Abstract
The presence of multiple homologs of the same yeast ATG genes endows an extra layer of complexity on the autophagic machinery in higher eukaryotes. The physiological function of individual homologs in the autophagy pathway remains poorly understood. Here we characterized the function of the two atg16 homologs, atg-16.1 and atg-16.2, in the autophagy pathway in C. elegans. We showed that atg-16.2 mutants exhibit a stronger autophagic defect than atg-16.1 mutants. atg-16.2; atg-16.1 double mutants display a much more severe defect than either single mutant. ATG-16.1 and ATG-16.2 interact with themselves and each other and also directly associate with ATG-5. atg-16.1 mutant embryos exhibit a wild-type expression and distribution pattern of LGG-1/Atg8, while LGG-1 puncta are markedly fewer in number and weaker in intensity in atg-16.2 mutants. In atg-16.2; atg-16.1 double mutants, the lipidated form of LGG-1 accumulates, but LGG-1 puncta are completely absent. ATG-16.2 ectopically expressed on the plasma membrane provides novel sites of LGG-1 puncta formation. We also demonstrated that the C-terminal WD repeats are dispensable for the role of atg-16.2 in aggrephagy (the degradation of protein aggregates by autophagy). Genetic epistasis analysis placed atg-16.2 upstream of atg-2, epg-6, and atg-18. Our study indicated that C. elegans ATG-16s are involved in specifying LGG-1 puncta formation and the two ATG-16 homologs have partially redundant yet distinct functions in the aggrephagy pathway.
Keywords: atg-16.1, atg-16.2, aggrephagy, C. elegans
Introduction
Macroautophagy (hereafter referred to as autophagy) is a lysosome-mediated degradation process, which involves de novo formation of a crescent-shaped membrane sac, called the phagophore, and its further expansion and closure to form a double-membrane structure, known as the autophagosome, and finally its delivery to the lysosome/vacuole for degradation.1,2 Yeast genetic studies have identified more than 18 autophagy-related (ATG) genes that are essential for autophagosome formation.1,2 The Atg proteins act in a hierarchical order in organizing the phagophore assembly site (PAS), from which autophagosomes originate.3,4 The autophagy pathway in higher eukaryotes involves more dynamic membrane remodeling events.5 Many autophagosomes can form simultaneously at multiple sites. The ER, Golgi, plasma membrane, and other membrane structures have been shown to contribute to the membrane source for autophagosome formation.6-8 The more elaborate autophagic machinery in higher eukaryotes involves highly conserved yeast Atg proteins and also metazoan-specific factors.9,10 The presence of multiple homologs of the same yeast Atg genes confers another layer of complexity. For example, mammals have at least seven Atg8 orthologs, which are divided into the MAP1LC3/LC3, GABARAP, and GABARAPL2/GATE-16 subfamilies.11 The single Atg4 in yeast has two C. elegans orthologs and four mammalian orthologs which exhibit differential processing activity toward Atg8 proteins.12,13 The physiological function of individual homologs in the autophagy pathway remains poorly understood.
Among Atg proteins, the two essential ubiquitin-like conjugation systems are required for elongation and closure of phagophores.1,2 The ubiquitin-like protein Atg8 is activated by the E1-like enzyme Atg7 and then transferred to the E2-like conjugating enzyme Atg3 and finally conjugated to phosphatidylethanolamine (PE).2 The ubiquitin-like molecule Atg12 is covalently conjugated to Atg5 via the sequential reactions of Atg7 and the E2-like enzyme Atg10.2 The Atg12–Atg5 conjugate further interacts noncovalently with Atg16, which self-oligomerizes to mediate the formation of a multimeric complex.14,15 One function of the Atg12–Atg5-Atg16 complex in autophagosome formation is to regulate Atg8–PE conjugation and its targeting to the PAS.3,4 The Atg12–Atg5 conjugate possesses an E3-like activity for Atg8 lipidation by promoting the transfer of Atg8 from Atg3 to PE.16 Atg16 is not essential for Atg8 lipidation in vitro, but is necessary for efficient Atg8–PE conjugation under starvation conditions in vivo.3
The two ubiquitin-like conjugation systems are highly conserved in higher eukaryotes. The mammalian Atg16 ortholog ATG16L1 mediates formation of the multimeric ATG12–ATG5-ATG16L1 complex.17 In addition to the N-terminal ATG5 binding domain and the self-oligomerization coiled-coil domain, ATG16L1 possesses seven WD repeats at the C terminus, whose role in autophagy remains unknown.17 In mammals, LC3 lipidation is abolished in atg5 and atg16l knockout cells.18,19 The ATG12–ATG5-ATG16L1 complex also specifies the site of LC3 lipidation.20,21 Another mammalian Atg16 homolog, ATG16L2, homo- and hetero-oligomerizes with ATG16L1 and also forms a multimeric complex with ATG12–ATG5.22 However, ATG16L2 is neither recruited to phagophores nor required for starvation-induced autophagy.22 The role of ATG16L2 and its interaction with ATG16L in the autophagy pathway under physiological conditions has yet to be determined.
We demonstrated previously that during C. elegans embryogenesis a variety of protein aggregates are selectively degraded by autophagy, a process termed aggrephagy.23 Using this genetic model, we performed genetic screens to identify essential autophagy genes.9 In addition to conserved and divergent homologs and orthologs of yeast Atg proteins, we isolated several autophagy genes conserved in higher eukaryotes but absent in yeast.9,10,24-27 Most Atg proteins have only one ortholog in C. elegans except for Atg4, Atg8, and Atg16, which have two orthologs. Here we characterized the role of the two atg16 homologs in the autophagy pathway. Loss of function of either atg-16.1 or atg-16.2 results in defective degradation of protein aggregates. atg-16.2; atg-16.1 double mutants exhibit much more severe autophagic defects than either single mutant. ATG-16.1 and ATG-16.2 self-interact and associate with each other and also with ATG-5. We showed that ATG-16 is not required for lipidation of LGG-1, but is essential for formation of LGG-1 punctate structures. Our study revealed that the two Atg16 homologs play partially redundant yet differential functions in the aggrephagy pathway.
Results
Two orthologs of yeast Atg16 are present in C. elegans
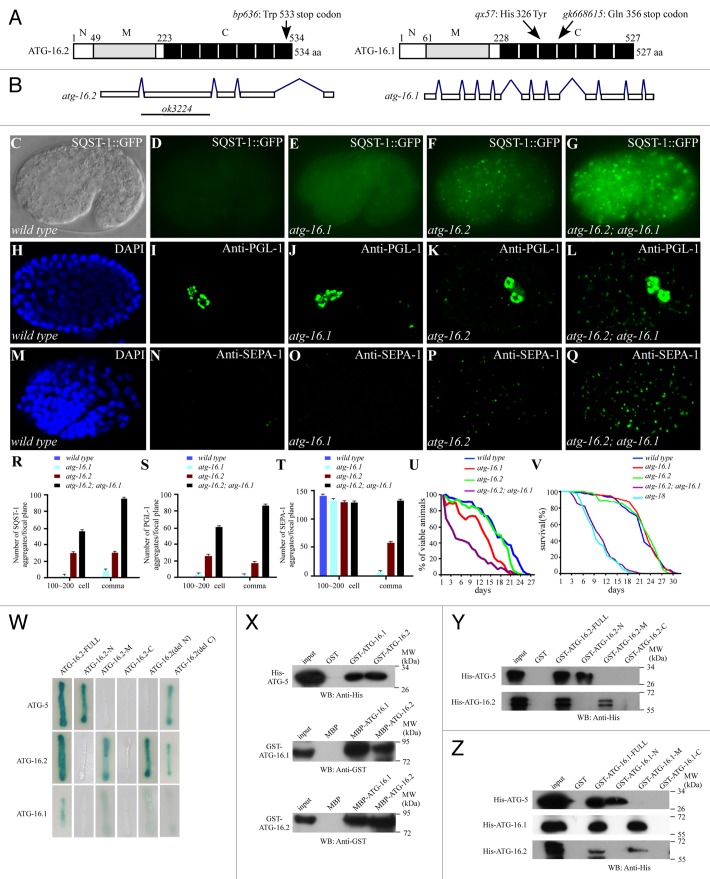
C. elegans has two atg16 homologs, atg-16.1 and atg-16.2, which encode proteins with 527 and 534 amino acids, respectively. ATG-16.1 and ATG-16.2 contain seven WD repeats at their C termini, which are also shared by mammalian ATG16L but are absent in yeast Atg16 (Fig. 1A). ATG-16.2 shares 28.8% identity and 48.3% similarity to human ATG16L1 (NP_060444.3); ATG-16.1 has 29.7% identity and 49.3% similarity to human ATG16L1. ATG-16.1 also exhibits 42.4% identity and 62.9% similarity to ATG-16.2 (Fig. S1A). Phylogenetic analysis revealed that ATG-16.1 and ATG-16.2 are more closely related to human ATG16L1 than to ATG16L2 (Fig. S1B).
Figure 1. Defective degradation of protein aggregates in atg-16.1 and atg-16.2 mutants. (A) Schematic structure of ATG-16s. Both ATG-16.1 and ATG-16.2 contain an N-terminal region (N), a coiled-coil domain (M, in gray) in the middle and a C-terminal motif with seven WD repeats (C, black boxes). (B) Gene structures of atg-16.1 and atg-16.2. atg-16.2 maps on linkage group II (LG II). Transgenes containing fosmid WRM0641aG04 or a PCR product containing a single gene K06A1.5 rescued the defective degradation of SQST-1 aggregates in atg-16.2(bp636) mutants. Intron-exon boundaries were confirmed by cDNA sequencing. atg-16.1 maps on linkage group X (LG X). (C and D) No SQST-1::GFP aggregates are detected in wild-type embryos. (C) Nomarski image of the embryo shown in (D). (E) SQST-1::GFP accumulates into a few aggregates in atg-16.1 mutants. (F) Many SQST-1::GFP aggregates are found in atg-16.2 mutants. (G) SQST-1::GFP accumulates into a large number of aggregates in atg-16.2; atg-16.1 mutants. (C–G) The embryos are at the comma stage. (H and I) In a wild-type embryo, P granules, labeled by anti-PGL-1, are exclusively localized in germline precursor cells Z2 and Z3. (H) DAPI image of the embryo shown in (I). (J–L) PGL granules are present in somatic cells in atg-16.1, atg-16.2, and atg-16.2; atg-16.1 mutant embryos. (H-L) The embryos were at about the 200-cell stage. (M and N) SEPA-1 aggregates are undetectable in wild-type embryos at the comma stage. (M). DAPI image of the embryo shown in (N). (O) SEPA-1 aggregates are absent in atg-16.1 mutant embryos at the comma stage. (P) SEPA-1 aggregates are present in a comma-stage atg-16.2 mutant embryo. (Q) Numerous SEPA-1 aggregates are formed in atg-16.2; atg-16.1 mutant embryos at the comma stage. (R–T) Average number of aggregates formed by SQST-1::GFP, PGL-1, and SEPA-1 per confocal image in various genotypes. Error bars represent the standard deviation (s.d.) of five confocal images. The number of SQST-1 and PGL-1 aggregates at the ~100 to 200-cell stage shows statistical significance among the samples (P < 0.05 and P < 0.001, respectively). The number of SQST-1, PGL-1, and SEPA-1 aggregates at the comma stage shows statistical significance among the samples (P < 0.001). (U) atg-16.1 and atg-16.2 mutants L1 larvae show a reduction in the survival under food-depletion conditions compared with wild type (log-rank test, P = 0.000). The median survival duration of wild type, atg-16.1, and atg-16.2 was 18.000 [95% confidence interval (CI) 16.851–19.149], 13.000 [95% CI 12.050–13.950], and 17.000 [95% CI 16.030–17.970] days. Moreover, atg-16.2; atg-16.1 double mutant L1 larvae have dramatically shortened survival in the absence of food compared with wild type, atg-16.1, and atg-16.2 mutants (log-rank test, P = 0.000). The median survival duration of atg-16.2; atg-16.1 double mutant was 4.000 [95% CI 2.826–5.174] days. At least 500 L1 larvae of wild type and atg-16 mutants were examined. (V) Survival curves of wild type, atg-18, and atg-16 mutants. The median survival time was 23 d for wild type, 24 d for atg-16.1 and atg-16.2 mutants, 11 d for atg-16.2; atg-16.1 double mutant, and 10 d for atg-18 mutant. Loss of function of atg-16.1 or atg-16.2 has no significant effect on the life span compared with wild type. However, atg-16.2; atg-16.1 double mutants show dramatic reduction in median survival time compared with wild-type animals (log-rank test, P = 0.000). (W) Pairwise interactions between ATG-16.1, ATG-16.2, and ATG-5 in yeast two-hybrid analysis using an X-gal assay. -N, -M, and -C in (W, Y, and Z) stand for different fragments of ATG-16.2 as shown in (A). ATG-16.2(del N) and ATG-16.2(del C) indicate mutant ATG-16.2 deleting the N and C termini, respectively. (X) In vitro pulldown assay showing pairwise interactions between ATG-16.1, ATG-16.2, and ATG-5. WB, western blot. (Y) The N-terminal region of ATG-16.2 interacts with ATG-5, while the coiled-coil domain mediates the self-interaction of ATG-16.2. (Z) ATG-16.1 directly interacts with ATG-5 through the N-terminal region. The coiled-coil domain mediates the interaction of ATG-16.1 with itself and ATG-16.2.
atg-16.1 and atg-16.2 act differentially in the autophagy pathway
In genetic screens to find mutants with defective degradation of the C. elegans SQSTM1/p62 ortholog SQST-1, we identified the bp636 mutation. Further genetic mapping and rescue experiments demonstrated that bp636 is an allele of atg-16.2, in which the tryptophan at amino acid 533 is mutated to a stop codon. atg-16.2(ok3224), obtained from the knockout consortium, deletes amino acids 125 to 299 and causes a frame-shift (Fig. 1B). atg-16.2(ok3224) was used in this study. The mutant allele of atg-16.1(qx57) was identified in genetic screens to isolate mutants that suppress the accumulation of LGG-1::GFP aggregates in tat-1 mutants. The histidine at amino acid 326 is mutated to tyrosine in atg-16.1(qx57) mutants (Fig. 1A). This residue is conserved in ATG-16.2 and human ATG16L1 (Fig. S2A). However, mutating the corresponding histidine at the residue 331 did not impair the function of atg-16.2 (Fig. S2B–S2E). atg-16.1(gk668615) contains a glutamine to stop codon mutation at amino acid 356 (Fig. 1A). atg-16.1(gk668615) caused a weaker autophagic defect than qx57 (Fig. S2H–S2M). atg-16.1(RNAi) caused no autophagic defect neither enhanced the defect in atg-16.1(qx57) mutants. Thus, atg-16.1(qx57) was used in this study.
We examined degradation of a variety of autophagy substrates in atg-16.1 and atg-16.2 single mutants and atg-16.2; atg-16.1 double mutants. In wild-type embryos, SQST-1::GFP is weakly expressed and diffusely localized in the cytoplasm (Fig. 1C and D).9 In atg-16.1 mutants, a few small SQST-1::GFP aggregates were observed in embryos (Fig. 1E and R). Compared with atg-16.1 mutants, more SQST-1 aggregates were formed in atg-16.2 mutants and the aggregates were present even in comma stage embryos (~500 cells) (Fig. 1F and R). Compared with single mutants, expression of SQST-1::GFP was stronger and the number of aggregates was much greater in atg-16.2; atg-16.1 double mutants (Fig. 1G and R). SQST-1 aggregates were detected in embryos at all stages and persisted in larvae in double mutants (Fig. 1G; Fig. S2F and S2G).
Other autophagy substrates, including PGL granule components PGL-1 and SEPA-1, were also examined in atg-16 mutants. In wild-type embryos, the germline-specific P granule component PGL-1 is exclusively restricted to germ blastomeres (Fig. 1H and I).23 PGL-1 granules were ectopically detected in somatic cells before the ~200-cell stage in atg-16.1 and atg-16.2 single mutants (Fig. 1J and K). atg-16.2 mutants contained more PGL-1 granules than atg-16.1 mutants. In atg-16.2; atg-16.1 double mutants, PGL-1 aggregates were much greater in number throughout embryogenesis and persisted to early larval stages (Fig. 1L and S). In wild-type embryos, SEPA-1 aggregates are detected in early-stage embryos and become undetectable from the comma stage onward due to their selective removal by autophagy (Fig. 1M and N).23 Loss of function of either atg-16 caused ectopic accumulation of SEPA-1 aggregates, as observed for SQST-1 and PGL-1, in comma-stage embryos. atg-16.2 mutants contained more SEPA-1 aggregates than atg-16.1 mutants (Fig. 1O, P, and T). In atg-16.2; atg-16.1 mutants, SEPA-1 aggregates were far more numerous and had greater fluorescence intensity, and persisted in embryos of all stages (Fig. 1Q and T).
We further compared the degradation of protein aggregates in atg-16 mutants with other autophagy mutants. atg-16.2; atg-16.1 mutants exhibited the same degree of defect as mutants of other essential autophagy genes with only one homolog such as atg-3 and atg-5 (Fig. S2N). Taken together, our data show that loss of either atg-16.1 or atg-16.2 activity causes defective degradation of protein aggregates. atg-16.2 mutants show a more severe defect than atg-16.1 mutants. Simultaneous depletion of atg-16.1 and atg-16.2 causes a much stronger phenotype than either single mutant, indicating that atg-16.1 and atg-16.2 have a redundant role in the aggrephagy pathway.
atg-16.1 and atg-16.2 act redundantly in other autophagy-regulated processes
We further determined whether atg-16 is involved in other autophagy processes in addition to aggrephagy. Autophagy activity is required for the survival of L1 larvae in the absence of food.28 Under food-depletion conditions, the median survival duration of wild-type L1 larvae is about 18 d. The median survival duration of atg-16.1 and atg-16.2 mutants was 13 and 17 d, respectively, and it was reduced to 4 d in atg-16.2; atg-16.1 double mutants (Fig. 1U).
Autophagy activity also regulates the life span of adult animals.28 The median survival times were not significantly affected by loss of function of atg-16.1 or atg-16.2 but were dramatically reduced in atg-16.2; atg-16.1 double mutants, which is similar to atg-18 mutants (Fig. 1V). Another characteristic of autophagy mutants is that they grow slowly and fewer mutant embryos develop into larvae (Fig. S2O and S2P). Both processes were much more severely affected in the double mutants. In summary, atg-16.1 and atg-16.2 function redundantly in multiple autophagy-regulated processes.
atg-16.1 and atg-16.2 are widely expressed
To determine the expression pattern of atg-16.2, we constructed a translational fusion reporter with GFP inserted at the C terminus of ATG-16.2. The atg-16.2::gfp reporter was functional in rescuing defective degradation of SQST-1::GFP aggregates in atg-16.2(ok3224) mutants. ATG-16.2::GFP was diffusely expressed in the cytoplasm in most, if not all, cells during embryogenesis (Fig. S3A and S3B). At post-embryonic stages, ATG-16.2::GFP was widely expressed, including in neurons, pharyngeal muscles, body wall muscle cells and intestinal cells (Fig. S3E and S3F; and data not shown). The functional translational fusion atg-16.1::gfp reporter exhibited the same expression pattern as ATG-16.2::GFP (Fig. S3C, S3D, S3G, S3H, S3K, and S3L). Expression of ATG-16.2::GFP and ATG-16.1::GFP remained unchanged in lgg-1 or atg-3 mutants (Fig. S3I and S3J; and data not shown).
The functional redundancy of atg-16.1 and atg-16.2 in the autophagy pathway prompted us to investigate whether loss of function of one gene results in compensatory upregulation of the other gene. Quantitative real-time PCR analysis indicated that levels of atg-16.1 mRNA in atg-16.2(ok3224) mutants and levels of atg-16.2 mRNA in atg-16.1 mutants were slightly increased compared with wild-type animals (Fig. S3M and S3N). We further crossed the atg-16.1::gfp transgene into atg-16.2(ok3224) mutants and found that the fluorescent intensity showed no obvious changes. Similarly, expression of the atg-16.2 reporter displayed no evident changes in atg-16.1 mutants (data not show). Therefore, loss of function of one atg-16 gene does not cause obvious compensatory upregulation of the other.
ATG-16.1 and ATG-16.2 interact with themselves, with each other and with ATG-5
We performed a yeast-two-hybrid assay to identify proteins that interact with ATG-16.2. Out of 34 isolated clones, 3 encoded ATG-5 and 2 encoded ATG-16.2 (Fig. 1W). In vitro pulldown assays confirmed the interaction between ATG-16.2 and ATG-5 and also self-interaction of ATG-16.2 (Fig. 1X). ATG-16.1 also self-associated and interacted with ATG-16.2 and ATG-5 in yeast-two-hybrid and pulldown assays (Fig. 1W and X; Fig. S3O).
By analyzing a series of ATG-16.2 deletions, we found that the N-terminal region of ATG-16.2 (amino acids 1 to 49) was sufficient to interact with ATG-5 (Fig. 1Y). HIS-ATG-16.2 was specifically pulled down by the coiled-coil domain (CCD), covering amino acids 50 to 223 of ATG-16.2, but not GST alone or other fragments of ATG-16.2 lacking the coiled-coil domain (Fig. 1Y). Consistent with the evidence that ATG-16.1 and ATG-16.2 share similar structures, the N-terminal region of ATG-16.1 (amino acids 1 to 61) interacted with ATG-5 and the CCD of ATG-16.1 (amino acids 56 to 228) self-associated (Fig. 1Z; Fig. S3O). The CCD also mediated the interaction between ATG-16.1 and ATG-16.2 (Fig. 1Z).
As ATG-16.1 and ATG-16.2 self-interact and also associate with each other and with ATG-5, we determined whether overexpression of atg-16.1 could rescue the defect in atg-16.2 mutants. An hs::atg-16.1 transgene, in which the expression of atg-16.1 is driven by the heat shock promoter, failed to rescue the defective degradation of PGL-1 aggregates in atg-16.2 mutants after the heat-shock-induced expression of atg-16.1, while the defect was completely rescued by an hs::atg-16.2 transgene (Fig. 2U–W and data not shown), suggesting that overexpression of atg-16.1 cannot substitute the function of atg-16.2.
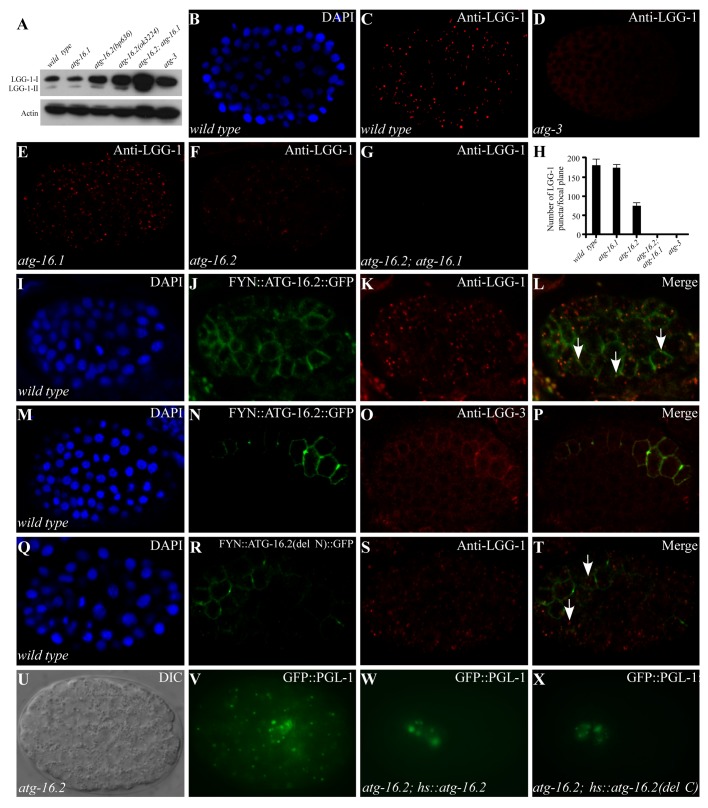
Figure 2. Expression of LGG-1 in atg-16 mutants. (A) Western blot analysis shows the levels of LGG-1-I and LGG-1-II in various autophagy mutants. (B and C) LGG-1 forms distinct punctate structures in a wild-type embryo at the ~100 to 200-cell stage. (B). DAPI image of the embryo shown in (C). (D) LGG-1 is diffusely localized and does not form aggregates in atg-3 mutants. (E) atg-16.1 mutant embryos show a wild-type pattern of LGG-1 puncta. (F and G) LGG-1 puncta are greatly reduced in atg-16.2 mutant embryos (F) and absent in atg-16.2; atg-16.1 mutant embryos (G). (H) Average number of LGG-1 puncta per confocal image in various mutants. Error bars represent the s.d. of five confocal images. (I–L) Localization of LGG-1 puncta to the plasma membrane in embryos expressing FYN::ATG-16.2::GFP (labeled by arrows). 60.5 ± 0.11% of LGG-1 puncta (n = 445) are located on the plasma membrane. (M–P) LGG-3, detected by anti-LGG-3 antibody, also forms a few puncta on the plasma membrane in embryos expressing FYN::ATG-16.2::GFP. (Q–T) LGG-1 puncta are largely dispersed in the cytoplasm (indicated by arrows) in embryos expressing FYN::ATG-16.2(del N)::GFP, in which the N-terminal ATG-5-interacting domain is deleted. Only 25.1 ± 0.03% of LGG-1 puncta (n = 490) are located on the plasma membrane. (U and V) GFP::PGL-1 granules accumulate in somatic cells in atg-16.2 mutant embryos. (U) Nomarski image of the embryo shown in (V). (W) A Phs::atg-16.2 transgene rescues the accumulation of PGL granules in somatic cells in atg-16.2 mutant embryos. GFP::PGL-1 is restricted to the germline precursor cells Z2 and Z3. (X) Ectopic accumulation of PGL granules in atg-16.2 mutant embryos is rescued by a Phs::atg-16.2(del C) transgene, in which the C-terminal WD repeats are deleted. (U–X). The embryos were at about the 200-cell stage.
atg-16.2 and atg-16.1 mutants have distinct effects on formation of LGG-1/Atg8 puncta
To further determine at which stage the autophagic process is impaired in atg-16 mutants, we used an anti-LGG-1 antibody to determine the expression and distribution of LGG-1, the C. elegans homolog of yeast Atg8. In wild-type embryos, both unlipidated (LGG-1-I) and lipidated (LGG-1-II) forms of LGG-1 are present, while LGG-1-II is undetectable in atg-3 mutants by immunoblotting assay (Fig. 2A).9 Levels of both LGG-1-I and LGG-1-II were unchanged in atg-16.1 embryos compared with wild type, but were elevated in atg-16.2(bp636) and atg-16.2(ok3224) mutants. Both forms of LGG-1 were dramatically increased by simultaneously depleting atg-16.1 and atg-16.2, indicating that neither atg-16 homolog is required for lipidation of LGG-1 (Fig. 2A; Fig. S3P and S3Q).
In wild-type embryos, LGG-1 forms punctate structures, mostly between the ~100- and ~200-cell stages (Fig. 2B and C). The number of aggregates dramatically decreases as development proceeds and only a few are detected in 4-fold stage embryos.9 The formation of LGG-1 puncta requires LGG-1 lipidation, as LGG-1 puncta are absent in atg-3 mutants (Fig. 2D).9 LGG-1 puncta had a wild-type distribution pattern in atg-16.1 mutants but were dramatically reduced in number and intensity in atg-16.2(ok3224) mutant embryos and were completely absent in atg-16.2; atg-16.1 double mutants (Fig. 2E–H). These results indicate that atg-16 activity is required for recruiting lipidated LGG-1 to form punctate structures.
We further investigated whether atg-5 is required for lipidation of LGG-1 and its accumulation into puncta. Two mutant alleles of atg-5, bp484, and bp546, were isolated in our genetic screens for defective degradation of protein aggregates. bp484 contains a G to A nucleotide substitution at the consensus “-1” position of the splice acceptor site in the first intron. atg-5(bp546) contains a glutamic acid to lysine change at amino acid 130. In atg-5 mutants, LGG-1-II was undetectable in the immunoblotting assay and no LGG-1 puncta were detected during embryogenesis (Fig. S3R–S3T),9 indicating that, unlike atg-16, atg-5 is essential for conjugation of PE to LGG-1. Double mutants of atg-5 and atg-16.2 were embryonic lethal, preventing us from examining their genetic interactions.
ATG-16.2 is sufficient to nucleate LGG-1 punctate structures
We next determined whether ATG-16.2 is involved in LGG-1 puncta formation. The plasma membrane localization motif of the FYN protein (MGCVQCKDKEATKLTE) was added to the N terminus of ATG-16.2::GFP. As expected, embryos expressing fyn::atg-16.2::gfp showed plasma membrane localization (Fig. 2I and J). Immunostaining with anti-LGG-1 antibody revealed that LGG-1 puncta seem to appear preferentially at the plasma membrane in cells expressing FYN::ATG-16.2::GFP (Fig. 2K and L). LGG-3 (the C. elegans homolog of yeast Atg12), detected by anti-LGG-3 antibody, displayed a diffuse expression pattern in wild-type embryos. In cells expressing fyn::atg-16.2::gfp, LGG-3 also appeared to be recruited to the plasma membrane and formed a few punctate structures (Fig. 2M–P). Embryos expressing fyn::atg-16.2(del N)::gfp, in which the N-terminal ATG-5 binding motif is deleted, showed that much less LGG-1 puncta were recruited to the plasma membrane (Fig. 2Q–T and data not shown), suggesting that interaction with ATG-5 is essential for the role of ATG-16.2 in nucleation of LGG-1 puncta.
The WD repeats are dispensable for the role of atg-16.2 in the aggrephagy pathway
Seven WD repeats are present at the C termini of C. elegans and mammalian Atg16 homologs. To determine whether the WD repeats are essential for the role of atg-16.2 in the autophagy pathway, we created the atg-16.2(del C) transgene, in which the WD repeats are deleted. This transgene rescued the defective degradation of SQST-1 aggregates and PGL-1 granules in atg-16.2 mutants (Fig. 2U–X; Fig. S3U and S3V). To exclude the possibility that the ATG-16.2(del C) mutant protein forms a complex with the LGG-3-ATG-5 conjugates via ATG-16.1, we crossed the same hs::atg-16.2(del C) transgene into atg-16.2; atg-16.1 double mutants. Defective degradation of SQST-1 aggregates and PGL-1 granules in atg-16.2; atg-16.1 mutants was also rescued (Fig. S3W and S3X; and data not shown). Therefore, the WD repeats are dispensable for the role of atg-16.2 in the aggrephagy pathway.
Genetic interactions of atg-16.2 with other autophagy genes
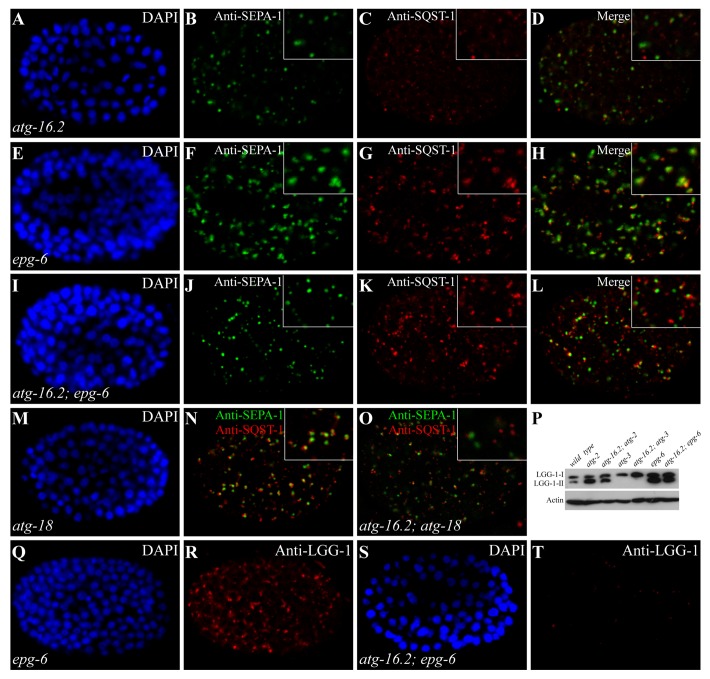
We showed previously that mutants of autophagy genes acting at discrete steps of autophagosome formation exhibited different autophagic phenotypes affecting the formation, morphology and distribution pattern of PGL granules, SQST-1 aggregates and LGG-1 puncta.9,10 Based on these phenotypes, we performed epistasis analysis to place autophagy genes in a hierarchical order in the aggrephagy pathway.10 We used the same approach to determine the genetic interactions of atg-16.2 with other autophagy genes. In atg-16.2 mutants, SQST-1 and SEPA-1 aggregates are small and spherical in shape and are separable from each other (Fig. 3A–D), but are enlarged and largely colocalized in epg-6 and atg-2 mutants (Fig. 3E–H and data not shown).10 In atg-16.2; epg-6 and atg-16.2; atg-2 mutants, SQST-1 and SEPA-1 aggregates were smaller in size and were largely separable, resembling those in atg-16.2 single mutants (Fig. 3I–L and data not shown). A majority of SQST-1 aggregates are closely aligned, but do not colocalize, with PGL granules in atg-18 mutants (Fig. 3M and N).10 In atg-16.2; atg-18 mutants, SQST-1 and SEPA-1 aggregates localized far apart from each other (Fig. 3O), just as in atg-16.2 single mutants, indicating that atg-16.2 functions upstream of epg-6, atg-2, and atg-18.
Figure 3. Epistasis analysis of atg-16.2 and other autophagy mutants. (A–D) In atg-16.2 mutants, SEPA-1 and SQST-1 aggregates are small and spherical and are separable from each other. (A). DAPI image of the embryo shown in (B). (E–H) SEPA-1 aggregates are enlarged and largely colocalize with SQST-1 aggregates in epg-6 mutants. Colocalization refers to two aggregates showing > 70% overlap in their fluorescence signals. (I-L) In atg-16.2; epg-6 double mutants, similar to atg-16.2 single mutants, SEPA-1 aggregates are small in size and separable from SQST-1 aggregates. (M–N) Small SQST-1 aggregates are closely aligned with SEPA-1 aggregates in atg-18 mutants. (M) DAPI image of the embryo shown in (N). (O) SQST-1 and SEPA-1 aggregates are largely separable in atg-16.2; atg-18 mutants. (P) LGG-1-II accumulates in atg-16.2; atg-2 and atg-16.2; epg-6 mutants, but is completely absent in atg-16.2; atg-3. (Q and R) Accumulation of LGG-1 puncta in epg-6 mutant embryos. (Q). DAPI image of the embryo shown in (R). (S and T) In atg-16.2; epg-6 mutants, LGG-1 puncta are absent, as in atg-16.2 single mutants shown in Figure 2F. (S). DAPI image of the embryo shown in (T).
We also examined the expression level and distribution pattern of LGG-1 when atg-16.2 and other autophagy genes were simultaneously depleted. In epg-6 and atg-2 mutants, levels of LGG-1-II dramatically increase and LGG-1 puncta are enlarged and accumulate in late-stage embryos (Fig. 3Q and R and data not shown).10 Simultaneous depletion of atg-16.2 in these mutants showed that LGG-1-II still accumulated, but formed only a few small puncta that were dispersed in the cytoplasm (Fig. 3P, S, and T), consistent with that atg-16.2 acts upstream of epg-6 and atg-2.
Discussion
In this study, we demonstrated that the two atg-16 homologs, atg-16.1 and atg-16.2, function differentially in the autophagy pathway in C. elegans. atg-16.1 mutants exhibit milder autophagy defects than atg-16.2 mutants. atg-16.2; atg-16.1 double mutants display much more severe defects than either single mutant. Both ATG-16.1 and ATG-16.2 show similar domain organization and exhibit similar spatial and temporal expression patterns.
In yeast and mammals, the Atg12–Atg5-Atg16 complex has two distinct functions in the Atg8–PE conjugation system, facilitating Atg8–PE conjugation and also targeting Atg8–PE to the PAS in yeast and LC3–PE to phagophores in mammalian cells.3,20,21 Here we demonstrate that loss of function of atg-5 abolishes LGG-1 lipidation and puncta formation. The role of Atg16 in Atg8 lipidation, however, appears to be different in C. elegans and other systems. Atg16 is required for efficient lipidation under starvation conditions in yeast.3 Mice deficient in atg16l show impaired LC3 lipidation.19 Lipidation of LGG-1 increases rather than reduces in atg-16.2; atg-16.1 mutants. As in other systems, ATG-16 is required for formation of LGG-1 puncta. In atg-16.2; atg-16.1 double mutants, LGG-1-II accumulates but LGG-1 puncta are completely absent, whereas forced expression of plasma membrane-localized ATG-16.2 is sufficient to nucleate ectopic LGG-1 puncta. Distinct from yeast Atg16, C. elegans and mammalian Atg16 orthologs possess WD repeats at their C termini. We showed here that the WD repeats are dispensable for the role of ATG-16.2 in the aggrephagy pathway. WD repeats may function in autophagy-independent processes; for example, ATG16L1 participates in cell secretion.29 ATG-16.1 and ATG-16.2 are capable of self-interaction and of associating with each other and ATG-5. The different functions of ATG-16.1 and ATG-16.2 in autophagy may result from interaction with additional factors. Formation of distinct ATG-16.1 and ATG-16.2 complexes may modulate the efficiency of autophagy during C. elegans development. Our results show that atg-16.1 and atg-16.2 have partially redundant functions in, but contribute differentially to, the autophagy pathway in C. elegans.
Materials and Methods
Strains
The following strains were used in this work: atg-16.1(qx57), atg-16.1(gk668615), atg-16.2(bp636), atg-16.2(ok3224), atg-3(bp412), atg-5(bp484), atg-5(bp546), atg-18(gk378), atg-2 (bp576), epg-1(bp414), epg-6(bp242), bpIs131(sepa-1::gfp), and bpIs151(sqst-1::gfp).
atg-2(bp576) has a tryptophan to stop codon mutation at amino acid 715, which causes a deletion of the C-terminal 1560 amino acids.10epg-6(bp242) contains an arginine to stop codon mutation at amino acid 161.10 They are likely to be null and were used in this study.
Isolation and identification of atg-16 homologs
Animals carrying sqst-1::gfp reporters were used for EMS mutagenesis. Embryos derived from F2 animals were screened for accumulation of SQST-1::GFP aggregates. Subsequent genetic and molecular analysis revealed that one mutation, bp636, is an allele of atg-16.2.
atg-16.1(qx57) was isolated from a forward genetic screen for mutations which suppress the intestinal GFP::LGG-1-labeled structures in tat-1(qx30) mutants. qx57 was mapped to the left arm of chromosome X by single nucleotide polymorphism (SNP) mapping. We amplified atg-16.1 cDNA and found that it encodes a protein that actually starts at the methionine at amino acid 52 of ATG-16.1 predicted by wormbase.
atg-16.2(ok3224) is likely to be a genetic null. atg-16.1(qx57) is probably not a null allele, as the mutation locates in the WD repeats.
Starvation assay
Embryos collected by the synchronization method were incubated in M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4 in 1 L water) at 20 °C. At each time point, an aliquot of each sample was placed on a plate seeded with OP50, a leaky uracil-requiring strain of E. coli. After 3 d at 20 °C, the number of animals surviving to L4 or adulthood was counted.
Life span
One hundred L4 larvae were maintained at 20 °C and transferred to fresh plates every day during the reproductive period and every 4 to 5 d thereafter. Animals were scored as dead if they failed to respond to a gentle tap on the head and tail with a platinum wire.
Reporter construction
The atg-16.1::gfp and atg-16.2::gfp reporters were constructed by a PCR-fusion based approach. One fragment contained ~2 kb promoter and the entire coding region of atg-16.1 and atg-16.2. The fusion PCR product was coinjected into wild-type animals with pRF4(rol-6). At least three stable transgenic lines were analyzed.
In vitro pull-down assay
cDNAs encoding ATG-16.1, ATG-16.2, ATG-5, and fragments of ATG-16.1 and ATG-16.2 were cloned into pAT-28a(+) (for His fusion), pGEX-6P-1 (for GST fusion) or pMAL-C2X (for MBP tagging). The pull-down assay was performed as previously described.10 Bound proteins were analyzed by western blot using an anti-GST and anti-His antibody.
Yeast two-hybrid assay
The yeast two-hybrid assay was performed using the ProQuest Two-Hybrid System (Invitrogen, 10835). cDNA encoding full length atg-16.2 was fused with the GAL4 DNA binding domain in the vector pPC97 then transformed into yeast strain Mav203 (Invitrogen, 10835) to screen a C. elegans cDNA library cloned in the GAL4 activation domain containing vector pPC86 (Invitrogen, 11288-016). Clones that grew on Leu-Trp-His- plates containing 25 mM 3AT were isolated for further analysis.
Indirect immunofluorescence
Permeabilization of embryos was performed by freeze-cracking methods.23 Freeze-cracked animals were fixed, blocked, and incubated with diluted primary antibody at room temperature for 2 to 4 h. The primary antibodies used in this study included anti-LGG-1 (rat or rabbit), anti-LGG-3 (rat), anti-SEPA-1 (rabbit), and anti-SQST-1 (rat) antibodies. The worms were then washed 3 times and incubated with Texas Red-conjugated goat anti-rat IgG (Jackson Labs, 112-075-003) or FITC-conjugated goat anti-rabbit IgG (Jackson Labs, 112-095-003). Slides were viewed using a confocal microscope (Zeiss LSM 510 Meta plus Zeiss Axiovert zoom, National Institute of Biological Sciences; Beijing). Five confocal images were analyzed for the quantification of aggregates.
RNA isolation and real-time RT-PCR
atg-16.1, atg-16.2, and wild-type animals were collected and total RNA was extracted with Trizol reagent (Sigma). Total isolated RNA (2 μg) from each sample was reverse transcribed using an Invitrogen Superscript III kit. Quantitative PCR reactions were performed as described previously.13atg-16.1 and atg-16.2 mRNA levels in mutants were normalized to the level of wild-type worms, which was set to 1. Error bars indicate the standard deviation (s.d.) of three independent experiments.
Supplementary Material
Acknowledgments
We thank Dr Isabel Hanson for editing the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40OD010440). This work was supported by the National Basic Research Program of China (2013CB910100, 2011CB910100) to HZ. The research of HZ was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute.
Glossary
Abbreviations:
- ATG
autophagy-related
- DAPI
4,6-diamino-2-phenyl indole
- epg
ectopic PGL granules
- GFP
green fluorescent protein
- lgg-1
LC3, GABARAP and GATE-16 family
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/26095
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/26095
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 5.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–65. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 6.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 7.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–69. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–55. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 10.Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovács AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–57. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Li Y, Wang F, Noda NN, Zhang H. Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem. 2012;287:29457–67. doi: 10.1074/jbc.M112.365676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–25. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 16.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 18.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 19.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishibashi K, Fujita N, Kanno E, Omori H, Yoshimori T, Itoh T, Fukuda M. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12–5-16L2 complex. Autophagy. 2011;7:1500–13. doi: 10.4161/auto.7.12.18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–21. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Tian E, Wang F, Han J, Zhang H. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009;5:608–15. doi: 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- 25.Yang P, Zhang H. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in C. elegans. Autophagy. 2011;7:159–65. doi: 10.4161/auto.7.2.14223. [DOI] [PubMed] [Google Scholar]
- 26.Sun T, Wang XW, Lu Q, Ren HY, Zhang H. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011;7:1308–15. doi: 10.4161/auto.7.11.17759. [DOI] [PubMed] [Google Scholar]
- 27.Liang Q, Yang P, Tian E, Han J, Zhang H. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy. 2012;8:1426–33. doi: 10.4161/auto.21163. [DOI] [PubMed] [Google Scholar]
- 28.Kovács AL, Zhang H. Role of autophagy in Caenorhabditis elegans. FEBS Lett. 2010;584:1335–41. doi: 10.1016/j.febslet.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.