Abstract
PINK1, a mitochondrial serine/threonine kinase, is the product of a gene mutated in an autosomal recessive form of Parkinson disease. PINK1 is constitutively degraded by an unknown mechanism and stabilized selectively on damaged mitochondria where it can recruit the E3 ligase PARK2/PARKIN to induce mitophagy. Here, we show that, under steady-state conditions, endogenous PINK1 is constitutively and rapidly degraded by E3 ubiquitin ligases UBR1, UBR2 and UBR4 through the N-end rule pathway. Following precursor import into mitochondria, PINK1 is cleaved in the transmembrane segment by a mitochondrial intramembrane protease PARL generating an N-terminal destabilizing amino acid and then retrotranslocates from mitochondria to the cytosol for N-end recognition and proteasomal degradation. Thus, sequential actions of mitochondrial import, PARL-processing, retrotranslocation and recognition by N-end rule E3 enzymes for the ubiquitin proteosomal degradation defines the rapid PINK1 turnover. PINK1 steady-state elimination by the N-end rule identifies a novel organelle to cytoplasm turnover pathway that yields a mechanism to flag damaged mitochondria for autophagic elimination.
Keywords: mitochondrial import, PARKIN, ubiquitin, PARL, mitophagy
Introduction
Two genes mutated in autosomal recessive forms of familial Parkinson disease, PINK1 and PARKIN have been suggested to play a role in mitochondrial quality control. Genetic and cell biological studies indicate that the mitochondrial kinase PINK1 acts in the same pathway as the cytosolic E3 ligase PARKIN1-3 by recruiting PARKIN to dysfunctional mitochondria to induce their elimination by autophagy.4 PINK1 signals mitochondrial damage by accumulating selectively on the outer mitochondrial membrane (OMM) of depolarized mitochondria.5-8 However, the expression of PINK1 in healthy mitochondria is barely detectable following import into the inner mitochondrial membrane (IMM) and sequential processing by the proteases MPP in the matrix and PARL in the IMM.9-13 Although many studies have focused on the subcellular and intramitochondrial localization of PINK1 under steady-state conditions,14-21 how it is eliminated is still unknown.
Results
PARL-cleaved PINK1 can form cytosolic aggregates with SQSTM1/p62
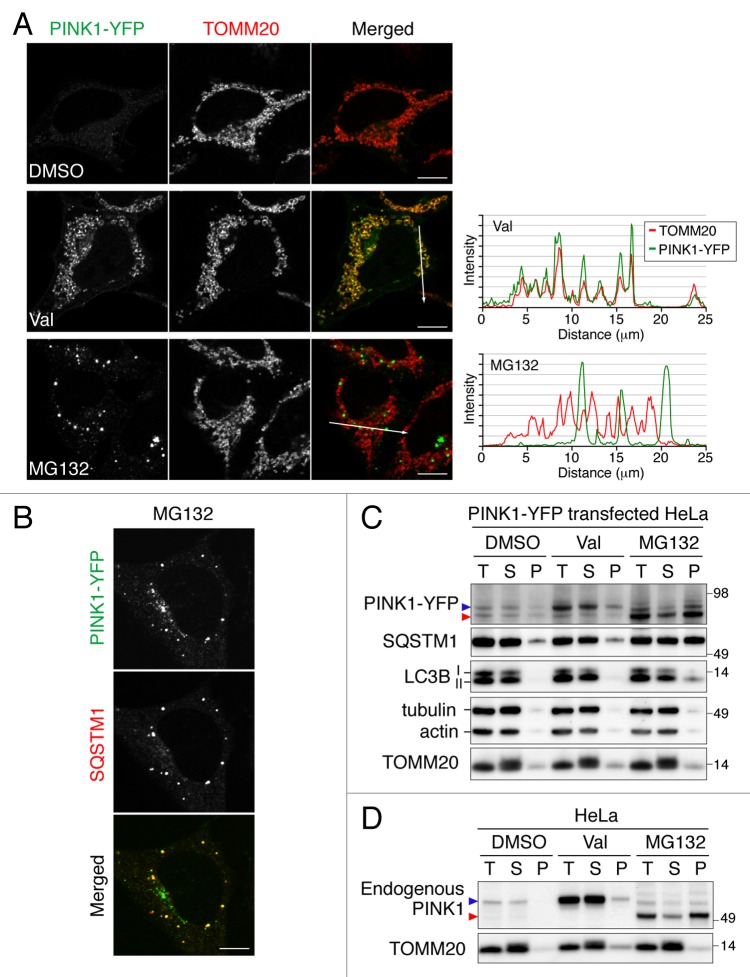
To understand PINK1 localization and stability, we treated HeLa cells expressing PINK1-YFP with dimethyl sulfoxide (DMSO), valinomycin or MG132 (Fig. 1A). Normally, PINK1-YFP expression in most cells is below the level of detection, consistent with models of rapid PINK1 turnover.6,7 However, exposure to valinomycin, which disrupts the mitochondrial inner membrane potential, induces PINK1 accumulation on mitochondria. Although treatment of cells with the proteasome inhibitor MG132 also enhances the PINK1-YFP signal, it is found in dot-like structures, which are not colocalized with TOMM20 but nicely merge with the cytosolic protein aggregate marker SQSTM1 (Fig. 1A and B). Similar results were also observed in HeLa and HCT116 cells stably expressing PINK1-YFP (Fig. S1). Immunoblotting analysis confirmed that PINK1-YFP forms aggregates upon MG132 treatment (Fig. 1C). In the absence of valinomycin or MG132, two weak bands of PINK1-YFP, the full-length and the PARL-cleaved forms, were recovered in the supernatant after solubilization with a detergent. The increased level of full-length PINK1-YFP generated by valinomycin was also collected in the soluble fraction. However, MG132 treatment specifically increased the PARL-cleaved form in a detergent-insoluble fraction. While tubulin, actin and TOMM20 were collected in the soluble fraction under all conditions tested, a fraction of the lipidated MAP1LC3B/LC3B (LC3-II) and SQSTM1 proteins were found in the detergent-insoluble fraction in cells treated with MG132 also supporting the microscopic observation that PINK1-YFP forms aggregates colocalized with SQSTM1. Notably, endogenous PINK1 showed an aggregate distribution pattern indistinguishable from that of PINK1-YFP (Fig. 1D). Taken together, these results indicate that proteasome inhibition selectively increases the level of the PARL-cleaved form of PINK1 as aggregates in the cytosol, but not in mitochondria.
Figure 1. PARL-cleaved PINK1 can form cytosolic aggregates. (A and B) Microscopic analysis of HeLa cells transiently expressing PINK1-YFP treated with DMSO, valinomycin (Val) or MG132 for 3 h. Cells were immunostained with anti-TOMM20 (A) and with anti-SQSTM1 (B) antibodies. Line scan plots (taken from the images along the white arrows in (A) are shown in the right panel. Scale bars: 10 μm. (C and D) Immunoblotting of total cell lysate (T), supernatant (S) and pellet (P) fractions after solubilization with 2% Triton X-100 followed by centrifugation. Blue and red arrowheads represent the full-length and PARL-cleaved forms, respectively.
N-end rule pathway governs PINK1 degradation
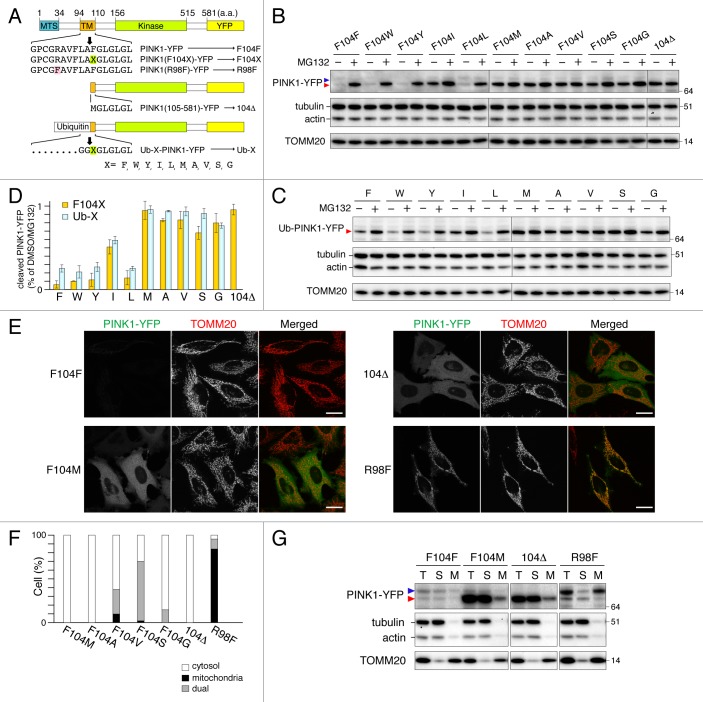
To mimic the PARL-cleaved PINK1 expression in the cytosol, we made truncated PINK1-YFP lacking the N-terminal 1 to 104 residues (104Δ) (Fig. 2A). In contrast to endogenous or ecotopic full-length PINK1-YFP, recombinant 104Δ was surprisingly stable in the cytosol without proteasome inhibition (Fig. 2B and E). One conceivable difference between recombinant expressed 104Δ and PARL-cleaved PINK1 is the first N-terminal amino acid residue. PARL cleaves between A103 and F104 of PINK1, thereby yielding a phenylalanine at the N terminus10 whereas 104Δ yields an N-terminal methionine from the start codon.
Figure 2. N-end rule pathway governs PINK1 degradation. (A) Scheme of PINK1-YFP variants. Bold downward arrows indicate the cleavage sites of PINK-YFP and Ub-PINK1-YFP by PARL and DUBs, respectively. (B and C) HeLa cells transiently expressing PINK1-YFP (B) or Ub-PINK1-YFP (C) variants were treated with DMSO or MG132 for 3 h. Total cell lysates were analyzed by immunoblotting. Tubulin, actin and TOMM20 were used as loading controls. (D) Proteasomal degradation efficiencies of cleaved forms of PINK1-YFP or Ub-PINK1-YFP variants prepared as in (B and C) were quantified. Data are the mean ± SD of three independent experiments. (E) Microscopy analysis of HeLa cells transiently expressing PINK1-YFP variants. Cells were immunostained with anti-TOMM20. Scale bars: 20 μm. (F) Quantification of localization of the PINK1-YFP mutated to stabilizing residues. Dual means that cells have a YFP signal both in mitochondria and in the cytosol. (G) Transfected cells as in (E) were fractionated. T, total fraction of post-nuclear supernatant; S, post-mitochondrial supernatant; M, mitochondrial fraction. Blue and red arrowheads denote full-length (or MPP-cleaved for R98F) and PARL-cleaved forms, respectively.
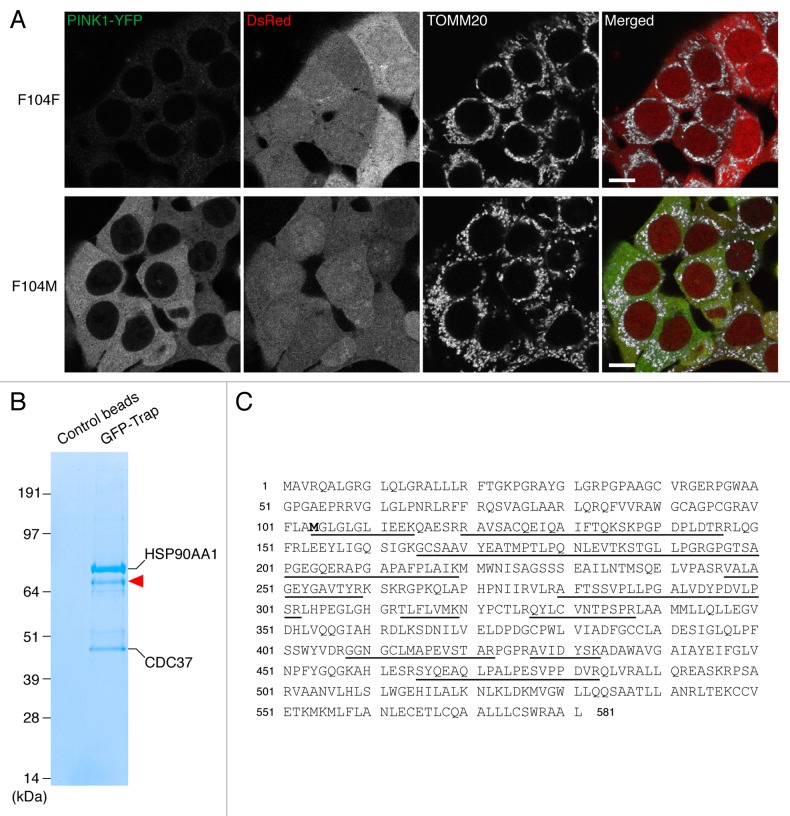
According to the N-end rule pathway, certain N-terminal amino acids function as signals for ubiquitin-mediated degradation of the proteins by the proteasome.22 The signal of N-end degradation (N-degron) in mammals is typically composed of N-terminal primary destabilizing amino acids (R, K and H for type-1, and bulky hydrophobic residues F, W, Y, I and L for type-2) and an internal lysine residue for polyubiquitination.23 To investigate if PARL-cleaved PINK1 is a substrate for the N-end rule pathway, we constructed PINK1-YFP mutants where residue F104 was replaced with other destabilizing residues (W, Y, I and L for type-2 degrons) and stabilizing residues24 (M, A, V, S and G) (Fig. 2A). Wild-type (WT) PINK1-YFP (PINK1F104F-YFP) was hard to detect both by fluorescent microscopy (Fig. 2E) and by immunoblotting (Fig. 2B) and was stabilized by MG132 treatment (Fig. 2B and D). Similar results were obtained for PINK1 F104 mutants that generated other type-2 degrons (Fig. 2B and D; Fig. S2). By contrast, when PINK1-YFP was mutated to yield stabilizing residues, their cleaved forms were stable without MG132 treatment (Fig. 2B and D; Fig. S2). Microscopy and subcellular fractionation assays indicated that PINK1-YFP cleaved to yield stabilizing residues were mostly localized in the cytosol while PINK1R98F-YFP, whose mutation abolishes processing by PARL,9 was localized inside mitochondria (Fig. 2E–G; Fig. S2). To rule out the possibility that PARL cleaves some of the PINK1 mutants at a false processing site, we also generated cleaved forms of PINK1-YFP by fusion to ubiquitin (Ub) (Fig. 2A). This Ub fusion technique25 enables the expression of PINK1-YFP cleaved by DUBs to generate the desired N-terminal amino acid variants in the cytosol. Immunoblotting of Ub-fusion generated PINK1 mutants showed that their sensitivities to proteasome degradation were very similar to those of the corresponding PARL-processed PINK1-YFP variants (Fig. 2C and D), strongly suggesting that PARL cleavage occurs after A103 in all mutants. Furthermore, in order to identify the PARL cleavage site of stabilizing mutant PINK1F104M by mass spectrometry, we made a stable HCT116 cell line expressing PINK1F104M-YFP. To compare the expression level of PINK1F104M-YFP with that of WT PINK1-YFP, we used the internal ribosome entry site (IRES)-DsRed system, which directs coexpression of PINK1-YFP and cytosolic DsRed under a single promoter. As shown in Figure 3A, fluorescent signals of PINK1F104M-YFP were clearly found in the cytosol while those of WT PINK1 were barely detectable, showing that F104M mutation increases protein stability in the cytosol. After affinity purification of PINK1F104M-YFP with beads conjugated with GFP-binding protein, three strong coomassie blue-labeled bands were observed on SDS-PAGE (Fig. 3B). Mass spectrometry identified the upper and lower bands as HSP90AA1/Hsp90 and CDC37, respectively, consistent with previous reports.20,26 Furthermore, several tryptic peptides corresponding to cleaved PINK1 were identified from a band labeled by the red arrowhead in Figure 3B, and identification of the peptide MGLGLGLIEEK indicates that PARL cleaves PINK1F104M-YFP between A103 and M104 (Fig. 3C). Therefore, all these results indicate that the new N-terminal amino acid generated by PARL processing defines PINK1 stability in the cytosol correlating with the N-end rule.
Figure 3. PARL cleaves PINK1F104M-YFP between A103 and M104. (A) HCT116 cells stably expressing wild-type (F104F) and the F104M mutant of PINK1-YFP-IRES-DsRed were immunostained with anti-TOMM20. Scale bars: 10 μm. (B) HCT116 cells stably expressing PINK1F104M-YFP were solubilized with 0.5% Triton X-100 and subjected to immunoprecipitation with GFP-Trap or control beads. Proteins were analyzed by SDS-PAGE followed by coomassie brilliant blue staining. Red arrowhead represents PARL-cleaved F104M mutant. Mass spectrometry identified HSP90AA1 and CDC37 as interacting partners of cleaved F104M mutant. (C) The protein band described by arrowhead in (B) was analyzed by LC/MS/MS, and the identified sequences of PINK1F104M moiety are underlined. Identifying the peptide corresponding to MGLGLGLIEEK indicates that PARL cleaves the PINK1F104M mutant between A103 and M104.
N-end stabilizing PINK1 mutant (F104M) can recruit PARKIN to depolarized mitochondria followed by mitophagy
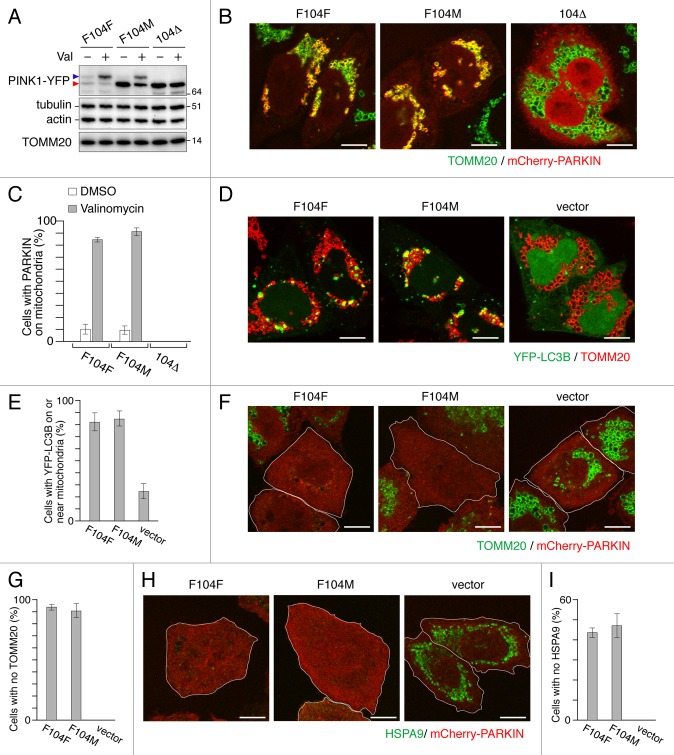
Uncoupling mitochondrial membrane potential shunts PINK1 from the IMM and PARL processing to the OMM where it accumulates as a full-length form.9 Although PARL-processed PINK1F104M-YFP was more stable than WT PINK1-YFP in the cytosol, the accumulation of full-length F104M mutant on mitochondria induced by valinomycin was comparable to that of WT PINK1-YFP (Fig. 4A) and, when expressed in pink1−/− MEFs, the F104M mutant could recruit PARKIN to depolarized mitochondria with an efficiency similar to that of WT PINK1-YFP (Fig. 4B and C). Therefore, the F104M mutation affects stability following PARL cleavage, but not the stability on the OMM of damaged mitochondria. In order to assess whether the PINK1F104M mutant can promote mitochondrial autophagy, we looked at LC3B localization by microscopy. For this purpose, we constructed pink1 KO HeLa cells by TALEN-mediated gene editing (See materials and methods for details). After 3 h valinomycin treatment, YFP-LC3B formed dot-like structures on or near mitochondria in pink1 KO HeLa cells stably expressing mCherry-PARKIN only when WT PINK1F104F or PINK1F104M was expressed (Fig. 4D and E). Furthermore, when we expressed WT PINK1F104F-YFP or PINK1F104M-YFP in pink1 KO cells and treated with valinomycin for 12 h, we observed that 80% and 40% of the cells completely lost immunostaining of TOMM20 and a mitochondrial matrix protein HSPA9/GRP75, respectively (Fig. 4F–I). These events were PINK1 dependent because pink1 KO HeLa cells with control vectors had neither LC3B translocation to mitochondria nor mitochondrial protein degradation. Thus, the N-end stabilizing PINK1F104M mutant has the ability to promote PARKIN-mediated mitophagy.
Figure 4. PINK1F104M mutant has the ability to recruit PARKIN to depolarized mitochondria followed by mitophagy. (A) Immunoblotting of HeLa cells expressing PINK1-YFP variants (F104F, F104M and 104Δ) treated with or without valinomycin (Val) for 3 h. Blue and red arrowheads represent the full-length and the cleaved forms of PINK1-YFP, respectively. (B and C) pink1−/− MEF cells were cotransfected with PINK1-YFP variants and mCherry-PARKIN. After 12 h of transfection, cells were treated with DMSO or valinomycin for 4 h. (B) Cells were then immunostained with anti-TOMM20. Scale bars: 10 μm. (C) Cells with mCherry-PARKIN on mitochondria were quantified. Error bars were calculated from three independent experiments. (D and E) pink1 KO HeLa cells stably expressing mCherry-PARKIN were cotransfected with YFP-LC3B and untagged either PINK1 WT(F104F) or PINK1(F104M) or pcDNA3.1(+) (vector). Cells were treated with valinomycin for 3 h, and immunostained with anti-TOMM20. (D) Scale bars: 10 μm. (E) Cells with YFP-LC3B on or near mitochondria were quantified. Error bars represent the results from three independent experiments. (F–I) pink1 KO HeLa cells stably expressing mCherry-PARKIN were transfected with WT PINK1-YFP (F104F), PINK1F104M-YFP or pEYFP-C1 (vector). Cells were treated with valinomycin for 12 h, and then immunostained with anti-TOMM20 or anti-HSPA9 antibodies. (F and H) YFP-positive cells are shown in white outlines. Scale bars: 10 μm. (G and I) Cells with complete degradation of TOMM20 or HSPA9 were quantified. Error bars represent the results from three independent experiments.
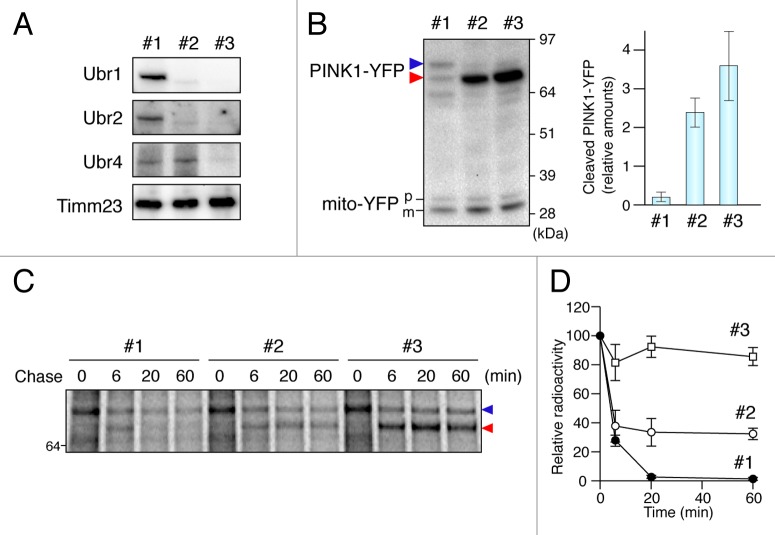
UBR1, UBR2 and UBR4 are responsible for PINK1 recognition
To confirm N-end rule degradation, we examined the role of 3 cytosolic E3 ubiquitin ligases involved in N-degron recognition. In mammals, E3 enzymes UBR1, UBR2 and UBR4 are important for recognition of type-2 degrons such as the phenylalanine motif on PARL-cleaved PINK1.27 PINK1-YFP was transiently expressed in ubr1ubr2 double KO MEF cells stably expressing firefly luciferase RNAi (ubr1−/−ubr2−/−LucRNAi) and the double KO MEF cells stably expressing a Ubr4 short-hairpin RNAi (ubr1−/−ubr2−/−Ubr4RNAi) (Fig. 5A). Strong and selective accumulation of the cleaved form of PINK1 was observed as compared with that in the corresponding WT cells (Fig. 5B). We also monitored the turnover and degradation of PINK1 by pulse-chase experiments (Fig. 5C and D). For this purpose, we utilized CCCP treatment of cells to uncouple mitochondria in order to transiently accumulate PINK1 on mitochondria. When CCCP was washed out, the accumulated full-length PINK1-YFP rapidly disappeared. On the other hand, the PARL-cleaved form of PINK1-YFP in ubr1−/−ubr2−/−Ubr4RNAi MEFs was significantly stabilized indicating that N-end rule E3 ubiquitin ligases in the cytosol mediate the final proteosomal degradation of PARL-cleaved PINK1.
Figure 5. UBR proteins are responsible for PINK1 recognition. (A) Immunoblotting of total cell lysates from wild type (#1), ubr1−/−ubr2−/−LucRNAi (#2) and ubr1−/−ubr2−/−Ubr4RNAi (#3) MEFs. (B) Total cell lysates from MEF cells cotransfected with PINK-YFP and mito-YFP at a 20:1 DNA ratio were analyzed by immunoblotting with anti-GFP antibody. In order to monitor any transfection efficiency discrepancies between the different cell lines, we used mito-YFP as an internal control, which was cotransfected with PINK1-YFP. The graph shows the relative amount of PARL-cleaved PINK1-YFP to mature mito-YFP, expressed as the mean ± SD of three independent experiments. Blue and red arrowheads denote full-length and PARL-cleaved PINK1-YFP. p and m denote precursor and mature forms of mito-YFP. (C) CCCP-treated MEF cells labeled with 35S-methionine/cysteine after PINK1-YFP transfection were subjected to CCCP-washout followed by chase for the indicated time periods. Immunoprecipitated PINK1-YFP was subjected to radioimaging. (D) Quantification of amounts of PARL-cleaved PINK1-YFP relative to full length PINK1-YFP at 0 time during the chase periods with the mean ± SD of three independent experiments. The amount of full-length PINK1-YFP at 0 time was set to 100%.
PARL-cleaved PINK1 retrotranslocates from mitochondria
Identifying mutants of PINK1 stabilized in the cytosol allowed us to explore how PINK1 escapes the mitochondrion. As the PINK1 precursor contains an N-terminal MTS, newly synthesized PINK1 is imported into mitochondria. Typically, this class of mitochondrial precursor proteins is imported via the TOM and TIMM23 complexes, the translocators of the outer and inner membranes, respectively, and is integrated into the IMM by a stop-transfer mechanism.28,29 To explore PINK1 retrotranslocation from mitochondria back to the cytosol, we performed in vitro mitochondrial import assays. Isolated mitochondria were incubated with 35S-radiolabeled PINK1 precursor. After import, mitochondrial pellet (pel) and supernatant (sup) fractions were separated by centrifugation. PARL-cleaved PINK1 was found not only in the mitochondrial fraction, but also in the supernatant (Fig. 6A). We also examined two control mitochondrial substrates, IMS soluble protein HTRA2/OMI and the matrix targeted Su9-DHFR (fusion protein between the mitochondrial presequence of Neurospora crassa subunit 9 of F0-ATPase and mouse dihydrofolate reductase). Mature forms of both HTRA2 and Su9-DHFR were exclusively found in the mitochondrial fraction (Fig. 6A). We performed PK protection assays to determine the localization of the cleaved PINK1 in the mitochondrial pellet. As shown in Figure 6B, the mitochondrial proteins MFN1 (a membrane integrated OMM protein), cytochrome c (Cyt.c) and HTRA2 (IMS soluble proteins) were not found in the supernatant. While MFN1 was entirely degraded by 10 μg/ml of PK, the IMS proteins were protected against any concentration of PK tested, confirming that PK accessibility is limited to the surface of the OMM. Although a high exposure image revealed that a very minor portion of cleaved PINK1 in mitochondria was resistant to PK, most of the cleaved PINK1 as well as nonimported full-length PINK1 in both mitochondrial and supernatant fractions were rapidly degraded by low concentrations of PK, suggesting that most of the PARL-cleaved PINK1 in the mitochondrial fraction became exposed to the cytosolic face.30 The folate analog MTX tightly stabilizes DHFR folding thereby inhibiting Su9-DHFR import through the narrow channel of the TOM complex in the OMM and proteolytic processing by MPP (Fig. S3). In contrast, the cleaved form of PINK1-DHFR was found in the supernatant even in the presence of MTX (Fig. S3), indicating that PARL processing of PINK1 on the IMM can occur while the large C-terminal kinase domain of PINK1 spans through the TOM complex in the OMM into the cytosol.
Figure 6. PINK1 precursor retrotranslocates from mitochondria to the cytosol after cleavage by PARL. (A) In vitro synthesized 35S-labeled proteins were incubated with mitochondria with or without valinomycin (Val). The mitochondrial pellet (pel) and the supernatant (sup) fractions were separated by centrifugation. 10% and 20% represent the amount of input of translation product. (B) PINK1 import samples were treated with PK and then the pel and sup were separated. The samples were subjected to radioimaging (low and high exposures) for PINK1 or immunoblotting (IB). (C) PINK1 import into parl−/− mitochondria. Blue, green and red arrowheads indicate the full-length, MPP-cleaved and PARL-cleaved PINK1, respectively. (D) Microscopic analysis of Parl+/+ and parl−/− MEF cells transiently expressing PINK1F104M-YFP. Cells were immunostained with anti-TOMM20. Scale bars: 10 μm. Right panel shows quantification of localization of PINK1-YFP wild-type (F104F), F104M and 104Δ mutants in Parl+/+ and parl−/− MEF cells. Dual means YFP signal both in mitochondria and in the cytosol. ND, not determined because of the low expression level. (E) Schematic representation of YFP-fusion proteins with N-terminal 155 residues of PINK1 or HTRA2. Arrows indicate the cleavage site by PARL for PINK1-YFP variants and by an IMS protease for HTRA2(1-155)-YFP. (F) Microscopic analysis of HeLa cells transiently expressing PINK1(1-155)-YFP, PINK1(1-155)F104M-YFP and HTRA2(1-155)-YFP. Cells were immunostained with anti-TOMM20. Scale bars: 10 μm. (G) The transfected cells as in (F) were treated with or without MG132 for 3 h. Total cell lysates were analyzed by immunoblotting with anti-GFP and anti-TOMM20 antibodies. Blue and red arrowheads represent precursor proteins and cleaved proteins, respectively.
To examine the effect of PARL on PINK1 retrotranslocation, we imported PINK1 into mitochondria isolated from parl−/− MEFs. The mitochondria yielded only MPP-cleaved PINK1 (Fig. 6C, green arrowhead), but this band was not found in the supernatant fraction. Microscopy observation also clearly revealed that PINK1F104M-YFP, whose cleaved form was stably localized in the cytosol of WT MEFs, was arrested in the mitochondria of parl−/− MEFs (Fig. 6D). These results show that cleavage within the TM domain by PARL is essential for PINK1 export from mitochondria. Otherwise the PINK1 precursor remains within the mitochondria. To determine the PINK1 region responsible for retrotranslocation, we next constructed a chimeric protein with the N-terminal 155 residues of PINK1 fused to YFP (Fig. 6E). Whereas it has a weak fluorescent signal on mitochondria (Fig. 6F), MG132 treatment increased expression of the PARL-cleaved form (Fig. 6G) similar to that of endogenous PINK1 (Fig. 1D). Furthermore, PINK1(1-155)F104M-YFP, a mutant with an N-end stabilizing residue was stably localized in the cytosol (Fig. 6F). However, YFP fused to the N-terminal 155 residues of HTRA2, whose topology is quite similar to that of PINK131 (Fig. 6E), was neither found in the cytosol nor responded by MG132 treatment (Fig. 6F and G). These results indicate that an N-terminal region (1-155 residues) of PINK1 is sufficient to yield export to the cytosol.
Discussion
We and others have previously found that newly synthesized PINK1 precursor accumulates on the outer membrane of depolarized mitochondria where it can recruit the E3 ubiquitin ligase PARKIN from the cytosol to mitochondria (Fig. 7). This event triggers elimination of damaged mitochondria by mitophagy. Here, we show that in healthy mitochondria, the N-end rule pathway has a prominent role in the rapid degradation of the cleaved PINK1. Although many N-end substrates have been discovered so far, PINK1 is the first substrate we are aware of identified to target an intracellular organelle for N-terminal processing and for subsequent degradation by the proteasome (Fig. 7). In this model, PARL contributes not only to the retrotranslocation of the PINK1 precursor to the cytosol by cleavage of the membrane spanning domain, but also to rapid proteasomal degradation by cleavage generation of an N-degron.
Figure 7. Schematic model of PINK1 trafficking to damaged or healthy mitochondria. Newly synthesized PINK1 precursor is accumulated on the outer membrane of damaged mitochondria, which can induce PARKIN recruitment from the cytosol to mitochondria. On the other hand, the N-terminal part of PINK1 precursor can be imported into the inner membrane of healthy mitochondria via TOM and TIMM23 translocator complexes. The N-terminal MTS and TM segment are cleaved by MPP and PARL, respectively. Subsequently, the cleaved PINK1 is released to the cytosol where the N-end rule specific E3 enzymes UBR1, UBR2 and UBR4 recognize the destabilizing N-terminal phenylalanine residue of cleaved PINK1 for proteasomal degradation.
Upon identification of N-end rule mediated turnover of PINK1, we were able to introduce mutations that prevent proteasomal degradation and, combined with in vitro and in vivo analysis, we could investigate the mechanism of PINK1 retrotranslocation to the cytosol. What is the conceivable mechanism of PINK1 retrotranslocation? So far, one other protein, yeast fumarase, is known to be released from mitochondria back to the cytosol.32 After cleavage of the presequence by MPP during cotranslational import, a population of the mature fumarase retrotranslocates from mitochondria to the cytosol using folding of the mature domain as a driving force for this movement. In contrast, PINK1 retrotranslocation does not necessarily depend on the folding of the C-terminal kinase domain as only the N-terminal 155 residues of PINK1 are sufficient to export YFP fused to the C terminus to the cytosol. Mitochondrial import of classical MTS-containing precursor proteins occurs at a contact site where the OMM and IMM are closely apposed and 50 to 60 amino acid residues of extended polypeptides are sufficient to span both mitochondrial membranes.33,34 Therefore, the extended C-terminal part of PINK1 including the kinase domain could be exposed to the cytosol when the TM of PINK1 is arrested in the TIMM23 complex. Many IMS or IMM-anchored mitochondrial precursor proteins can laterally diffuse away from the contact site between the TOM and the TIM23 complexes soon after the TIM23 complex recognizes the hydrophobic TM segment, thereby pulling the C-terminal domain into the IMS using lateral diffusion as a driving force. Thus, the TM segment of PINK1 may be cleaved very rapidly by PARL after release from or perhaps while within the TIM23 complex allowing the cleaved PINK1 to be released back to the cytosol with minimal or no force driving export to the cytosol. Alternatively, other mitochondrial proteins such as CLPX/ClpXP or m-AAA proteases may coordinate with PARL to facilitate PINK1 retrotranslocation and/or to preclude PINK1 import further into IMS.13
The ephemeral life of PINK1 enables this sophisticated sensor to monitor and regulate dysfunctional mitochondrial through a well-orchestrated proteolytic cascade.
Materials and Methods
Cell culture and transfection
HeLa and MEF cells were cultured in Dulbecco’s modified Eagles medium (DMEM) (GIBCO, 31053028) supplemented with 10% (v/v) fetal calf serum, 10 mM HEPES buffer, 1 mM sodium pyruvate, 1 mM glutamine and nonessential amino acids. HCT116 cells were cultured in McCoy’s 5A medium (GIBCO, 16600082) supplemented with 10% (v/v) fetal calf serum, 1 mM glutamine and nonessential amino acids. Cells were cultured at 37°C in a 5% CO2 incubator. To transfect HeLa and parl−/− MEF cells for immunoblotting and microscopy, plasmids were mixed with Fugene HD (Promega, E2311) at a 1:3 ratio in Opti-MEM (GIBCO, 31985070). After 10 min incubation, the complex was added to each well containing cells and medium. For pink1−/− MEFs, Lipofectamine LTX (Invitrogen, 15338100) was used according to the manufacturer’s instruction. Valinomycin, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), and MG132 were used at final concentrations of 10 μM, 20 μM and 10 μM, respectively. Stable cell lines were established by recombinant retrovirus infection as follows. DNA fragments encoding wild-type or F104M mutant PINK1-YFP were cloned into NotI/ClaI sites of a pRetroX-IRES-DsRed-Express vector. Vector particles were produced in HEK293T cells grown in poly-lysine coated plates by cotransfection with Gag-Pol, VSV-G and the aforementioned plasmid. After 12 h of transfection, the media were changed and the cells were further incubated for 24 h. The viral supernatants were then infected into HCT116 or HeLa cells grown in 6-well plates with 8 μg/ml polybrene (Sigma, 107689).
Antibodies
The following antibodies were used for immunoblotting: rabbit anti-GFP (Invitrogen, A-11122), mouse anti-SQSTM1 (clone 2C11, Novus Biologicals, H00008878-M01), rabbit anti-LC3B (Sigma, L7543), mouse anti-α-Tubulin (clone B-5-1-2, Invitrogen, 32-2500) and mouse anti-Actin (clone AC-40, Sigma, A4700), rabbit anti-TOMM20 (Santa Cruz Biotechnology, Inc., sc-11415), rabbit anti-PINK1(Novus Biologicals, BC100-494), mouse anti-TIMM23 (clone 32, BD Transduction Laboratories, 611222), mouse anti-Cytochrome c (clone 7H8.2C12, BD Transduction Laboratories, 556433), rabbit anti-HTRA2/Omi (R&D Systems, AF1458), rabbit anti-MFN1 (generated as described previously35), rabbit anti-UBR4 (abcam, ab86738), rabbit anti-UBR1 and chicken anti-UBR2 (kind gifts from Dr Yong Tae Kwon36,37). The following antibodies were used for immunostaining: guinea pig anti-SQSTM1 (Progen, GP62-C), rabbit anti-HSPA9/GRP75 (Cell Signaling, 3593) and rabbit anti-TOMM20 (Santa Cruz Biotechnology, Inc., sc-11415).
Human PINK1 plasmid construction
Wild-type PINK1-YFP, PINK1R98F-YFP, and PINK1F104W-YFP plasmids were described previously.7,9 Other PINK1 mutations (F104X)-YFP (X = Y, I, L, M, A, V, S, and G) were introduced by PCR-based site-directed mutagenesis using appropriate primer pairs. PINK1(105-581)-YFP was made as follows. A DNA fragment encoding PINK1(105-581) and 5′-portion of EYFP was amplified by PCR using Hind-PINK1(105)-F (5′-ggccAAGCTT gccaccATGG GGCTAGGGCT GGGCCTCATC GA -3′) and Long-EGFP-Rv (5′-GTGGCCGTTT ACGTCGCCGT CCAGCTCGAC CAG -3′) as primers and PINK1-YFP as a template. The amplified PCR products were treated with HindIII and BamHI and introduced into the same sites of EYFP-N1. Ub-fusion plasmids were constructed as follows. The amplified DNA fragments encoding PINK1(105-581) by using EcoR-PINK1(105)-F (5′-GGCCgaattc GGGCTAGGGC TGGGCCTCAT-3′) and Long-EGFP-Rv as primers were treated with EcoRI and BamHI and inserted into EcoRI/BamHI sites of EYFP-N1 to make EcoRI-PINK1(105-581)-YFP. Ubiquitin DNA fragments amplified by PCR using primers HindIII-Ub-Fw (5′-ggccAAGCTT gccaccATGC AGATCTTCGT GAAGACTCT-3′) and Ub-EcoRI-Rv (5′-GGCCgaattc CCCACCTCT GAGACGGAGT A-3′) were inserted into HindIII/EcoRI sites of EcoRI-PINK1(105-581)-YFP. Finally, Ub-X (X = F, W, Y, I, L, M, A, V, S, and G) mutations were introduced by PCR-based site-directed mutagenesis using appropriate primer pairs. Introduction of mutations was confirmed by DNA sequencing. For in vitro protein synthesis, the appropriate DNA fragments were cloned into pTNT vector.
Construction of pink1 KO HeLa cell line
pink1 KO HeLa cell was generated by TALEN technology with targeting site as following: GTTGCTTCCA GGGAGAGGCC CAggtaccAG TGCACCAGGA GAAGGGCAGG AGCGAG (underlined sequences and lower-case sequences indicate the target sites for left and right TALEs and KpnI restriction enzyme site). 16-mer left and right TALEs were assembled according to Huang et al.38 and cloned into a final TALEN vector modified from Miller et al.39 0.8 μg of left and right TALEN constructs were cotransfected with 0.4 μg pEYFP-C1 vector in HeLa cells and YFP-positive cells were FACS sorted and plated into 96-well plates two days after transfection. Single colonies were picked and screened by PCR with KpnI digestion. Finally pink1 KO of positive clones were confirmed by immunoblotting with anti-PINK1 antibody.
Immunocytochemistry and confocal imaging
Cells grown on 2-well coverglass chamber slides were fixed with 4% paraformaldehyde in PBS for 25 min at room temperature. Fixed cells were permeabilized with 0.15% Triton X-100 in PBS for 15 min, and blocked with 10% BSA in PBS for 30 min. Indicated primary antibodies and corresponding secondary antibodies (Alexa Fluor 594 goat anti-rabbit IgG (A11012), Alexa Fluor 647 goat anti-rabbit IgG (A21244) and Alexa Fluor 647 goat anti-guinea pig IgG (A21450) purchased from Invitrogen) were added for immunostaining. The images of immunostained cells were captured using an inverted confocal microscope (LSM510 Meta, Carl Zeiss) with a 63× /1.4 NA oil differential interference contrast Plan-Apochromat objective lens. For image analysis, Volocity (PerkinElmer) and/or Photoshop (Adobe) software were used.
Separation of soluble and insoluble proteins
Transfected cells treated with DMSO, valinomycin, or MG132 were solubilized with 2% TX-100 buffer (25 mM HEPES-KOH pH7.5, 300 mM NaCl, 2%(v/v) Triton X-100, protease inhibitor cocktail [PIC] [Roche, 04693159001]) on ice for 15 min, and then subjected to centrifugation at 20,800 × g for 10 min at 4°C to separate soluble supernatants and insoluble pellet fractions. The supernatants were precipitated with trichloroacetic acid (TCA).
Mitochondrial isolation and in vitro protein import assay
HCT116 cells grown in two 150 mm cell culture dishes were homogenized using a Teflon pestle (Thomas Scientific) in 4 ml of 20 mM HEPES-KOH pH7.4, 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 2 mg/ml BSA and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Cell homogenates were then centrifugated at 800 × g at 4°C for 10 min. The supernatants were subjected to centrifugation at 10,000 × g at 4°C for 20 min to obtain mitochondria-rich pellets. Radiolabeled precursor proteins were synthesized in vitro by coupled transcription/translation rabbit reticulocyte lysates (Promega, L2080) in the presence of [35S]methionine/cysteine protein-labeling mix (PerkinElmer, NEG072007MC). After incubation at 30°C for 90 min, radiolabeled translation products were incubated with isolated mitochondria in import buffer (250 mM sucrose, 20 mM HEPES-KOH pH7.4, 80 mM KCl, 5 mM ATP, 10 mM sodium succinate, 5 mM MgCl2, 1 mM methionine and 2 mM dithiothreitol [DTT]) at 24°C for the indicated times. Mitochondria and supernatants were separated by centrifugation at 15,000 × g at 4°C for 5 min. Mitochondrial pellets were washed once with SH buffer (250 mM sucrose and 20 mM HEPES-KOH pH7.4) and the supernatants were precipitated with TCA. Proteins were analyzed by SDS-PAGE and detected by Phosphorimager (STORM 840, Amersham Biosciences). To disrupt the mitochondrial membrane potential in vitro, 1 μM valinomycin was added to the import buffer prior to import reactions. For protease treatment, samples were incubated on ice for 15 min with indicated concentrations of Proteinase K (Sigma, P6556), and the reactions were stopped by addition of 1 mM PMSF. To fold the DHFR moiety with methotrexate (MTX) (Sigma, M9929), in vitro synthesized DHFR fusion proteins were preincubated with 10 μM MTX and 10 μM NAPDH (Sigma, N5130) at 24°C for 10 min, and then import reactions were performed in the presence of 10 μM MTX and 10 μM NAPDH.
Immunoblotting
For preparation of total cell lysate, cells were washed twice with cold PBS and lysed with NuPAGE LDS sample buffer (Invitrogen, NP0008) supplemented with 80 mM DTT and PIC. For Figure 5A, collected cells were solubilized with 2% CHAPS buffer (25 mM HEPES-KOH pH7.5, 300 mM NaCl, 2%(w/v) CHAPS, PIC) on ice for 30 min and then protein concentrations were determined. Soluble fractions precipitated with TCA were lysed with sample buffer. The appropriate amounts of proteins were applied and separated on 4–12% Bis-Tris SDS-PAGE (Invitrogen, NP0322BOX and NP0323BOX) with MES or MOPS SDS running buffer (Invitrogen, NP0001 or NP0002). After transfer to PVDF membranes, blocking and incubation with primary antibodies, proteins were detected using horseradish peroxidase-coupled secondary antibodies (GE Healthcare Life Sciences, NA9340V and NA9310V) and ECL Plus western blotting detection reagents (GE Healthcare Life Sciences, RPN2132).
Subcellular fractionation
Transfected HeLa cells cultured in 100 mm dishes were washed with PBS twice before harvest. Cells were homogenized in 1 ml of 20 mM HEPES-KOH pH7.4, 220 mM mannitol, 70 mM sucrose, 500 mM NaCl, 1 mM EDTA and 0.5 mM PMSF. Cell homogenates were centrifugated at 800 × g at 4°C for 10 min. The post-nuclear supernatants were then split in half and one half was TCA-precipitated as a total fraction. The other half was further centrifuged at 10,000 × g at 4°C for 20 min. The supernatant and pellet fractions were used as a post-mitochondrial supernatant and mitochondria-rich fractions, respectively.
Pulse-chase of PINK1-YFP
PINK1-YFP transfected MEF cells grown in 6-well plates were preincubated for 20 min in DMEM lacking methionine and cysteine (GIBCO, 21013024) supplemented with 10% (v/v) dialyzed fetal calf serum, 10 mM HEPES buffer, 1 mM sodium pyruvate, 1 mM glutamine, nonessential amino acids, and 20 μM CCCP and labeled with Expre35S35S protein labeling mix (PerkinElmer, NEG072007MC) for 60 min at a concentration of 4 MBq/ml. The cells were washed with 1 ml of normal DMEM three times, and then incubated in DMEM during the chase time. After washing twice with cold PBS, cells were solubilized with 500 μl of 0.5% lysis buffer (10 mM Tris-HCL pH7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5%(v/v) Triton X-100, PIC) on ice for 20 min. After centrifugation at 20,000 × g at 4°C for 10 min, the supernatant, whose volume was adjusted to 1 ml with wash buffer (10 mM Tris-HCL pH7.5, 150 mM NaCl, 0.5 mM EDTA, PIC) were subjected to immunoprecipitation. Ten microliters of 50%(v/v) equilibrated GFP-Trap beads (Allele Biotechnology, ACT-CM-GFA0050) were added to each sample and mixed gently for 1 h at 4°C. The beads were washed three times with 1 ml of wash buffer and then resuspended with 40 μl of 2× LDS sample buffer. The immunoprecipitated proteins were dissociated by heating at 99°C for 10 min, and analyzed by SDS-PAGE and radioimaging.
Mass spectrometry
The excised protein bands were alkylated and then incubated with 10 μl of 12.5 ng/μl trypsin in 50 mM NH4HCO3 overnight at 37°C. The resultant tryptic peptides were extracted from the gel by successive incubation with (1) 50% CH3CN/5% trifluoroacetic acid and (2) 75% CH3CN /0.1% trifluoroacetic acid. The extracts from each step were pooled and dried in a SpeedVac. For LC/MS/MS experiments we used a LTQ XL ion trap mass spectrometer (Thermo Fisher Scientific, USA) with an ADVANCE ion max source (Michrom Bioresources) coupled to Surveyor HPLC system with a Micro AS autosampler (Thermo Finnigan). A reverse-phase capillary column (Magic C18, 0.2 × 150 mm) purchased from Michrom BioResources was used to separate the peptides injected. The MS spectra and MS/MS spectra data were collected using Xcalibur software. The data were searched against NCBInc. Human database using Sequest software.
Supplementary Material
Acknowledgments
We thank C Wang for construction of the pink1 KO HeLa cell line, DP Sideris and the other members of the Youle laboratory for valuable discussions and comments, Y Li for help with mass spectrometry, YT Kwon for the kind gift of ubr1−/−ubr2−/−LucRNAi and ubr1−/−ubr2−/−Ubr4RNAi MEF cells and of anti-UBR1 and anti-UBR2 antibodies, B. De Strooper for parl−/− MEFs and Z Zang for pink1−/− MEFs. This research is supported by Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (to KY) and by the National Institute of Neurological Disorders and Stroke intramural program.
Glossary
Abbreviations:
- PINK1
PTEN-induced putative kinase 1
- PARK2/PARKIN
Parkinson protein 2
- UBR
ubiquitin protein ligase E3 component n-recognin
- PARL
presenilin-associated rhomboid-like
- MPP
matrix processing peptidase
- DUBs
deubiquitinating enzymes
- MEF
mouse embryonic fibroblast
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- MTS
mitochondrial targeting sequence
- TOM
translocator of the outer membrane
- TIMM23/TIM23
translocase of inner mitochondrial membrane 23 homolog (yeast)
- DHFR
dihydrofolate reductase
- PK
proteinase K
- MTX
methotrexate
- TM
transmembrane
- OMM
outer mitochondrial membrane
- IMM
inner mitochondrial membrane
- IMS
intermembrane space
- knockout
KO
- TALEN
transcription activator-like effector nuclease
- HTRA2/OMI
HtrA serine peptidase 2
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24633
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24633
References
- 1.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 3.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 4.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 6.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–42. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–79. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, et al. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum Mol Genet. 2011;20:1966–74. doi: 10.1093/hmg/ddr077. [DOI] [PubMed] [Google Scholar]
- 12.Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–67. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 13.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–85. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–92. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 15.Muqit MM, Abou-Sleiman PM, Saurin AT, Harvey K, Gandhi S, Deas E, et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J Neurochem. 2006;98:156–69. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 16.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, et al. The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat Cell Biol. 2007;9:1243–52. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–7. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takatori S, Ito G, Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett. 2008;430:13–7. doi: 10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Weihofen A, Ostaszewski B, Minami Y, Selkoe DJ. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum Mol Genet. 2008;17:602–16. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 21.Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–74. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 23.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–89. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lévy F, Johnsson N, Rümenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci U S A. 1996;93:4907–12. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–99. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 26.Moriwaki Y, Kim YJ, Ido Y, Misawa H, Kawashima K, Endo S, et al. L347P PINK1 mutant that fails to bind to Hsp90/Cdc37 chaperones is rapidly degraded in a proteasome-dependent manner. Neurosci Res. 2008;61:43–8. doi: 10.1016/j.neures.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–36. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 30.Becker D, Richter J, Tocilescu MA, Przedborski S, Voos W. Pink1 kinase and its membrane potential (Deltaψ)-dependent cleavage product both localize to outer mitochondrial membrane by unique targeting mode. J Biol Chem. 2012;287:22969–87. doi: 10.1074/jbc.M112.365700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadomatsu T, Mori M, Terada K. Mitochondrial import of Omi: the definitive role of the putative transmembrane region and multiple processing sites in the amino-terminal segment. Biochem Biophys Res Commun. 2007;361:516–21. doi: 10.1016/j.bbrc.2007.07.053. [DOI] [PubMed] [Google Scholar]
- 32.Yogev O, Naamati A, Pines O. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 2011;278:4230–42. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- 33.Gaume B, Klaus C, Ungermann C, Guiard B, Neupert W, Brunner M. Unfolding of preproteins upon import into mitochondria. EMBO J. 1998;17:6497–507. doi: 10.1093/emboj/17.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamano K, Kuroyanagi-Hasegawa M, Esaki M, Yokota M, Endo T. Step-size analyses of the mitochondrial Hsp70 import motor reveal the Brownian ratchet in operation. J Biol Chem. 2008;283:27325–32. doi: 10.1074/jbc.M805249200. [DOI] [PubMed] [Google Scholar]
- 35.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol Cell Biol. 2007;21:8007–21. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–71. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 39.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.