Abstract
Autophagy is a cellular defense response to stress conditions, such as nutrient starvation. The type III phosphatidylinositol (PtdIns) 3-kinase, whose catalytic subunit is PIK3C3/VPS34, plays a critical role in intracellular membrane trafficking and autophagy induction. PIK3C3 forms multiple complexes and the ATG14-containing PIK3C3 is specifically involved in autophagy induction. Mechanistic target of rapamycin (MTOR) complex 1, MTORC1, is a key cellular nutrient sensor and integrator to stimulate anabolism and inhibit catabolism. Inactivation of TORC1 by nutrient starvation plays a critical role in autophagy induction. In this report we demonstrated that MTORC1 inactivation is critical for the activation of the autophagy-specific (ATG14-containing) PIK3C3 kinase, whereas it has no effect on ATG14-free PIK3C3 complexes. MTORC1 inhibits the PtdIns 3-kinase activity of ATG14-containing PIK3C3 by phosphorylating ATG14, which is required for PIK3C3 inhibition by MTORC1 both in vitro and in vivo. Our data suggest a mechanistic link between amino acid starvation and autophagy induction via the direct activation of the autophagy-specific PIK3C3 kinase.
Keywords: MTOR, autophagy, PIK3C3, ATG14, BECN1
Introduction
Macroautophagy, hereafter referred to as autophagy, is a catabolic process in which double-membrane autophagosomes sequester organelles or portions of cytosol and fuse with lysosomes for degradation. Autophagy plays important roles in a wide range of biological processes, including cell survival under stress conditions, removal of altered and damaged intracellular proteins and organelles, and cellular defense against pathogen infection. Efforts have been taken in recent years to uncover the mechanisms of autophagosome formation, which is mainly based on the autophagy-related (ATG) gene products initially found in yeast followed by the identification of orthologs in higher eukaryotes.1 These ATG proteins form several functional units during autophagy, including the ATG1/unc-51-like kinase (ULK) complex, the type III PtdIns 3-kinase PIK3C3/VPS34 complex I, two ubiquitin-like protein conjugation systems (ATG12–ATG5-ATG16L1 and the mammalian ortholog of yeast Atg8, MAP1LC3 (LC3), ATG9 and the ATG2-ATG18 complex, to promote the autophagy progression.
PIK3C3 is the catalytic subunit of the sole class III phosphatidylinositol-3-kinase that specifically phosphorylates phosphatidylinositol to generate phosphatidylinositol-3-phosphate (PtdIns3P).2 Early work has implicated PtdIns3P produced by PIK3C3 in several aspects of endosome-to-lysosome trafficking,3 while later studies have discovered that PIK3C3, in addition to its role in endosomal trafficking,4 has a key role in autophagy.5 In yeast, there are two distinct PIK3C3 complexes: complex I (composed of Vps34, Vps15, Atg6, and Atg14) that is involved in autophagy, and complex II composed of (Vps34, Vps15, Atg6, and Vps38) that functions in endosome-to-Golgi complex retrograde trafficking.5 In mammals, the orthologs of Atg6 and Vps34, BECN1/Beclin 1 and PIK3C3, also form a core complex and regulate cellular activity ranging from vesicle trafficking to autophagy. The mammalian ortholog of Atg14 has been identified to bind to BECN1-PIK3C3 and form an autophagy-specific complex.6,7 Mammals have at least two other stable PIK3C3 complexes, containing either UVRAG (UV irradiation resistance-associated gene, putative ortholog of yeast Vps38) or UVRAG and KIAA0226/Rubicon, a negative regulator of autophagy.7,8 Notably, interaction of ATG14 or UVRAG with the BECN1-PIK3C3 core complex is mutually exclusive.6,9 It is considered that by forming distinct protein complexes, the localization and activity of PIK3C3 are orchestrated to regulate different biological process, including autophagy and retrograde transport. Recently, Kim et al. have reported a role of AMPK in the differential regulation of different PIK3C3 complexes response to energy starvation.10 AMPK activation by glucose starvation stimulates the proautophagy PIK3C3 complex by phosphorylating BECN1 and inhibits the nonautophagy PIK3C3 complex by phosphorylating PIK3C3, thus revealing a mechanism of PIK3C3 regulation by energy starvation. Because autophagy is also induced by acute amino acid starvation, in which AMPK is not involved, it is of great interest to determine how different PIK3C3 complexes are regulated in response to nutrient stress.
The serine/threonine protein kinase MTOR regulates intracellular homeostasis by coordinating anabolic and catabolic processes with nutrient, energy and growth factor signaling.11 MTOR resides in two distinct multiprotein complexes: MTOR complex 1 (MTORC1) and 2 (MTORC2). Although several other associated proteins exist, MTORC1 and MTORC2 are distinguished by two unique accessory subunits, a regulatory-associated protein of MTOR (RPTOR) and a rapamycin-insensitive companion of MTOR (RICTOR), respectively. As a critical nutrient sensor, MTORC1 has been known for a long time to be tightly linked to autophagy regulation. In yeast, activation of TORC1 hyperphosphorylates Atg13, preventing its interaction with Atg1.12,13 Although the mammalian ATG13 can also be phosphorylated by MTORC1, this phosphorylation apparently does not disrupt the interaction between MTOR and ATG13. However, MTORC1 phosphorylates ULK1 (mammalian ortholog of Atg1) and ATG13, through which it sequesters ULK1-ATG13-RB1CC1 (RB1-inducible coiled-coil 1)/FIP200 complex in an inactive state.14–16 Moreover, MTORC1 inhibits ULK1 activation via a direct phosphorylation.17 Recently MTOR has also been found to play a positive role in the termination of autophagy upon prolonged starvation and help the restoration of functional lysosomes.18 However, little is known about whether MTOR also regulates other components/complexes in the autophagy process. It has been reported that TOR signaling is required for starvation-induced recruitment of PtdIns3P to autophagosomes in Drosophila,19 although it is not clear whether this is due to a direct regulation of PIK3C3 activity by TOR. In the present work, we studied the regulation of PIK3C3 complexes by MTOR and showed that MTORC1 specifically inhibits the ATG14-containing PIK3C3 complex through phosphorylating ATG14 at multiple sites. This phosphorylation in ATG14 is important for autophagy inhibition under nutrient sufficiency.
Results
MTOR inhibition activates the autophagy-related PIK3C3 complex
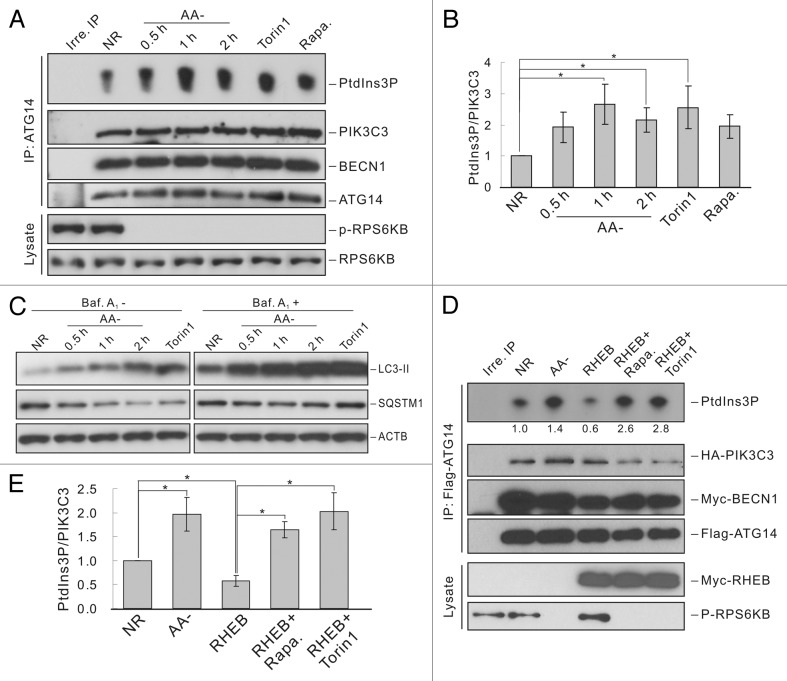
Association of ATG14 with BECN1 is required for PIK3C3 localization to the phagophore and autophagosome formation.6–9 To study whether autophagy-related PIK3C3 complex is regulated by nutrient conditions, ATG14-containing PIK3C3 complexes were immunoprecipitated from mouse embryonic fibroblasts (MEFs) that were cultured in nutrient-rich or amino acid starvation conditions and PIK3C3 activity was tested by an in vitro lipid kinase assay using PtdIns as a substrate. Amino acid starvation had no effect on the interaction between ATG14 and PIK3C3 or BECN1, but significantly increased ATG14-containing PIK3C3 complex activity in a time-dependent manner, in which 1 h starvation resulted in the highest PIK3C3 activity (Figure 1A and B). Moreover, treatment with rapamycin and Torin1, two MTOR-specific inhibitors, also increased ATG14-containing PIK3C3 activity, although rapamycin showed a milder effect compared with amino acid starvation or Torin1 treatment (Figure 1A and B). These data suggest that inhibition of MTORC1 by amino acid starvation may be responsible for activation of the ATG14-containing PIK3C3. We also tested LC3-II accumulation and SQSTM1/p62 degradation, two markers of autophagy, under amino acid starvation. Conversion of LC3-I to LC3-II was induced upon amino acid starvation (Figure 1C). Similarly, SQSTM1 levels were decreased in a time-dependent manner in response to amino acid starvation, indicating proper autophagy induction under this condition (Figure 1C). Notably, cotransfection of RHEB, an activator of MTOR, significantly reduced the activity of the ATG14-containing PIK3C3 complex, while further treatment with rapamycin or Torin1 reversed this effect (Figure 1D and E). It should be pointed out that PI3KR4/VPS15, a regulatory subunit required for PIK3C3 activity, was not included in the cotransfection for the sake of reducing the complexity of transfection. Because we purposely expressed the epitope-tagged PIK3C3 at a low level similar to endogenous protein in order to preserve the physiological regulation of PIK3C3, epitope-tagged PIK3C3 was fully activated by endogenous PI3KR4 (Fig. S1). These data show that MTOR negatively regulates the activity of autophagy-related PIK3C3 complex in response to nutrient deficiency.
Figure 1. MTOR is required for autophagy-related PIK3C3 complex regulation. (A) ATG14 containing PIK3C3 is activated under conditions of MTOR inhibition. HEK293 cells were cultured in following conditions: nutrient-rich (NR), amino acid free (AA-, indicated time), Torin1 (100 nM, 1 h) or rapamycin (50 nM, 1 h) treatment. ATG14-containing PIK3C3 complex was immunoprecipitated with ATG14 antibody and used for lipid kinase assay. (B) Quantification of (A). (C) Induction of autophagy by MTOR inhibition. HEK293 cells were treated similarly as in (A) with or without bafilomycin A1 (Baf A1) (100 nM) treatment for 1 h before harvesting cells. LC3 and SQSTM1 level were detected using specific antibodies. (D) RHEB inhibits ATG14 containing PIK3C3 via activation of MTOR. HEK293 cells were transfected with Flag-ATG14, Myc-BECN1 and HA-PIK3C3 together with pcDNA3 or RHEB. ATG14-containing PIK3C3 complex was immunoprecipitated with Flag antibody and used for lipid kinase assay. (E) Quantification of (D). The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a statistically significant difference.
AMPK is reported to be an important regulator of autophagy upon glucose starvation. Therefore, we also tested whether AMPK is involved in amino acid-induced PIK3C3 regulation. The AMPKα wild-type (WT) and Ampkα knockout (KO) MEFs exhibited similar activation of ATG14-containing PIK3C3 complex in response to amino acid starvation or rapamycin treatment (Fig. S2), suggesting that AMPK is not involved in amino acid starvation-induced PIK3C3 activation. Notably, Ampkα KO MEFs showed higher PIK3C3 activity than WT MEFs although the reason is unknown.
RRAG GTPases-MTOR pathway mediates the autophagy response to amino acid starvation
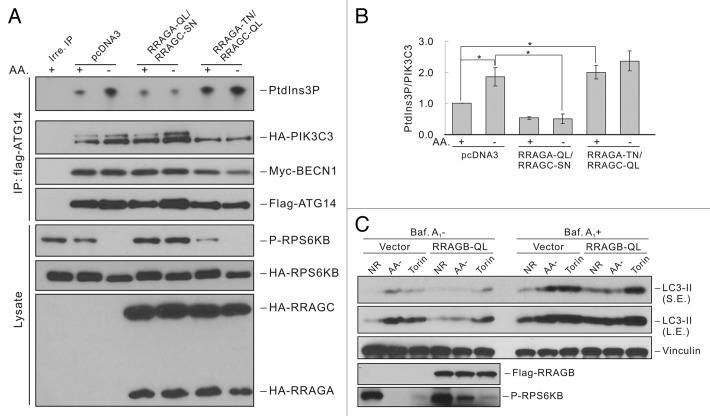
MTORC1 is regulated by amino acid signaling through Rag GTPases.20,21 To further confirm that MTOR mediates nutrient regulation on ATG14-containing PIK3C3 complexes, we tested the effect of RRAGAQ66L (QL, constitutively GTP-bound)/RRAGCS75N (SN, constitutively GDP-bound) or RRAGAT21N (TN, constitutively GDP-bound)/RRAGCQ120L (QL, constitutively GTP-bound) mutants. We found that ATG14-containing PIK3C3 complex had low activity and could not be activated by amino acid starvation when coexpressed with RRAGA-QL and RRAGC-SN, which constitutively activates MTORC1 even in the absence of amino acids (Figure 2A and B). Moreover, expression of RRAGA-TN and RRAGC-QL, which dominantly inhibited MTORC1, led to significant activation of PIK3C3 complex even under nutrient-rich conditions (Figure 2A and B). To further examine the function of Rag GTPases in autophagy, we stably expressed the constitutively active form of RRAGB (RRAGBQ99L) in 293A cells. Amino acid starvation-induced autophagy flux was suppressed as indicated by the reduced LC3-II accumulation, while Torin1-induced autophagy was not affected (Figure 2C) since Torin1 directly inhibits MTOR. These data indicate that MTOR mediates the amino acid signal to regulate ATG14-associated PIK3C3 activity and autophagy.
Figure 2. Rag GTPases-MTOR pathway mediates the autophagy response to amino acid starvation. (A) Rag GTPases suppress ATG14-PIK3C3 activation upon amino acid starvation. HEK293 cells were transfected with Flag-ATG14, Myc-BECN1 and HA-PIK3C3 together with pcDNA3, RRAGA-QL and RRAGC-SN, or RRAGA-TN and RRAGC-QL. ATG14-containing PIK3C3 complex was immunoprecipitated with Flag antibody and subjected to lipid kinase assay. (B) Quantification of (A). (C) Constitutively active RRAGB blocks autophagy via MTORC1 activation. HEK293A cells stably expressing control vector or RRAGBQ99L were subjected to amino acid starvation or Torin1 (100 nM) treatment with or without bafilomycin A1 (100 nM) treatment for 1 h before harvesting cells. LC3 levels were detected by immunoblot. LE: long exposure; SE: short exposure. The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a statistically significant difference.
Different PIK3C3 complexes are differentially regulated by MTOR
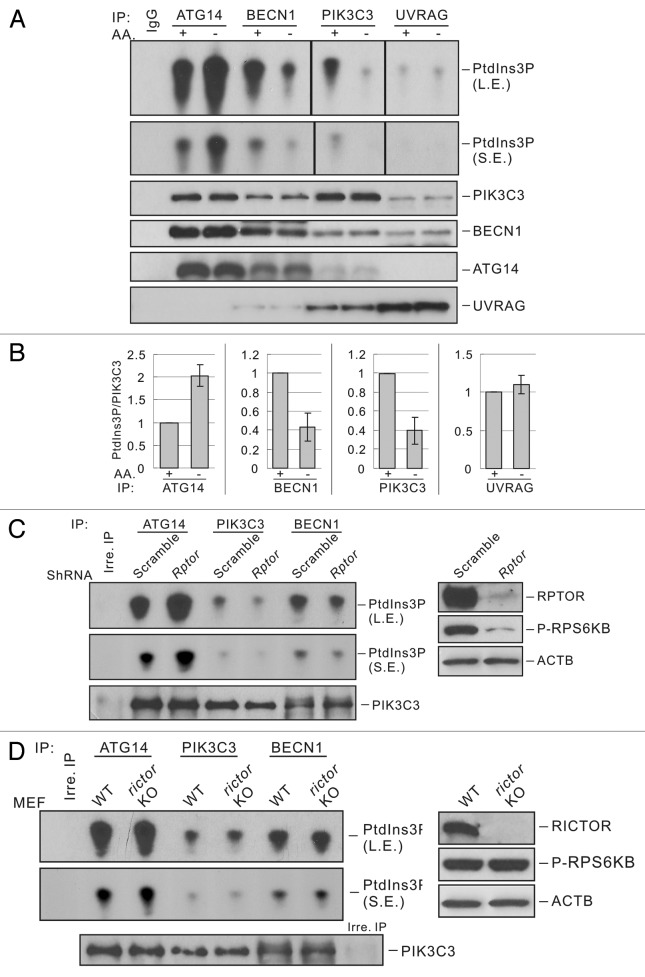
PIK3C3 can form multiple complexes, which are involved in the regulation of endosomal trafficking and autophagy. It has been reported that autophagy and non-autophagy PIK3C3 complexes are differentially regulated by AMPK upon glucose starvation.10 To determine whether these PIK3C3 complexes are also regulated by different nutrient stresses, we measured the lipid kinase activity of autophagic PIK3C3 complexes (prepared by ATG14 and UVRAG immunoprecipitation) and non-autophagy PIK3C3 complexes (prepared by BECN1 and PIK3C3 immunoprecipitation). We found that ATG14-associated PIK3C3 was activated upon amino acid starvation. In contrast, the nonautophagy PIK3C3 complexes (BECN1-PIK3C3 or PIK3C3 alone) were inhibited by a similar treatment (Figure 3A and B). These results are consistent with the regulation of PIK3C3 complexes upon glucose starvation. However, the activity of UVRAG-associated PIK3C3 was not affected by amino acid starvation, which is different from glucose starvation-induced regulation of UVRAG-PIK3C3. These observations show that different PIK3C3 complexes are indeed differentially regulated by amino acid starvation. Interestingly, the composition of each complex was not affected by nutrient starvation (Figure 3A and B), suggesting that the regulation of PIK3C3 activity is not due to the disruption of complexes but possibly by post-translational modifications. We also tested whether AMPK is involved in the regulation of different PIK3C3 complexes in response to amino acid starvation. We found that AMPKα WT and Ampkα KO MEFs displayed similar regulation of each PIK3C3 complexes (Fig. S3). Therefore, AMPK appears to play no direct role in PIK3C3 regulation in response to amino acid starvation.
Figure 3. MTORC1, but not MTORC2, is involved in the regulation of different PIK3C3 complexes. (A) Amino acid starvation only activates the ATG14-containing PIK3C3 lipid kinase activity. Different PIK3C3 complexes were immunoprecipitated from MEFs that were cultured in nutrient-rich or AA-free medium for 1 h, and subjected to lipid kinase assay and immunoblot. (B) Quantification of (A). (C) RPTOR knockdown activates ATG14-PIK3C3. Different PIK3C3 complexes from scrambled or Rptor KD MEFs were immunoprecipitated with corresponding antibodies and subjected to lipid kinase assay. The right panel indicates RPTOR protein levels in control or Rptor KD MEFs. (D) Rictor knockout has no effect on ATG14-PIK3C3 activity. Different PIK3C3 complexes from RICTOR WT or Rictor KO MEFs were immunoprecipitated with corresponding antibodies and subjected to lipid kinase assay. The right panel indicates RICTOR protein levels in WT or Rictor KO MEFs.
To further confirm that amino acid starvation-induced differential regulation of PIK3C3 complexes is through MTOR pathway, we tested the effect of RHEB or RRAGA and RRAGC. Amino acid starvation activated the ATG14-containing PIK3C3 complex, while this effect was compromised by overexpression of RHEB or RRAGA-QL and RRAGC-SN (Fig. S4A and S4B). Moreover, amino acid starvation-induced inhibition of the BECN1-PIK3C3 or PIK3C3 pool was also reversed by overexpression of RRAGA-QL and RRAGC-SN or RHEB (Fig. S4A and S4B). These data indicate that MTOR is involved in both the activation and inhibition of different PIK3C3 complexes in response to amino acids, and constitutive activation of MTOR by RHEB or Rag GTPases suppresses the effects of amino acid starvation.
MTORC1, but not MTORC2, regulates PIK3C3
MTOR exists in two complexes, MTORC1 or MTORC2. These two complexes differ in subunit composition, regulation and function. Our data indicate that MTORC1 may be involved in PIK3C3 regulation because rapamycin only inhibits MTORC1 that is regulated by amino acids (Figures 1 and 2). To demonstrate the function of MTORCs in PIK3C3 regulation, we determined the activity of PIK3C3 in Rptor knockdown (KD) or Rictor KO MEFs, which represent the loss of function of MTORC1 or MTORC2, respectively. Rptor KD resulted in an activation of ATG14-containing PIK3C3 complex but an inhibition of BECN1-PIK3C3 (Figure 3C). Therefore, Rptor knockdown generated an effect similar to that caused by nutrient starvation. Rptor KD also resulted in higher basal autophagy flux as indicated by LC3-II accumulation (Fig. S5A). These data support a role of MTORC1 in PIK3C3 regulation. In contrast, Rictor KO cells exhibited similar activity in each PIK3C3 pool compared with WT cells (Figure 3D), suggesting that MTORC2 is not involved in PIK3C3 regulation. We also tested the response of Rptor KD or Rictor KO MEFs to nutrient starvation. It was noted that Rptor KD cells did not show substantial response to nutrient conditions (Fig. S5B), while Rictor KO cells still displayed normal responses as WT cells (Fig. S5C).
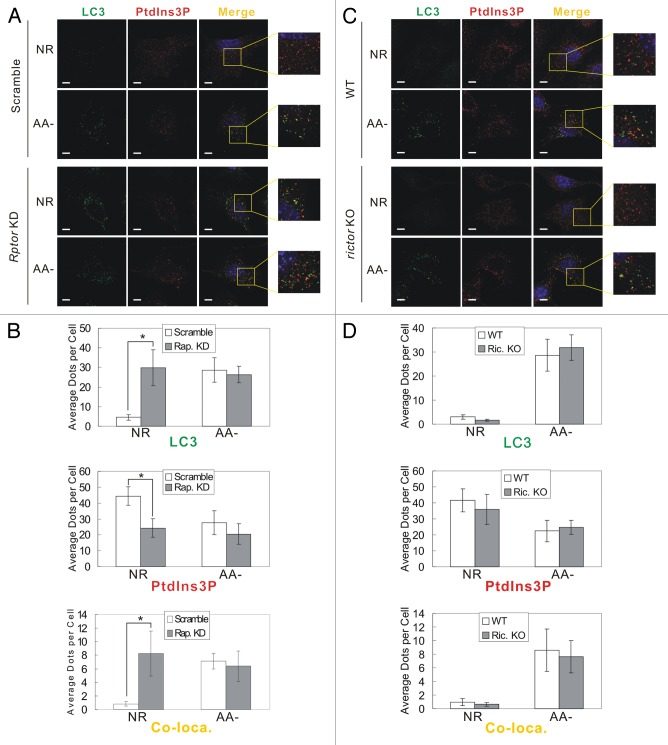
To further investigate the regulation of PIK3C3 activity and autophagy by MTOR complexes, we tested cellular PtdIns3P and LC3 puncta formation by immunostaining. Cellular PtdIns3P was stained with GST-2XFYVE domain fusion protein, which specifically binds to PtdIns3P.22 Total PtdIns3P dots were reduced upon amino acid starvation in control cells, consistent with previous report that total PtdIns3P is decreased by nutrient starvation (Figure 4A and B).23,24 However, PtdIns3P costaining with LC3 puncta (a marker of autophagosomes) was significantly increased upon amino acid starvation (Figure 4A and B), consistent with the results of in vitro PIK3C3 kinase assay that the autophagy-specific ATG14-PIK3C3 complex is activated under starvation. Notably, only a portion of LC3 puncta (about 25%) showed colocalization with PtdIns3P. A possible explanation is that after the recruitment of LC3 PtdIns3P might be dephosphorylated by PtdIns3-phosphatases in mature autophagosomes.25,26 Interestingly, Rptor KD cells showed strong induction of LC3 puncta formation and suppression of PtdIns3P dots compared with control cells under nutrient-rich conditions, which is quite similar to the staining pattern observed under amino acid starvation (Figure 4A and B). In addition, there was a significant increase of colocalization of LC3 puncta and PtdIns3P in Rptor KD cells. In contrast, Rictor KO cells exhibited a similar staining pattern of LC3 and PtdIns3P as WT cells under both nutrient-rich and nutrient-stress conditions (Figure 4C and D). Collectively, the above data demonstrate that MTORC1, but not MTORC2, is involved in PIK3C3 regulation and autophagy induction in response to amino acid starvation.
Figure 4. MTORC1, but not MTORC2, regulates autophagy-specific PIK3C3 activity. (A) RPTOR knockdown enhances basal autophagy and autophagy-specific PIK3C3 activity. Scrambled or Rptor KD MEFs were cultured in either nutrient-rich or amino acid-free medium for 1 h. Cells were fixed and subjected to immunostaining with LC3 antibody and GST-FYVE probe. (B) Quantification of the staining in (A). (C) Rictor knockout has not effect on autophagy induction by amino acid starvation. WT and Rictor KO MEFs were treated and stained similarly as in (A). (D) Quantification of the staining in (C). The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a statistically significant difference.
MTORC1 inhibits ATG14 complexes in vitro
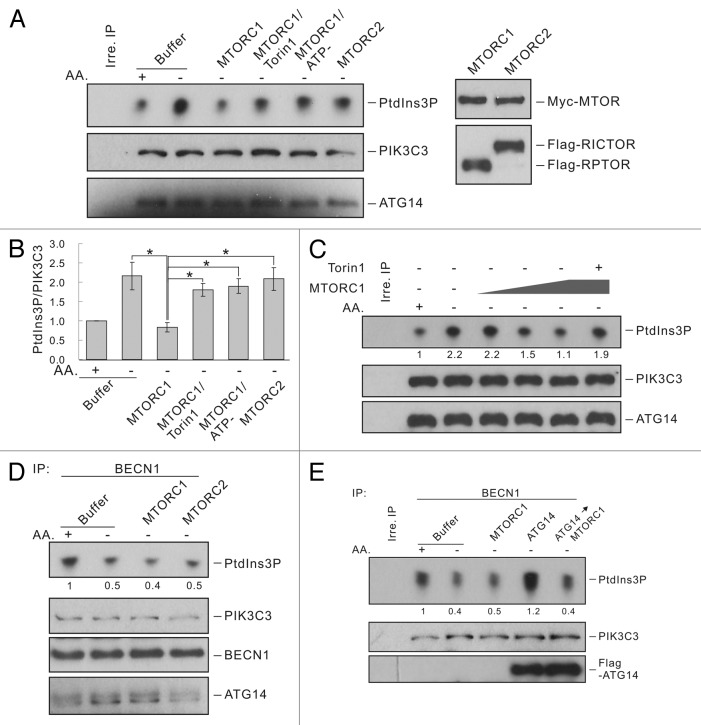
To test whether MTORC1 directly regulates PIK3C3 complexes, we immunoprecipitated ATG14-containg PIK3C3 complexes and treated the sample in vitro with MTORC1 or MTORC2 immunopurified from HEK293 cells. Interestingly, in vitro treatment with MTORC1 significantly decreased the lipid kinase activity of ATG14-PIK3C3 complex prepared from nutrient-starved cells (Figure 5A and B). In contrast, MTORC2 treatment did not show any effect on the lipid kinase activity of the ATG14-containing PIK3C3 complex. This data are consistent with the observations described in the previous sections that MTORC1 regulates ATG14-containing PIK3C3. MTORC1 inhibited ATG14-PIK3C3 in vitro in a dose-dependent manner (Figure 5C). Moreover, the inhibition of MTOR kinase by Torin1 blocked the inhibitory effect of MTORC1 on PIK3C3 activity (Figure 5A). Similarly, MTORC1 could not inhibit ATG14-PIK3C3 when ATP was omitted in the reaction. These data show that MTORC1 directly inhibits ATG14-PIK3C3 complex in a manner dependent on MTOR kinase activity.
Figure 5. MTORC1, but not MTORC2, regulates the ATG14-containing PIK3C3 complex in vitro. (A) MTORC1 inhibits ATG14-PIK3C3 activity in vitro. ATG14-containing complexes were immunoprecipitated with ATG14 antibody and treated with MTORC1 or MTORC2 purified from HEK293 cells in vitro. For MTORC1 treatment, reactions in the presence of Torin1 (50 nM) or without ATP were set up as controls. The immune-complexes were then subjected to lipid kinase assay. Right panels show the relative amount of MTORC1 and MTORC2 used in the assay. (B) Quantification of (A). The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a significant difference. (C) Dose-dependent inhibition of ATG14-PIK3C3 by MTORC1. ATG14-containing complexes were treated with increasing amount of MTORC1 in vitro and subjected to lipid kinase assay. The presence of Torin1 is indicated (last lane). (D) The BECN1-PIK3C3 complex is not regulated by MTORC1 or MTORC2. BECN1-PIK3C3 complex were treated with MTORC1 or MTORC2 in vitro and then subjected to lipid kinase assay. (E) ATG14 increased the basal PIK3C3 activity and also confers inhibition by MTORC1. BECN1-PIK3C3 complex were immunoprecipitated with BECN1 antibody and incubated with Flag-ATG14 protein (50 ng) for 10 min. After washing, the immune-complexes were further treated with MTORC1 and finally subjected to lipid kinase assay. Immunoblot shows Flag-ATG14 bound to the BECN1-PIK3C3 complex. The numbers under top panels in (C–E) represent the relative density of each dot normalized to PIK3C3 levels.
Contrary to ATG14-PIK3C3, the activity of the BECN1-PIK3C3 complex is downregulated upon nutrient starvation. Thus, we tested whether this complex is regulated by MTORC1. Interestingly, the activity of the BECN1-PIK3C3 complex was not directly affected by treatment with either MTORC1 or MTORC2 in vitro (Figure 5D). These data suggest that the presence of ATG14 may confer PIK3C3 inhibition by MTORC1. To test this hypothesis, we incubated purified ATG14 with the BECN1-PIK3C3 complex. After washing, the complex was further treated with MTORC1. As expected, binding of ATG14 increased PIK3C3 lipid kinase activity. Remarkably, MTORC1 strongly inhibited the lipid kinase activity of BECN1-PIK3C3 only when purified ATG14 was bound (Figure 5E). Therefore, addition of ATG14 likely converted the ATG14 free BECN1-PIK3C3 to ATG14 containing complex, which was then inhibited by MTORC1.
MTOR phosphorylates ATG14 at multiple sites
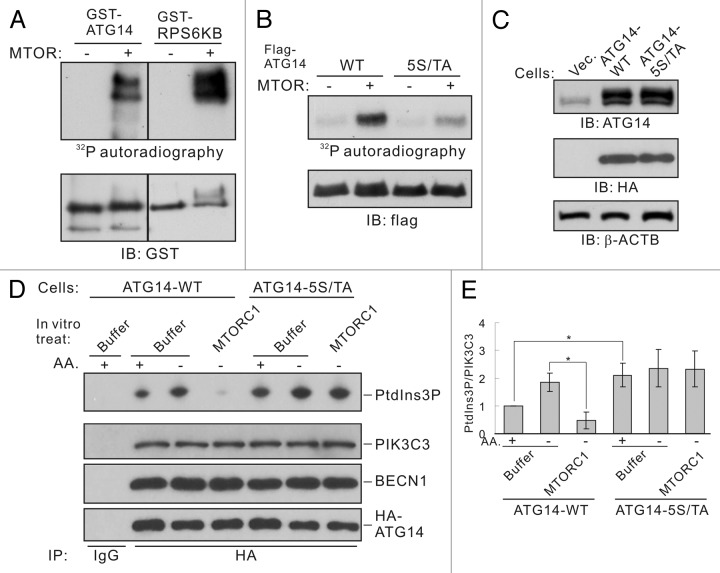
Previous data strongly indicate that MTOR may regulate the autophagy-related PIK3C3 complex through targeting ATG14. Therefore, we made efforts to determine whether ATG14 is a phosphorylation substrate of MTOR. In vitro kinase assay showed that GST-fused recombinant ATG14 was directly phosphorylated by MTOR (Figure 6A). We then identified phosphorylation sites by mass spectrometry. Five serine/threonine sites (Ser3, Ser223, Thr233, Ser383, and Ser440) were found to be phosphorylated by MTOR in vitro (Fig. S6). Except Ser223, all other four sites fit the MTOR consensus motif.27 Further experiments demonstrated that mutation of all the five sites to alanine was able to eliminate most of the phosphorylation potential of ATG14 by MTOR (Figure 6B; Fig. S7A). Moreover, reversion of any of the alanine residues back to serine or threonine could not fully restore MTOR phosphorylation indicating that ATG14 is phosphorylated on multiple serine and threonine sites (Fig. S7B).
Figure 6. MTORC1 inhibits ATG14-PIK3C3 complex activity by phosphorylating ATG14. (A) Phosphorylation of ATG14 by MTOR. Recombinant GST-tagged ATG14 or RPS6KB were expressed and purified from E. coli, and used as substrates for in vitro MTOR kinase assay. Phosphorylation was determined by 32P-autoradiograph and the protein levels were determined by immunoblot. GST-RPS6KB was included as a positive control for MTOR kinase assay. (B) Mutation of the five phosphorylation residues abolishes ATG14 phosphorylation by MTOR in vitro. HEK293 cells were transfected with Flag-ATG14-WT or 5S/TA mutant as well as Myc-BECN1 and HA-PIK3C3. Flag-ATG14 proteins were purified by immunoprecipitation with Flag antibody and subjected to in vitro MTOR kinase assay. (C) HEK293 cells stably expressing HA-ATG14-WT or HA-ATG14-5S/TA. The HA-tagged ATG14 run slightly slower than the endogenous ATG14 (top panel). (D) Amino acid starvation activates ATG14-WT but not the ATG14-5S/TA associated PIK3C3 activity. ATG14-containing PIK3C3 complexes were immunoprecipitated from HEK293 stable cells under either nutrient-rich or -starvation conditions using HA antibody. The immune-complexes were treated with MTORC1 and then subjected to lipid kinase assay. (E) Quantification of (D). The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a statistically significant difference.
To investigate the function of these phosphorylation sites, we made HEK293 cells that stably express ATG14-WT or ATG14-5S/TA mutant proteins (Figure 6C). PIK3C3 lipid kinase activity and regulation were tested in these stable cells. When treated with MTORC1 in vitro, the PIK3C3 complex containing ATG14-WT was inhibited (Figure 6D and E). In contrast, the PIK3C3 complex containing ATG14-5S/TA was not inhibited by MTORC1 treatment in vitro. Notably, the PIK3C3 complex containing ATG14-5S/TA also showed elevated basal activity under nutrient-rich conditions. These results indicate that phosphorylation of ATG14 is important for inhibition of the ATG14-containing PIK3C3 by MTORC1.
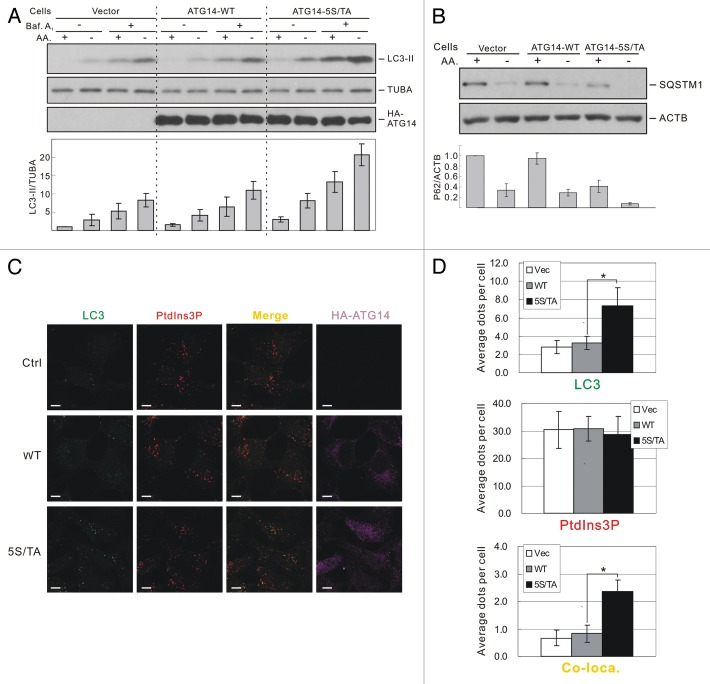
MTOR is a negative regulator for autophagy; therefore, the phosphorylation of ATG14 by MTOR would be predicted to exert an inhibitory effect on autophagy. To test this hypothesis, we checked autophagy flux in HEK293 stable cells. Cells expressing the ATG14-5S/TA mutant protein had a significantly higher level of autophagy flux indicated by both LC3-II accumulation (Figure 7A) and SQSTM1 degradation (Figure 7B). Consistently, more LC3 puncta formation as well as colocalization of LC3 puncta and PtdIns3P were observed in ATG14-5S/TA cells compared with ATG14-WT cells under nutrient-rich conditions (Figure 7C and D). Interestingly, overall PtdIns3P level did not show a statistical difference between the two cell lines. Therefore, expression of ATG14-5S/TA selectively increased PtdIns3P at the autophagosomes. Under amino acid starvation conditions, however, both LC3 puncta formation and colocalization of LC3 and PtdIns3P were increased to similar levels in ATG14-WT and ATG14-5S/TA cells (Fig. S8). Collectively, these data indicate an important role for TORC1 in the direct inhibition of the autophagy-specific PIK3C3 kinase complex at times of nutrient sufficiency.
Figure 7. ATG14-5S/TA mutant promotes autophagy level. (A) Expression of ATG14-5S/TA enhances LC3-II. HEK293 cells stably expressing control vector, ATG14-WT or ATG14-5S/TA were treated with either nutrient-rich or AA-free medium for 1 h with or without presence of bafilomycin A1 (100 nM). LC3 levels were detected by immunoblot. The histogram under the blots represents quantification of each LC3-II band normalized by tubulin. (B) ATG14-5S/TA enhances SQSTM1 degradation. The histogram under the blots represents quantification of each SQSTM1 band normalized by actin. (C) ATG14-5S/TA promotes autophagy. HEK293A stable cells stably expressing control vector, ATG14-WT or ATG14-5S/TA were cultured in nutrient-rich medium for 1 h. Cells were fixed and subjected to immunostaining with GST-FYVE probe (PtdIns3P) and antibodies against HA and LC3. Scale bar: 10 μm. (D) Quantification of the staining in (C). The error bars represent the standard error of the mean from independent experiments within same treatment group. Stars indicate a statistically significant difference.
Discussion
It is well established that MTOR negatively regulates autophagy. Nutrient starvation induces autophagy by inactivating MTOR. Supporting a critical role of MTOR in autophagy, inhibition of MTOR by rapamycin is sufficient to induce autophagy even in the presence of nutrients. One mechanism of MTOR-mediated inhibition of autophagy is by phosphorylation and inhibition of the ULK1 kinase. In this report, we demonstrated that the catalytic subunit of class III PtdIns3K, PIK3C3, particularly the autophagy-specific and ATG14-containing complex, is directly regulated by MTOR in response to nutrient starvation.
During autophagy initiation, PIK3C3 complex is recruited to the phagophore and phosphorylates PtdIns to generate PtdIns3P, which further recruits downstream effectors (e.g., WIPI1,WIPI2 and ZFYVE1/DFCP1) and promotes phagophore formation.28 This study provides a biochemical basis for the selective activation of the autophagy specific PIK3C3 complex in response to amino acid starvation. Our results demonstrated that the ATG14-containing PIK3C3 complex is significantly activated upon MTORC1 inhibition by pharmacological inhibitors or amino acid starvation. Accordingly, constitutively high MTORC1 activity suppresses activation of the autophagy-specific and ATG14-associated PIK3C3 complex. Consistent with our model, it has been reported that ATG14 localizes to the endoplasmic reticulum (ER) and recruits a subset of PIK3C3 to form a platform for autophagosome formation.29 This study provides a molecular mechanism for MTORC1 in the regulation of the ATG14-associated and autophagy-specific PIK3C3 upon amino acid starvation.
The inhibition of ATG14-PIK3C3 by MTORC1 appears to be a direct phosphorylation event, as the immunopurified ATG14-PIK3C3 can be inhibited by MTORC1 treatment in vitro. Inhibition of MTOR kinase activity by Torin1 or omission of ATP in the kinase reaction abolishes the inhibitory effect of MTORC1 on ATG14-PIK3C3. ATG14 appears to play a critical role in this regulation. MTORC1 could not inhibit PIK3C3 in the absence of ATG14. Addition of ATG14 confers the ability of PIK3C3 to be inhibited by MTORC1 in vitro. These results indicate that MTORC1 phosphorylates ATG14 or another component in the PIK3C3 complex to inhibit the lipid kinase activity.
ATG14 is a direct substrate of MTORC1 and its phosphorylation is important for inhibition of the associated PIK3C3 by MTORC1. Mutation of the phosphorylation sites to alanine renders the ATG14-5S/TA-containing PIK3C3 complex resistant to in vitro inhibition by MTORC1 treatment. Overexpression of the ATG14-5S/TA mutant protein significantly elevated the basal autophagy level under nutrient-rich conditions, indicating the ATG14-5S/TA is constitutively active, consistent with the insensitivity of the ATG14-5S/TA to inhibition by MTORC1 in vivo. It is notable that amino acid starvation further induces autophagy response in cells expressing ATG14-5S/TA. This observation strongly argues that release from MTOR inhibition of the ATG14-PIK3C3 is not enough to fully activate the autophagic machinery. Additional regulation or signaling pathways may be required for full autophagy induction under nutrient stress. Interestingly, it is reported that ULK1 is inhibited through phosphorylation by MTOR under nutrient-rich conditions.17 This (ULK1 activation) may provide another link from nutrient starvation to autophagy regulation.
A recent report from our laboratory has revealed a mechanism of differential regulation of different PIK3C3 complexes under glucose starvation, in which AMPK plays an essential role.10 AMPK inhibits the nonautophagy PIK3C3 complexes by phosphorylating PIK3C3 to suppress normal cellular activity whereas activates the proautophagy PIK3C3 complexes by phosphorylating BECN1 to induce autophagy. Although ATG14 is not the direct substrate of AMPK, its binding to BECN1-PIK3C3 promotes BECN1 phosphorylation and inhibits PIK3C3 phosphorylation by AMPK. However, AMPK is not directly regulated by amino acid starvation, which also induces autophagy. The current work demonstrates that MTORC1 regulates the autophagy-specific ATG14-PIK3C3 complex by targeting ATG14 in response to amino acid starvation. The MTORC1-dependent phosphorylation of ATG14 inhibits the pro-autophagy PIK3C3 complex activity, thereby suppressing autophagy. We also observed that the nonautophagy PIK3C3 complexes are inhibited by amino acid starvation, which is similar to the phenomenon induced by glucose starvation.10 However, this inhibition seems not to be due to a direct effect of MTOR as in vitro MTOR treatment did not inhibit the nonautophagy PIK3C3 complexes, indicating a different mechanism responsible for this inhibition. Taken together, we propose a model that AMPK and MTOR, two key cellular nutrient sensors, coordinately regulate autophagy by phosphorylating different components of PIK3C3 complexes.
In summary, this study establishes an essential role of MTORC1 in regulation of the autophagy-specific, ATG14-associated PIK3C3 complex in response to amino acid starvation. We propose that this mechanism of regulation is important for autophagy induction. Moreover, we demonstrated an obligatory role of ATG14 to support the PIK3C3 complex regulation by MTORC1. Finally, our study showed that MTORC1 inhibits ATG14-containing complex via direct phosphorylation of ATG14. PIK3C3 regulation is likely to be rather complex given the complex biology of autophagy. Besides MTORC1, PIK3C3 is likely to be regulated by other upstream components in the autophagy pathway, such as ULK1/Atg1. Elucidation of the molecular mechanism of PIK3C3 regulation will significantly advance our understanding of autophagy biology.
Materials and Methods
Antibodies and plasmids
Anti-PIK3C3 (#4263 for WB), BECN1 (#3738 for WB), phospho-RPS6KB (#9205), RPS6KB (#9202), RPTOR (#2280), RICTOR (#2114), and LC3 (#2775) for WB antibodies were purchased from Cell Signaling Technology. Anti-UVRAG (M160-3), ATG14 (PD026 for immunoprecipitation), and LC3 (PM036 for immunostaining) antibodies were obtained from MBL. Anti-PIK3C3 (Echelon, ZR015) and BECN1 (Bethyl, A302-567A) antibodies were used for immunoprecipitation. Anti-vinculin (V9264), TUBA/tubulin, α (T9026), Flag (F3165), and ATG14 (A6358 for WB) antibodies were obtained from SIGMA. The ACTB/actin, β (sc-47778) antibody was from Santa Cruz. Anti-HA (MMS-101P) and Myc (MMS-150P) antibodies were from Covance. Anti-SQSTM1 (GP62-C) antibody was purchased from Progen Biotechnik.
Cell culture, transfection, lentiviral, and retroviral infection
HEK293 or HEK293A cells were cultured in DMEM (Invitrogen, 11965118) containing 10% FBS (Thermo Scientific, SH30910.03) 50 μg/mL penicillin/streptomycin, 1 mM sodium pyruvate (Invitrogen, 11360070) and 0.1 mM MEM non-essential amino acid (Invitrogen, 11140050). Immortalized MEF cells were cultured in the same medium as above plus 55 μM β-mercaptoethanol. Transfections were performed with polyethylenimine. For amino acid starvation, cells were incubated with amino-acid free DMEM containing 10% dialyzed FBS, 50 μg/mL P/S and 1 mM sodium pyruvate for the indicated time.
To establish control and Rptor knockdown stable cells in WT MEF cells, A lentiviral construct pLKO.1 containing the scrambled sequence or shRNA targeting the mouse Rptor gene (provided by Dr David Sabatini, MIT)30 was cotransfected with viral packaging plasmids (psPAX2 and pMD2.G) into HEK293T cells. Viral supernatant was harvested after 48 h and filtered through 0.45-μm filter. Target cells were infected in the presence of 8 μg/ml Polybrene. Stable pools were obtained in the presence of 2 μg/ml puromycin (InvivoGen, ant-pr-5). To establish HEK293A cells stably expressing RRAGBQ99L mutant, lentiviral constructs of pLJM1 empty vector or pLJM1-RRAGBQ99L (David Sabatini, Addgene plasmid 19315)21 was cotransfected with psPAX2 and pMD2.G and virus infection was performed as described above. To establish HEK293 cells stably expressing HA-tagged ATG14-WT or ATG14-5S/TA mutant protein, retroviral constructs of pQCXIH control vector, pQCXIH-HA-ATG14-WT or -5S/TA was transfected into HEK293P cells. Virus infection was performed as described above. Stable pools were obtained in the presence of 100 μg/ml hygromycin B (Invitrogen, 10687-010).
Immunoprecipitation and western blot
For immunoprecipitation of MTOR-containing complexes, cells were lysed in CHAPS lysis buffer (50 mM HEPES [pH 7.4], 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 10 mM NaF, 100 mM NaCl, 0.3% CHAPS, and protease inhibitors).30 Immune complexes conjugated to beads were also washed with the same buffer. For other immunoprecipitations, cells were lysed with mild lysis buffer (MLB; 10 mM Tris at pH 7.5, 2 mM EDTA, 100 mM NaCl, 1% NP-40, 50 mM NaF, 1 mM Na3VO4 and protease inhibitor cocktail [Roche, 5056489001]). Indicated antibodies were coupled with protein A-Sepharose beads in MLB for 2 h or overnight at 4 °C. The immune complex was then added to cell lysate and incubated at 4 °C for 2 h. The resulting beads were washed with MLB three times before subjected to western blot analysis. For LC3 analysis, cells were harvested with Laemmli sample buffer and subjected to western blot analysis. Densitometric analysis of western blot bands was performed with Quantity One software (Bio-Rad).
Immunofluorescent staining
Cells were cultured on fibronectin-coated coverslips to the appropriate density and treated as indicated in experiments. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 50 μg/ml digitonin for 10 min. After blocking in 2% BSA/2% goat serum for 30 min, slides were incubated with primary antibodies diluted in blocking buffer for 1 h at room temperature. After washing with PBS, slides were incubated with Alexa Fluor 488- (A11008), Alexa Fluor 546- or Alexa Fluor 647-conjugated secondary antibodies (Invitrogen, A10036 and A20990) diluted in blocking buffer for 1 h. The slides were then washed and mounted. Images were taken on an Olympus FV1000 confocal microscope (CMME, UC San Diego, La Jolla, CA).
Purification of MTOR complexes
The method for MTOR complexes purification has been described with some modifications.30 Briefly, Myc-MTOR was cotransfected with either Flag-RPTOR or Flag-RICTOR into HEK293 cells. Cells were lysed in CHAPS lysis buffer. MTORC1 and MTORC2 were immunoprecipitated with anti-Flag M2 Affinity Gel (Sigma, A2220). Purified MTOR complexes were eluted with 200 μg/ml 3 × FLAG peptide in 50 mM HEPES, pH 7.4 and 100 mM NaCl. Excessive flag peptide was removed by desalting with desalting spin column (Thermo Scientific, 89883).
In vitro treatment on PIK3C3 complexes and in vitro kinase assay
The PIK3C3 immune-complex was washed three times with MLB and once with MTOR kinase buffer (25 mM HEPES, pH 7.4, 50 mM KCl and 10 mM MgCl2). Beads of immununoprecipitates were then incubated with MTORC1 or MTORC2 in MTOR kinase buffer supplemented with 4 mM MnCl2 and 50 μM ATP at 30 °C for 15 min. After the reaction, the beads were washed once with MLB containing 0.05% SDS and twice with MLB and then continue to in vitro PIK3C3 lipid kinase assay. For MTOR kinase assay, GST fusion proteins or immune-complex purified from HEK293 cells were used as substrates and reaction was performed as described above except that 3 uCi of 32P was added to the reactions.
In vitro PIK3C3 lipid kinase assay
PIK3C3 immune-complex was washed three times with MLB and once with KA buffer (20 mM HEPES, pH7.4, 1 mM EGTA, 0.4 mM EDTA, 5 mM MgCl2, and 0.05 mM DTT). A fraction (1/5) of the beads of immununoprecipitates was aliquoted for western blot. Residual beads was incubated with 50 μl of KA buffer containing 0.1 mg/ml phosphatidylinositol (Sigma, p0639), 50 μM cold ATP, 5 μCi 32P-ATP, 5 mM MnCl2, and 50 μM DTT at 37 °C for 30 min. Reactions were terminated by adding 1/5 vol. of 1N HCl and the lipid was extracted by 2 vol. of CHCl3:MeOH (1:1). Same volume of lipid product was loaded on a thin-layer-chromatography (TLC) plate (Whatman, 4865-821) and separated by TLC buffer of CHCl:MeOH:NH4OH:Water (129:100:4.29:24). 32P-PtdIns3P was subjected to autoradiography on an X-ray film.
Mass spectrometry
Excised Coomassie-stained SDS-PAGE gel bands were subjected to in-gel proteolytic digestion with trypsin. Following gel slice digestion the digestion products were desalted using C18-micro ZipTips (Millipore, ZTC18S960) per the manufacturer’s instructions and dried by vacuum centrifugation. The resulting peptide sample was resuspended in 7 μL of 0.1% formic acid and 5 μL was analyzed by LC/ESI MS with a 2D Nano-HPLC (Eksigent) coupled to a LTQ-OrbiTrapXL (Thermo Scientific) mass spectrometer using an LC MS ion source configuration. In-line de-salting was accomplished using an IntegraFrit trap column (75 μm × 25 mm; New Objective, TRC-25) packed with reverse phase Magic C18AQ (5-μm, 200 Å resin; Michrom Bioresources, CN5/61270/00) followed by peptide separations on a PicoFrit column (75 μm × 200 mm; New Objective, PF75##-200) packed with reverse phase Magic C18AQ (5-μm, 100 Å resin; Michrom Bioresources, CN5/66170/00) directly mounted on the electrospray ion source. A 40 min nonlinear gradient was used starting at 2% acetonitrile with 0.1% formic acid. The percentage of acetonitrile was increased to 40% over 40 min, increased to 90% over 0.1 min and then held at 90% for 5 min, then decreased to 2% over 0.1 min and held at 2% for 15 min. A flow rate of 400 nL/min was used for chromatographic separations and a spray voltage of 2500 V was applied to the electrospray tip. The LTQ-OrbiTrap instrument was operated in a data-dependent mode, switching automatically between MS survey scans in the OrbiTrap (AGC target value 1,000,000, resolution 60,000, and injection time 0.01 s) with MS/MS spectra acquisition in the linear ion trap (AGC target value of 10,000 and injection time 0.1 s). The 5 most intense ions from the Fourier-transform (FT) full scan were selected for fragmentation in the linear ion trap by collision-induced dissociation. Normalized collision energy of 35% and an isolation width of 2.0 were used for MS2 events and selected ions were dynamically excluded for 30 s. Charge state screening was enabled to exclude unassigned, +1, and greater than +4 charge states from being selected for analysis. The protein database search algorithm X!Tandem was used to identify peptides from a custom protein database in which the GST-ATG14 amino acid sequences was appended to an E. coli database to which common protein contaminants had been added. Variable monoisotopic mass modifications of 15.9949 on methionine (oxidation) and 79.9663 on serine, threonine, and tyrosine (phosphorylation), and a static modification of 57.02146 on cysteine (carbamidomethylation) were used. Peptide false discovery rates were measured using Peptide Prophet and results were stored and analyzed in the Computational Proteomics Analysis System (CPAS). Peptides were filtered with a minimum Peptide Prophet probability that produced a false discovery rate of approximately 5% and the resulting peptides were grouped into proteins.
Statistical analysis
Results of western blots and lipid kinase assay were quantified by densitometry analysis using Quantity One software (Bio-Rad). The data was analyzed by ANOVA and the pairwise comparisons were done by Bonferroni post-hoc test. Experiments were repeated at least three times for quantification. Immunostaining results were quantified by counting puncta formation and the statistical significance of differences between mean values (P < 0.05) was evaluated using the unpaired Student t test. At least three different visual fields containing at least 50 cells were counted for each condition. All data were expressed as the mean ± standard deviation.
Supplementary Material
Acknowledgments
We wish to thank Dr Joungmok Kim for critical discussion and suggestions on this study. Phosphosite identification by mass spectrometry was performed by the Proteomics Facility at the Fred Hutchinson Cancer Research Center. This work is supported by grants from NIH (KLG).
Glossary
Abbreviations:
- AA
amino acid
- Baf A1
bafilomycin A1
- KD
knockdown
- KO
knockout
- LC3
MAP1LC3
- MEFs
mouse embryonic fibroblasts
- MTOR
mechanistic target of rapamycin
- NR
nutrient-rich
- PIK3C3
type III phosphatidylinositol (PtdIns) 3-kinase, catalytic subunit
- PtdIns3P
phosphatidylinositol-3-phosphate
- RICTOR
RPTOR independent companion of MTOR, complex 2
- RPTOR
regulatory-associated protein of MTOR, complex 1
- ULK1
unc-51-like kinase
- UVRAG
UV radiation resistance associated
- WT
wild-type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/26058
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/26058
Reference
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–48. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 4.Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–54. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 9.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–6. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49:379–87. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–66. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–88. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–65. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–43. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–78. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 26.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–58. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.