skeleton.92 Multiple measurements of autophagy in sumf1−/− chondrocytes reveal severe lysosomal vacuolization and an increased number of autophagosomes compared with wild-type chondrocytes. Similarly to cells treated with an inhibitor of lysosomal acidification (bafilomycin A1), sumf1−/− chondrocytes accumulate autophagic vacuoles and present low ATP content, suggesting that the decreased survival of the sumf1−/− chondrocytes is directly linked to the defective digestion of the autophagic cargo.93
Pathogenesis of the MPS
Although the pathogenic mechanisms are still unclear, defective autophagy is a generalized phenomenon occurring in many LSDs; cells and tissues isolated from patients and mouse models of LSDs display higher numbers of autophagosomes compared with controls, most likely the result of a defective lysosome-autophagosome fusion (Table 1). As a consequence, autophagic substrates, such as polyubiquitinated proteins and dysfunctional mitochondria, are significantly elevated in LSD cell and tissue samples.94,95 Providing a direct insight into the biochemical defect, sumf1−/− mice exhibit a significant accumulation of cholesterol in lysosomal membranes. As a consequence, there is impaired distribution of the SNARE (soluble NSF attachment protein receptors) that are key components of the cellular membrane fusion machinery.96 Interestingly, wild-type cells “loaded” with cholesterol in vitro mirror the fusion defects observed in LSD cells. Conversely, lowering cholesterol levels restores normal lysosomal function. From this perspective, the lysosome-autophagosome fusion defect in LSDs reflects the abnormal lysosomal membrane lipid composition.96
Autophagy and the Pathogenesis of Osteoarthritis (OA)
Pathogenesis of osteoarthritis
OA is among the most prevalent aging-related diseases and the most prevalent joint disease.97 The two main risk factors for OA are aging and mechanical load. Excessive mechanical loading can occur in the acute setting of joint injury or can be chronically due to abnormal joint shape, malalignment, or as a result of occupational and recreational activities.98 The earliest changes in cartilage are enzymatic degradation of GAGs and cartilage proteins, and loss of chondrocytes in the superficial zone; this region is exposed to shear and compressive forces during movement.99 With depletion of many of the original chondrocytes, there is emergence of clusters of densely packed cells, which are phenotypically distinct from the original cartilage cells. Cell activation in OA cartilage has been interpreted as an unsuccessful attempt at tissue repair and, as the condition progresses, it leads to further cartilage defects.100
Changes in autophagy protein expression and activation in aging and OA
Conceptually, autophagy in normal adult articular cartilage is an important mechanism for cellular homeostasis. Thus, cells in the superficial zone display a robust expression of autophagy proteins BECN1, ATG5, and MAP1LC3.101 When MAP1LC3 is tagged with GFP, the highest GFP signal is observed in cells present in the superficial and middle zones of the knee articular cartilage (Fig. 1). Few cells in the deep cartilage zone exhibit detectable levels of GFP-MAP1LC3 signal. As with other tissues, starvation increases the number of autophagosomes in chondrocytes.14
During the aging process in mouse and human knee articular cartilage, there is a decrease in ULK1 (unc-51 like autophagy activating kinase 1), MAP1LC3, and BECN1 protein expression. The reduction of these key regulators of autophagy is accompanied by increased apoptosis.14 Using a rapidly progressing experimental mouse model of OA, a time-dependent reduction in these autophagy proteins was noted.101 Since this reduction is observed in relatively young mice it is apparently not a consequence of aging-related events. However, for both surgical OA and mechanically injured cartilage, the increase in cell death suggests that autophagy may contribute to survival mechanisms (Table 1).
In contrast to the reduction in autophagic proteins in nonproliferating chondrocytes, the cell clusters in OA cartilage express high levels of these proteins101 thereby confirming an earlier study that suggested that chondrocytes in OA cartilage display numerous autophagic MAP1LC3 puncta.102 Levels of MAP1LC3-II are also increased in the superficial and middle zones in a rat model of OA.14 When full-thickness cartilage explants are subjected to high-impact mechanical compression, there are immediate matrix changes and a low level of cell death, accompanied by a short and transient increase in the levels of MAP1LC3-II, and a marked reduction in ULK1, MAP1LC3, and BECN1.101 Thus, during the development of OA, increased autophagy may reflect an adaptive stress response. Furthermore, failure to mount an autophagic response may lead to further degeneration.
Consequences of dysregulated autophagy in OA
The reduction in autophagy protein levels and activity lends strong support to the hypothesis that basal autophagic activity decreases with age, thus contributing to the accumulation of damaged organelles and macromolecules and susceptibility to aging-related diseases.103 Indeed, prior studies demonstrated mitochondrial dysfunction in various animal models and in human OA.104 In addition, mitochondrial DNA mutations increase in OA chondrocytes.105 Damaged mitochondria, producing high levels of ROS, promote pro-inflammatory signaling, as they initiate formation of inflammasomes and activation of other inflammatory pathways.106 In knee chondrocytes, IL1- or nitric oxide-dependent increase in expression of MAP1LC3 and BECN1 activates autophagy.107 Furthermore, autophagy activation prevents IL1-mediated suppression of cartilage matrix degradation while reducing the levels of MMP13, ADAMTS5 (ADAM metalloproteinase with thrombospondin type 1 motif, 5), and ROS. Given that one of the cytoprotective functions of autophagy is removal of damaged mitochondria,108 the IL1–induced OA-like gene expression changes might possibly occur through reduction in the intracellular ROS level via elimination of damaged mitochondria.
As discussed above, HIF1A-like proteins have recently been linked to the regulation of autophagy in chondrocytes.85 With respect to HIF1A, in the superficial cartilage zone, there is a moderate level of protein expression; both expression and autophagy are increased in OA cells.102 It is likely that HIF1A upregulation due to the hypoxic state of the tissue serves to promote chondrocyte autophagy. With respect to EPAS1/HIF2A (endothelial PAS domain protein 1), in young animals, the highest expression level is in the superficial zone. In this case, upregulation of EPAS1 lowers intracellular ROS levels by promoting the activities of the dismutating proteins, CAT (catalase) and SOD (superoxide dismutase); from this perspective, it can be regarded as cytoprotective. In OA cartilage, EPAS1 expression is induced in the earlier stage of OA, and it is downregulated at later stages.109,110
A second aspect of EPAS1 is that it increases expression of RUNX2 (runt-related transcription factor 2), a transcription factor, which induces expression of proteins associated with chondrocyte hypertrophy [collagen X, MMP13, and VEGFA (vascular endothelial growth factor A)].111 As RUNX2 and its target genes are overexpressed in OA cartilage, it has been suggested that chondrocytes express a differentiation program that is more characteristic of a hypertrophic state.112 On this basis, increased EPAS1-induced RUNX2 activation and hypertrophic differentiation may promote OA. EPAS1 overexpression promotes cartilage destruction and conversely, the severity of experimental OA is reduced in Epas1 mutant mice.110,113 Indeed, when Epas1 is silenced, IL1B-induced expression of ADAMTS4, MMP1, MMP3, MMP9, MMP12, and MMP13 is significantly decreased. Thus, HIF1A and EPAS1 have overlapping but differing roles: whereas EPAS1 appears to be cytoprotective, HIF1A promotes autophagy while sensitizing cells to local apoptogens. Put another way, EPAS1 regulates the extent of the autophagic response and can be viewed as acting as a brake to the accelerator function of HIF1A-like proteins. Finally, it should be noted that stress- or cytokine-induced activation of EPAS1 could outweigh the homeostatic effects of HIF1A, and promote chondrocyte hypertrophy, cell death, and matrix degradation.102 The marked inhibition of autophagy would have a negative impact on chondrocyte survival and differentiation.
Therapeutic Targets for Skeletal Disease
This review highlighted the importance of the autophagic response in relationship to the pathogenesis of diseases affecting bone and cartilage. The authors suspect that aside from disorders discussed here in some detail, there are hints that autophagic dysfunction may influence other diseases of the skeletal tissues. An incomplete list would include growth disorders, dysregulated endocrine function, osteoporosis, fibrous dysplasia, rheumatoid arthritis, and degenerative disc disease.
A priori, before delineating therapies directed at removing dysfunctional organelles and accumulated aggregated proteins, the underlying autophagic “defects” need to be defined with care. From what is already known of the autophagic process, it is clear that unless the nature of the defect is established, promotion of autophagic flux may well trigger the induction of apoptosis or even senescence. In this case, for many skeletal tissues where cell numbers are normally low and longevity is extended, this type of treatment would exacerbate the disease state. In contrast, when the site of aberrant dysfunction is known, modulation of the autophagy response may be a rational approach for treatment of conditions like the MPS.
While caution is urged, there have been some attempts to modify skeletal cell function through modulators of the autophagic response. Of the drugs examined, in OA tissues, the autophagy activator rapamycin prevents cell death and GAG loss, maintains cartilage cellularity, and decreases the expression of ADAMTS5, an important enzyme in cartilage extracellular matrix degradation.114 Likewise, on the basis of p-MTOR and or p-RPS6KB1 expression, there is evidence indicating that over 50% of chordomas, tumors of the intervertebral disc, are responsive to MTOR inhibitors.115 Since this kinase is the nexus for signals from the AKT-PtdIns3P, MAPK1/3, and TP53 signaling pathways, as well as nutrient (amino acid) levels and sensors of the oxygen and energy status, there are likely to be multiple sites for control of these skeletal tumors. Undoubtedly, molecular delineation of the autophagic process in skeletal tissues will provide unique functional insights, while at the same time uncover new approaches to treating the host of diseases that affect the axial and appendicular skeleton.
Acknowledgments
This work was supported by NIH grants AR050087 and AR055655 (IMS), AG007996 and AR056026 (ML), Arthritis Research UK (17655)(CW) and (18865) (RL), and Italian Telethon grant S12008TELB (CS). The authors thank Bradley Snyder for the art work.
Glossary
Abbreviations:
- ADAMTS
ADAM metallopeptidase with thrombospondin type 1 motif
- AKT
v-akt murine thymoma viral oncogene homolog
- ALS
amyotrophic lateral sclerosis
- PRKAA2
protein kinase, AMP-activated, alpha 2 catalytic subunit
- ATF4
activating transcription factor 4
- ATG
autophagy-related (protein)
- ATG
autophagy-related (gene)
- BECN1
Beclin 1, autophagy-related
- BID
BH3 interacting domain death agonist
- BMP
bone morphogenetic protein
- BNIP3
BCL2/adenovirus E1B 19 kDa interacting protein 3
- CALCOCO2/NDP52
calcium binding and coiled-coil domain 2
- cAMP
cyclic adenosine monophosphate
- CSF1
colony stimulating factor 1 (macrophage)
- CTSK
cathepsin K
- DVL2
dishevelled segment polarity protein 2
- EGLN1
egl-9 hypoxia-inducible factor 1
- EPAS1
endothelial PAS domain protein 1
- FOXO
forkhead box O
- GAG
glycosaminoglycans
- HIF1A
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
- KEAP1
kelch-like ECH-associated protein 1
- KIR
KEAP1-interacting region
- LAMP2
lysosomal-associated membrane protein 2
- MAP1LC3
microtubule-associated protein 1 light chain 3
- LIR
LC3-interacting region
- LSDs
lysosomal storage disorders
- MMP
matrix metallopeptidase
- MPS
mucopolysaccharidoses
- MTOR
mechanistic target of rapamycin
- NBR1
neighbor of BRCA1 gene 1
- NF1
neurofibromin 1
- NFKB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NFE2L2/NRF2
nuclear factor, erythroid 2-like 2
- OA
osteoarthritis
- OPTN
optineurin
- PB1
Phox and Bem1p domain
- PDB
Paget disease of bone
- Ptdlns3P
phosphatidylinositol-3-phosphate
- PTH
parathyroid hormone
- PTHLH
parathyroid hormone-like hormone
- TNFSF11/RANKL
tumor necrosis factor (ligand) superfamily, member 11
- ROS
reactive oxygen species
- RUNX2
runt-related transcription factor 2
- S1P
sphingosine 1-phosphate
- SNARE
soluble NSF attachment protein receptor
- SOST
sclerostin
- SQSTM1
sequestosome 1
- SUMF1
sulfatase modifying factor 1
- TLR4
toll-like receptor 4
- TP53
tumor protein p53
- TRAF6
TNF receptor-associated factor 6, E3 ubiquitin protein ligase
- UBA
ubiquitin-like modifier activating enzyme ubiquitin-associated (domain)
- VCP
valosin containing protein
- VEGFA
vascular endothelial growth factor A
- WDFY3/ALFY
WD repeat and FYVE domain containing 3
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/26679
References
- 1.Romer AS. The “ancient history” of bone. Ann N Y Acad Sci. 1963;109:168–76. doi: 10.1111/j.1749-6632.1963.tb13466.x. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481:314–20. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce BF, Xing L, Shakespeare W, Wang Y, Dalgarno D, Iuliucci J, Sawyer T. Regulation of bone remodeling and emerging breakthrough drugs for osteoporosis and osteolytic bone metastases. Kidney Int Suppl. 2003;85:S2–5. doi: 10.1046/j.1523-1755.63.s85.2.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Guo YF, Liu YZ, Liu YJ, Xiong DH, Liu XG, Wang L, Yang TL, Lei SF, Guo Y, et al. Pathway-based genome-wide association analysis identified the importance of regulation-of-autophagy pathway for ultradistal radius BMD. J Bone Miner Res. 2010;25:1572–80. doi: 10.1002/jbmr.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagao M, Feinstein TN, Ezura Y, Hayata T, Notomi T, Saita Y, Hanyu R, Hemmi H, Izu Y, Takeda S, et al. Sympathetic control of bone mass regulated by osteopontin. Proc Natl Acad Sci U S A. 2011;108:17767–72. doi: 10.1073/pnas.1109402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanagita M. BMP antagonists: their roles in development and involvement in pathophysiology. Cytokine Growth Factor Rev. 2005;16:309–17. doi: 10.1016/j.cytogfr.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Yang W, Tyler MA, Gao X, Duan C, Kim SO, Aronson JF, Popov V, Takahashi H, Saito H, et al. Noggin attenuates cerulein-induced acute pancreatitis and impaired autophagy. Pancreas. 2013;42:301–7. doi: 10.1097/MPA.0b013e31825b9f2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 9.Waters S, Marchbank K, Solomon E, Whitehouse CA. Autophagic receptors Nbr1 and p62 coregulate skeletal remodeling. Autophagy. 2010;6:981–3. doi: 10.4161/auto.6.7.13155. [DOI] [PubMed] [Google Scholar]
- 10.Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, Xavier GM, Kashima TG, Cobourne MT, Richards GO, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci U S A. 2010;107:12913–8. doi: 10.1073/pnas.0913058107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–25. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandri M, Barberi L, Bijlsma AY, Blaauw B, Dyar KA, Milan G, Mammucari C, Meskers CG, Pallafacchina G, Paoli A, et al. Signalling pathways regulating muscle mass in ageing skeletal muscle. The role of the IGF1-Akt-mTOR-FoxO pathway. Biogerontology. 2013;14:303–23. doi: 10.1007/s10522-013-9432-9. [DOI] [PubMed] [Google Scholar]
- 13.Almeida M. Unraveling the role of FoxOs in bone--insights from mouse models. Bone. 2011;49:319–27. doi: 10.1016/j.bone.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caramés B, Kiosses WB, Akasaki Y, Brinson DC, Eap W, Koziol J, Lotz MK. Glucosamine activates autophagy in vitro and in vivo. Arthritis Rheum. 2013;65:1843–52. doi: 10.1002/art.37977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279:47109–14. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- 16.Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–51. doi: 10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahm AM, Bohensky J, Adams CS, Shapiro IM, Srinivas V. Bone cell autophagy is regulated by environmental factors. Cells Tissues Organs. 2011;194:274–8. doi: 10.1159/000324647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia J, Yao W, Guan M, Dai W, Shahnazari M, Kar R, Bonewald L, Jiang JX, Lane NE. Glucocorticoid dose determines osteocyte cell fate. FASEB J. 2011;25:3366–76. doi: 10.1096/fj.11-182519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia X, Kar R, Gluhak-Heinrich J, Yao W, Lane NE, Bonewald LF, Biswas SK, Lo WK, Jiang JX. Glucocorticoid-induced autophagy in osteocytes. J Bone Miner Res. 2010;25:2479–88. doi: 10.1002/jbmr.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teitelbaum SL. The osteoclast and its unique cytoskeleton. Ann N Y Acad Sci. 2011;1240:14–7. doi: 10.1111/j.1749-6632.2011.06283.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Shao J, Chen W, Li YP. Osteoclast differentiation and gene regulation. Front Biosci. 2007;12:2519–29. doi: 10.2741/2252. [DOI] [PubMed] [Google Scholar]
- 22.Pavlos NJ, Ng PY. “Fusion and fission” unveils remarkable insights into osteoclast plasticity. Calcif Tissue Int. 2012;91:157–8, author reply 159. doi: 10.1007/s00223-012-9620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pederson L, Ruan M, Westendorf JJ, Khosla S, Oursler MJ. Regulation of bone formation by osteoclasts involves Wnt/BMP signaling and the chemokine sphingosine-1-phosphate. Proc Natl Acad Sci U S A. 2008;105:20764–9. doi: 10.1073/pnas.0805133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii M, Kikuta J. Sphingosine-1-phosphate signaling controlling osteoclasts and bone homeostasis. Biochim Biophys Acta. 2013;1831:223–7. doi: 10.1016/j.bbalip.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Lee SH, Lee SY, Lee YS, Kim BJ, Lim KH, Cho EH, Kim SW, Koh JM, Kim GS. Higher circulating sphingosine 1-phosphate levels are associated with lower bone mineral density and higher bone resorption marker in humans. J Clin Endocrinol Metab. 2012;97:E1421–8. doi: 10.1210/jc.2012-1044. [DOI] [PubMed] [Google Scholar]
- 26.Arnett TR, Gibbons DC, Utting JC, Orriss IR, Hoebertz A, Rosendaal M, Meghji S. Hypoxia is a major stimulator of osteoclast formation and bone resorption. J Cell Physiol. 2003;196:2–8. doi: 10.1002/jcp.10321. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Chen G, Zhang W, Xu N, Zhu JY, Jia J, Sun ZJ, Wang YN, Zhao YF. Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1α/BNIP3 signaling pathway. J Cell Physiol. 2012;227:639–48. doi: 10.1002/jcp.22768. [DOI] [PubMed] [Google Scholar]
- 28.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966–74. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung YH, Yoon SY, Choi B, Sohn DH, Yoon KH, Kim WJ, Kim DH, Chang EJ. Microtubule-associated protein light chain 3 regulates Cdc42-dependent actin ring formation in osteoclast. Int J Biochem Cell Biol. 2012;44:989–97. doi: 10.1016/j.biocel.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC. Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62’s role in neurodegenerative disease. J Biomed Biotechnol. 2006;2006:62079. doi: 10.1155/JBB/2006/62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isakson P, Holland P, Simonsen A. The role of ALFY in selective autophagy. Cell Death Differ. 2013;20:12–20. doi: 10.1038/cdd.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hocking LJ, Mellis DJ, McCabe PS, Helfrich MH, Rogers MJ. Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochem Biophys Res Commun. 2010;402:543–8. doi: 10.1016/j.bbrc.2010.10.076. [DOI] [PubMed] [Google Scholar]
- 33.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–79. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung PY, Van Hul W. Paget’s disease of bone: evidence for complex pathogenetic interactions. Semin Arthritis Rheum. 2012;41:619–41. doi: 10.1016/j.semarthrit.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Ralston SH, Layfield R. Pathogenesis of Paget disease of bone. Calcif Tissue Int. 2012;91:97–113. doi: 10.1007/s00223-012-9599-0. [DOI] [PubMed] [Google Scholar]
- 36.Chamoux E, Couture J, Bisson M, Morissette J, Brown JP, Roux S. The p62 P392L mutation linked to Paget’s disease induces activation of human osteoclasts. Mol Endocrinol. 2009;23:1668–80. doi: 10.1210/me.2009-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon LR. Billiard-player’s fingers: an unusual case of Paget’s disease of bone. Br Med J. 1979;1:931. doi: 10.1136/bmj.1.6168.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lever JH. Paget’s disease of bone in Lancashire and arsenic pesticide in cotton mill wastewater: a speculative hypothesis. Bone. 2002;31:434–6. doi: 10.1016/S8756-3282(02)00833-5. [DOI] [PubMed] [Google Scholar]
- 39.Michou L, Collet C, Morissette J, Audran M, Thomas T, Gagnon E, Launay JM, Laplanche JL, Brown JP, Orcel P. Epidemiogenetic study of French families with Paget’s disease of bone. Joint Bone Spine. 2012;79:393–8. doi: 10.1016/j.jbspin.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Roodman GD. Insights into the pathogenesis of Paget’s disease. Ann N Y Acad Sci. 2010;1192:176–80. doi: 10.1111/j.1749-6632.2009.05214.x. [DOI] [PubMed] [Google Scholar]
- 41.Morales-Piga AA, Rey-Rey JS, Corres-González J, García-Sagredo JM, López-Abente G. Frequency and characteristics of familial aggregation of Paget’s disease of bone. J Bone Miner Res. 1995;10:663–70. doi: 10.1002/jbmr.5650100421. [DOI] [PubMed] [Google Scholar]
- 42.Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum Mol Genet. 2002;11:2735–9. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- 43.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 44.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–44. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 45.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285:16782–8. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–6. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem. 2006;281:18015–24. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 48.Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22:1645–54. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–46. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–89. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 51.Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, Layfield R. Loss of ubiquitin-binding associated with Paget’s disease of bone p62 (SQSTM1) mutations. J Bone Miner Res. 2005;20:619–24. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- 52.Cavey JR, Ralston SH, Sheppard PW, Ciani B, Gallagher TR, Long JE, Searle MS, Layfield R. Loss of ubiquitin binding is a unifying mechanism by which mutations of SQSTM1 cause Paget’s disease of bone. Calcif Tissue Int. 2006;78:271–7. doi: 10.1007/s00223-005-1299-6. [DOI] [PubMed] [Google Scholar]
- 53.Najat D, Garner T, Hagen T, Shaw B, Sheppard PW, Falchetti A, Marini F, Brandi ML, Long JE, Cavey JR, et al. Characterization of a non-UBA domain missense mutation of sequestosome 1 (SQSTM1) in Paget’s disease of bone. J Bone Miner Res. 2009;24:632–42. doi: 10.1359/jbmr.081204. [DOI] [PubMed] [Google Scholar]
- 54.Rea SL, Walsh JP, Ward L, Magno AL, Ward BK, Shaw B, Layfield R, Kent GN, Xu J, Ratajczak T. Sequestosome 1 mutations in Paget’s disease of bone in Australia: prevalence, genotype/phenotype correlation, and a novel non-UBA domain mutation (P364S) associated with increased NF-kappaB signaling without loss of ubiquitin binding. J Bone Miner Res. 2009;24:1216–23. doi: 10.1359/jbmr.090214. [DOI] [PubMed] [Google Scholar]
- 55.Hocking LJ, Whitehouse C, Helfrich MH. Autophagy: a new player in skeletal maintenance? J Bone Miner Res. 2012;27:1439–47. doi: 10.1002/jbmr.1668. [DOI] [PubMed] [Google Scholar]
- 56.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inomata M, Niida S, Shibata K, Into T. Regulation of Toll-like receptor signaling by NDP52-mediated selective autophagy is normally inactivated by A20. Cell Mol Life Sci. 2012;69:963–79. doi: 10.1007/s00018-011-0819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–31. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colleran A, Ryan A, O’Gorman A, Mureau C, Liptrot C, Dockery P, Fearnhead H, Egan LJ. Autophagosomal IkappaB alpha degradation plays a role in the long term control of tumor necrosis factor-alpha-induced nuclear factor-kappaB (NF-kappaB) activity. J Biol Chem. 2011;286:22886–93. doi: 10.1074/jbc.M110.199950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Mariño G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 2011;30:4908–20. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niso-Santano M, Criollo A, Malik SA, Michaud M, Morselli E, Mariño G, Lachkar S, Galluzzi L, Maiuri MC, Kroemer G. Direct molecular interactions between Beclin 1 and the canonical NFκB activation pathway. Autophagy. 2012;8:268–70. doi: 10.4161/auto.8.2.18845. [DOI] [PubMed] [Google Scholar]
- 62.Salminen A, Hyttinen JM, Kauppinen A, Kaarniranta K. Context-dependent regulation of autophagy by IKK-NF-kappaB signaling: impact on the aging process. Int J Cell Biol. 2012;2012:849541. doi: 10.1155/2012/849541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70:1582–8. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falchetti A, Di Stefano M, Marini F, Ortolani S, Ulivieri MF, Bergui S, Masi L, Cepollaro C, Benucci M, Di Munno O, et al. GenePage Project Genetic epidemiology of Paget’s disease of bone in italy: sequestosome1/p62 gene mutational test and haplotype analysis at 5q35 in a large representative series of sporadic and familial Italian cases of Paget’s disease of bone. Calcif Tissue Int. 2009;84:20–37. doi: 10.1007/s00223-008-9192-8. [DOI] [PubMed] [Google Scholar]
- 65.Michou L, Morissette J, Gagnon ER, Marquis A, Dellabadia M, Brown JP, Siris ES. Novel SQSTM1 mutations in patients with Paget’s disease of bone in an unrelated multiethnic American population. Bone. 2011;48:456–60. doi: 10.1016/j.bone.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Collet C, Michou L, Audran M, Chasseigneaux S, Hilliquin P, Bardin T, Lemaire I, Cornélis F, Launay JM, Orcel P, et al. Paget’s disease of bone in the French population: novel SQSTM1 mutations, functional analysis, and genotype-phenotype correlations. J Bone Miner Res. 2007;22:310–7. doi: 10.1359/jbmr.061106. [DOI] [PubMed] [Google Scholar]
- 67.Daroszewska A, van ’t Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, Rose L, Rose K, Ralston SH. A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget’s disease-like disorder in mice. Hum Mol Genet. 2011;20:2734–44. doi: 10.1093/hmg/ddr172. [DOI] [PubMed] [Google Scholar]
- 68.Azzam E, Helfrich MH, Hocking LJ. Paget’s disease-causing mutations in Sequestosome-1 impair autophagic protein degradation. J Bone Miner Res. 2011;26:Abstr 1081. [Google Scholar]
- 69.Fecto F, Siddique T. UBQLN2/P62 cellular recycling pathways in amyotrophic lateral sclerosis and frontotemporal dementia. Muscle Nerve. 2012;45:157–62. doi: 10.1002/mus.23278. [DOI] [PubMed] [Google Scholar]
- 70.Rubino E, Rainero I, Chiò A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, et al. TODEM Study Group SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–62. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–27. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–88. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Badadani M, Nalbandian A, Watts GD, Vesa J, Kitazawa M, Su H, Tanaja J, Dec E, Wallace DC, Mukherjee J, et al. VCP associated inclusion body myopathy and paget disease of bone knock-in mouse model exhibits tissue pathology typical of human disease. PLoS One. 2010;5:e13183. doi: 10.1371/journal.pone.0013183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albagha OM, Visconti MR, Alonso N, Langston AL, Cundy T, Dargie R, Dunlop MG, Fraser WD, Hooper MJ, Isaia G, et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat Genet. 2010;42:520–4. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Obaid R, Wani S, Ralston SH, Albagha OM. OPTN negatively regulates osteoclast formation in vitro. Bone. 2012;50(Suppl 1):S92–3. doi: 10.1016/j.bone.2012.02.276. [DOI] [Google Scholar]
- 76.Wilsman NJ, Bernardini ES, Leiferman E, Noonan K, Farnum CE. Age and pattern of the onset of differential growth among growth plates in rats. J Orthop Res. 2008;26:1457–65. doi: 10.1002/jor.20547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson HC, Garimella R, Tague SE. The role of matrix vesicles in growth plate development and biomineralization. Front Biosci. 2005;10:822–37. doi: 10.2741/1576. [DOI] [PubMed] [Google Scholar]
- 78.Álvarez-García Ó, García-López E, Loredo V, Gil-Peña H, Mejía-Gaviria N, Rodríguez-Suárez J, Ordóñez FÁ, Santos F. Growth hormone improves growth retardation induced by rapamycin without blocking its antiproliferative and antiangiogenic effects on rat growth plate. PLoS One. 2012;7:e34788. doi: 10.1371/journal.pone.0034788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shapiro IM, Adams CS, Freeman T, Srinivas V. Fate of the hypertrophic chondrocyte: microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res C Embryo Today. 2005;75:330–9. doi: 10.1002/bdrc.20057. [DOI] [PubMed] [Google Scholar]
- 80.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 81.Srinivas V, Bohensky J, Zahm AM, Shapiro IM. Autophagy in mineralizing tissues: microenvironmental perspectives. Cell Cycle. 2009;8:391–3. doi: 10.4161/cc.8.3.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15:2865–76. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shapiro IM, Srinivas V. Metabolic consideration of epiphyseal growth: survival responses in a taxing environment. Bone. 2007;40:561–7. doi: 10.1016/j.bone.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–14. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 85.Bohensky J, Leshinsky S, Srinivas V, Shapiro IM. Chondrocyte autophagy is stimulated by HIF-1 dependent AMPK activation and mTOR suppression. Pediatr Nephrol. 2010;25:633–42. doi: 10.1007/s00467-009-1310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terkhorn SP, Bohensky J, Shapiro IM, Koyama E, Srinivas V. Expression of HIF prolyl hydroxylase isozymes in growth plate chondrocytes: relationship between maturation and apoptotic sensitivity. J Cell Physiol. 2007;210:257–65. doi: 10.1002/jcp.20873. [DOI] [PubMed] [Google Scholar]
- 88.Schröder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–76. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 89.Muenzer J. Overview of the mucopolysaccharidoses. Rheumatology (Oxford) 2011;50(Suppl 5):v4–12. doi: 10.1093/rheumatology/ker394. [DOI] [PubMed] [Google Scholar]
- 90.Cosma MP, Pepe S, Annunziata I, Newbold RF, Grompe M, Parenti G, Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–56. doi: 10.1016/S0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 91.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–96. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 92.Settembre C, Annunziata I, Spampanato C, Zarcone D, Cobellis G, Nusco E, Zito E, Tacchetti C, Cosma MP, Ballabio A. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci U S A. 2007;104:4506–11. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Settembre C, Arteaga-Solis E, McKee MD, de Pablo R, Al Awqati Q, Ballabio A, Karsenty G. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22:2645–50. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shimada Y, Klionsky DJ. Autophagy contributes to lysosomal storage disorders. Autophagy. 2012;8:715–6. doi: 10.4161/auto.19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–30. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fraldi A, Annunziata F, Lombardi A, Kaiser HJ, Medina DL, Spampanato C, Fedele AO, Polishchuk R, Sorrentino NC, Simons K, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–20. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51:241–8. doi: 10.1016/j.bone.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen QT, Wong BL, Chun J, Yoon YC, Talke FE, Sah RL. Macroscopic assessment of cartilage shear: effects of counter-surface roughness, synovial fluid lubricant, and compression offset. J Biomech. 2010;43:1787–93. doi: 10.1016/j.jbiomech.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hollander AP, Pidoux I, Reiner A, Rorabeck C, Bourne R, Poole AR. Damage to type II collagen in aging and osteoarthritis starts at the articular surface, originates around chondrocytes, and extends into the cartilage with progressive degeneration. J Clin Invest. 1995;96:2859–69. doi: 10.1172/JCI118357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lotz MK, Otsuki S, Grogan SP, Sah R, Terkeltaub R, D’Lima D. Cartilage cell clusters. Arthritis Rheum. 2010;62:2206–18. doi: 10.1002/art.27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Almonte-Becerril M, Navarro-Garcia F, Gonzalez-Robles A, Vega-Lopez MA, Lavalle C, Kouri JB. Cell death of chondrocytes is a combination between apoptosis and autophagy during the pathogenesis of Osteoarthritis within an experimental model. Apoptosis. 2010;15:631–8. doi: 10.1007/s10495-010-0458-z. [DOI] [PubMed] [Google Scholar]
- 102.Bohensky J, Terkhorn SP, Freeman TA, Adams CS, Garcia JA, Shapiro IM, Srinivas V. Regulation of autophagy in human and murine cartilage: hypoxia-inducible factor 2 suppresses chondrocyte autophagy. Arthritis Rheum. 2009;60:1406–15. doi: 10.1002/art.24444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 104.Blanco FJ, Rego I, Ruiz-Romero C. The role of mitochondria in osteoarthritis. Nat Rev Rheumatol. 2011;7:161–9. doi: 10.1038/nrrheum.2010.213. [DOI] [PubMed] [Google Scholar]
- 105.Kim J, Xu M, Xo R, Mates A, Wilson GL, Pearsall AW, 4th, Grishko V. Mitochondrial DNA damage is involved in apoptosis caused by pro-inflammatory cytokines in human OA chondrocytes. Osteoarthritis Cartilage. 2010;18:424–32. doi: 10.1016/j.joca.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 106.Tschopp J. Mitochondria: Sovereign of inflammation? Eur J Immunol. 2011;41:1196–202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- 107.Sasaki H, Takayama K, Matsushita T, Ishida K, Kubo S, Matsumoto T, Fujita N, Oka S, Kurosaka M, Kuroda R. Autophagy modulates osteoarthritis-related gene expression in human chondrocytes. Arthritis Rheum. 2012;64:1920–8. doi: 10.1002/art.34323. [DOI] [PubMed] [Google Scholar]
- 108.Kim J, Xu M, Xo R, Mates A, Wilson GL, Pearsall AW, 4th, Grishko V. Mitochondrial DNA damage is involved in apoptosis caused by pro-inflammatory cytokines in human OA chondrocytes. Osteoarthritis Cartilage. 2010;18:424–32. doi: 10.1016/j.joca.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 109.Saito T, Fukai A, Mabuchi A, Ikeda T, Yano F, Ohba S, Nishida N, Akune T, Yoshimura N, Nakagawa T, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med. 2010;16:678–86. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 110.Yang S, Kim J, Ryu JH, Oh H, Chun CH, Kim BJ, Min BH, Chun JS. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat Med. 2010;16:687–93. doi: 10.1038/nm.2153. [DOI] [PubMed] [Google Scholar]
- 111.Solomon LA, Bérubé NG, Beier F. Transcriptional regulators of chondrocyte hypertrophy. Birth Defects Res C Embryo Today. 2008;84:123–30. doi: 10.1002/bdrc.20124. [DOI] [PubMed] [Google Scholar]
- 112.Wu L, Huang X, Li L, Huang H, Xu R, Luyten W. Insights on biology and pathology of HIF-1α/-2α, TGFβ/BMP, Wnt/β-catenin, and NF-κB pathways in osteoarthritis. Curr Pharm Des. 2012;18:3293–312. doi: 10.2174/1381612811209023293. [DOI] [PubMed] [Google Scholar]
- 113.Yang S, Kim J, Ryu JH, Oh H, Chun CH, Kim BJ, Min BH, Chun JS. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat Med. 2010;16:687–93. doi: 10.1038/nm.2153. [DOI] [PubMed] [Google Scholar]
- 114.Li D, Wu Z, Duan Y, Hao D, Zhang X, Luo H, Chen B, Qiu G. TNFα-mediated apoptosis in human osteoarthritic chondrocytes sensitized by PI3K-NF-κB inhibitor, not mTOR inhibitor. Rheumatol Int. 2012;32:2017–22. doi: 10.1007/s00296-011-1929-4. [DOI] [PubMed] [Google Scholar]
- 115.Tamborini E, Virdis E, Negri T, Orsenigo M, Brich S, Conca E, Gronchi A, Stacchiotti S, Manenti G, Casali PG, et al. Analysis of receptor tyrosine kinases (RTKs) and downstream pathways in chordomas. Neuro Oncol. 2010;12:776–89. doi: 10.1093/neuonc/noq003. [DOI] [PMC free article] [PubMed] [Google Scholar]