Abstract
The paramyxoviruses represent a diverse virus family responsible for a wide range of human and animal diseases. In contrast to other viruses, such as HIV and influenza virus, which use a single glycoprotein to mediate host receptor binding and virus entry, the paramyxoviruses require two distinct proteins. One of these is an attachment glycoprotein that binds receptor, while the second is a fusion glycoprotein, which undergoes conformational changes that drive virus-cell membrane fusion and virus entry. The details of how receptor binding by one protein activates the second to undergo conformational changes have been poorly understood until recently. Over the past couple of years, structural and functional data have accumulated on representative members of this family, including parainfluenza virus 5, Newcastle disease virus, measles virus, Nipah virus and others, which suggest a mechanistic convergence of activation models. Here we review the data indicating that paramyxovirus attachment glycoproteins shield activating residues within their N-terminal stalk domains, which are then exposed upon receptor binding, leading to the activation of the fusion protein by a ‘provocateur’ mechanism.
Introduction
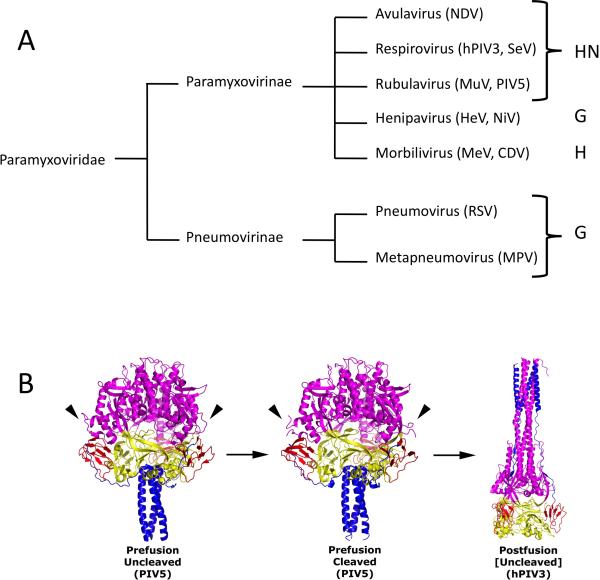
The Paramyxoviridae are enveloped, negative strand RNA viruses that infect both humans and animals [1]. The family is divided into two major subfamilies, the Paramyxovirinae and the Pneumovirinae (Figure 1) and includes, among others mumps virus, measles virus, respiratory syncytial virus (RSV), metapneumovirus (MPV), parainfluenza viruses (PIV) 1-5, and Newcastle disease virus (NDV). The paramyxoviruses are associated with significant health and economic burdens worldwide.
Figure 1. The attachment and fusion glycoproteins of the Paramyxoviridae.
(A) Pseudo phylogenetic tree of examples of the Paramyxoviridae. No evolutionary distance is implied by the lines. Genera are listed to the right and associated attachment glycoprotein abbreviations are indicated to the far right. NDV, Newcastle disease virus; HeV, Hendra virus; NiV, Nipah virus; MeV, measles virus; CDV, canine distemper virus; MuV, mumps virus; PIV5, parainfluenza virus 5; SeV, Sendai virus; PIV3, parainfluenza virus 3; RSV, respiratory syncytial virus; MPV, metapneumovirus. (B) Structure of distinct conformational states of the F protein ectodomain. Side-view ribbon diagrams of prefusion PIV5 F protein in its uncleaved (PDB ID code: 2B9B) [67] and protease-cleaved states (PDB ID code 4GIP) [7], as well as the uncleaved postfusion structure of the hPIV3 F ectodomain (PDB ID code: 1ZTM) [68]. Arrowheads indicate the position of protease cleavage sites in the prefusion structures for two of the three chains (the third is hidden from view). Arrows indicate the progression of F through its various conformational states. An exception for the uncleaved postfusion hPIV3 F structure is noted by square brackets as cleavage activation is a biological requirement for fusion activity, although not for refolding of the F ectodomain into the postfusion form. Each structure is color-coded by domain (DI, yellow; DII, red; DIII, magenta; HRB, blue). Reproduced from Welch and coworkers (2012) [7].
Paramyxovirus entry into cells requires the fusion of the virion envelope with a host cell membrane. For nearly all paramyxoviruses, membrane fusion is triggered at the cell surface in a receptor-dependent, pH-independent manner [1-3] and two viral glycoproteins mediate this process – an attachment protein referred to as HN, H, or G (HN/H/G), depending on the virus, and the fusion (F) glycoprotein [1,2]. The F proteins form trimers and are class I viral fusion proteins [1-6]. F initially folds to a metastable, pre-fusion conformation, and upon activation, it refolds to catalyze membrane fusion, juxtaposing the target cell and viral membranes (Figure 1). Proteolytic cleavage of F is required for fusion activity, but does not induce any major structural changes in the prefusion structure (Figure 1) [7].
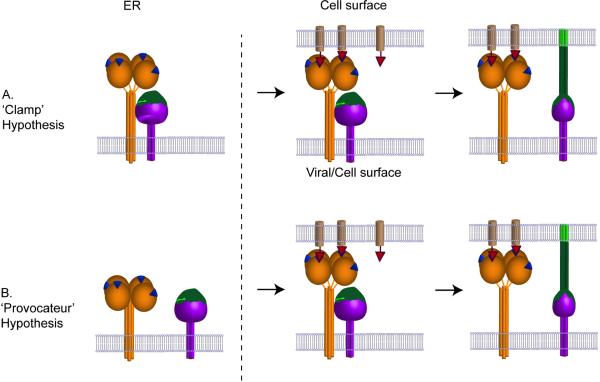
Most paramyxoviruses, excluding the Pneumovirinae subfamily (RSV and MPV) (Figure 1), require both F and the receptor binding attachment protein for membrane fusion [8-13]. Early studies suggested that F and the receptor binding protein have to physically interact for fusion to occur [9,12,14-16]. Two distinct models have arisen for the membrane fusion triggering mechanism, explaining how F is regulated to refold at the right time and right place.
In the “dissociation” or “clamp” model (Figure 2) the attachment protein retains F in its metastable, prefusion form. F and HN/H/G associate intracellularly, arriving at the cell surface as a complex to be incorporated into virions. When HN/H/G binds to its receptor, F is released and triggered to refold and drive membrane merger [15,17,18] (Figure 2).
Figure 2. Schematic diagram illustrating the ‘Clamp’ hypothesis and the ‘Provocateur’ hypothesis.
(A) In the ’Clamp’ hypothesis the attachment protein HN, (H or G) and the F protein interact during their transport through the exocytic pathway and at the cell or viral surface dissociate upon receptor engagement with the attachment protein, allowing the F protein to trigger in a manner analogous to a loaded spring. (B) In the ‘Provocateur’ hypothesis, the attachment protein and the F proteins do not associate inside the cell but do associate at the cell or viral surface, and dissociate after the attachment protein engages with the receptor. The protein-protein interaction between HN (H, G) and F triggers the F protein. The receptor binding protein is illustrated in orange as a globular head linked, via a flexible linker, to a four helix bundle stalk. The receptor binding site is shown as a blue triangle. The F protein is illustrated as a purple trimer with the domain that refolds in green. The receptor molecule is illustrated as a light brown cylinder with a red triangle as the attachment point. Potential changes in the HN/H/G stalk on receptor engagement are not illustrated.
In the second model, known as the “association” or “provocateur” model [19], HN/H/G actively triggers the metastable F protein by destabilizing it after receptor binding. In this model, HN/H/G is not required to stabilize the prefusion F conformation. HN/H/G could preassociate with, but not activate, F or alternatively, HN/H/G may only associate with F after receptor binding. A major difference between the clamp and provocateur models is that in the clamp model HN/H/G stabilizes prefusion F whereas in the provocateur model HN/H/G destabilizes prefusion F (Figure 2).
The interactions of F and HN/H/G have been extensively investigated. For measles virus and henipaviruses, mutant analyses have indicated an inverse correlation between fusion promotion and the strength of the F-H/G interaction [20-22]. These results have been interpreted to support the clamp model, although the data may be consistent with alternative explanations. For NDV, certain mutations that decrease fusion also decreased F–HN interactions [23]. However, other mutations in both measles virus and NDV attachment glycoproteins decrease fusion while retaining F binding [24-26]. To accommodate these somewhat diverse observations, it has been suggested that paramyxoviruses that use proteinaceous receptors have a different triggering mechanism from those that use sialic acid as a receptor.
One of the principal tenets of the provocateur hypothesis is that F can be expressed in its prefusion form without HN/H/G. This has been demonstrated by using monoclonal antibodies (mAbs) that specifically recognize prefusion or postfusion PIV5 F [19,27]. In addition, elevated temperatures convert prefusion PIV5 F to postfusion F in the absence of HN expression, with heat providing a surrogate trigger, in support of the provocateur hypothesis [19,27]. Recent data show that measles virus, canine distemper virus and henipavirus F are all expressed in the prefusion form in the absence of H or G and that heat converts prefusion to postfusion F [24,28,29]. These data indicate that the prefusion F conformation can be maintained in the absence of a clamp, supporting a generalizable provocateur model for paramyxoviruses.
Although the majority of available data supports a coalescence of activation models, two recent publications suggest that hPIV3 F is stabilized in its pre-triggered state as long as the HN interaction is prolonged [30,31]. However, the hPIV3 data do show that when F was coexpressed with influenza virus HA as an irrelevant receptor binding protein to tether effector and target cell membranes, over 70% the level of fusion of HN/F coexpression was observed [31]. This result raises the question of what triggered hPIV3 F: spontaneous activation at 37°C or interaction with an unknown receptor? In any case, the data demonstrate that functional, prefusion hPIV3 F is expressed without HN, consistent with the provocateur model.
Paramyxovirus attachment proteins show diversity in receptor binding.
The two major subfamilies of the Paramyxoviridae, the Paramyxovirinae and the Pneumovirinae (Figure 1), also delineate a structural division in the attachment proteins. The Paramyxovirinae subfamily attachment glycoproteins, HN/H/G, are structurally related, but engage different cellular receptors. Three genera (Rubulaviruses, Respiroviruses and Avulaviruses), which include mumps virus, PIV5, hPIV1-4 and NDV, encode a hemagglutinin-neuraminidase (HN) protein (Figure 1). HN both binds to and cleaves sialic acid receptors, promoting virus attachment during entry and budding after infection, respectively. Viruses in the morbillivirus (e.g. measles virus) and henipavirus (Nipah/Hendra) genera encode attachment proteins, H and G respectively, that are structurally related to HN, but which bind to protein receptors rather than sialic acid. By contrast, the Pneumovirinae, which includes RSV and MPV, encode a distinct attachment glycoprotein (G), which bears no apparent structural relationship to those in the Paramyxovirinae subfamily (Figure 1).
The Paramyxovirinae HN/H/G proteins are all type II integral membrane proteins, consisting of a membrane proximal stalk and a C-terminal globular receptor binding domain (RBD). The RBDs adopt a six-blade beta propeller fold characteristic of sialidases, with individual structural adaptations and variations (Figure 3). The glycoproteins form tetramers and share common functional characteristics [32-42]. Receptor engagement by the C-terminal RBD is thought to initiate conformational changes, leading to F activation, with F specificity encoded in the N-terminal stalk. Since the Pneumovirinae G proteins are not structurally related to the Paramyxovirinae HN/H/G proteins and are also not required for F activation, the Pneumovirinae subfamily entry mechanism appears to differ substantively.
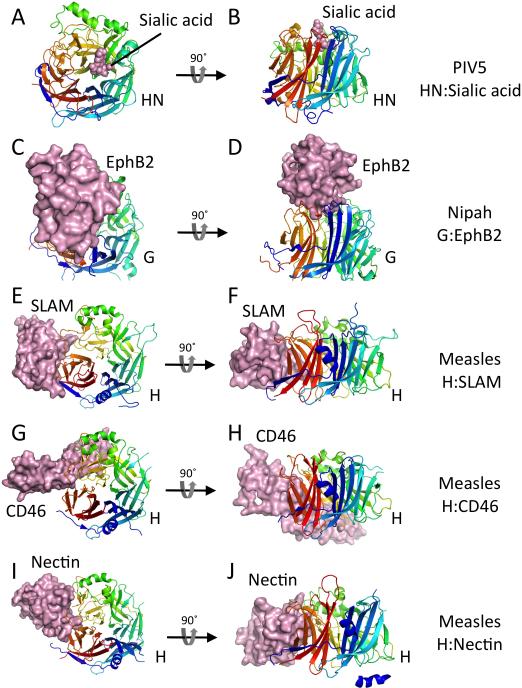
Figure 3. Receptor complexes of paramyxovirus attachment glycoprotein receptor binding domains.
For all panels, the receptor binding domains (RBDs) are shown in cartoon format, colored in a rainbow from N (blue) to C (red). The receptors are shown in solid surface format colored light pink. The RBDs all show a characteristic 6-bladed beta-propeller domain that is observed in sialidases and neuraminidases, although the Nipah virus G and measles virus H proteins do not bind sialic acid as their receptors. Pairs of panels are shown with two views of each receptor complex, related by a 90° rotation as indicated. (A, B) Two views of the PIV5 HN:sialylalactose complex (PDB ID code 1Z4X). (C, D) Two views of the Nipah virus G:EphB2 complex (PDB ID code 2VSM). (E, F) Two views of the measles virus H:SLAM complex (PDB ID code 3ALZ). (G, H) Two views of the measles virus H:CD46 complex (PDB ID code 3INB). (I, J) Two views of the measles virus H:nectin complex (PDB ID code 4GJT).
Crystal structures of receptor complexes of the RBDs from PIV5 HN, measles virus H and Nipah/Hendra virus G have been determined (Figure 3) [32,37,38,41-43]. HN binds sialic acid in a central pocket at one face of the beta propeller barrel, within an active site that conserves essential catalytic residues found in other neuraminidases. HN catalytic activity is reduced at neutral pH, enabling attachment and entry, but is activated at lower pH, enabling sialic acid removal during biosynthesis and budding. The Nipah virus G receptor is ephrin B2/3 (EphB2/3). The Nipah virus RBD also engages receptor in the central pocket of the beta propeller domain, using vestiges of the sialidase active site [32,37]. Measles virus H utilizes SLAM or nectin-4 as physiological receptors. A subset of measles virus variants adapted to using CD46 to trigger entry, likely as a result of passaging virus in tissue culture. In contrast to HN and henipavirus G, measles virus H engages all three protein receptors to the side of the beta propeller domain (Figure 3) [41-43].
The relationship between receptor binding and F activation is incompletely understood, but the receptor structures indicate that any general features of the F activation mechanism should be consistent with the different types of receptors (protein or carbohydrate), the different affinities of receptor engagement and the different geometries of the RBD:receptor complexes. Comparisons of unbound and bound RBDs indicate only minor structural changes upon engaging receptors. Measles virus entry can be reengineered by inserting a 6His tag into the RBD and utilizing an anti-his tag antibody as the entry receptor, or by appending other binding domains to H [44-46]. Thus any model for receptor dependent entry must take into account the ability of the attachment glycoproteins to recognize diverse input signals and convert these to a common output to activate F.
Structures of HN ectodomains indicate that RBDs shield the N-terminal stalk region involved in F activation.
Considerable evidence indicates that the stalk domains of paramyxovirus HN/H/G proteins directly interact with F [14,21,23,26,30,47-55]. However, until recently, no structural information was available on these domains.
Crystal structures of the NDV HN ectodomain [56], the PIV5 stalk domain by itself [47] and the PIV5 ectodomain [57] have provided insights into the stalk region, and unanticipated interactions with the globular RBDs (Figure 4). All three crystal structures show that the stalk forms a parallel 4-helix bundle (4HB). The PIV5 stalk and ectodomain structures show adjacent segments of undecad (11-mer) and heptad (7-mer) hydrophobic repeat regions, with a kink at the junction of the distinct coiled coil regions [47]. The stalk structures contain key residues defining an F binding and activation site (Figure 4) surrounding the kink. Mutations that block F activation map to the surface of the stalk and a subset have been shown to disrupt binding to F. Other stalk mutations block F activation, but retain F binding, suggesting that binding is necessary but insufficient for activation and that additional interactions between the stalk and F may be required for F triggering. These observations suggest that F activation may involve two steps, F binding followed by F activation (reviewed in ref. [13]).
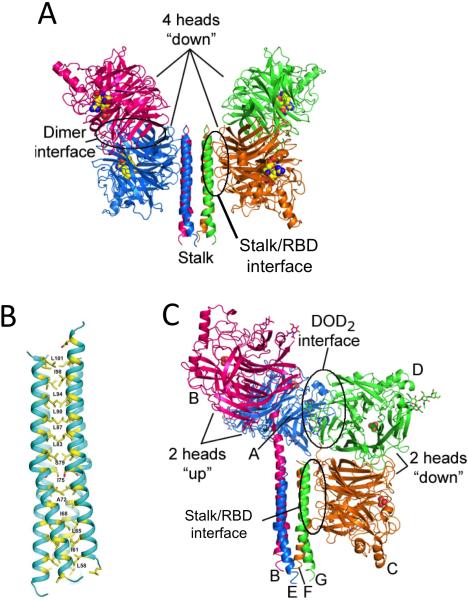
Figure 4. Structures of the NDV and PIV5 HN ectodomain and stalk regions.
(A) The NDV HN crystal structure (PDB ID: 3TIE) [56] representing the “4-heads-down” conformation is shown in cartoon format. Active site residues (E400, R415, and Y525) are shown as yellow, blue and red spheres. The locations of the second sialic acid binding sites are not shown. The stalk/RBD interface is circled and labeled. (B) Structure of the PIV5 HN 4HB stalk domain encompassing residues 56-108 (PDB ID code 3TSI) [47] . Residues in ‘a’ ‘d’ and ‘h’ positions of the hydrophobic core of the 4-helix bundle (yellow) are numbered. (C) Structure of the “2-heads-up/2-heads-down” conformation revealed by the PIV5-HN ectodomain crystal structure (PDB ID code 4JF7) [57]. The two dimers of RBD head domains (2-heads up and 2-heads down) are indicated.
The NDV and PIV5 HN crystal structures also revealed interactions between the RBD and stalk (Figure 4) [56,57]. In both, an extensive interface is observed between the base of an RBD beta propeller domain and the 4HB, partially covering residues implicated in F binding and activation. Pairs of HN RBDs associate as dimers, with only one of the RBDs interacting with the stalk (Figure 4). The arrangement of the NDV HN protein is 2-fold symmetric about the stalk, with RBD dimers flanking the 4HB, forming a ‘4-heads down’ state. The receptor binding sites point away from the stalk-RBD interface. By contrast the PIV5 HN structure reveals a hybrid, asymmetric conformation, with one pair of the RBDs arranged above the 4HB stalk, and the other pair arranged as in NDV HN (Figure 4). The PIV5 pair of RBDs in the ‘up’ conformational state reorient their receptor binding sites away from the stalk and TM domains, consistent with a reorientation to bind sialic acid receptors on target cells. The PIV5 HN conformation thus appears to represent a potential intermediate along the pathway from a pre-receptor bound ‘4-heads down’ conformation to a post receptor bound ‘4-heads up’ conformation. These structures suggested that receptor engagement could reorient the RBDs away from the stalk 4HB, thereby facilitating F engagement and activation in a receptor dependent manner. The conversion of the attachment glycoprotein from a ‘4-heads down’ to a ‘4-heads up’ conformation does not necessarily require receptor induced conformational changes within the globular RBD, but the reorientation away from the N-terminal stalk may be achieved by optimizing multivalent receptor interactions within the tetramer. This may explain the apparent flexibility in receptor specificity and binding site location on the RBD, highlighted by the variety of naturally occurring receptors, as well as retargeting studies with engineered measles virus H [44-46].
The stalk regions constitutively activate F mediated membrane fusion.
The HN ectodomain crystal structures raised the possibility that HN initially adopts an autoinhibited prefusion state, with inhibitory RBD-stalk interactions, suggesting that a truncated, ‘headless’ stalk lacking the RBDs might be constitutively activating. Consistent with this hypothesis, we showed that the PIV5 HN stalk (residues 1-117) is sufficient to activate F for fusion, indicating that the RBDs are dispensable for PIV5 F triggering [58]. The PIV5 HN stalk does not activate other non-cognate F proteins and mutants with N-linked carbohydrate sites on the outer surface of the PIV5 HN stalk ablate fusion, consistent with the inhibition of direct F-stalk interactions [58].
Two lines of evidence indicate that the expression of PIV5 HN stalk, in comparison to wt HN, more readily overcomes an energy barrier to fusion activation. First, the stalk triggers fusion to a greater extent at 33°C than wt HN at 33°C. Second, the stalk triggers fusion of the hypofusogenic mutant F-P22L [59], orders of magnitude more readily than wt HN. This suggests a distinct energy requirement of the PIV5 HN, possibly for the RBDs to undergo structural rearrangements. The HN stalk may be able to better activate F-P22L because of an unlimited dwell time for the interaction with F.
It has recently been shown that F mediated fusion can be similarly triggered by ‘headless’ stalk constructs from measles virus H [60], Nipah virus G [61], mumps virus HN and NDV HN (Bose et al., 2014 in preparation). Thus, constitutive stalk activation is a general property across HN/H/G receptor binding proteins. It remains unclear why only specific lengths of stalk activate F [58]; [60,61], why in some cases the stalk requires stabilization [60] and why the introduction of disulfide bonds either inhibits fusion or can stabilize the stalk [49,62,63], (Bose et al., 2014 in preparation).
Little is known concerning the region of F that interacts with the HN/H/G stalk, but recent studies implicate residues near the base of the prefusion measles virus F head [64]. Chimeric F proteins generated between PIV5 F and simian virus 41 F have also been used to map the HN interaction site [65], but the identified region is relatively large. Finally, multiple hydrophobic residues in the PIV5 F immunoglobulin-like (Ig-like) domain II may potentially interact with HN [66] and some of these residues map to the same general location on PIV5 F as identified for measles virus F [64,66].
A comprehensive stalk exposure model for fusion activation.
We previously proposed a simple model for paramyxovirus fusion activation [58], in which exposure of the F-activating stalk region is triggered by receptor binding. In this model, the attachment glycoprotein RBDs adopt a prefusion ‘4-heads down’ state and protect the F-activating stalk region. On binding receptor, the heads move to attain ‘up’ positions, fully exposing the stalk, allowing F to interact and triggering fusion as anticipated by the provocateur model.
The generality of this stalk exposure model is supported by three consistent sets of observations across multiple paramyxoviruses: (1) Two (NDV and PIV5) crystal structures show analogous RBD-stalk interactions near the F-interaction sites that could restrict F binding, (2) F proteins of many paramyxoviruses adopt a prefusion conformation in the absence of their attachment glycoproteins, and (3) ‘headless’ stalks from the attachment glycoproteins of many paramyxoviruses can constitutively activate their respective F proteins. These accumulating data are most consistent with a ‘provocateur’ model for F activation, but at odds with the ‘clamp’ model initially proposed for measles virus and henipaviruses. Importantly, mutagenesis studies have revealed that F binding can be separated from F activation, indicative of a two-step process for initiating membrane fusion, involving a step of F binding followed by a subsequent step of F activation mediated by distinct residues in the attachment glycoprotein stalk.
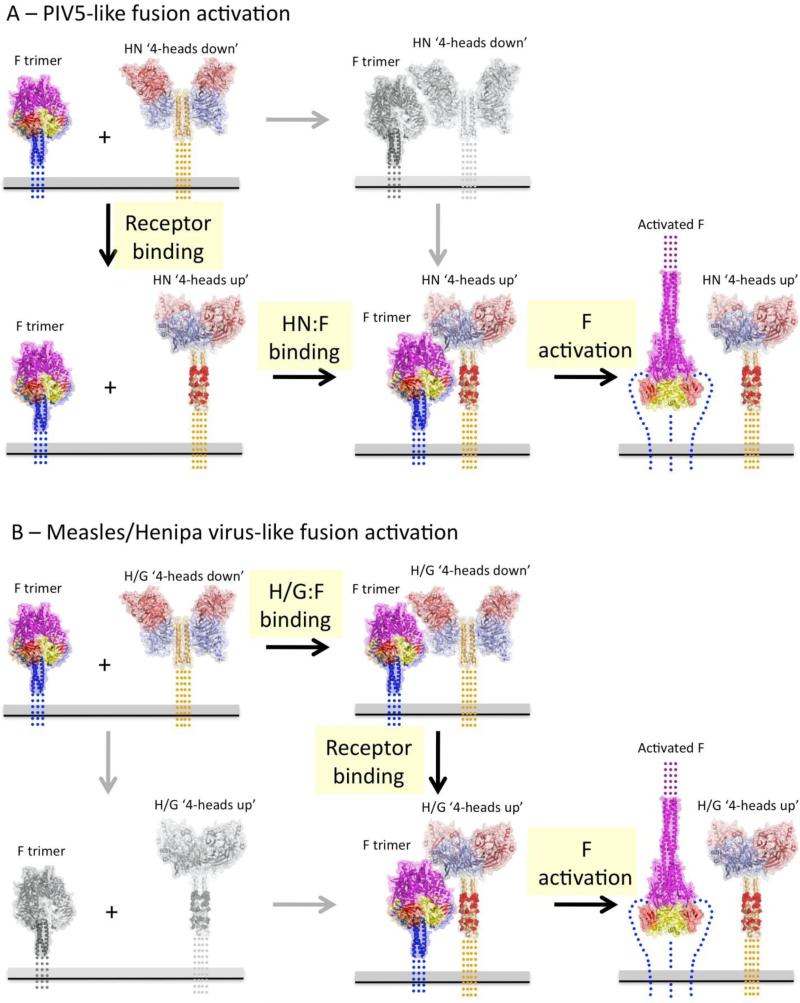
A possible reconciliation of the different models associated with different paramyxoviruses is shown in Figure 5. PIV5-like paramyxoviruses may exhibit no HN-F complex formation prior to receptor binding, which triggers exposure of the HN stalk region enabling the two steps of F binding and activation (Figure 5A). Measles or henipa-like viruses form H/G:F complexes prior to receptor binding, but these complexes may not trigger until RBDs reorient and fully reveal key activating residues of the stalk, following a distinct but related two-step pathway for membrane fusion (Figure 5B). In both of these model scenarios, engagement of the fully exposed stalk instigates the refolding of F, and this could conceivably involve stalk conformational changes as well. In summary, a convergence of structural and functional data indicate that receptor-binding by the paramyxovirus attachment glycoproteins leads to the exposure of N-terminal stalk residues that can constitutively bind and activate refolding of F to trigger membrane fusion, consistent with a ‘provocateur’ role for the stalk region that is regulated by interactions with the receptor binding domains.
Figure 5. Models for receptor-dependent fusion activation for paramyxoviruses within the Paramyxovirinae subfamily.
Current experiments indicate that the attachment glycoproteins from many paramyxoviruses within the Paramyxovirinae subfamily adhere to a general set of features. The attachment glycoproteins are structurally related and have a globular, C-terminal RBD that can interact with various receptors with diverse complex architectures. The attachment glycoprotein specificity for F activation lies within the N-terminal, tetrameric stalk and the ‘headless’ stalk domains constitutively activate F. The F proteins fold to a functional, prefusion state in the absence of the attachment proteins, indicating that a stabilizing clamp is not required for their function. The structures of two different attachment glycoprotein ectodomains show a stalk-RBD interaction that occludes residues implicated in F binding and activation. Stalk residues that are separately responsible for F protein binding and F protein activation have been identified, suggesting two distinct steps in F activation, which could be energetically different in different viruses. These data indicate that F binding is not absolutely required for F stabilization, disfavoring a clamp model, and that F binding Is necessary but insufficient for F activation. The data appear most consistent with a provocateur model, where residues in the HN/H/G stalk domain are specifically involved in destabilizing the prefusion F protein to undergo conformational changes that drive membrane fusion. Two related models that capture these features for PIV5-like and Henipa/measles virus-like activation pathways are shown in panels (A) and (B). The grey panels and arrows indicate low probability pathway steps based on available functional data. Red atoms in the post-receptor bound HN/H/G stalk represent F interacting and activating residues. ‘Activated” prefusion F is indicated by CPK atoms for residues that undergo large conformational changes to the post-fusion conformation (far right panels). (A) PIV5-like fusion activation involves weak or no interaction between prefusion F and prefusion HN proteins. Binding to receptors on target cells would fully expose the stalk region, allow F to bind and then to be activated at the appropriate time and place. (B) Henipa/measles virus-like fusion activation involves preformed complexes between prefusion F and H/G proteins that does not expose activating residues with the H/G stalk to F. Receptor binding would reorient the H/G RBDs, allowing full binding to the stalk and subsequent destabilization of prefusion F. The two pathways show significant functional conservation, as revealed by extensive comparative data that has been collected. Differences in the nature and stability of the prefusion HN/H/G complexes with F may reflect evolutionary differences in the strength of these interactions but these differences are not in and of themselves indicative of fundamentally different activation mechanisms or pathways. On the contrary, the accumulating data suggest an overall conserved mechanism of receptor-dependent activation across the Paramyxovirinae subfamily of the paramyxoviruses.
Highlights.
Competing ‘clamp’ and ‘provocateur’ models have been proposed as Paramyxovirus entry mechanisms.
Paramyxovirus attachment proteins contain receptor-binding (RBD) and stalk domains, which activate the fusion (F) protein.
Attachment protein RBD and stalk domains can interact, potentially regulating F activation in a receptor-dependent manner.
Headless stalk constructs of multiple attachment proteins, lacking RBDs, constitutively activate F mediated fusion.
Accumulating data favor a general ‘provocateur’ model for F protein activation.
Acknowledgements
We thank Dr. Sayantan Bose for helpful discussions. Work in the authors’ laboratories is supported in part by National Institutes of Health research grants AI-23173 (to R.A.L) and GM-61050 (to T.S.J.). R.A.L is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Sixth Edition Vol. 1. Lippincott Williams & Wilkins; 2013. pp. 957–995. [Google Scholar]
- 2.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell CJ, Luque LE. The structural basis of paramyxovirus invasion. Trends Microbiol. 2006;14:243–246. doi: 10.1016/j.tim.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 7.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl. Acad. Sci. USA. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison T, McQuain C, McGinnes L. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J. Virol. 1991;65:813–822. doi: 10.1128/jvi.65.2.813-822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Q, Hu X, Compans RW. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 1997;71:650–656. doi: 10.1128/jvi.71.1.650-656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heminway BR, Yu Y, Galinski MS. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 1994;31:1–16. doi: 10.1016/0168-1702(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 13.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future virology. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bousse T, Takimoto T, Gorman WL, Takahashi T, Portner A. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology. 1994;204:506–514. doi: 10.1006/viro.1994.1564. [DOI] [PubMed] [Google Scholar]
- 15.Sergel T, McGinnes LW, Peeples ME, Morrison TG. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 16.Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 17.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 18.Bossart KN, Fusco DL, Broder CC. Paramyxovirus entry. Adv. Exp. Med. Biol. 2013;790:95–127. doi: 10.1007/978-1-4614-7651-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J. Virol. 2009;83:10857–10868. doi: 10.1128/JVI.01191-09. [Description of the provocateur hypothesis. For PIV5 MAbs recognize prefusion F in the absence of HN expression. Heat used as a surrogate for HN triggering.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Betrolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 2006;80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop KA, Hickey AC, Khetawat D, Patch JR, Bossart KN, Zhu Z, Wang LF, Dimitrov DS, Broder CC. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J. Virol. 2008;82:11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 2002;76:5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Brindley MA, Takeda M, Plattet P, Plemper RK. Triggering the measles virus membrane fusion machinery. Proc. Natl. Acad. Sci. USA. 2012;109:E3018–E3027. doi: 10.1073/pnas.1210925109. [Stalk requires structural flexibility for activation of measles virus F. Only one dimer of H needs to bind receptor to trigger fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirza AM, Iorio RM. A mutation in the stalk of the NDV HN protein prevents triggering of the F protein despite allowing efficient HN-F complex formation. J. Virol. 2013;87:8813–8815. doi: 10.1128/JVI.01066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J. Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. USA. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Ader N, Brindley M, Avila M, Orvell C, Horvat B, Hiltensperger G, Schneider-Schaulies J, Vandevelde M, Zurbriggen A, Plemper RK, et al. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J. Virol. 2013;87:314–326. doi: 10.1128/JVI.01826-12. [Monoclonal antibodies that recognize CDV prefusion F recognize F at the cell surface in the absence of H expression. Heat can be used as a surrogate for H activation of fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Chan YP, Lu M, Dutta S, Yan L, Barr J, Flora M, Feng YR, Xu K, Nikolov DB, Wang LF, et al. Biochemical, conformational and immunogenic analysis of soluble trimeric forms of henipavirus fusion glycoproteins. J. Virol. 2012;86:11457–11471. doi: 10.1128/JVI.01318-12. [Monoclonal antibodies recognize Hendra and Nipah virus prefusion F when F is expressed without G. Heat can be used as a surrogate for G activation of fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porotto M, Devito I, Palmer SG, Jurgens EM, Yee JL, Yokoyama CC, Pessi A, Moscona A. Spring-loaded model revisited: paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 2011;85:12867–12880. doi: 10.1128/JVI.05873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porotto M, Salah ZW, Gui L, Devito I, Jurgens EM, Lu H, Yokoyama CC, Palermo LM, Lee K, Moscona A. Regulation of paramyxovirus fusion activation: the hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pre-triggered state. J. Virol. 2012;86:12838–12848. doi: 10.1128/JVI.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 33.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 34.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 35.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. USA. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 37.Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan P, Thompson T, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virius 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:1–13. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J. Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan P, Paterson RG, Leser GP, Lamb RA, Jardetzky TS. Structure of the Ulster strain Newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence. PLoS Pathog. 2012;8:e1002855. doi: 10.1371/journal.ppat.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat. Struct. Mol. Biol. 2013;20:67–72. doi: 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 43.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 44.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paraskevakou G, Allen C, Nakamura T, Zollman P, James CD, Peng KW, Schroeder M, Russell SJ, Galanis E. Epidermal growth factor receptor (EGFR)-retargeted measles virus strains effectively target EGFR- or EGFRvIII expressing gliomas. Mol. Ther. 2007;15:677–686. doi: 10.1038/sj.mt.6300105. [DOI] [PubMed] [Google Scholar]
- 46.Allen C, Vongpunsawad S, Nakamura T, James CD, Schroeder M, Cattaneo R, Giannini C, Krempski J, Peng KW, Goble JM, et al. Retargeted oncolytic measles strains entering via the EGFRvIII receptor maintain significant antitumor activity against gliomas with increased tumor specificity. Cancer Res. 2006;66:11840–11850. doi: 10.1158/0008-5472.CAN-06-1200. [DOI] [PubMed] [Google Scholar]
- 47.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four- helix bundle and the role of the stalk in fusion promotion. J. Virol. 2011;85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maar D, Harmon B, Chu D, Schulz B, Aguilar HC, Lee B, Negrete OA. Cysteines in the stalk of the Nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J. Virol. 2012;86:6632–6642. doi: 10.1128/JVI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk J, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 2012;287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. Probing the spatial organization of measles virus fusion complexes. J. Virol. 2009;83:10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 52.Deng R, Wang Z, Mahon PJ, Marinello M, Mirza A, Iorio RM. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology. 1999;253:43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 53.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stone-Hulslander J, Morrison TG. Mutational analysis of heptad repeats in the membrane- proximal region of Newcastle disease virus HN protein. J. Virol. 1999;73:3630–3637. doi: 10.1128/jvi.73.5.3630-3637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. USA. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [Atomic structure of HN ectodomain (heads plus stalk) of Newcastle disease virus show heads to be a dimer of dimers and the stalk is a 4-helix bundle. Two of the four head domains, one from each dimer, make contacts with the stalk. Head arrangement known as ‘four heads down’ conformation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 2013;9:e1003534. doi: 10.1371/journal.ppat.1003534. [Atomic structure of a PIV5 ectodomain in the ‘two heads up/two heads down’ conformation. One chain of tetrameric stalk shows connection with its head domain. The structure supports a model in which the heads of HN transition from down to up upon receptor binding thereby releasing steric constraints and facilitating the interaction between critical HN-stalk residues and F.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. USA. 2012;109:E2625–E2634. doi: 10.1073/pnas.1213813109. [The headless HN stalk that lacks the receptor binding domain, activates fusion. Provides evidence that expression of the HN stalk lowers the activation barrier for fusion. Proposes a simple model for paramyxovirus activation in which the receptor-binding protein stalk domain provides the primary trigger in fusion and the globular head domains undergo structural changes to regulate activation of fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 60**.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J. Virol. 2013;87:11693–11703. doi: 10.1128/JVI.01945-13. [The headless measles virus H protein activates F for fusion. Only some lengths of stalk activate F and they need to be stabilized by addition of a tetramerization domain. This leads to the suggestion that the 4-HB stalk changes conformation for fusion activation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides-Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS. Pathog. 2013:e1003770. doi: 10.1371/journal.ppat.1003770. [The headless Nipah virus G protein activates F for fusion. MAb reactivity data suggest two head conformational changes are required for exposure of the stalk.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Apte-Sengupta S, Navaratnarajah CK, Cattaneo R. Hydrophobic and charged residues in the central segment of the measles virus hemagglutinin stalk mediate transmission of the fusion-triggering signal. J. Virol. 2013;87:10401–10404. doi: 10.1128/JVI.01547-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J. Biol. Chem. 2012;287:38543–38551. doi: 10.1074/jbc.M112.410563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W. Cattaneo R: Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J. Biol. Chem. 2012;287:33026–33035. doi: 10.1074/jbc.M112.373308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsurudome M, Nakahashi M, Matsushima Y, Ito M, Nishio M, Kawano M, Komada H, Nosaka T. Full conversion of the hemagglutinin-neuraminidase specificity of the parainfluenza virus 5 fusion protein by replacement of 21 amino acids in its head region with those of the simian virus 41 fusion protein. J. Virol. 2013;87:8342–8350. doi: 10.1128/JVI.03549-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bose S, Heath CM, Shah PA, Alayyoubi M, Jardetzky TS, Lamb RA. Mutations in the parainfluenza virus 5 fusion (F) protein reveal domains important for fusion triggering and metastability. J. Virol. 2013;87:13520–13531. doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin H-S, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]