Abstract
Exposure to polycyclic aromatic hydrocarbons (PAHs) in the food chain is the major human health hazard associated with the Deepwater Horizon oil spill. Phenanthrene is a representative PAH present in crude oil, and it undergoes biological transformation, photooxidation, and chemical oxidation to produce its signature oxygenated derivative, phenanthrene-9,10-quinone. We report the downstream metabolic fate of phenanthrene-9,10-quinone in HepG2 cells. The structures of the metabolites were identified by HPLC–UV–fluorescence detection and LC–MS/MS. O-mono-Glucuronosyl-phenanthrene-9,10-catechol was identified, as reported previously. A novel bis-conjugate, O-mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol, was discovered for the first time, and evidence for both of its precursor mono conjugates was obtained. The identities of these four metabolites were unequivocally validated by comparison to authentic enzymatically synthesized standards. Evidence was also obtained for a minor metabolic pathway of phenanthrene-9,10-quinone involving bis-hydroxylation followed by O-mono-sulfonation. The identification of 9,10-catechol conjugates supports metabolic detoxification of phenanthrene-9,10-quinone through interception of redox cycling by UGT, COMT, and SULT isozymes and indicates the possible use of phenanthrene-9,10-catechol conjugates as biomarkers of human exposure to oxygenated PAH.
Introduction
The Deepwater Horizon oil spill was the largest release of crude oil in U.S. history.1,2 Over 200 million gallons of crude oil were released from the Macondo well in the Gulf of Mexico.3−5 Crude oil is a complex mixture and contains thousands of hydrocarbons of various types. Among them, polycyclic aromatic hydrocarbons (PAHs) are recognized as one of the most toxic and persistent components.6
Phenanthrene is one of the most abundant petrogenic PAHs based on the compositional analysis of Macondo well oil.7 Biological transformation, photooxidation, and chemical oxidation of phenanthrene produce its signature oxygenated derivative, phenanthrene-9,10-quinone.8−11 Contamination of the food chain with PAHs is a great concern related to human health.12 Filter-feeding bivalves near contaminated sediments are the first invertebrate targets for exposure to PAHs and can accumulate PAHs because they have a very poor metabolic clearance for PAHs. PAHs can then move through the food chain via predation. Teleost fish have a well-developed capacity to metabolize PAHs, and the metabolites of PAHs can be more harmful than the parent PAHs. Thus, both shell fish and fin fish can be contaminated by PAHs and oxygenated PAH,13 and fin fish may be contaminated by PAH metabolites as well. If contaminated sea food is consumed, then a human health hazard could exist.
The toxicity of representative oxygenated PAH phenanthrene-9,10-quinone has been associated with the generation of reactive oxygen species (ROS) via the process of redox cycling.14,15 Phenanthrene-9,10-quinone can be enzymatically and nonenzymatically reduced back to the catechol and can establish futile redox cycles that result in the amplification of ROS until cellular reducing equivalents (e.g., NADPH) are depleted.14,15 The enzyme responsible for the one-electron reduction of phenanthrene-9,10-quinone to the semiquinone radical is NADPH:cytochrome P450 oxidoreductase (POR), whereas the enzymes responsible for the two-electron reduction of phenanthrene-9,10-quinone to the catechol include NAD(P)H:quinone oxidoreductase 1 (NQO1), aldo-keto reductases (AKRs), and carbonyl reductase (CBR).14−19 It has been also reported that the redox cycling of phenanthrene-9,10-quinone is terminated in human A549 cells through transporting a metabolite of phenanthrene-9,10-quinone, an O-mono-glucuronosyl catechol conjugate, to extracellular space.20 These findings suggest that investigating the downstream metabolism of phenanthrene-9,10-quinone is key to understanding the detoxification of this oxygenated PAH.
Human liver cells are major sites of ingestion exposure to phenanthrene-9,10-quinone and generation of its metabolites. However, to our knowledge, information relevant to the liver cell-based metabolism of phenanthrene-9,10-quinone is lacking. The objective of this study was to elucidate the metabolic fate of phenanthrene-9,10-quinone in human hepatoma (HepG2) cells as a model for metabolism by human hepatocytes.
We found that the pathways of phenanthrene-9,10-quinone metabolism involved the reduction to the catechol followed by O-mono-glucuronidation, O-mono-sulfonation, O-mono-methylation, and O-mono-methylation-O-mono-sulfonation as well as bis-hydroxylation followed by O-mono-sulfonation. We conclude that quinone reduction followed by phase II conjugation is a potential detoxification pathway of phenanthrene-9,10-quinone following ingestion. As catechol conjugates were major metabolites, they could be used as biomarkers of human exposure to phenanthrene-9,10-quinone and oxygenated PAH in general.
Materials and Methods
Caution:
These chemicals are dangerous. All PAHs are potentially hazardous and should be handled in accordance with the NIH Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Reagents
Cell culture medium and reagents were all obtained from Invitrogen Co. (Carlsbad, CA) except for fetal bovine serum (FBS), which was purchased from Hyclone (Logan, UT). Human recombinant uridine 5′-diphospho-glucuronosyltransferases (UGT) 2B7 Supersomes (microsomes from baculovirus-infected insect cells expressing UGTs) were obtained from BD Biosciences (San Jose, CA). Human recombinant sulfotransferases (SULT) 1A1 and human recombinant catechol-O-methyltransferase (COMT) were expressed and purified according to published methods.21,22 Phenanthrene, phenanthrene-9,10-quinone, dithiothreitol, uridine-5′-diphosphoglucuronic acid (UDPGA), adenosine 3′-phosphate 5′-phosphosulfate (PAPS), and S-(5′-adenosyl)-l-methionine (AdoMet) chloride were purchased from Sigma-Aldrich Co. (St. Louis, MO). All other chemicals used were of the highest grade available, and all solvents were HPLC grade.
Cell Culture
HepG2 (human hepatocellular carcinoma) cells were obtained from American Type Culture Collection and were maintained in minimum essential medium (MEM) with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and were passaged every week at a 1:7 dilution. Cultured cells with a passage number of 10–20 were used in the experiments to reduce variability resulting from cell culture conditions.
Detection and Identification of Phenanthrene-9,10-quinone Metabolites in HepG2 Cells
The cells (∼5 × 106) were treated with phenanthrene-9,10-quinone (1 μM, 0.2% DMSO) in MEM (without phenol red) containing 10 mM glucose as an energy source. The culture media were collected at 0 and 24 h and were subsequently acidified with 0.1% formic acid before extraction twice with a 1.5-fold volume of cold ethyl acetate saturated with H2O. The acidification is an essential step because it leads to the neutralization of sulfate and glucuronide conjugates that can then be extracted into the organic phase. The organic phases from the extracted culture media were combined and dried under vacuum. The residue was redissolved in 150 μL of methanol. No analysis was performed on the resultant aqueous phase.
For UV and fluorescence studies, a 10 μL aliquot was analyzed on a tandem Waters Alliance 2695 chromatographic system (Waters Corporation, Milford, MA) with a Waters 2996 photodiode array (PDA) detector and a Waters 2475 multi λ fluorescence (FLR) detector. Separations were accomplished on a reversed-phase (RP) column (Zorbax-ODS C18, 5 μm, 4.6 mm × 250 mm) (DuPont Co., Wilmington, DE) with a guard column at ambient temperature. The mobile phase consisted of 5 mM ammonium acetate and 0.1% trifluoroacetic acid (TFA) (v/v) in H2O (solvent A) and 5 mM ammonium acetate and 0.1% TFA in acetonitrile (solvent B) and was delivered at a flow rate of 0.5 mL/min. The linear gradient elution program was as follows: 5 to 95% B over 30 min followed by an isocratic hold at 95% B for another 10 min. At 40 min, B was returned to 5% in 1 min, and the column was equilibrated for 19 min before the next injection. The total run time for each analysis was 60 min. Eluants from the column were introduced sequentially into the PDA detector and the FLR detector. Excitation (λex) and emission (λem) wavelengths for the FLR detector were set at 252 and 365 nm, respectively, based on the spectral properties of phenanthrene.
For MS analysis, a 10 μL aliquot was analyzed on a Waters Alliance 2690 HPLC system (Waters Corporation, Milford, MA) coupled to a Finnigan LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The column, mobile phase, flow rate, and linear gradient elution program were the same as described above. During LC–MS/MS analysis, up to 10 min of the initial flow was diverted away from the mass spectrometer before evaluation of the eluants. The mass spectrometer was operated in both the positive and negative ion modes with an electrospray ionization (ESI) source. Eluants were monitored on the LTQ using product ion scan (MS2), subsequent MS/MS/MS (MS3), and pseudo-selected reaction monitoring (SRM) modes. The mass spectrometry parameters, including spray voltage (4 kV in positive ion mode; 4.5 kV in negative ion mode), sheath gas flow rate (35 arbitrary units in both ion modes), auxiliary gas flow rate (18 arbitrary units in both ion modes), capillary temperature (220 °C in both ion modes), capillary voltage (27 V in positive ion mode; −5 V in negative ion mode), and tube lens (110 V in positive ion mode; −22.05 V in negative ion mode), were automatically optimized with a phenanthrene-9,10-quinone standard solution in methanol. An isolation width of three bracketed around the m/z of interest, an activation Q of 0.25, and an activation time of 30 ms were used for data acquisition. Xcalibur version 2.0 software (Thermo Fisher Scientific) was used to control the LC–MS/MS system and to process the data.
In some instances, another 5 μL aliquot was analyzed on a nano-Acquity ultra-performance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Separations were accomplished on an analytical column (C18, 1.7 μm BEH130, 150 μm × 100 mm) (Waters Corporation, Milford, MA) at 50 °C. The mobile phase consisted of 0.1% formic acid (v/v) in H2O (solvent A) and 0.1% formic acid (v/v) in acetonitrile (solvent B) and was delivered at a flow rate of 1.6 μL/min. The linear gradient elution program was as follows: an isocratic hold at 5% B for 5 min and then 5 to 95% B over 30 min followed by an isocratic hold at 95% B for another 10 min. At 46 min, B was returned to 5% in 2 min, and the column was equilibrated for 12 min before the next injection. The total run time for each analysis was 60 min. The mass spectrometer was operated in the positive and negative ion modes with a nanoelectrospray ionization (nano-ESI) source after accurate calibration with the manufacturer’s calibration mixture. The ionization voltage was set to 1.5 kV, and the capillary temperature was set to 200 °C. Full-scan spectra were acquired with a resolving power of 60 000 full-width half-maximum (FWHM) in a mass range from m/z 100 to 800.
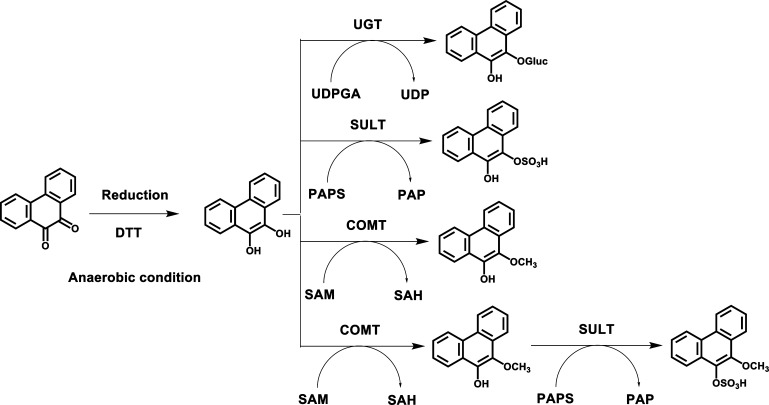
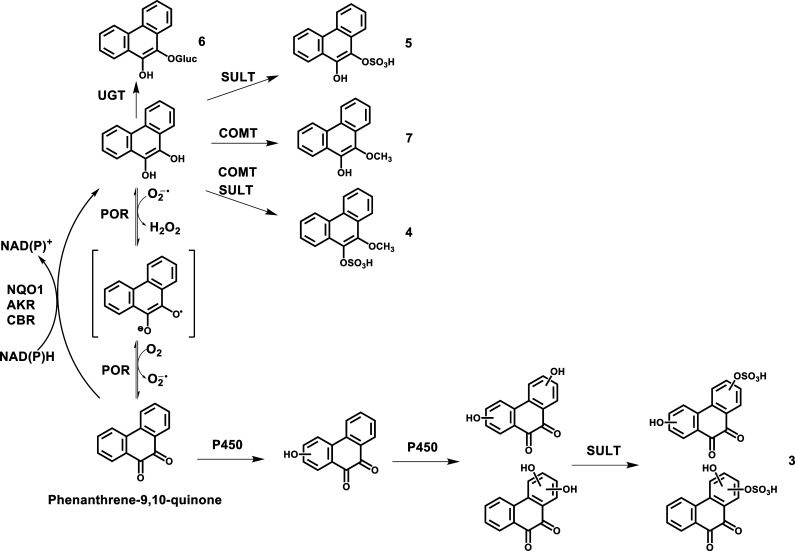
Synthesis of Four Phenanthrene-9,10-catechol Conjugates
Synthetic routes of four phenanthrene-9,10-catechol conjugates are shown in Scheme 1. Experiments were conducted anaerobically in a glovebox purged with argon as previously described so that phenanthrene-9,10-quinone could be reduced with dithiothreitol to the catechol.21−23 Anaerobic incubation is required because phenanthrene-9,10-catechol is air-sensitive and can autooxidize in air back to the quinone. All solutions were degassed by freeze–pump–thaw cycling five times and were stored in sealed containers filled with argon. The reactions were performed in 1.5 mL amber glass vials with polytetrafluoroethylene/silicone septa closures. Once the catechol conjugates are formed, they are no longer air sensitive. UGT2B7 and SULT1A1 were selected as the conjugating enzymes because previous studies showed that they could conjugate benzo[a]pyrene-7,8-catechol.21,23
Scheme 1. Synthetic Routes of Four Phenanthrene-9,10-catechol Conjugates.
For glucuronidation of phenanthrene-9,10-catechol, the reaction system consisted of 10 mM phosphate buffer (pH 7.4), 0.025 mg/mL of alamethicin, 1.0 mM dithiothreitol, 5.0 mM MgCl2, 1 mM UDPGA, 30 μg of human recombinant UGT2B7 microsomes, and 10 μM phenanthrene-9,10-quinone in a final volume of 0.2 mL. For sulfonation of phenanthrene-9,10-catechol, the reaction system consisted of 10 mM phosphate buffer (pH 7.4), 1.0 mM dithiothreitol, 5.0 mM MgCl2, 20 μM PAPS, 1 μg of human recombinant SULT1A1, and 10 μM phenanthrene-9,10-quinone in a final volume of 0.2 mL. For methylation of phenanthrene-9,10-catechol, the reaction system consisted of 10 mM phosphate buffer (pH 7.8), 1.0 mM dithiothreitol, 1.0 mM MgCl2, 50 μM AdoMet, 1 μg of human recombinant COMT, 10 μM phenanthrene-9,10-quinone in a final volume of 0.2 mL. The reaction systems for mono-conjugation were incubated at 37 °C for 1 h.
For bis-conjugation, methylation of phenanthrene-9,10-catechol was conducted first at 37 °C for 1 h followed by sulfonation of O-mono-methyl phenanthrene-9,10-catechol at 37 °C for another 1 h, which was initiated by the addition of 20 μM PAPS and 1 μg of human recombinant SULT1A1 into the methylation system. The negative controls of the four reactions involved initiating the reaction in the absence of the respective cofactors. All of the reactions were quenched by the addition of 50 μL of ice-cold 1% formic acid and were chilled on ice.
The reaction mixtures were extracted twice with a 2-fold volume of cold ethyl acetate saturated with H2O. The combined ethyl acetate layer was backwashed with 0.2 mL of 1% formic acid by vigorous vortexing and centrifuged at 16 000 g. The organic phases were combined and dried under vacuum. The residue was dissolved in 100 μL methanol and was subsequently analyzed by HPLC–UV–FLR detection and LC–MS/MS in the same manner as described above. The identity of phenanthrene-9,10-catechol conjugates was validated using MS2, subsequent MS3, and pseudo-SRM modes.
Time Course of Phenanthrene-9,10-quinone Consumption and Its Metabolite Formation in HepG2 Cells
The cells (∼5 × 106) were treated with phenanthrene-9,10-quinone (1 μM, 0.2% DMSO), and the culture media were collected at 0, 3, 8, 24, 48, and 72 h. To each sample of medium was added 1 μL phenanthrene stock solution in DMSO as an internal standard (30 ng/mL), which was subsequently extracted and analyzed by HPLC–UV–FLR in the same manner as described above. Peak area ratios of each analyte and internal standard were calculated and plotted against the incubation time using Microsoft Excel. Data points are the mean of three measurements ± SD.
Results
Strategy
HepG2 cells were treated with phenanthrene-9,10-quinone to detect and identify the potential metabolites using HPLC–UV–FLR and ion trap LC–MS/MS. The combination of UV and FLR detection was used to confirm the number of metabolites of interest because most PAH metabolites show strong UV and fluorescence signals. The peak areas on the UV and FLR chromatograms provided the metabolite profiles with regard to their relative quantitation. The structural information of the metabolites was obtained on the basis of the corresponding MS2 and MS3 spectra from ion trap LC–MS/MS. The identities of the metabolites were subsequently validated by comparison to authentic enzymatically synthesized standards.
Because phenanthrene-9,10-quinone does not show a fluorescence signal, phenanthrene was selected to obtain the optimal pair of λex and λem wavelengths (Figure S1) to detect the metabolites of phenanthrene-9,10-quinone. Pilot studies with phenanthrene-9,10-quinone in this cell line indicated that the metabolite profile from the organic phase of the extracted media at 24 h was the most comprehensive among the time points studied (data not shown). Thus, in our studies, we elected to detect metabolites following a 24 h treatment with phenanthrene-9,10-quinone. Few, if any, metabolites were observed in the cell pellets, and these were not processed further.
We considered three possible routes for phenanthrene-9,10-quinone metabolism to detect and identify metabolites by ion trap LC–MS/MS. First, we predicted that reduction of phenanthrene-9,10-quinone to the catechol followed by formation of phase II conjugates could be the major metabolic pathway of phenanthrene-9,10-quinone. Because phenanthrene-9,10-catechol is symmetric and planar, no regioisomers or stereoisomers of its phase II metabolites are allowed. Second, we predicted that hydroxylation of phenanthrene-9,10-quinone followed by formation of phase II conjugates could be another metabolic pathway. Because phenanthrene-9,10-quinone is symmetric, four regioisomers of its mono-hydroxylation metabolites could be generated at most. In terms of bis-hydroxylation, the two hydroxyl groups could either be both located on the same terminal ring or they could be located on the two different terminal rings. Third, a combination of these metabolic pathways was also predicted. On the basis of these predictions, the potential metabolites of phenanthrene-9,10-quinone can be detected, identified, and validated subsequently using authentic synthesized standards.
Detection of Phenanthrene-9,10-quinone Metabolites in HepG2 Cells by HPLC–UV–FLR
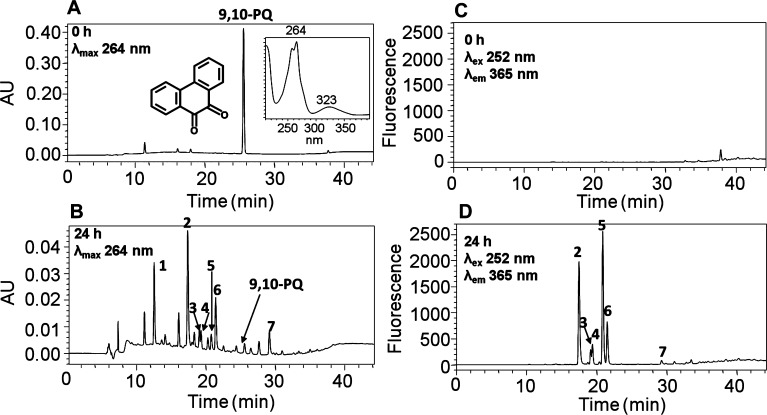
Comparison of UV chromatograms at λmax 264 nm of the 0 (Figure 1A) and 24 h chromatographic runs (Figure 1B) showed that seven metabolites of phenanthrene-9,10-quinone were detected in the organic phase of the ethyl acetate-extracted media after acidification of the media from HepG2 cells. The corresponding UV spectra of these seven metabolites were extracted from the UV chromatogram and are shown in Figure S2. The peak with a retention time of 25.57 min was attributed to phenanthrene-9,10-quinone but was barely detectable at 24 h and had a much lower intensity than the peak at 0 h, suggesting that phenanthrene-9,10-quinone is rapidly metabolized by HepG2 cells over this time course.
Figure 1.
HPLC detection of phenanthrene-9,10-quinone metabolites in HepG2 cells. (A) UV chromatogram at λmax 264 nm of the 0 h chromatographic run. (B) UV chromatogram at λmax 264 nm of the 24 h chromatographic run. (C) FLR chromatogram at λex 252 nm and λem 365 nm of the 0 h chromatographic run. (D) FLR chromatogram at λex 252 nm and λem 365 nm of the 24 h chromatographic run. HepG2 cells (∼5 × 106) were treated with phenanthrene-9,10-quinone (1 μM, 0.2% (v/v) DMSO) in MEM (without phenol red) containing 10 mM glucose. The cell media were collected at 0 and 24 h and subsequently acidified with 0.1% formic acid before extraction with ethyl acetate. The extracts were analyzed by HPLC–UV–FLR. 9,10-PQ = phenanthrene-9,10-quinone.
Comparison of FLR chromatograms at λex 252 nm and λem 365 nm of the 0 (Figure 1C) and 24 h chromatographic runs (Figure 1D) showed that there were six fluorescence peaks in Figure 1D, corresponding to metabolites 2–7 in Figure 1B, validating that these peaks were derived from phenanthrene-9,10-quinone and were now fully aromatic. The peak corresponding to metabolite 1 with a retention time of 12.55 min was detected only in the UV chromatogram (Figure 1B) but was not detected in the FLR chromatogram (Figure 1D), suggesting the loss of the phenanthrene fluorophore.
Identification of O-Mono-glucuronosyl-phenanthrene-9,10-catechol
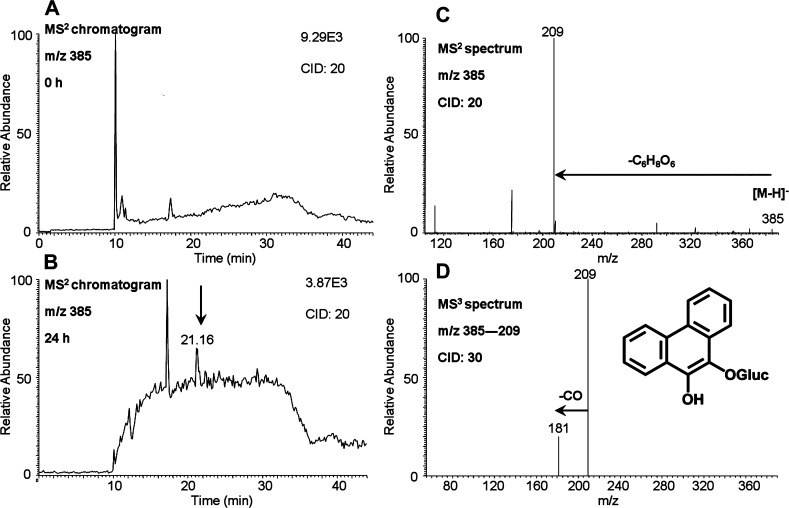
O-Mono-glucuronosyl-phenanthrene-9,10-catechol was detected in the culture media from HepG2 cells following treatment with 1 μM phenanthrene-9,10-quinone for 24 h. One peak with the retention time of 21.16 min was detected by monitoring the MS2 chromatograms (m/z 385) at 0 (Figure 2A) and 24 h (Figure 2B) in the negative ion mode. The corresponding MS2 spectra (m/z 385) of this metabolite showed the loss of glucuronide (176 amu) from the deprotonated molecular ion (Figure 2C). The MS3 spectra (m/z 385 → 209 →) of this metabolite showed loss of one CO group (Figure 2D). Because phenanthrene-9,10-catechol is symmetric and planar, there is only one possibility for the structure of this metabolite and thus the specific position of the glucuronide group can be assigned. Comparison of the retention time of O-mono-glucuronosyl-phenanthrene-9,10-catechol with metabolite 6 on HPLC–UV–FLR in Figure 1 showed an excellent concordance.
Figure 2.
O-Mono-glucuronosyl-phenanthrene-9,10-catechol detected in HepG2 cells. (A) MS2 chromatogram at 0 h. (B) MS2 chromatogram at 24 h. (C) MS2 spectrum. (D) MS3 spectrum. HepG2 cells (∼5 × 106) were treated with phenanthrene-9,10-quinone (1 μM, 0.2% (v/v) DMSO) in MEM (without phenol red) containing 10 mM glucose. The cell media were collected at 0 and 24 h and were subsequently acidified with 0.1% formic acid before extraction with ethyl acetate. The extracts were analyzed on an ion trap LC–MS/MS.
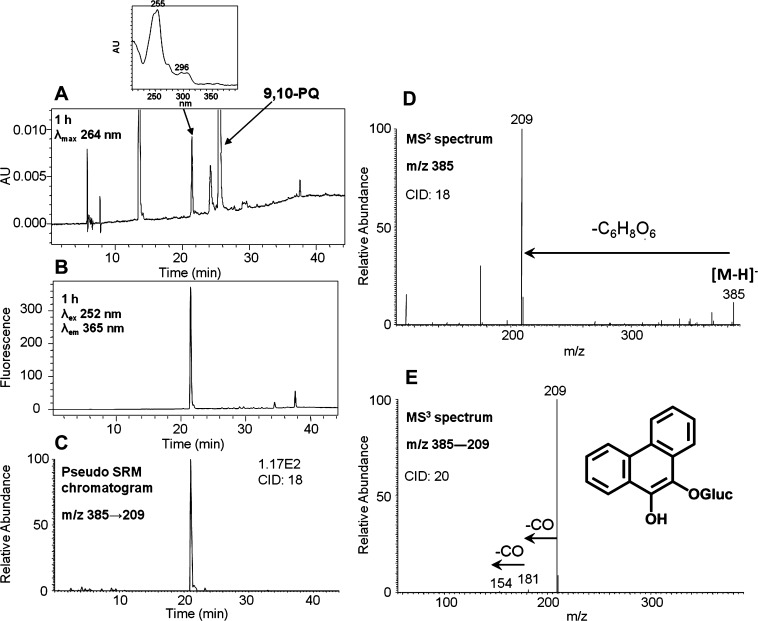
To generate the O-mono-glucuronosyl-phenanthrene-9,10-catechol standard, phenanthrene-9,10-quinone was reduced by dithiothreitol and reacted with UGT2B7 plus UDPGA. This reaction generated a light yellow solid after extraction and solvent evaporation. Subsequent HPLC–UV–FLR analysis showed that phenanthrene-9,10-quinone was not completely consumed, probably because of the low activity of microsomal UGTs. However, a significant peak that was absent in the negative control was observed that eluted earlier than phenanthrene-9,10-quinone (Figure 3A,B) and that had the same retention time of metabolite 6 in Figure 1. This synthetic product showed a deprotonated molecule [M – H]− at m/z 385 under Q1 full-scan negative ion mode, corresponding to the molecular weight of a deprotonated O-mono-glucuronosyl catechol. The fragmentation pattern of this synthetic product obtained from the MS2 and MS3 spectra (Figure 3D,E) was essentially identical to metabolite 6 and confirmed the metabolite to be the deprotonated O-mono-glucuronosyl catechol (m/z 385). The FLR chromatogram (Figure 3B) and pseudo-SRM chromatogram (Figure 3C) further confirmed the formation of only one product and thus its definite structure can be unequivocally ascertained.
Figure 3.
Characterization of synthetic O-mono-glucuronosyl-phenanthrene-9,10-catechol. (A) UV chromatogram at λmax 264 nm. (B) FLR chromatogram at λex 252 nm and λem 365 nm. (C) Extracted ion chromatogram of the pseudo-SRM transition. (D) MS2 spectrum. (E) MS3 spectrum. 9,10-PQ = phenanthrene-9,10-quinone. The product profiles were obtained after a 1 h incubation of phenanthrene-9,10-catechol with UGT2B7 and UDPGA.
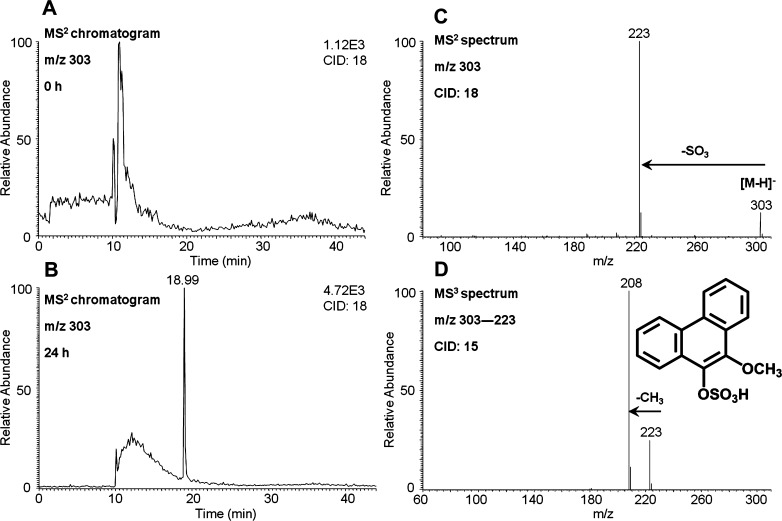
Identification of O-Mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol
O-Mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol was detected in the culture media from HepG2 cells following treatment with 1 μM phenanthrene-9,10-quinone for 24 h. One peak with a retention time of 18.99 min was detected by monitoring the MS2 chromatograms (m/z 303) at 0 (Figure 4A) and 24 h (Figure 4B) in the negative ion mode. The corresponding MS2 spectra (m/z 303) of this metabolite showed the loss of sulfate (80 amu) from the deprotonated molecular ion (Figure 4C). The MS3 spectra (m/z 303 → 223 →) of this metabolite showed the characteristic loss of CH3 (Figure 4D). Because phenanthrene-9,10-catechol is symmetric and planar, there is only one possibility for the structure of this metabolite and thus the specific positions of the methyl group and the sulfate group can be assigned. Comparison of the retention time of O-mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol with metabolite 4 on HPLC–UV–FLR in Figure 1 showed an excellent concordance.
Figure 4.
O-Mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol detected in HepG2 cells. (A) MS2 chromatogram at 0 h. (B) MS2 chromatogram at 24 h. (C) MS2 spectrum. (D) MS3 spectrum. HepG2 cells (∼5 × 106) were treated with phenanthrene-9,10-quinone (1 μM, 0.2% (v/v) DMSO) in MEM (without phenol red) containing 10 mM glucose. The cell media were collected at 0 and 24 h and were subsequently acidified with 0.1% formic acid before extraction with ethyl acetate. The extracts were analyzed on an ion trap LC–MS/MS.
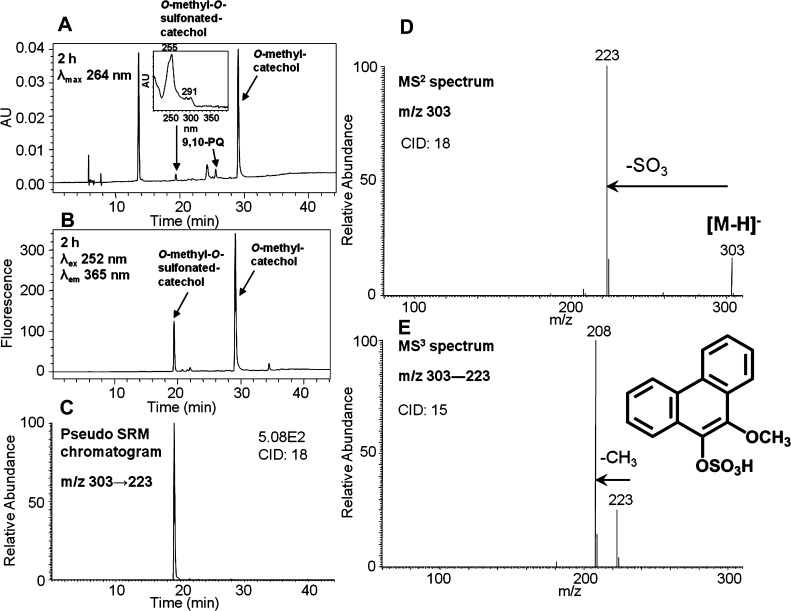
To generate the O-mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol standard, phenanthrene-9,10-quinone was reduced by dithiothreitol followed by sequential phase II conjugation with COMT plus AdoMet and SULT1A1 plus PAPS. This reaction generated a light yellow solid after extraction and solvent evaporation. HPLC–UV–FLR analysis showed that phenanthrene-9,10-quinone was completely consumed, and a peak was observed that eluted later than phenanthrene-9,10-quinone (Figure 5A,B), corresponding to O-mono-methyl catechol. Another significant peak that was absent in the negative control was also observed that eluted earlier than phenanthrene-9,10-quinone (Figure 5A,B) and that had the same retention time as metabolite 4 in Figure 1. This polar synthetic product showed a deprotonated molecule [M – H]− at m/z 303 under Q1 full-scan negative ion mode, corresponding to the molecular weight of a deprotonated O-mono-methyl-O-mono-sulfonated catechol. The fragmentation pattern of this synthetic product obtained from the MS2 and MS3 spectra (Figure 5D,E) confirmed the structure of the deprotonated O-mono-methyl-O-mono-sulfonated catechol (m/z 303). The pseudo-SRM chromatogram (Figure 5C) further confirmed the formation of only one product and thus the definite structure can be unequivocally ascertained.
Figure 5.
Characterization of synthetic O-mono-methyl-O-mono-sulfonated-phenanthrene-9,10-catechol. (A) UV chromatogram at λmax 264 nm. (B) FLR chromatogram at λex 252 nm and λem 365 nm. (C) Extracted ion chromatogram of the pseudo-SRM transition. (D) MS2 spectrum. (E) MS3 spectrum. The product profiles were obtained at 2 h, which included a 1 h incubation of phenanthrene-9,10-catechol with COMT and AdoMet followed by an additional 1 h incubation with SULT1A1 and PAPS.
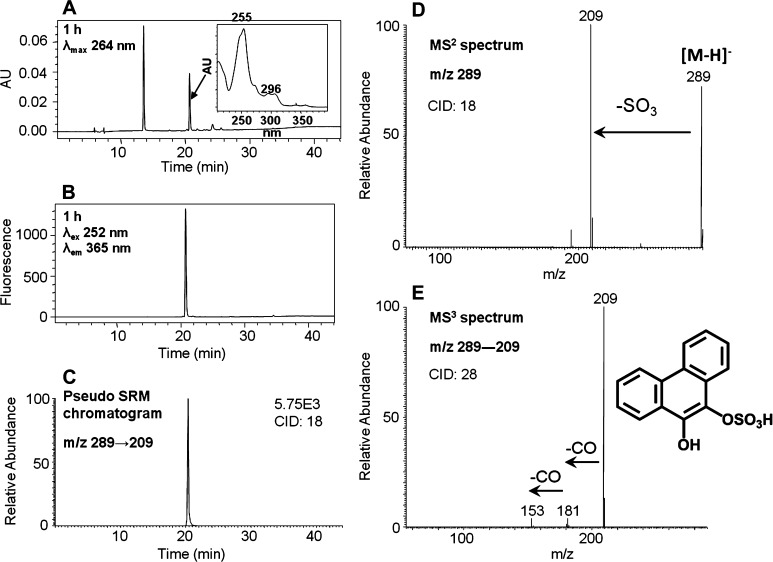
Identification of O-Mono-sulfonated-phenanthrene-9,10-catechol
The identification of metabolite 5 proved to be difficult because it was present in low UV abundance and had weak ionization upon ion trap LC–MS/MS. To resolve this issue, we proposed that this metabolite could be an O-mono-sulfonated-phenanthrene-9,10-catechol. We enzymatically synthesized this conjugate as follows. Phenanthrene-9,10-quinone was reduced by dithiothreitol followed by phase II conjugation catalyzed by SULT1A1 in the presence of PAPS. This reaction generated a light yellow solid after extraction and solvent evaporation. Subsequent HPLC–UV–FLR analysis showed that phenanthrene-9,10-quinone was completely consumed, and a significant peak that was absent in the negative control was observed that eluted earlier than phenanthrene-9,10-quinone (Figure 6A,B) and that had the same retention time and UV spectrum as metabolite 5 in Figure 1. This synthetic product showed a deprotonated molecule [M – H]− at m/z 289 under Q1 full-scan negative ion mode, corresponding to the molecular weight of a deprotonated O-mono-sulfonated catechol. The MS2 spectra (m/z 289) of this synthetic product showed the loss of sulfate (80 amu) from the deprotonated molecular ion (Figure 6D). The MS3 spectra (m/z 289 → 209 →) of this synthetic product showed the subsequent loss of two CO groups (Figure 6E). Because phenanthrene-9,10-catechol is symmetric and planar, there is only one possibility for the structure of this synthetic product and thus the specific position of the sulfate group can be assigned. The FLR (Figure 6B) and pseudo-SRM (Figure 6C) chromatograms further confirmed the formation of only one product and thus its definite structure can be unequivocally ascertained.
Figure 6.
Characterization of synthetic O-mono-sulfonated-phenanthrene-9,10-catechol. (A) UV chromatogram at λmax 264 nm. (B) FLR chromatogram at λex 252 nm and λem 365 nm. (C) Extracted ion chromatogram of the pseudo-SRM transition. (D) MS2 spectrum. (E) MS3 spectrum. The product profiles were obtained after 1 h incubation of phenanthrene-9,10-catechol with SULT1A1 and PAPS.
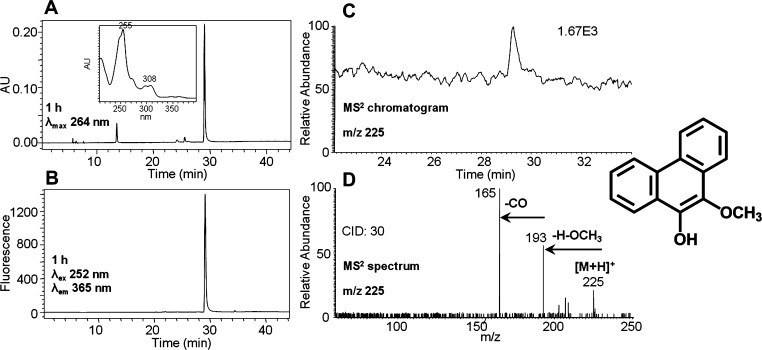
Identification of O-Mono-methyl-phenanthrene-9,10-catechol
Metabolite 7 was found to be O-mono-methyl-phenanthrene-9,10-catechol. This metabolite, although detected by the ion trap LC–MS/MS, generated a weak signal. However, subsequent high-resolution MS data obtained on the Orbitrap gave the accurate mass for this metabolite. Furthermore, the identity was ascertained by comparison to an authentic standard synthesized enzymatically. The reduction of phenanthrene-9,10-quinone followed by phase II conjugation catalyzed by COMT in the presence of AdoMet generated a light yellow solid after extraction and solvent evaporation. Subsequent HPLC–UV–FLR analysis showed that phenanthrene-9,10-quinone was completely consumed, and a significant peak that was absent in the negative control was observed that eluted later than phenanthrene-9,10-quinone (Figure 7A,B) and that had the same retention time and UV spectrum as metabolite 7 in Figure 1. This synthetic product showed a protonated molecule [M + H]+ at m/z 225 under Q1 full-scan positive ion mode, corresponding to the molecular weight of a protonated O-mono-methyl catechol. The MS2 spectra (m/z 225) of this synthetic product showed the loss of OCH3 plus H and subsequent loss of CO from the protonated molecular ion (Figure 7D), which is consistent with the fragmentation pattern of O-mono-methyl-benzo[a]pyrene-7,8-catechol generated in three human lung cell lines after treatment with benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione).24 Because phenanthrene-9,10-catechol is symmetric and planar, there is only one possibility for the structure of this synthetic product and thus the specific position of the methyl group can be assigned. The FLR (Figure 7B) and MS2 (Figure 7C) chromatograms further confirmed the formation of only one product and thus its definite structure can be unequivocally ascertained.
Figure 7.
Characterization of synthetic O-mono-methyl-phenanthrene-9,10-catechol. (A) UV chromatogram at λmax 264 nm. (B) FLR chromatogram at λex 252 nm and λem 365 nm. (C) MS2 chromatogram. (D) MS2 spectrum. The product profiles were obtained after 1 h incubation of phenanthrene-9,10-catechol with COMT and AdoMet.
Identification of O-Mono-sulfonated Bis-phenols of Phenanthrene-9,10-quinone
O-Mono-sulfonated bis-phenols of phenanthrene-9,10-quinone were detected in the culture media from HepG2 cells following treatment with 1 μM phenanthrene-9,10-quinone for 24 h. Two peaks with retention times of 15.88 (minor regioisomer) and 18.65 min (major regioisomer) were detected by monitoring the pseudo-SRM chromatograms (m/z 319 → 239) at 0 and 24 h in the negative ion mode (Figure S3). Pseudo-SRM transition 319 → 239 showed an increase of 80 amu over that for the bis-phenol (m/z 239), strongly indicating the occurrence of O-mono-sulfonation of the bis-phenol. However, the specific positions of two hydroxyl groups and the sulfate group could not be assigned on the basis of mass spectrometry data only. The retention time of the major regioisomer (18.65 min) showed a good concordance with metabolite 3 on HPLC–UV–FLR in Figure 1.
Identification of Other Metabolites of Phenanthrene-9,10-quinone by Nano-UPLC-Orbitrap-MS
A single isomer of a mono-hydroxylated-O-mono-glucuronosyl-phenanthrene-9,10-quinone was detected in HepG2 cells by monitoring the extracted ion chromatograms of Orbitrap full scan at 0 and 24 h in the negative ion mode (Figure S4). The specific position of the glucuronosyl group could not be assigned on the basis of mass spectrometry data only. A single isomer of a mono-hydroxylated-O-mono-methyl-phenanthrene-9,10-catechol was detected in HepG2 cells by monitoring the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the positive ion mode (Figure S5). The specific position of the hydroxyl group could not be assigned on the basis of mass spectrometry data only. Evidence for an O-mono-methyl-O-mono-glucuronosyl-phenanthrene-9,10-catechol metabolite in HepG2 cells was also obtained by monitoring the extracted ion chromatograms of the Orbitrap full scan at 0 and 24 h in the negative ion mode (Figure S6). Because phenanthrene-9,10-catechol is symmetric and planar, there is only one possibility for the structure of this metabolite and thus the specific positions of the methyl group and the glucuronosyl group can be assigned.
Time Course of Phenanthrene-9,10-quinone Consumption and Its Metabolite Formation in HepG2 Cells
The quantitation of phenanthrene-9,10-quinone, metabolites 1 and 7 was based on the UV peak area ratio of the analyte and phenanthrene (internal standard), whereas the quantitation of metabolites 2–6 was based on the fluorescence peak area ratio of the analyte and phenanthrene (internal standard). As shown in Figure S7, phenanthrene-9,10-quinone disappeared in HepG2 cells rapidly within the first 3 h followed by a slow decline. The metabolite profiles formed at different time points exhibited similar trends in HepG2 cells, with a slight increase over 72 h. On the basis of this semiquantitation, we find that metabolite 5 > metabolite 2 > metabolites 4–6. Thus, one of the major conjugates formed was O-mono-sulfonated-phenanthrene-9,10-catechol.
Discussion
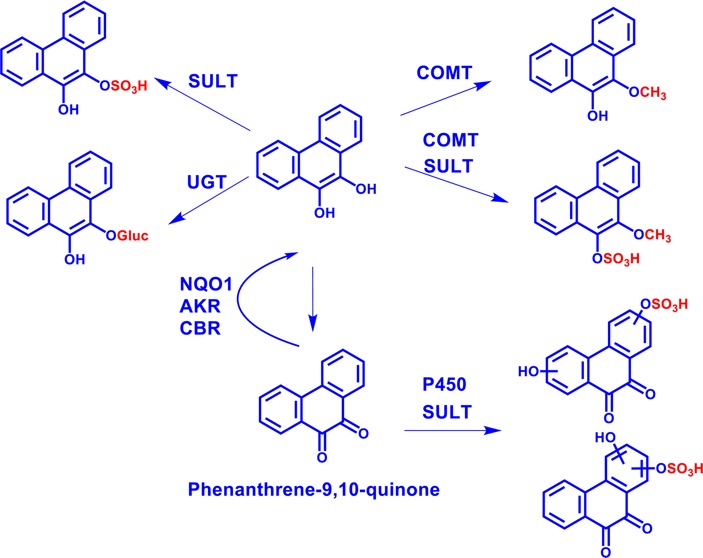
This study provides a comprehensive account of the metabolism of phenanthrene-9,10-quinone in a human liver cell line, HepG2. Phenanthrene-9,10-quinone is a signature oxygenated derivative of phenanthrene present in crude oil, but information about its downstream cellular metabolism is limited. Phenanthrene-9,10-quinone may enter the food chain after the oil has weathered, and understanding its metabolism in liver cells is critical in determining its human toxicity. Phenanthrene-9,10-quinone metabolites were detected and identified from the hepatoma cell culture media by HPLC–UV–FLR and LC–MS/MS (Table 1). Metabolites 3–7 in Figure 1 were identified as O-mono-sulfonated-bis-phenol, O-mono-methyl-O-mono-sulfonated-catechol, O-mono-sulfonated-catechol, O-mono-glucuronosyl-catechol, and O-mono-methyl-catechol in the order of their elution, respectively. Metabolites 1 and 2 in Figure 1 remain unassigned.
Table 1. Mass Spectrometric Properties for Phenanthrene-9,10-quinone Metabolites in HepG2 Cells.
| no.a | phenanthrene-9,10-quinone metabolites | retention time (min) | ion mode | m/z |
|---|---|---|---|---|
| 6 | O-mono-glucuronosyl catechol | 21.16 | negative | 385 [M – H]−, 209 [M – H-C6H8O6]−, 181 [M – H-C6H8O6-CO]−, 154 [M – C6H8O6-2CO]− |
| 5 | O-mono-sulfonated catechol | 20.40 | negative | 289 [M – H]−, 209 [M – H-SO3]−, 181 [M – H-SO3-CO]−, 153 [M – H-SO3-2CO]− |
| 7 | O-mono-methyl catechol | 29.29 | positive | 225 [M + H]+, 193 [M – OCH3]+, 165 [M – OCH3-CO]+ |
| 4 | O-mono-methyl-O-mono-sulfonated catechol | 18.99 | negative | 303 [M – H]−, 223 [M – H-SO3]−, 208 [M – H-SO3-CH3]− |
| 3 | O-mono-sulfonated bis-phenols | 15.88, 18.65 | negative | 319 [M – H]−, 239 [M – H-SO3]− |
no. corresponds to the metabolite in the UV and fluorescence chromatograms shown in Figure 1.
The major metabolic pathway of phenanthrene-9,10-quinone in HepG2 cells involves reduction to the catechol. Previous studies have shown that the reduction of phenanthrene-9,10-quinone is catalyzed by NQO1, AKRs, and, to a less extent, by CBR in a two-electron reduction step. In addition, phenanthrene-9,10-quinone can be reduced by two one-electron reduction steps by POR.14−19 Once formed, the catechol can be conjugated by O-mono-glucuronidation, O-mono-sulfonation, O-mono-methylation, and O-mono-methylation-O-mono-sulfonation (Scheme 2). Minor metabolic pathways involve bis-hydroxylation of the quinone followed by O-mono-sulfonation (Scheme 2), mono-hydroxylation of the quinone followed by O-mono-glucuronidation, and mono-hydroxylation of the O-mono-methyl-catechol (not shown in Scheme 2). Previous studies on the conjugation of benzo[a]pyrene-7,8-catechol showed evidence for O-mono-glucuronidation, O-mono-sulfonation, and O-mono-methylation.21−23 Thus, these pathways may be shared by both non-K-region and K-region catechols.
Scheme 2. Proposed Metabolic Pathway of Phenanthrene-9,10-quinone in HepG2 Cells.
The numbers for each metabolite correspond to the metabolites labeled in the UV and fluorescence chromatograms in Figure 1.
Among the metabolites of phenanthrene-9,10-quinone, the identities of four catechol conjugates, namely, O-mono-glucuronosyl catechol, O-mono-methyl-O-mono-sulfonated catechol, O-mono-sulfonated catechol, and O-mono-methyl catechol, were validated by comparison to authentic standards synthesized enzymatically. Because phenanthrene-9,10-catechol is symmetric and planar, there is only one structure that is possible for each phase II catechol conjugate. Therefore, the definite structures of these four catechol conjugates were unequivocally elucidated. Identification of O-mono-glucuronosyl catechol as a metabolite of phenanthrene-9,10-quinone in human A549 cells has been reported previously.20
A unique feature of the present work was the detection and identification of a phase II catechol bis-conjugate, namely, O-mono-methyl-O-mono-sulfonated catechol. We predicted that O-mono-methyl-O-mono-glucuronosyl catechol and O-mono-sulfonated-O-mono-glucuronosyl catechol might form as well. These two metabolites were not easily detected because of their trace amounts in HepG2 cells. However, O-mono-methyl-O-mono-glucuronosyl catechol was detected using the Orbitrap. By contrast, O-bis-glucuronosyl catechol, O-bis-sulfonated catechol, and O-bis-methyl catechol were not detected.
Because two regioisomers of O-mono-sulfonated bis-phenols were detected in HepG2 cells, mono-phenols and bis-phenols were also expected. These two intermediates were not detectable, probably because of their trace amounts in HepG2 cells. However, a single isomer of mono-hydroxylated-O-mono-glucuronosyl-phenanthrene-9,10-quinone and a single isomer of mono-hydroxylated-O-mono-methyl catechol were detected. The formation of mono-hydroxylated-O-mono-methyl catechol provides evidence that both catechol conjugation and mono-hydroxylation can occur, but the exact order of these biotransformations could not be ascertained. The specific positions at which phase I mono-hydroxylation and bis-hydroxylation of phenanthrene-9,10-quinone and the subsequent phase II conjugation occur were not assignable by LC–MS/MS. NMR spectroscopy would be needed to characterize the definite structures of these metabolites, but the approach is not feasible with the limited material isolated from cells.
Reduction of phenanthrene-9,10-quinone to the catechol followed by formation of phase II conjugates represents detoxification pathways of phenanthrene-9,10-quinone that result in loss of its redox activity. In contrast, other metabolites, such as the isomers of O-mono-sulfonated bis-phenols of the quinone, are still capable of redox cycling to produce oxidative stress and oxidative DNA damage.
Knowledge of the major metabolic pathways of phenanthrene-9,10-quinone in human liver cells could be used to identify biomarkers of phenanthrene-9,10-quinone exposure by measuring their presence in human urine and plasma. The present study suggests that O-mono-glucuronosyl catechol and O-mono-sulfonated catechol may be reasonable biomarkers of oxygenated PAH exposure because they are major metabolites and appear to be stable.
The phase II isozymes responsible for the formation of phenanthrene-9,10-catechol conjugates in HepG2 cells remain to be completely identified. However, the presence of catechol conjugates suggests that the redox cycling of phenanthrene-9,10-quinone is being intercepted. Detection of O-mono-sulfonated bis-phenols indicates bis-hydroxylation by cytochrome P450 followed by mono-sulfonation by SULTs in HepG2 cells. On the basis of the profiles of the metabolites synthesized enzymatically, UGT2B7 and SULT1A1 may be involved in the phase II conjugation of phenanthrene-9,10-catechol in HepG2 cells. Previous studies indicated that both of these enzyme isoforms are expressed in HepG2 cells.23,25 The other UGTs and SULTs isoforms expressed in HepG2 cells have not been examined in this study for their ability to conjugate phenanthrene-9,10-catechol.
The approach to detect and identify metabolites in this study was to employ MS2 followed by MS3, in combination with pseudo-SRM. MS2 followed by MS3 provides more structural information, but the sensitivity to detect potential metabolites is limited, which could result from the relatively low substrate concentration used (i.e., 1 μM) and the high background signal from the cell media matrices. Pseudo-SRM offers significantly higher sensitivity and selectivity and enables a wide range of potential transitions to be targeted as a result of the rapid cycle times, but less structural information is obtained. MS2 followed by MS3, in combination with pseudo-SRM are both based on the knowledge of the predicted metabolites and their likely MS/MS fragmentation patterns. Therefore, this approach was unable to detect and identify unexpected and unusual metabolites, which explains why the structures of metabolites 1 and 2 remain unassigned. Another explanation is that polar metabolites 1 and 2 were eluted with a higher percentage of the aqueous mobile phase, resulting in lower evaporation and lower ionization.
In summary, we have conducted the metabolism studies of phenanthrene-9,10-quinone in HepG2 cells. The major metabolic pathway of phenanthrene-9,10-quinone involves the reduction to the catechol followed by phase II mono-conjugation or bis-conjugation. In particular, we obtained evidence of the formation of phase II catechol bis-conjugates. Another minor metabolic pathway involves bis-hydroxylation followed by O-mono-sulfonation. Among these metabolic pathways, reduction to the catechol followed by phase II conjugation is a potential detoxification pathway of phenanthrene-9,10-quinone. The definite structures of four catechol conjugates were unequivocally elucidated and could be used as biomarkers of human exposure to this oxygenated PAH.
Glossary
Abbreviations
- PAPS
adenosine 3′-phosphate 5′-phosphosulfate
- AdoMet
S-(5′-adenosyl)-l-methionine
- AKR
aldo-keto reductase
- B[a]P-7,8-dione
benzo[a]pyrene-7,8-dione
- CBR
carbonyl reductase
- COMT
catechol-O-methyltransferase
- P450
cytochrome P450
- ESI
electrospray ionization
- FBS
fetal bovine serum
- HPLC–UV–FLR
high-performance liquid chromatography–ultraviolet–fluorescence
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- NQO1
NAD(P)H:quinone oxidoreductase 1
- PAH
polycyclic aromatic hydrocarbon
- POR
NADPH:cytochrome P450 oxidoreductase
- QR
quinone reductase
- ROS
reactive oxygen species
- SULTs
sulfotransferases
- UDPGA
uridine-5′-diphosphoglucuronic acid
- UGTs
uridine 5′-diphospho-glucuronosyltransferases
Supporting Information Available
Excitation wavelength and emission wavelength spectra of phenanthrene; UV spectra of metabolites 1–7 in HepG2 cells; extracted ion chromatograms of pseudo-SRM transitions for O-mono-sulfonated bis-phenols of phenanthrene-9,10-quinone detected in HepG2 cells; extracted ion chromatograms of Orbitrap full scan for mono-hydroxylated-O-mono-glucuronosyl-phenanthrene-9,10-quinone, mono-hydroxylated-O-mono-methyl-phenanthrene-9,10-catechol, and O-mono-methyl-O-mono-glucuronosyl-phenanthrene-9,10-catechol detected in HepG2 cells; and time course of phenanthrene-9,10-quinone (9,10-PQ) consumption and its metabolite formation in HepG2 cells. This material is available free of charge via the Internet at http://pubs.acs.org.
This publication was made possible by Deepwater Horizon Research Consortia grant no. U01/U19 ES020676-03 and by National Institute of Environmental Health Sciences (NIEHS), NIH, DHHS grant no. P30 ES013508 (T.M.P.). The contents of this article are the responsibility solely of the authors and do not necessarily represent the official views of the NIEHS or NIH.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Joye S. B.; MacDonald I. R.; Leifer I.; Asper V. (2011) Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat. Geosci. 4, 160–164. [Google Scholar]
- Atlas R. M.; Hazen T. C. (2011) Oil biodegradation and bioremediation: A tale of the two worst spills in US history. Environ. Sci. Technol. 45, 6709–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy C. M.; Arey J. S.; Seewald J. S.; Sylva S. P.; Lemkau K. L.; Nelson R. K.; Carmichael C. A.; McIntyre C. P.; Fenwick J.; Ventura G. T.; Van Mooy B. A. S.; Camilli R. (2012) Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. U.S.A. 109, 20229–20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt M. K.; Camilli R.; Crone T. J.; Guthrie G. D.; Hsieh P. A.; Ryerson T. B.; Savas O.; Shaffer F. (2012) Review of flow rate estimates of the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. U.S.A. 109, 20260–20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryerson T. B.; Camilli R.; Kessler J. D.; Kujawinski E. B.; Reddy C. M.; Valentine D. L.; Atlas E.; Blake D. R.; de Gouw J.; Meinardi S.; Parrish D. D.; Peischl J.; Seewald J. S.; Warneke C. (2012) Chemical data quantify Deepwater Horizon hydrocarbon flow rate and environmental distribution. Proc. Natl. Acad. Sci. U.S.A. 109, 20246–20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. D.; Osofsky H. J.; Lichtveld M. Y. (2011) The Gulf oil spill. N. Engl. J. Med. 364, 1334–1348. [DOI] [PubMed] [Google Scholar]
- National Institute of Standards and Technology (2012) Standard Reference Material (SRM) 2779, Gulf of Mexico Crude Oil, National Institute of Standards and Technology Certificate of Analysis, pp 1–7, Gaithersburg, MD.
- Boyland E.; Sims P. (1962) Metabolism of polycyclic compounds. 21. The metabolism of phenanthrene in rabbits and rats: Dihydrodihydroxy compounds and related glucosiduronic acids. Biochem. J. 84, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David B.; Boule P. (1993) Phototransformation of hydrophobic pollutants in aqueous-medium 0.1. PAHs adsorbed on silica. Chemosphere 26, 1617–1630. [Google Scholar]
- Barbas J. T.; Sigman M. E.; Dabestani R. (1996) Photochemical oxidation of phenanthrene sorbed on silica gel. Environ. Sci. Technol. 30, 1776–1780. [Google Scholar]
- Holt J.; Hothem S.; Howerton H.; Larson R.; Sanford R. (2005) 9,10-Phenanthrenequinone photoautocatalyzes its formation from phenanthrene, and inhibits biodegradation of naphthalene. J. Environ. Qual. 34, 462–468. [PubMed] [Google Scholar]
- Ylitalo G. M.; Krahn M. M.; Dickhoff W. W.; Stein J. E.; Walker C. C.; Lassitter C. L.; Garrett E. S.; Desfosse L. L.; Mitchell K. M.; Noble B. T.; Wilson S.; Beck N. B.; Benner R. A.; Koufopoulos P. N.; Dickey R. W. (2012) Federal seafood safety response to the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. U.S.A. 109, 20274–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera F.; Mendez J.; Pasaro E.; Laffon B. (2010) Review on the effects of exposure to spilled oils on human health. J. Appl. Toxicol. 30, 291–301. [DOI] [PubMed] [Google Scholar]
- Matsunaga T.; Arakaki M.; Kamiya T.; Endo S.; El-Kabbani O.; Hara A. (2009) Involvement of an aldo-keto reductase (AKR1C3) in redox cycling of 9,10-phenanthrenequinone leading to apoptosis in human endothelial cells. Chem.–Biol. Interact. 181, 52–60. [DOI] [PubMed] [Google Scholar]
- Rodriguez C. E.; Sobol Z.; Schiestl R. H. (2008) 9,10-Phenanthrenequinone induces DNA deletions and forward mutations via oxidative mechanisms in the yeast Saccharomyces cerevisiae. Toxicol. In Vitro 22, 296–300. [DOI] [PubMed] [Google Scholar]
- Taguchi K.; Fujii S.; Yamano S.; Cho A. K.; Kamisuki S.; Nakai Y.; Sugawara F.; Froines J. R.; Kumagai Y. (2007) An approach to evaluate two-electron reduction of 9,10-phenanthraquinone and redox activity of the hydroquinone associated with oxidative stress. Free Radical Biol. Med. 43, 789–799. [DOI] [PubMed] [Google Scholar]
- Matsunaga T.; Shinoda Y.; Inoue Y.; Shimizu Y.; Haga M.; Endo S.; El-Kabbani O.; Hara A. (2011) Aldo-keto reductase 1C15 as a quinone reductase in rat endothelial cell: Its involvement in redox cycling of 9,10-phenanthrenequinone. Free Radical Res. 45, 848–857. [DOI] [PubMed] [Google Scholar]
- Chesis P. L.; Levin D. E.; Smith M. T.; Ernster L.; Ames B. N. (1984) Mutagenicity of quinones: Pathways of metabolic activation and detoxication. Proc. Natl. Acad. Sci. U.S.A. 81, 1696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz C. A.; Quinn A. M.; Park J. H.; Harvey R. G.; Bolton J. L.; Maser E.; Penning T. M. (2011) Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase, and carbonyl reductases to redox-cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-o-quinone. Chem. Res. Toxicol. 24, 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K.; Shimada M.; Fujii S.; Sumi D.; Pan X.; Yamano S.; Nishiyama T.; Hiratsuka A.; Yamamoto M.; Cho A. K.; Froines J. R.; Kumagai Y. (2008) Redox cycling of 9,10-phenanthraquinone to cause oxidative stress is terminated through its monoglucuronide conjugation in human pulmonary epithelial A549 cells. Free Radical Biol. Med. 44, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Huang M.; Blair I. A.; Penning T. M. (2012) Detoxication of benzo[a]pyrene-7,8-dione by sulfotransferases (SULTs) in human lung cells. J. Biol. Chem. 287, 29909–29920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Jin Y.; Chen M.; Huang M.; Harvey R. G.; Blair I. A.; Penning T. M. (2011) Detoxication of structurally diverse polycyclic aromatic hydrocarbon (PAH) o-quinones by human recombinant catechol-O-methyltransferase (COMT) via O-methylation of PAH catechols. J. Biol. Chem. 286, 25644–25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Huang M.; Blair I. A.; Penning T. M. (2013) Interception of benzo[a]pyrene-7,8-dione by UDP glucuronosyltransferases (UGTs) in human lung cells. Chem. Res. Toxicol. 26, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.; Liu X.; Basu S. S.; Zhang L.; Kushman M. E.; Harvey R. G.; Blair I. A.; Penning T. M. (2012) Metabolism and distribution of benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione) in human lung cells by liquid chromatography tandem mass spectrometry: Detection of an adenine B[a]P-7,8-dione adduct. Chem. Res. Toxicol. 25, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerink W. M.; Schoonen W. G. (2007) Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol. In Vitro 21, 1592–1602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.