Abstract
The role of histidine in channel-forming transmembrane (TM) helices was investigated by comparing the transmembrane helices from Virus protein ‘u’ (Vpu) and the M2 proton channel. Both proteins are members of the viroporin family of small membrane proteins that exhibit ion channel activity, and have a single TM helix that is capable of forming oligomers. The transmembrane helices from both proteins have a conserved tryptophan towards the C-terminus. Previously, alanine 18 of Vpu was mutated to histidine in order to artificially introduce the same HXXXW motif that is central to the proton channel activity of M2. Interestingly, the mutated Vpu TM resulted in an increase in helix tilt angle of 11° in lipid bilayers compared to the wild-type Vpu TM. Here, we find the reverse, when histidine 37 of the HXXXW motif in M2 was mutated to alanine, it decreased the helix tilt by 10° from that of wild-type M2. The tilt change is independent of both the helix length and the presence of tryptophan. In addition, compared to wild-type M2, the H37A mutant displayed lowered sensitivity to proton concentration. We also found that the solvent accessibility of histidine-containing M2 is greater than without histidine. This suggests that the TM helix may increase the solvent exposure by changing its tilt angle in order to accommodate a polar/charged residue within the hydrophobic membrane region. The comparative results of M2, Vpu and their mutants demonstrated the significance of histidine in a transmembrane helix and the remarkable plasticity of the function and structure of ion channels stemming from changes at a single amino acid site.
Keywords: Vpu, M2, lipid bilayers, OS solid-state NMR
Introduction
Histidine serves important roles in the transmembrane (TM) domains of membrane proteins, including pH sensing, ligand binding, and metal transport (Stewart et al. 2007, Rehwald et al. 1999, Larson et al. 2010). It has a significant impact on both the structure and function of the proteins, but the details of its mechanisms are far from being fully understood. Perhaps the best-characterized histidine-containing TM helix is that of the M2 proton channel from Influenza A virus (Pinto et al, 1992, Rossman and Lamb, 2011, Wang et al. 2011a, Cross et al. 2012). It is a type III integral phosphoprotein that plays a critical role in virus replication. The polypeptide has a single TM helix, which tetramerizes to form the channel. The channel opens upon acidification (below pH 6.5) allowing the flow of protons from endosome into the virion. This subsequently leads to the release of its genetic material into the cytoplasm. As part of a conserved HXXXW motif, the histidine residue (H37) within the TM domain serves as the gating mechanism for the proton flow (Hu et al. 2006, Takeuchi et al. 2003).
Previously, we have described studies of the structure and function of another small viral membrane protein that forms an ion channel, Virus protein ‘u’ (Vpu) from HIV-1 (Ma et al. 2002, Marassi et al. 1999, Park et al. 2006, Park et al. 2003, Park and Opella 2007, Skasko et al. 2011, Skasko et al. 2012). Vpu is a type I integral phosphoprotein and contains a single TM helix, which oligomerizes and is permeable to monovalent cations Na+ and K+ with marginal permeability to divalent cations (Schubert et al. 1996). Its TM domain also directly interacts with that of BST2/Tetherin, an interferon induced host factor that restricts the release of mature virus particles, and antagonizes the restriction function (Rong et al. 2009, Skasko et al. 2012). While the wild-type Vpu TM does not contain a histidine, it does have a conserved tryptophan residue (W22) towards the C-terminus, corresponding to the tryptophan in the M2 TM. By mutating alanine 18 to histidine, four residues away from the tryptophan, Vpu was able to acquire characteristics of M2, including susceptibility to the channel blocker, rimantadine, which specifically targets the M2 channel (Hout et al. 2006). Moreover, in our previous structural studies of wild-type Vpu TM and its A18H mutant, we found that their helix tilt angles, in the same phospholipid bilayer environment, differed by 11°.
In order to find out whether this large tilt change can also be observed in the M2 TM, we expressed in bacteria and purified wild-type M2 TM and its H37A mutant. The sequence for the wild-type M2-TM is shown in Figure 1, and it is aligned with that of Vpu-TM using the conserved tryptophan as the reference point. The lysine residues were added to the C-terminus to facilitate purification and to minimize aggregation. They are unstructured and mobile, and thus do not interfere with the properties of the TM domain (Park et al. 2006). The helix tilt and rotation of the polypeptides in phospholipid bilayers are determined using oriented sample (OS) solid-state NMR on 15N labeled proteins, and the results are compared with the previous data obtained from the corresponding Vpu constructs. To further characterize the effects of histidine, we mapped the response to proton concentration by observing the chemical shift perturbation of the backbone amides induced by pH change. In addition, to address the question of whether the tryptophan is important for the tilt change, the W41 residue was mutated to an alanine.
Figure 1.

Sequence comparison of the wild-type Vpu TM and wild-type M2 TM polypeptides with sites of mutation highlighted in bold.
Materials and Methods
Protein expression and purification
Expression and purification procedures for the protein constructs have been described (Park et al. 2006). Briefly, the proteins were expressed with ketosteriod isomerase (KSI) as a fusion within the pET31b (+) vector (Novagen, www.emdmillipore.com). Vectors were transformed into OverExpress™ C41 (DE3) cells and grown in M9 medium with the nitrogen provided by 15N ammonium sulfate (Cambridge Isotope Laboratories, www.isotope.com). The overexpressed polypeptide was isolated initially as inclusion bodies and then subjected to Nickel affinity chromatography. The target protein was cleaved from the fusion protein, KSI, by adding excess cyanogen bromide and incubating at room temperature for 3 hours. The final purified protein was obtained by precipitating the KSI in 1:4 (v/v) HFIP:dichloromethane, and filtering the supernatant through a 0.2 μm PTFE syringe filter (Millipore, www.millipore.com).
NMR sample preparation
Samples for solution NMR spectroscopy were prepared by dissolving lyophilized protein powder in an aqueous solution containing 150 mM 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), 90% H2O, and 10% D2O. The pH was adjusted by adding aliquots from a 1 M stock solution of either NaOH or HCl. Samples for solid-state NMR were prepared by first dissolving 2 mg of protein powder in an aqueous solution of DHPC and then adding the solution to 20 mg of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) powder. The final DMPC/DHPC molar ratio (q) was 3.2, and the total lipid content was 15% by weight. The mixture was temperature cycled between 45°C and 0°C with vigorous vortexing until it became transparent, and then the final pH was adjusted by adding either NaOH or HCl. Mn-EDTA chelate was prepared as previously described (Cook and Opella 2011) and then directly added to the protein-containing micelle solution at a final concentration of 2 mM.
NMR data acquisition and processing
The solution NMR data were acquired on a Bruker 600 MHz spectrometer equipped with a 1H/13C/15N triple-resonance cryoprobe. 1H/15N HSQC spectra were acquired at 50°C with 2048 t2 points and 256 t1 points. The solid-state data were acquired on a Bruker 700 MHz spectrometer equipped with a home-built 1H/15N double-resonance probe with a 5 mm solenoid coil and a strip-shield to minimize sample heating (Wu et al. 2009). One-dimensional 15N NMR spectra were acquired at 42°C using spin-lock cross-polarization with 512 points, and the two-dimensional 1H/15N dipolar coupling/15N chemical shift spectra separated local field (SLF) spectra were acquired at 42°C using the SAMPI4 pulse sequence (Nevzorov and Opella 2003) with 512 t2 points and 54 t1 points. The spectra were processed with NMRPipe (Delaglio et al. 1995) and displayed using NMRView (Johnson 2004). Changes in the chemical shifts (Δδ) in the solution NMR spectra were calculated using the equation below.
In the Mn-EDTA paramagnetic relaxation enhancement experiments, the intensities of the 1H/15N HSQC resonances were measured using NMRViewJ (One Moon Scientific, www.onemoonscientific.com/nmrviewj). The intensities were normalized to the largest resonance signal that is fully resolved from a spectrum that was recorded before (reference) and after the addition of 2mM Mn-EDTA. The percent reduction in relative intensity (%CRI) is calculated according to the equation below using the normalized intensity.
Structure Calculation
All four structures were calculated with the same protocol using Xplor-NIH version 2.26 (Schwieters et al. 2003). The calculations were processed in torsion space for 10 ps with orientational restraints obtained from the NMR experiments described above. Conventional simulated annealing protocol is employed to start from 1500 K, end at 50 K with 12.5 K step-size, followed by Cartesian minimization. Ideal dihedral angels (ϕ=-61°, ψ=-45°) are assigned to the TM residues with ±10 degrees of variation. Dipolar coupling and chemical shift anisotropy (CSA) restraints were applied through the whole calculation, with force constants 0.5 kcal•mol-1rad-2 and 0.02 kcal•mol-1ppm-2 respectively at high temperature, then ramped to 5 kcal•mol-1rad-2 and 0.2 kcal•mol-1ppm-2 during annealing. For all non-Gly residues, the principal CSA values of 55.3, -97.7 and 42.3 were used. And the order parameter was estimated to be 0.9 for all four cases and applied to Da in the calculation.
Results
We used established OS solid-state NMR techniques to study the TM polypeptides in magnetically aligned phospholipid bilayers (bicelles) (De Angelis et al. 2005, De Angelis and Opella 2007, Opella and Marassi 2004). The solid-state NMR spectra obtained from samples of uniformly and selectively 15N labeled proteins reveal details on the tilt angle and rotation of transmembrane helices relative to the bilayer normal through the orientationally-dependent frequencies of the resonances. Two-dimensional 1H/15N dipolar coupling/15N chemical shift SLF spectra for the wild-type M2 TM and the H37A M2 TM polypeptides are presented in Figure 2A and B, respectively. In order to facilitate direct comparisons, our previously published SLF spectra of wild-type Vpu TM and the A18H Vpu TM are also presented in Figure 2D and C, respectively (Park et al. 2006). The signals in all of the spectra have line widths of ~ 2 ppm in the 15N chemical shift dimension and ~ 300 Hz in the heteronuclear dipolar coupling dimension. These relatively narrow line widths indicate that the helices are well aligned along with the phospholipid bilayers by the magnetic field, and that each polypeptide has a single conformation. All of the spectra have the wheel-like pattern characteristic of a TM helix that is tilted with respect to the bilayer normal. The dotted lines were calculated based on the tilt angle that provided the best fit to the experimental data, assuming ideal helical secondary structure. In all cases, the patterns observed in the experimental spectra are well simulated by a single polarity index slant angle (PISA) wheel (Marassi and Opella, 2000; Wang et al, 2000). The helix tilt angles with respect to the bilayer normal are 30° and 28° for wild-type Vpu TM and H37A M2 TM, respectively. With the histidine residue present to form the HXXXW motif, the tilt angle increases by ~10° in both cases relative to the same sequence without the histidine. Specifically, the tilt angles are 41° and 38° for A18H Vpu TM and for wild-type M2 TM, respectively. The positions of the histidine/alanine and tryptophan resonances are indicated on the spectra (Figure 2).
Figure 2.
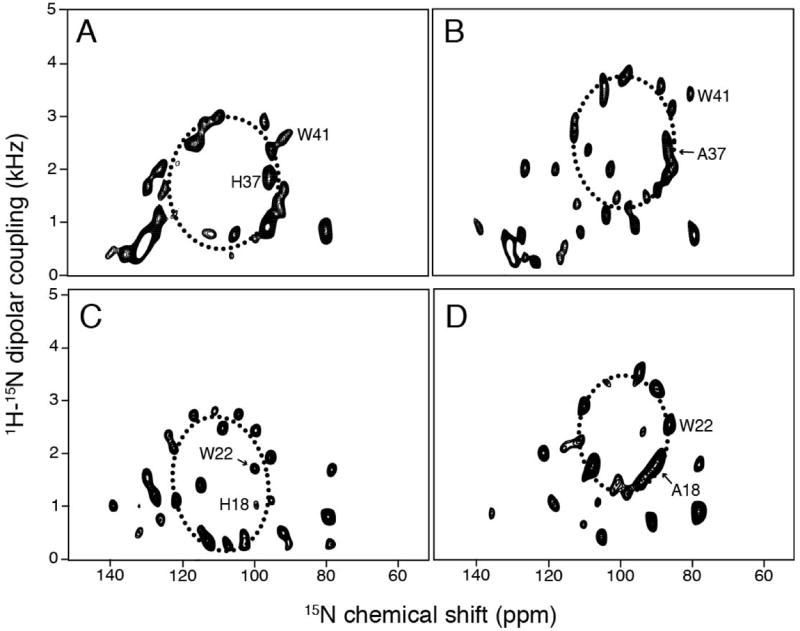
Two-dimensional 1H/15N dipolar coupling/15N chemical shift SLF solid-state NMR spectra of uniformly 15N-labeled samples in magnetically aligned bicelles. (A) wild-type M2 TM. (B) H37A M2 TM. (C) wild-type Vpu TM. (D) A18H Vpu TM. PISA wheel simulations are superimposed onto the spectra as dashed ellipses. Helix tilt angles for (A), (B), (C), and (D) are 38°, 28°, 41°, and 30°, respectively.
Resonances were assigned by obtaining the same type of spectra from samples of selectively 15N-labeled proteins. Two-dimensional SLF spectra of 15N leucine labeled proteins are shown in Figures 3A and B for wild-type M2 TM and H37A M2 TM, respectively. This enables the determination of the relative locations of residues on a helical wheel representation of the polypeptide since the PISA wheel spectra have the same periodic pattern as the secondary structure. The relative positions of the leucines on the helical wheel demonstrates that the helix azimuthal rotations differ by 15°-20° between the wild-type M2 TM and H37A M2 TM. The rotation angle of the helical wheels was confirmed by analysis of the spectra of 15N isoleucine labeled samples (data not shown). Interestingly, the helix rotations for the M2 constructs are similar to those for the Vpu constructs when the conserved tryptophan residue is used as the reference point in the sequence alignment. By fitting the selective labeling data to ideal helical wheels, resonance assignments could be made for the M2 constructs. The assignments of the leucine signals were further confirmed by D2O exchange experiment in which the samples contained 90% D2O (not shown). The disappearance of the resonances corresponding to L26 and L46 indicate that these two residues are at the ends of the helix and more accessible to the solvent than those on the interior of the helix.
Figure 3.
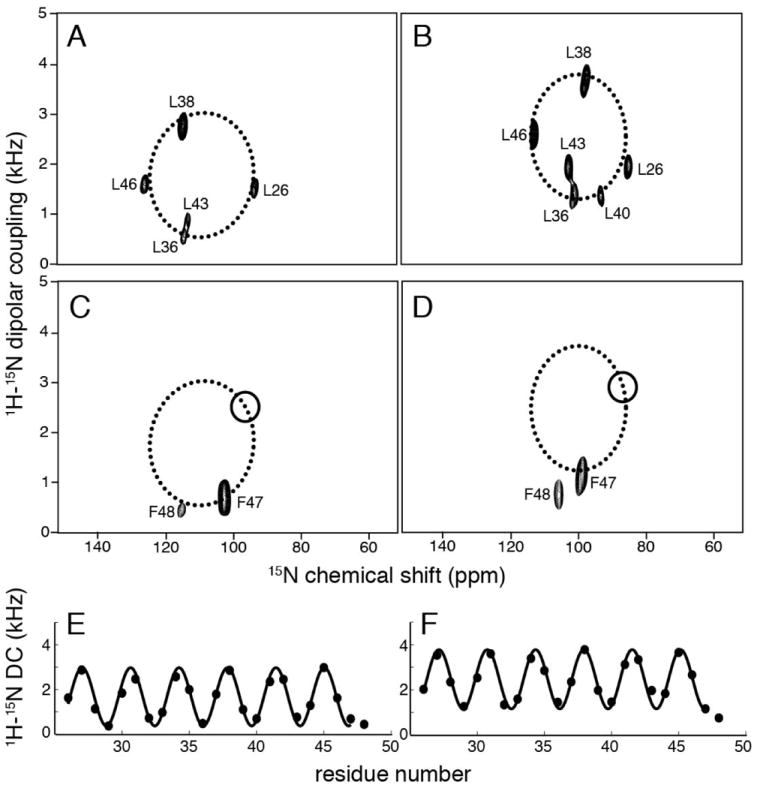
Two-dimensional 1H/15N dipolar coupling/15N chemical shift SLF solid-state NMR spectra of selectively leucine (A and B) and phenylalanine (C and D) 15N-labeled samples in magnetically aligned bicelles. (A) wild-type M2 TM. (B) H37A M2 TM. (C) wild-type Vpu TM. (D) A18H Vpu TM. For (C) and (D), circles indicate the expected position of F48 resonance on the PISA wheel if it were part of the transmembrane helix. Dipolar waves fitting are shown for (E) wild-type M2 TM and (F) H37A M2 TM.
Previous data on Vpu TM constructs display a difference in helix lengths between the wild-type and the A18H mutant (Park and Opella 2007). For the M2 constructs, the helix starts at residue 25, a proline which is a known helix breaker, especially at terminal regions (Senes et al. 2004). The C-terminal end is expected to occur at the two phenylalanines. In order to determine the exact C-terminal residue of the helix, selectively 15N phenylalanine labeled samples were prepared, and their two-dimensional SLF spectra are shown in Figure 3C for wild-type M2 TM and in Figure 3D for H37A M2 TM. In both cases F47 falls within the expected region of the PISA wheel, while the frequencies of F48 significantly deviate from a position indicated by the circle. This is a clear indication that the C-terminus of the TM helix of M2 stops at residue F47. The overall results for both wild-type and mutant M2 showed that they have a 22-residue TM helix that spans from L26 to F47. In contrast, previous studies on Vpu TM constructs showed that the helix spans 18 residues for wild-type Vpu TM and 22 residues for Vpu A18H TM. Dipolar waves for wild-type M2 TM and H37A M2 TM are shown in Figure 3E and 3F. In these plots, the measured values of the dipolar couplings are plotted as a function of residue number. Dipolar waves provide a direct and reliable assessment of the length, tilt angle, and rotation angle of the helices because they reflect the regularity of the secondary structure. Sinusoids with a period of 3.6 residues per turn fitted well to the dipolar coupling values plotted as a function of residue number, indicating that the constructs have no kinks or other deviations from an ideal helix (Mesleh et al. 2002). Similarly, the previously plotted dipolar waves for wild-type Vpu TM and A18H Vpu-TM both were consistent in showing a single helix without significant distortions.
Histidine is responsible for sensing pH change by the M2 transmembrane helix. To characterize the proton sensitivity of the TM helices, we used solution NMR experiments to characterize them in DHPC micelles at various pH values. The two-dimensional HSQC spectra of all of the uniformly 15N labeled constructs were fully resolved and assigned as described previously (Park and Opella 2007). This enabled changes associated with individual residues to be monitored through chemical shift perturbations of the backbone amide resonances. Spectra were recorded at pH 4 and pH 7.5 and the chemical shift differences between the frequencies measured for each resonance are plotted as a function of residue number for wild-type M2 TM (Figure 4A), H37A M2 TM mutant (Figure 4B), wild-type Vpu TM (Figure 4C), and A18H Vpu TM mutant (Figure 4D). Notably, the crucial residues for M2 proton transport, H37 and W42, had large chemical shift changes from low to high pH. In addition, residues flanking H37 and near the C-terminus also experienced significant changes in chemical shift frequencies. A18H Vpu TM displayed similar changes, except that the tryptophan residue appears somewhat less affected. In comparison, chemical shift changes for the H37A M2 TM were confined to the C-terminus. In particular, the backbone amide signals from A37 and W41 experienced little effect from the pH change, and similar results were observed for the corresponding residues of wild-type Vpu TM. Studies have shown that the histidine residue is the key for proton selectivity of M2 (Wang et al. 1995, Okada et al. 2001), and wild-type Vpu with an alanine at the corresponding position is unable to perform the proton transport function of M2 (Betakova 2010). The chemical shift perturbation plots are consistent with these biological functional data. Moreover, the results support a connection between the histidine and tryptophan residues in the proton transport activity of the HXXXW motif. In both wild-type M2 TM and A18H Vpu TM, the tryptophan residue displayed larger chemical shift changes as a result of changing the pH change compared to constructs that do not contain the histidine residue.
Figure 4.
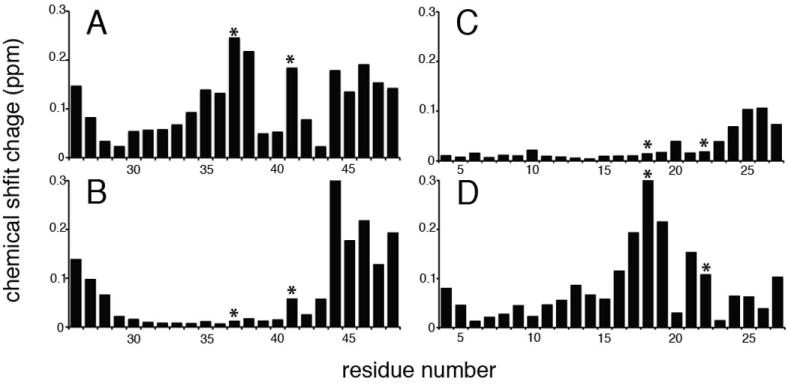
Chemical shift difference between 1H/15N HSQC spectra taken at pH 4 and pH 7.5 plotted against residue number. (A) wild-type M2 TM. (B) H37A M2 TM (C) wild-type Vpu TM. (D) A18H Vpu TM. Histidine/alanine and tryptophan residues from the H/AXXXW motifs are indicated with asterisks.
The tryptophan residue is known to function as the ion channel ‘gate-keeper’ for M2 (Tang et al. 2002) and serves as a membrane anchor for many TM helices in membrane proteins (De Planque et al. 2003). It is also possible that it has a role in concert with the histidine residue in determining the tilt angle of the TM helices. To investigate whether the HXXXW motif is required to maintain a relative large tilt angle of ~ 40°, the W41L M2 TM was prepared. Since leucine also has a bulky hydrophobic side-chain, it should result in minimal perturbation of the occupied volume. The one-dimensional spectra are shown in Figure 5 for uniformly 15N labeled aligned samples of wild-type M2 TM (Figure 5A) and W41L M2 TM (Figure 5B). The anisotropic 15N chemical shift region spans between 60 ppm and 140 ppm for both constructs, and this indicates that the tilt angles for both constructs are approximately the same. However, the resonances from W41L M2 TM have generally broader line widths, and this suggests the possibility of a less well defined molecular conformation, multiple conformations with small differences among them, or possibly increased motion. While the tryptophan residue is not required to maintain the tilt change induced by the histidine, it is may play a role in stabilizing the TM as a membrane anchor.
Figure 5.
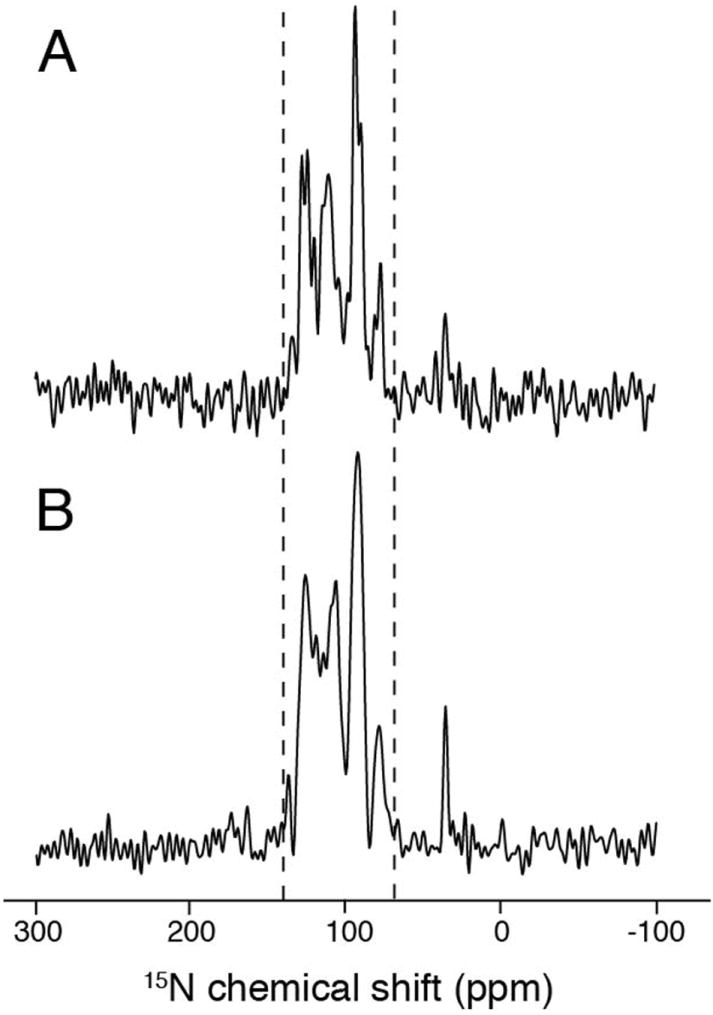
Comparison of one-dimensional 15N chemical shift spectra of samples in magnetically aligned bicelles. (A) wild-type M2 TM. (B) W41L M2 TM mutant. Similarity in tilt angle is indicated by the good overlap of the signal region between the dashed lines.
In addition, we performed all-atom molecular dynamics (MD) simulations in Xplor-NIH to correlate the experimental findings on the histidine residue in the polypeptides. The simulations were performed on the monomer of each construct. 10 structures are chosen for analysis from each calculation, the backbone RMSDs are 0.77±0.15Å and 0.91±0.27Å for wild type M2 TM and H37A M2 TM, respectively, and 0.81±0.24Å and 0.89±0.22Å, for wild-type Vpu TM and A18H Vpu TM, respectively. The results also showed significant differences in tilt angles between the same helices with and without histidine. The simulated structures are shown in Figure 6. Overall, the tilt angles correlate well with the experimental data. The discrepancies can be attributed to a number of factors including experimental error for PISA wheel analysis, and other intrinsic properties of the protein constructs that were not included in the simulation. Importantly, for both M2 and Vpu, the wild-type helix and its mutant also display significant differences in tilt angles consistent with the solid-state NMR data. This reinforces the conclusion that the presence of histidine increases the helix tilt angle. The change in tilt angle appears to be slightly larger for the case of Vpu which shows a difference of 14° in comparison to M2 which shows a difference of 8°. Their tilt angles, extracted from coordinates, are: 40.0° and 32.0° for wild-type and the mutant H37A M2-TM, 31.5° and 45.6° for wild-type and the mutant A18H Vpu-TM (Figure 6).
Figure 6.
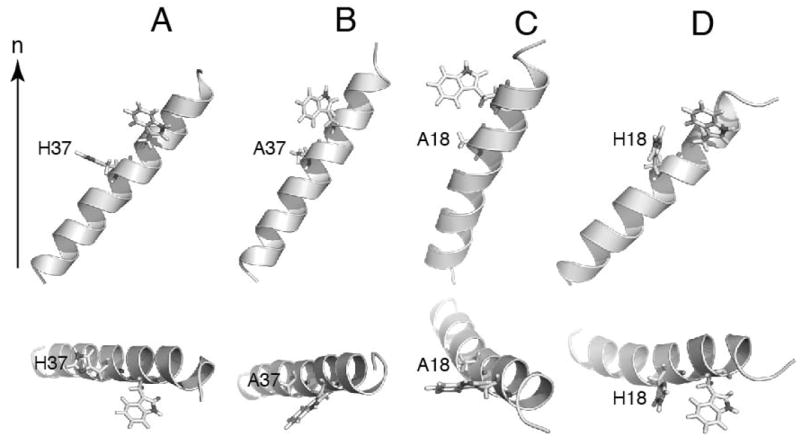
Orientation of transmembrane helix of the wild-type and mutants of Vpu and M2. Structures are refined against dipolar coupling and chemical shift anisotropy restraints. The alignments are achieved based on dipolar coupling and chemical shift anisotropy restraints as well. (A) wild-type M2 TM. (B) H37A M2 TM. (C) wild-type Vpu TM. (D) A18H Vpu TM. The orientations in membrane bilayers are show as relative to the membrane normal (n). The top view of each structure is shown at the bottom.
Discussion
Polypeptides corresponding to wild-type M2 TM of Influenza and its H37A mutant were studied in DHPC micelles by solution NMR and in DMPC/DHPC q=3.2 bicelles by solid-state NMR. The results are compared with our previous studies of wild-type Vpu TM and A18H Vpu TM (Park et al. 2006, Park et al. 2003, Park and Opella 2007). There have been a number of pertinent studies comparing the properties of these viroporins due to their similar structural and functional features (Cook et al. 2011, Sharma et al. 2011, Wang et al. 2011b). The results and interpretations described here rely on direct comparisons, therefore the same experimental conditions and methods were used throughout. This is a crucial feature of the study, since the observed functional and structural properties of membrane proteins, especially single TM channel proteins depend upon experimental conditions such as protein concentration, types of lipids, temperature, etc. The solid-state NMR results showed that the TM constructs containing a histidine residue, wild-type or prepared by mutation, have significantly higher helix tilt angles than those with an alanine instead of a histidine at that location in the sequence.
Several factors have been shown to affect the tilt angle of a transmembrane helix. Hydrophobic mismatch between bilayer thickness and helical length can result in tilt angle adjustment to optimize the membrane incorporation of the TM helix (Park and Opella 2005). However, in this study, the bilayer thickness is the same in all samples, since same phospholipid, i.e. DMPC, is used to form the bilayers for all of the TM helices. While the wild-type Vpu TM was observed to be shorter than its A18H mutant in our previous study (Park and Opella, 2007), the helical lengths are the same for wild-type M2 TM and its H37A mutant. Therefore, the exact natures of the bilayer and helices are not likely to be major factors in the observed tilt angle differences. Interfacial tryptophan residues have also been shown to directly contribute to tilt angle (Vostrikov and Koeppe 2011). We investigated this effect by mutating the tryptophan residue in wild-type M2-TM to a leucine, and the results show that the tilt angle is unchanged. Thus, while tryptophan plays the role of helix anchor, it does not make a significant contribution to the large tilt angle observed in the histidine-containing construct. There is some evidence that the tryptophan residue affects the structural stability of the TM helix.
The side chain of histidine has a pKa of approximately 6, and its role as a pH-sensing residue for the M2 proton channel is well established. This is clearly demonstrated in the chemical shift perturbation profile of the backbone amide. The histidine amide resonance displayed the largest chemical shift change when the pH was increased from 4 to 7.4 for both the wild-type M2 TM and A18H Vpu TM, and the replacement of histidine by alanine abolishes this perturbation. Depending on the pH, the histidine side chain can be positively charged or polar, and neither condition is favorable for insertion into a hydrophobic membrane environment. It has been suggested that the TM helix can accommodate this by increasing the solvent exposure of the unfavorable side chain by altering the helix tilt angle and shifting the membrane insertion of the residue. This may explain some of the large tilt angle variations observed in the solid-state NMR studies of transmembrane helices. Addition of the chelate Mn-EDTA to micelle samples of wild-type M2 TM and H37A M2 TM resulted in selective line broadening due to the paramagnetic property of manganese. The intensities of the resonances are reduced for amide sites in relatively close proximity (< 30 Å) with the chelate and/or exchanging with the solvent protons that have enhanced relaxation from the chelate. The calculation of ‘%Change in Relative Intensity’ (%CRI) is described in detail in the Material & Methods section, and this value is plotted against residue number in Figure 7. In principle, the sign of %CRI partially reflects the solvent exposure for backbone amides with the positive sign being less exposed and negative sign being more exposed. The two constructs display mostly similar profiles with the resonances from residues in terminal regions experiencing intensity reduction as expected. Residues 29 to 35 have positive values and thus are less exposed to solvent, likely due to the higher degree of hydrophobic incorporations and intra chain hydrogen bonds in the interior of the lipid acyl chain environment. The key difference lies within the region between residues 36 and 42. In particular, residues 41 and 38 have more negative %CRI for wild-type M2 TM than for H37A M2 TM, indicating that they are more exposed to the solvent. We cannot definitively differentiate between the lipids becoming more loosely packed around the region or the C-terminal region having more obtrusion into the solvent. It is likely to be former case since the signal intensity reduction of the 36-42 region does not follow a uniform trend as in the case of the terminal regions. However, it is certain that the packing of the lipids around the TM helix differs between the two constructs, and this would be expected to translate into differences in lipid incorporation for the bilayer samples as well. Since the bilayer is less flexible in terms of packing around the helix, it would be energetically favorable for the helix to increase the tilt angle to achieve more solvent exposure for the polar/charged residue(s).
Figure 7.
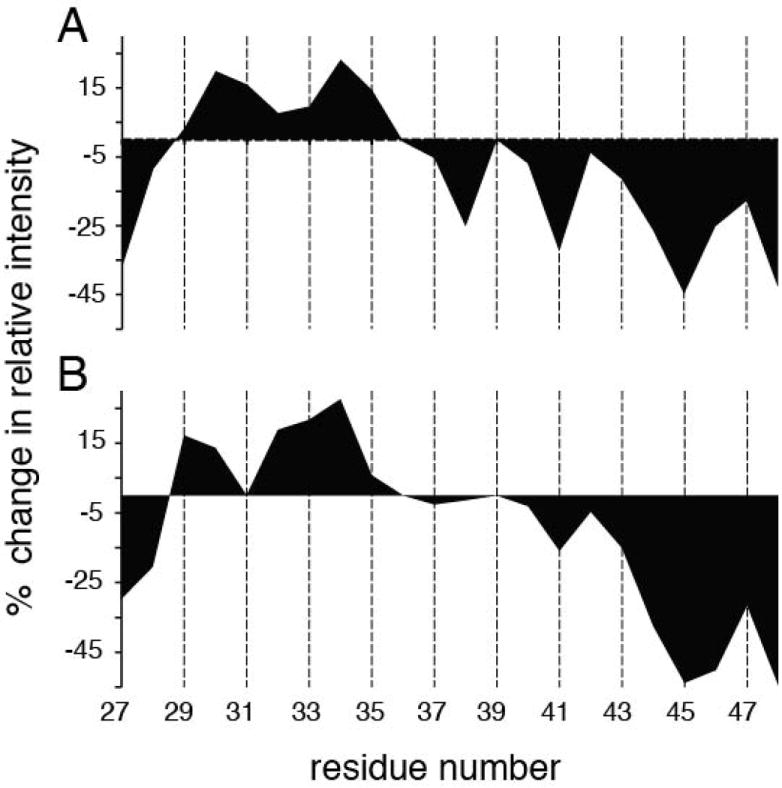
1H/15N HSQC signal intensity change due to paramagnetic relaxation enhancement of Mn-EDTA in solvent for individual residues of (A) wild-type M2-TM and (B) H37A M2 TM. Positive values suggest that the corresponding amides are less solvent accessible while the negative values suggest the opposite. Intensities for residues 31 and 39 of H37A M2 TM could not be properly extracted due to overlapping resonances.
Other residues have also been shown to change the tilt angles of TM helices. Previously, Koeppe and coworkers showed that substituting either an arginine or a lysine for a leucine within the GWALP type TM peptides also causes an increase in the helix tilt angle based on results of solid-state NMR and molecular dynamics simulations (Gleason et al. 2013, Vostrikov et al. 2010). This suggests that introducing a charged residue can have a significant impact on the helix tilt. However, we observed no significant difference in the tilt angle when the solid-state NMR experiments were performed at pH 7.4 instead of 4 (data not shown). Moreover, the result on the W41L mutant of M2 TM indicates that the tilt angle is not directly affected by the tryptophan residue, but it still serves as a stabilizing force. This suggests that within the pH range tested, the charge state of the histidine also did not have significant contribution to the observed large tilt change.
The mutations performed in this work also have biological implications. Studies have shown that the H37A mutation enables M2 to become permeable to Na+ and K+ ions, compared to the strict selectivity for H+ of the wild-type protein (Venkataraman et al. 2005). In contrast, wild-type Vpu forms an ion channel that is permeable to Na+ and K+ ions. The proton transport function is localized to the cooperative function of the histidine and tryptophan residues of M2. This is also found in the Vpu A18H mutant, but the magnitude of the chemical shift perturbation suggests that this effect may be weaker than in the wild-type M2. Vpu itself is unable to acidify a cellular compartment as M2 does (Betakova 2010), and it has been shown that the A18H Vpu TM was confined to the ER resulting in impaired viral release (Skasko et al. 2011). This suggests that the mutation may be disruptive to the protein localization process. Overall, the changes resulting from this single mutation appear to be detrimental for the proper cellular functions of these two viroporins (Hout et al. 2006, Park and Opella 2007, Takeuchi et al. 2003, Wang et al. 1995).
Acknowledgments
We thank Roger E. Koeppe II for providing helpful discussions and insights. We also thank Chris Grant and Albert Wu for assistance with instrumentation. The research was supported by Grants RO1GM099986, RO1GM066978, and R21GM075917 from the National Institutes of Health. It utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at the University of California, San Diego supported by grant P41EB002031.
Footnotes
Declaration of Interest
The authors declare that there are no conflicts of interest.
References
- Betakova T. Human immunodeficiency virus 1 Vpu protein does not affect the conversion of influenza A virus hemagglutinin to its low-pH conformation in an acidic trans-Golgi compartment. Acta Virol. 2010;54(3):197–203. doi: 10.4149/av_2010_03_197. [DOI] [PubMed] [Google Scholar]
- Cook GA, Opella SJ. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim Biophys Acta. 2011;1808(6):1448–1453. doi: 10.1016/j.bbamem.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GA, Zhang H, Park SH, Wang Y, Opella SJ. Comparative NMR studies demonstrate profound differences between two viroporins: p7 of HCV and Vpu of HIV-1. Biochim Biophys Acta. 2011;1808(2):554–560. doi: 10.1016/j.bbamem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Dong H, Sharma M, Busath DD, Zhou HX. M2 protein from influenza A: from multiple structures to biophysical and functional insights. Curr Opin Virol. 2012;2(2):128–133. doi: 10.1016/j.coviro.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis AA, Jones DH, Grant CV, Park SH, Mesleh MF, Opella SJ. NMR experiments on aligned samples of membrane proteins. Methods Enzymol. 2005;394:350–382. doi: 10.1016/S0076-6879(05)94014-7. [DOI] [PubMed] [Google Scholar]
- De Angelis AA, Opella SJ. Bicelle samples for solid-state NMR of membrane proteins. Nat Protoc. 2007;2(10):2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- De Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, 2nd, Separovic F, Watts A, Killian JA. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42(18):5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Gleason NJ, Vostrikov VV, Greathouse DV, Koeppe RE., 2nd Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc Natl Acad Sci U S A. 2013;110(5):1692–1695. doi: 10.1073/pnas.1215400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout DR, Gomez LM, Pacyniak E, Miller JM, Hill MS, Stephens EB. A single amino acid substitution within the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein renders simian-human immunodeficiency virus (SHIV(KU-1bMC33)) susceptible to rimantadine. Virology. 2006;348(2):449–461. doi: 10.1016/j.virol.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci U S A. 2006;103(18):6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Larson CA, Adams PL, Blair BG, Safaei R, Howell SB. The role of the methionines and histidines in the transmembrane domain of mammalian copper transporter 1 in the cellular accumulation of cisplatin. Mol Pharmacol. 2010;78(3):333–339. doi: 10.1124/mol.110.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002;11(3):546–557. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi FM, Ma C, Gratkowski H, Straus SK, Strebel K, Oblatt-Montal M, Montal M, Opella SJ. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc Natl Acad Sci U S A. 1999;96(25):14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144(1):150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesleh MF, Veglia G, Desilva TM, Marassi FM, Opella SJ. Dipolar waves as NMR maps of protein structure. J Am Chem Soc. 2002;124(16):4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov AA, Opella SJ. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J Magn Reson. 2003;164(1):182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza a virus. Biochemistry. 2001;40(20):6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104(8):3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006;91(8):3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol. 2003;333(2):409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- Park SH, Opella SJ. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J Mol Biol. 2005;350(2):310–318. doi: 10.1016/j.jmb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Park SH, Opella SJ. Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs. Protein Sci. 2007;16(10):2205–2215. doi: 10.1110/ps.073041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69(3):517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Rehwald M, Neuschafer-Rube F, De Vries C, Puschel GP. Possible role for ligand binding of histidine 81 in the second transmembrane domain of the rat prostaglandin F2alpha receptor. FEBS Lett. 1999;443(3):357–362. doi: 10.1016/s0014-5793(99)00007-1. [DOI] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol. 2009;83(15):7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398(1):12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Senes A, Engel DE, Degrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Sharma M, Li C, Busath DD, Zhou HX, Cross TA. Drug sensitivity, drug-resistant mutations, and structures of three conductance domains of viral porins. Biochim Biophys Acta. 2011;1808(2):538–546. doi: 10.1016/j.bbamem.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Tokarev A, Chen CC, Fischer WB, Pillai SK, Guatelli J. BST-2 is rapidly down-regulated from the cell surface by the HIV-1 protein Vpu: evidence for a post-ER mechanism of Vpu-action. Virology. 2011;411(1):65–77. doi: 10.1016/j.virol.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Wang Y, Tian Y, Tokarev A, Munguia J, Ruiz A, Stephens EB, Opella SJ, Guatelli J. HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem. 2012;287(1):58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AK, Kurschat CE, Burns D, Banger N, Vaughan-Jones RD, Alper SL. Transmembrane domain histidines contribute to regulation of AE2-mediated anion exchange by pH. Am J Physiol Cell Physiol. 2007;292(2):C909–918. doi: 10.1152/ajpcell.00265.2006. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Okada A, Miura T. Roles of the histidine and tryptophan side chains in the M2 proton channel from influenza A virus. FEBS Lett. 2003;552(1):35–38. doi: 10.1016/s0014-5793(03)00781-6. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277(42):39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280(22):21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- Vostrikov VV, Hall BA, Greathouse DV, Koeppe RE, 2nd, Sansom MS. Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J Am Chem Soc. 2010;132(16):5803–5811. doi: 10.1021/ja100598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov VV, Koeppe RE., 2nd Response of GWALP transmembrane peptides to changes in the tryptophan anchor positions. Biochemistry. 2011;50(35):7522–7535. doi: 10.1021/bi2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69(4):1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Nishimra K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J Magn Reson. 2000;144(1):162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- Wang J, Qiu JX, Soto C, Degrado WF. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr Opin Struct Biol. 2011a;21(1):68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta. 2011b;1808(2):510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Grant CV, Cook GA, Park SH, Opella SJ. A strip-shield improves the efficiency of a solenoid coil in probes for high-field solid-state NMR of lossy biological samples. J Magn Reson. 2009;200(1):74–80. doi: 10.1016/j.jmr.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


