Abstract
Background
New drug regimens of greater efficacy and shorter duration are needed for tuberculosis (TB) treatment. The identification of accurate, quantitative, non-culture based markers of treatment response would improve the efficiency of Phase 2 TB drug testing.
Methods
In an unbiased biomarker discovery approach, we applied a highly multiplexed, aptamer-based, proteomic technology to analyze serum samples collected at baseline and after 8 weeks of treatment from 39 patients with pulmonary TB from Kampala, Uganda enrolled in a Centers for Disease Control and Prevention (CDC) TB Trials Consortium Phase 2B treatment trial.
Results
We identified protein expression differences associated with 8-week culture status, including Coagulation Factor V, SAA, XPNPEP1, PSME1, IL-11 Rα, HSP70, Galectin-8, α2-Antiplasmin, ECM1, YES, IGFBP-1, CATZ, BGN, LYNB, and IL-7. Markers noted to have differential changes between responders and slow-responders included nectin-like protein 2, EphA1 (Ephrin type-A receptor 1), gp130, CNDP1, TGF-b RIII, MRC2, ADAM9, and CDON. A logistic regression model combining markers associated with 8-week culture status revealed an ROC curve with AUC=0.96, sensitivity=0.95 and specificity=0.90. Additional markers showed differential changes between responders and slow-responders (nectin-like protein), or correlated with time-to-culture-conversion (KLRK1).
Conclusions
Serum proteins involved in the coagulation cascade, neutrophil activity, immunity, inflammation, and tissue remodeling were found to be associated with TB treatment response. A quantitative, non-culture based, five-marker signature predictive of 8-week culture status was identified in this pilot study.
Keywords: Tuberculosis, treatment response, biomarkers, proteomics, multiplex analysis, SOMAscan, logistic regression model
Introduction
A number of new chemical entities and repurposed antibiotics are under clinical development that when combined and optimized in dosing may lead to shorter, safer, and more effective TB regimens [1]. Establishing the efficacy of newer regimens, however, is a significant challenge [2–6]. Eight week sputum culture status is the most frequently used surrogate marker of treatment outcome. However, its ability to distinguish efficacy of regimens has come under scrutiny recently [7–12]. The discovery of a robust, quantitative, non-culture based biomarker or “biomarker signature” of treatment effect that can be measured early in treatment could speed up clinical development of new regimens and could potentially also be helpful for individual monitoring of patients on treatment [13, 14].
Using an unbiased, aptamer-based platform, we sought to identify and quantify protein markers associated with sputum culture status after 8 weeks of rifamycin-based combination therapy for active TB that might be a measure of treatment response. To this end, we analyzed data and specimens made available by a Phase 2B CDC-funded TB Trials Consortium (TBTC) clinical trial, Study 29, comparing the efficacy of rifampin 10 mg/kg/day versus rifapentine 10 mg/kg/day as part of the intensive phase of treatment. We previously cataloged and described our biomarker discovery efforts using these serum samples via the SOMAscan proteomics technology [15].
The SOMAscan assay uses slow off-rate modified aptamers (SOMAmers), which are ssDNA aptamers that contain pyrimidine residues carrying hydrophobic entities at their 5- position [16] and have been selected for slow dissociation rates. The affinity of these reagents is, on average, over an order of magnitude higher compared to simple RNA or DNA aptamers [17–19]. Measurements of proteins in SOMAscan have been previously validated by ELISA and by SOMAmer-based pull-down followed by LC-MS/MS [17]. Performance criteria have also been determined for all SOMAmers used in our study, including binding affinity constants, serum and plasma titrations, intra- and interrun %CV, and limits of quantitation [17, 18].
In this study, we incorporated all demographic, clinical, radiographic and microbiologic data available from the selected patients from Study 29 in addition to the measured 1030 human proteins to ascertain which markers and which marker combinations measured at baseline and at 8 weeks are closely associated with two established intermediate microbiologic endpoints currently used in Phase 2 clinical trials, namely culture status after 8 weeks of treatment and time-to-culture-conversion.
METHODS
Study Population
Patients were enrolled from CDC TBTC Study 29, a prospective, multicenter, open-label Phase 2B clinical trial (ClinicalTrials.gov Identifier NCT00694629) comparing efficacy and safety of standard TB therapy comprised of rifampin, isoniazid, pyrazinamide and ethambutol with rifapentine replacing rifampin [20]. The 39 participants included in this pilot project were all from the TBTC site based in Kampala, Uganda, were 18 years or older, sputum smear positive and HIV-uninfected. Author PN selected for inclusion in this study samples from patients free of significant co-morbidities reported at enrollment (December, 2008 to July, 2009), and who had relatively normal renal, hepatic and hematologic function. The study participants had signed a written informed consent and agreed to HIV testing. IRB approval for TBTC Study 29 was obtained from all participating institutions and from the Center for Disease Control and Prevention (CDC). Additionally, this pilot project was also approved by the Committee on Human Research of the University of California, San Francisco (H45279-34102-02A).
Study Design and Data Analysis
Serum was collected, processed and stored at baseline (time of enrollment), and after 8 weeks (40 doses) of intensive phase therapy using a standardized protocol. Efficacy of the regimens was assessed through determination of sputum culture status on both Lowenstein-Jensen (LJ) solid media and BACTEC Mycobacterial Growth Indicator Tube (MGIT, Becton Dickinson and Co., Franklin Lakes, NJ) liquid media with the MGIT 960 system. Of the 39 participants included in this case-control study, 19 were culture negative at completion of 8 weeks of treatment on both media types and were classified as “responders” (controls), whereas 20 participants who remained culture positive on either (or both) of the culture media were classified as “slow responders” (cases). Additionally, 25 of the 39 participants had been randomly assigned to the rifapentine arm and 14 to the rifampin arm, out of whom 4 received between 3–5 days of therapy prior to enrollment. All 39 pairs of baseline and end of intensive phase treatment (8-week) serum samples, respectively, were included in the proteomic assay to measure 1030 proteins. The investigators were blinded to all participant clinical, radiographic and microbiologic data until after all proteomic measurements were completed and submitted to the CDC. Additional information including total duration of treatment and end of treatment cure status was retrieved from patient charts by author GM in Kampala, Uganda.
Proteomic Methods
Proteomic measurements were performed at SomaLogic Inc., (Boulder, CO) in a single SOMAscan™ assay run that was performed as previously described [19, 21]. The SOMAmer reagents used for SOMAscan™ assay had been generated in vitro by a process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment) via multiple rounds of selection, partitioning, and amplification [17]. The version 2 SOMAscan assay used serum at three different concentrations (5%, 0.3%, 0.01%) to ensure the precise measurement of low-, medium-, and high-abundant proteins within the dynamic range of the assay. Established procedures to monitor known sample handling artifacts [22] were followed to assess the integrity of the provided serum samples. The SOMAmer-based proteomic assay is based on equilibrium binding in solution of fluorophore-tagged SOMAmers and proteins, automated capture of the SOMAmers that are in complexes with their cognate proteins [17], and hybridization to an antisense probe array (Agilent Technologies, Santa Clara, CA, USA). The fluorescent signal generated in the hybridization step is captured, and protein concentrations are reported in relative fluorescence units (RFU).
Statistical analysis
Matlab™ (www.mathworks.com/matlab) and the R environment for statistical computing (http://www.r-project.org/) were used for statistical analysis. Fisher’s exact test [23] was used to compare the proportions of radiographic findings in responders and non-responders. Linear regression analysis [24] was used to assess the association between protein levels measured in log RFU and culture conversion times. The non-parametric Kolmogorov-Smirnov (KS) test [23] was applied for unpaired comparisons of the protein distributions in rapid and slow-responders at baseline and week 8 separately. The KS statistic is an unsigned quantity, though we report a “signed” value to convey the directionality in the differential expression, that is, positive or negative KS distances for increased or decreased protein levels, respectively, in a given comparison of interest. The Wilcoxon rank sum test [23] was used to identify proteins with paired (within-subject) differential response between baseline and week 8 in the responders and slower responders respectively. Multiple comparison corrections were performed using the false discovery rate (FDR) methodology [25]. For each statistic we report both p-values and the associated FDR corrected “q-values” computed with the R package q-value [26]. Given the modest sample size in this pilot study we used a 30% false discovery rate threshold for reporting findings. Stability selection [27] using the randomized LASSO applied to a logistic regression model was used to identify proteins that distinguish responders from slow-responders with high probability over a wide range of regularization parameters. A final logistic regression model was re-fit (without regularization) to the markers with highest selection probability, and the resulting sensitivity and specificity when classifying subjects by treatment response was estimated using stratified cross-validation.
RESULTS
Basic demographic parameters and their correlation with treatment response
Ages of the 39 study participants ranged from 19 to 53 years, and 28 were males. The average body mass index (BMI) was 19.3 kg/m2, and 22 (56%) participants had cavitary disease, three of whom had bilateral cavities at baseline. The severity of disease had been assessed in detail from chest radiographs, reporting CXR class (1, absent; 2, < 4 cm; 3, > 4 cm) and CXR extent (A, limited; B, moderate; C, extensive) [15, 20].
Younger age was significantly associated with enhanced TB treatment response in this study (Figure 1A). Responders were younger than slow-responders (median age 25.7 yrs vs. 31.8 yrs, p=0.01). Though responders had slightly higher BMI than slow-responders (BMI of 19.7 vs 18.8), the difference was not significant (p>0.1), nor was the difference in quantitative culture analysis at baseline, determined as number of days to detection in liquid culture (p>0.1). Among radiographic parameters used to assess the degree or severity of TB infection, chest radiograph (CXR) extent differed between responders and slow-responders (p= 0.02), with twice as many responders having CXR extent B (moderate) and twice as many slow-responders having CXR extent C (extensive), as shown in Figure 1A.
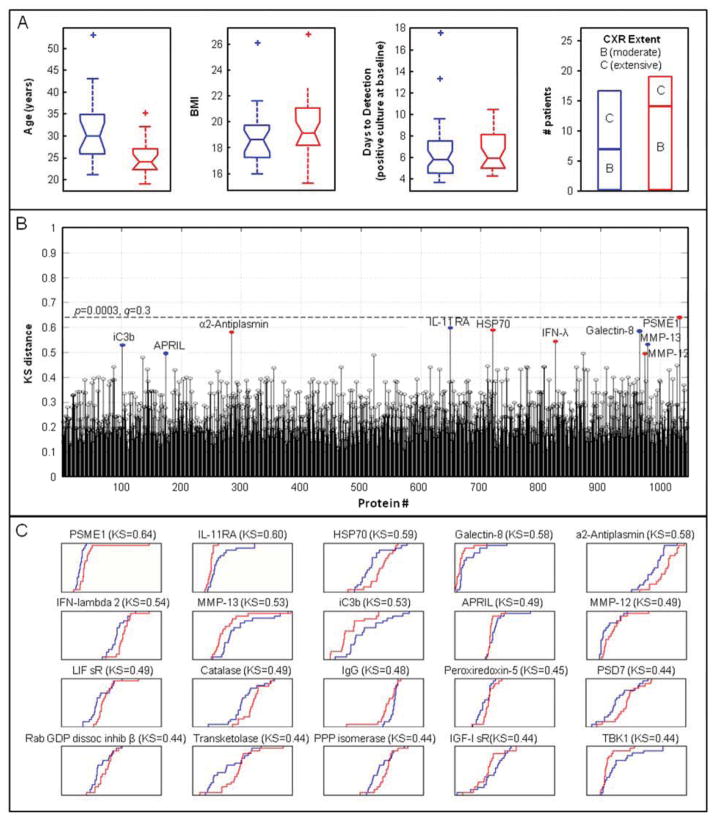
Figure 1.
Assessment of correlation of markers measured at baseline with treatment response. A, Box plots of patient demographic parameters in responders (red) and slow-responders (blue), depicting quartiles, medians, and outliers. B, KS distance plots of 1030 proteins measured in baseline samples from responders versus slow-responders. Colored dots mark the top ten proteins that are higher in responders (red) or slow-responders (blue). The dashed line indicates a 30% false discovery rate. C, Cumulative distribution function (CDF) plots of the most differentially expressed proteins in responders (red) versus slow-responders (blue) at baseline. Axis labels and scales for RFU (x axis) and for cumulative fraction of all samples within each group (y axis) were omitted for clarity.
Protein markers at baseline based on treatment response
Proteins that distinguish responders from slow-responders at baseline were identified using the Kolmogorov-Smirnov (KS) test. The KS distances were calculated for all 1030 proteins and are depicted in Figure 1B. Figure 1C shows plots of the empirical cumulative distribution functions of the relative fluorescence units (RFU) for the top 20 proteins with the largest KS distances between responders (red) and slow-responders (blue). Table 1 (upper half) shows the top ten proteins with the largest KS distances for the comparison between responders and slow-responders among all 1030 proteins. The top five proteins have a 29% false discovery rate indicating that we may expect one or two of these five to be false discoveries. Responders had higher levels than slow-responders of proteasome activator complex subunit 1 (PSME1), heat shock 70 kDa cognate protein 8 (HSP 70), α2-antiplasmin, interferon lambda 2 (IFN-λ), and matrix metalloproteinase 12 (MMP-12). In turn, slow responders had higher levels of interleukin 11 receptor antagonist (IL-11 Rα), Galectin-8, matrix metalloproteinase 13 (MMP-13), iC3b, and a proliferation inducing ligand of the TNF ligand family (APRIL) as compared to rapid responders.
Table 1.
Top 10 proteins distinguishing responders from slow-responders in comparisons of samples at baseline and at 8 weeks, ranked according to their KS distance.
| Analysis | Protein | SwissProt | Signed KSa | p-value | q-value |
|---|---|---|---|---|---|
| Baseline | |||||
| 1 | PSME1 | Q06323 | 0.639 | 0.000307 | 0.29 |
| 2 | IL-11 Rα | Q14626 | −0.597 | 0.00094 | 0.29 |
| 3 | HSP 70 | P11142 | 0.589 | 0.00115 | 0.29 |
| 4 | Galectin-8 | O00214 | −0.584 | 0.00131 | 0.29 |
| 5 | α2-Antiplasmin | P08697 | 0.582 | 0.0014 | 0.29 |
| 6 | IFN-lambda 2 | Q8IZJ0 | 0.545 | 0.00342 | 0.60 |
| 7 | MMP-13 | P45452 | −0.532 | 0.00463 | 0.64 |
| 8 | iC3b | P01024 | −0.529 | 0.00492 | 0.64 |
| 9 | APRIL | O75888 | −0.495 | 0.0104 | 0.99 |
| 10 | MMP-12 | P39900 | 0.495 | 0.0104 | 0.99 |
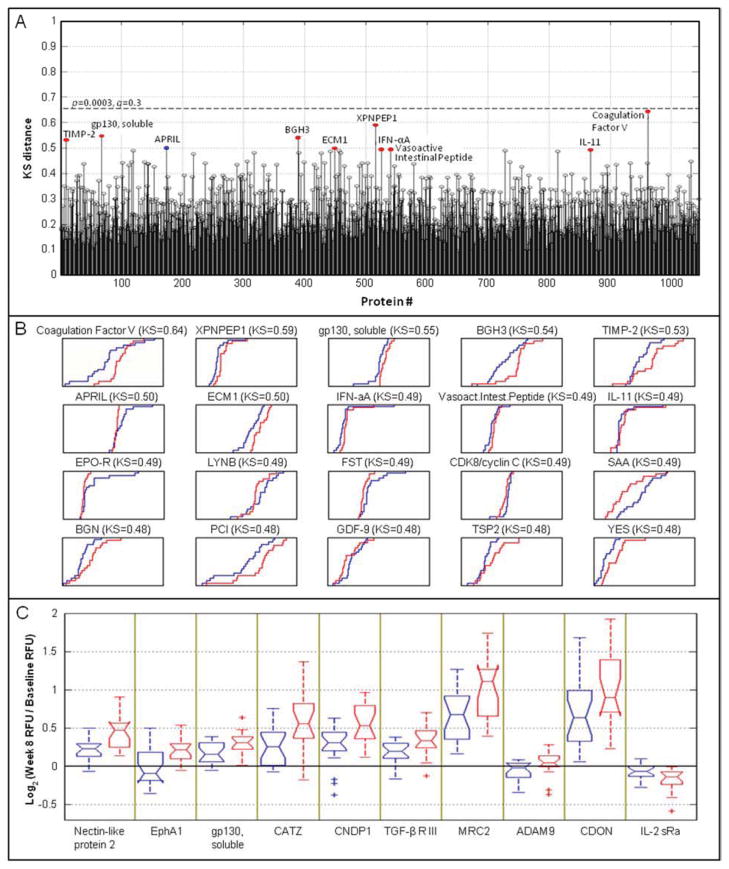
| 8 weeks | |||||
| 1 | Coagulation Factor V | P12259 | 0.645 | 0.000266 | 0.28 |
| 2 | XPNPEP1 | Q9NQW7 | 0.589 | 0.00115 | 0.60 |
| 3 | gp130, soluble | P40189 | 0.547 | 0.00321 | 0.80 |
| 4 | BGH3 | Q15582 | 0.539 | 0.00386 | 0.80 |
| 5 | TIMP-2 | P16035 | 0.532 | 0.00463 | 0.80 |
| 6 | APRIL | O75888 | −0.500 | 0.00932 | 0.80 |
| 7 | ECM1 | Q16610 | 0.500 | 0.00932 | 0.80 |
| 8 | IFN-αA | P01563 | 0.495 | 0.0104 | 0.80 |
| 9 | Vasoactive Intestinal Peptide | P01282 | 0.495 | 0.0104 | 0.80 |
| 10 | IL-11 | P20809 | 0.492 | 0.011 | 0.80 |
Positive KS values indicate higher protein levels in responders than slow-responders.
Protein markers at 8 weeks based on treatment response
The KS distances between responders and slow-responders were determined for all proteins measured in the 8-week samples (Figure 2A), and cumulative distribution function plots (Figure 2B) are shown for the top 20 proteins with the largest KS distances between responders (red) and slow-responders (blue). Table 1 (bottom half) show the proteins that exhibited the largest differential expression between responders and slow-responders at week 8 based on the KS distances. Coagulation Factor V showed the most significant difference and was elevated in responders compared to slow-responders. Xaa-Pro aminopeptidase 1 (XPNPEP1), soluble gp130, transforming growth factor-beta-induced protein ig-h3 (BGH3), metalloproteinase inhibitor 2 (TIMP-2), extracellular matrix protein 1 (ECM-1), vasoactive intestinal peptide (VIP), interferon alpha-2 (IFN-αA), IL-11 were also elevated in responders compared to slow-responders. Of the other proteins listed in that section of Table 1, only tumor necrosis factor ligand superfamily member 13 (APRIL) was lower in responders than slow-responders.
Figure 2.
Protein markers at 8 weeks based on treatment response. A, KS distance plots of all 1030 proteins measured in 8-week samples from responders versus slow-responders. Colored dots mark the top ten proteins that are higher in responders (red) or slow-responders (blue). The dashed line indicates a 30% false discovery rate. B, Cumulative distribution function (CDF) plots of the most differentially expressed proteins in responders (red) versus slow-responders (blue) at 8 weeks of TB treatment. Axis labels and scales for RFU (x axis) and for cumulative fraction of all samples within each group (y axis) were omitted for clarity. C, Box plots for the log2 ratio of week 8 to baseline signal in responders (red) and slow-responders (blue).
A paired analysis of responders and slow-responders was conducted using the log-ratio of within-subject week 8-to-baseline response as a “fold change” metric (Figure 2C). This analysis targets proteins that exhibit differential change between the two time points in the responders (red) compared to slow-responders (blue). Proteins were subsequently ranked using the Wilcoxon rank sum test to identify those with different median fold changes in the responders and slow-responders. The ten proteins with the most differential change are listed in Table 2. For the first nine of these proteins, the fold-change in signal from baseline to week 8 was larger in responders than in slow responders. These features were nectin-like protein 2, EphA1 (Ephrin type-A receptor 1), gp130, CATZ, CNDP1, TGF-b RIII, MRC2, ADAM9, and CDON. IL-2 sRa was the only protein that decreased in both groups, but decreased to a greater extent in responders compared to slow-responders.
Table 2.
Top 10 proteins distinguishing responders from slow-responders in comparisons of samples at baseline, using the 8 weeks-to-baseline log2-ratios, ranked according to their Rank Sum Z-score.
| Rank | Protein | SwissProt | Rank Sum Z score b | p-value | q-value |
|---|---|---|---|---|---|
| 1 | Nectin-like protein 1 | Q9BY67 | 3.301 | 0.00096 | 0.37 |
| 2 | EphA1 | P21709 | 3.077 | 0.0021 | 0.65 |
| 3 | gp130, soluble | P40189 | 2.852 | 0.0044 | 0.65 |
| 4 | CATZ | Q9UBR2 | 2.768 | 0.0057 | 0.65 |
| 5 | CNDP1 | Q96KN2 | 2.627 | 0.0086 | 0.65 |
| 6 | TGF-β R III | Q03167 | 2.599 | 0.0094 | 0.65 |
| 7 | MRC2 | Q9UBG0 | 2.515 | 0.012 | 0.65 |
| 8 | ADAM 9 | Q13443 | 2.459 | 0.014 | 0.76 |
| 9 | CDON | Q4KMG0 | 2.459 | 0.014 | 0.76 |
| 10 | IL-2 sRα | P01589 | −2.430 | 0.015 | 0.76 |
Positive rank sum values indicate a larger differential change between the two time points in the responders compared to slow-responders.
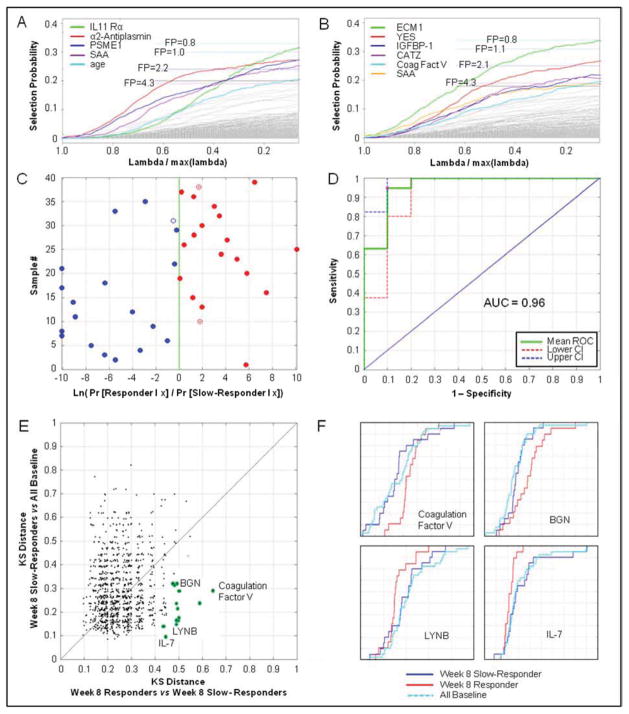
The association of treatment response with combinations of markers
In this pilot study we explored several strategies for generating signatures of treatment response, specifically measuring the association of serum protein measurements with the culture status at 8 weeks. In a first approach we sought the best combination of protein measurements and clinical covariates to classify slow-responders from responders. Stability selection was used to identify a subset of covariates from the set of 1030 protein measurements combined with age, gender, BMI, smear status, CXR class, CXR extent of disease, and time to detection after inoculation in liquid culture. The most stable predictive markers at baseline were IL-11 Rα, α2-Antiplasmin, PSME1, SAA, and subject age (Figure 3A). Three of the proteins are among those with large KS distances between responders and slow-responders as mentioned above, and the associated q-values suggested on average at least 1 of these was falsely discovered. This assessment is consistent with the average number of false discoveries expected by stability selection at different selection probabilities (Figure 3A, dashed lines). At eight weeks, the most highly associated stable markers with treatment response were ECM1, YES, IGFBP1, CATZ, Coagulation Factor V, and SAA (Figure 3B).
Figure 3.
Models and algorithms to “predict” treatment response at week 8. A, Stability paths for L1-regularized logistic regression using randomized lasso (weakness=0.25, W=0.9) applied to combination of baseline measurements and clinical covariates to classify responders from non-responders. Dashed lines indicate number of false positive (FP) discoveries at different selection probability thresholds computed from class-randomized observations. B, Stability paths for L1-regularized logistic regression of 8-week measurements and clinical covariates to classify responders from non-responders. C, Training sample classification based on (log) odds ratio produced by logistic regression model using five markers (IL-11 Rα, α2-Antiplasmin, PSME1, SAA, and subject age) measured at baseline. Red solid dots represent true positive classifications (responders), blue solid dots are true negative classifications (slow-responders); open dots are false positive or false negative results. D, ROC curve and point-wise 95% CI for training samples, showing AUC=0.96 and bootstrapped 95% CI (0.88, 0.99). E, Scatter plot of the KS distances of slow-responders (week 8) to baseline (all) versus responders (week 8) to slow-responders (week 8). Potential treatment response markers fall into the lower right area. F, CDF plots of representative candidate treatment response markers identified via KS distance; this metric illustrates that week 8 responder samples are distinctly different from week 8 slow-responder samples and baseline samples.
As an example of a five-marker signature to associate with treatment response, baseline measurements of IL-11 Rα, α2-Antiplasmin, PSME1, and SAA were combined with subject age in a logistic regression model. The corresponding sample classification (Figure 3C) and resulting ROC curve (Figure 3D) show the performance of this model on the training data. Since there were too few samples in this pilot study to withhold an independent “test set”, we used 5-fold stratified cross-validation to estimate model performance. Under cross-validation, the estimated AUC was 0.8±0.06 with sensitivity 0.8±0.11 and specificity 0.8±0.07. Similar performance was observed from a naive Bayes model constructed using baseline measurements of the first 5 proteins in Table 1 with the best KS distances (PSME1, IL-11 Rα, HSP70, Galectin-8, and α2-Antiplasmin).
The final analysis looked at markers that changed over the course of active therapy only in the responders, while their levels in the slow-responders remained the same as at baseline. In a scatter plot of the KS distances for the comparison of week-8 slow-responders to all baseline samples vs. the KS distances for the comparison of week-8 responders to slow-responders, markers associated with treatment response congregate in the lower right area (Figure 3E). Many of the proteins identified above were confirmed, such as Coagulation Factor V, XPNPEP1, YES, vasoactive intestinal peptide, and ECM1, but additional markers were found that distinguished responders from slow-responder in this analysis, including BGN (matrix proteoglycan), LYNB (tyrosine kinases), and IL-7. Empirical cumulative distribution functions for these markers illustrate that the levels of these proteins track closely together in baseline and week-8 slow-responders, but are clearly different in week-8 responders (Figure 3F).
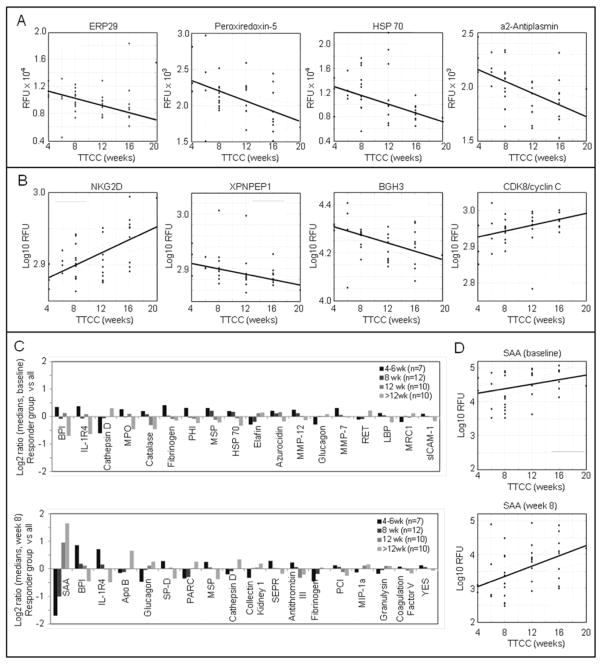
Markers associated with time-to-culture-conversion
Toward the identification of surrogate markers for treatment response, meta data and serum protein data were analyzed with regard to time-to-culture-conversion (TTCC), defined as the first of at least two consecutive time-points where negative cultures (solid and liquid) were obtained. Among the 39 participants there were six responder groups, with TTCC of 4 weeks (n=3), 6 weeks (n=4), 8 weeks (n=12), 12 weeks (n=10), 16 weeks (n=9), and 20 weeks (n=1). TTCC did not correlate with clinical data obtained at baseline, such as smear/culture results, chest X-ray classifications, presence of cavities, smoking status, or BMI (data not shown). Univariate regression analysis of baseline and week-8 protein measurements (log RFU) on TTCC was performed and sorted by statistical significance (Table 3). At baseline, lower levels of ERP29, peroxiredoxin-5, HSP-70, and α2-antiplasmin were associated with longer TTCC (Figure 4A). At 8 weeks, NKG2D (KLRK1) and CDK8 showed increased levels in samples from participants with longer TTCC, while XPNPEP1, and BGH3 (TGFBI) levels were lower (Figure 4B). Comparison of the medians of all 8-week measurements within the different responder groups showed a large number of proteins associated with neutrophil function (Figure 4C). BPI (bactericidal permeability-increasing factor) and IL-1 R4 were higher in fast-responders compared to slow-responders, and cathepsin D was lower in fast-responders compared to slow-responders. The largest differences between the responder groups were for SAA measured at 8 weeks, and a more detailed regression analysis using baseline data and week-8 data showed that signals dropped from baseline to week 8 by almost 10-fold, but a much sharper decrease was observed in samples from the fast-responders (Figure 4D).
Table 3.
Top 10 proteins at baseline and at 8 weeks that correlate with TTCC in univariate regression of log10 RFU on TTCC (measured in weeks), ranked by R2.
| Analysis | Protein | SwissProt | Slope a | R2 | p-value | q-value |
|---|---|---|---|---|---|---|
| Baseline | ||||||
| ERP29 | P30040 | −0.012 | 0.30 | 0.00037 | 0.39 | |
| Peroxiredoxin-5 | P30044 | −0.0078 | 0.24 | 0.0018 | 0.69 | |
| HSP 70 | P08107 | −0.016 | 0.23 | 0.002 | 0.69 | |
| α2-Antiplasmin | P08697 | −0.0058 | 0.21 | 0.0035 | 0.90 | |
| RANTES | P13501 | 0.0059 | 0.19 | 0.0051 | 1.00 | |
| IgG | P01857 | 0.0063 | 0.19 | 0.0091 | 1.00 | |
| Transketolase | P29401 | −0.01 | 0.17 | 0.0095 | 1.00 | |
| NKG2D (KLRK1) | P26718 | 0.0041 | 0.17 | 0.0096 | 1.00 | |
| Coagulation Factor V | P12259 | −0.0089 | 0.16 | 0.012 | 1.00 | |
| Coagulation Factor IX | P00740 | −0.0056 | 0.15 | 0.014 | 1.00 | |
| 8 weeks | ||||||
| NKG2D (KLRK1) | P26718 | 0.0043 | 0.26 | 0.00081 | 0.65 | |
| XPNPEP1 | Q9NQW7 | −0.0024 | 0.32 | 0.0012 | 0.65 | |
| BGH3 (TGFBI) | Q15582 | −0.0085 | 0.22 | 0.0026 | 0.71 | |
| CDK8/cyclin C | P49336 | 0.0041 | 0.23 | 0.0027 | 0.71 | |
| SAA | P02735 | 0.076 | 0.19 | 0.0051 | 0.83 | |
| Coagulation Factor V | P12259 | −0.0069 | 0.19 | 0.0053 | 0.83 | |
| YES | P07947 | −0.0038 | 0.19 | 0.0059 | 0.83 | |
| PARC | P55774 | 0.019 | 0.17 | 0.0095 | 0.83 | |
| CD39 | P49961 | 0.0031 | 0.17 | 0.011 | 0.83 | |
| LRIG3 | Q6UXM1 | −0.0055 | 0.16 | 0.011 | 0.83 | |
Linear model coefficient gives change in log RFU signal per week.
Figure 4.
Association of serum protein levels with TTCC. A, Regression of baseline protein data (log10 RFU) on TTCC. B, Regression of week-8 protein data (log RFU) on TTCC. C, Differential expression of proteins based on the medians of the responder groups at baseline (top) and at 8 weeks (bottom). D, Regression of SAA data (log10 RFU) on TTCC at baseline (top) and at 8 weeks (bottom).
DISCUSSION
In this pilot study, we identified a number of proteins that differed between treatment slow-responders and responders, at baseline or after 8 weeks of TB treatment. Serum amyloid A (SAA) protein was strongly associated with treatment response in multiple analyses performed. Many proteins involved in innate and adaptive immunity were differentially expressed, including gp-130, TNF pathway molecules, complement components, catalase, IgG, IFN-λ, PSME, and PSD7. At baseline, the strongest association of a marker with treatment response was PSME, an IFN-γ-inducible component of the immunoproteasome. Levels of this protein are increased under conditions of intensified immune response and are important for efficient antigen processing [28]. IL-11 Rα is a receptor for IL-11 and uses the high affinity gp130-transducing domain, which also appeared in both the baseline and week 8 data, and both are acute phase response proteins [29]. APRIL [30], a TNF family ligand, is involved in TGF-β signaling, and has been shown to have a role in the response to pathogens [31]. Both TGF-β and TNF are important cytokines in the immune response to TB [10, 32, 33]. APRIL has also been shown to be involved in promoting T-cell proliferation and survival [34]. MMP-12 and MMP-13 were differentially expressed at baseline. The major substrate for MMP-12 is elastin, an important constituent of lung connective tissue. MMP-12 appears to have a role in progression of lung diseases in which there is turnover of extracellular matrix components, and MMPs have been shown to be key mediators in TB pathology [35, 36]. Matrix proteoglycan (BGN) and BGH3 may also be involved in extracellular matrix and tissue remodeling [37]. Proteins involved in amyloids/fibrils (BGH3) and potentially HSP70 deserve greater attention and may have to do with the makeup of the TB lesions and can change with therapy. XPNPEP1 is a metalloaminopeptidase involved in the degradation of neuropeptides and the finding of VIP in these analyses is intriguing. Coagulation Factor V was the strongest differentially expressed marker at 8 weeks between responders and slow-responders and may suggest either better protein calorie nutrition in responders [38] or that tissue remodeling, changes in fibrinolysis and the resolution of pulmonary TB lesions has connections with the coagulation cascade that have not been described previously [39, 40].
In this study, we explored several mathematical models for the prediction of 8 week culture status. A logistic regression model using four features obtained during our measurements of serum protein levels at baseline together with subject age performed accurately in sample classification and resulted in an ROC curve with AUC=0.96. Similar performance was observed for a model containing the top five serum protein markers at baseline based on KS distances. Separately, we also selected the top markers at 8 weeks based on large KS distances (≥0.5), and constructed a five-feature (Coagulation Factor V, XPNPEP1, gp130, TIMP-2 and ECM1) naïve Bayes classifier to “predict” treatment response, which revealed an ROC curve with an AUC was 0.8±0.06. Given the small sample size of this pilot, such models are at risk for “over-fitting” and they need to be tested in properly designed validation studies using independent sample test sets to confirm their performance. Our attempt to correlate serum protein measurements with time to culture conversion showed limited significance, but corroborated some of the markers found in previous analyses and several other proteins linked to neutrophil function. While the presence of bilateral cavitary disease, or high sputum bacillary load as assessed by rapid time-to-detection on initial sputum cultures are reported predictors of a longer conversion time [41, 42], we did not find such a correlation in our sample set.
Our study has several limitations. We did not have additional time points or samples obtained earlier during treatment (i.e., at weeks 2, 4 or 6) and hence may have missed early changes in biomarker concentrations that may have stabilized by the 8-week time point. In addition, we recognize that culture status at 8 weeks is a less than perfect surrogate marker for treatment response or durable cure [7]. Patients who ultimately relapse can be culture negative at 2 months [12, 47, 48] and many of those who are culture positive at 2 months are ultimately cured, as was observed with all 39 patients in our study. We also cannot comment on markers previously linked with TB but not present on our array, and these include serum CA-125 [43, 44] and transthyretin [45]. Lastly, given the small sample size, false discovery rates exceed 25% in all analyses. Such high rates are not uncommon in small pilot studies –rather they are indicative of the challenges associated with large-scale statistical hypothesis testing in the presence of small effect sizes. It remains to be seen if serum protein signatures identified in this work will improve upon simple clinical measures of treatment response like extent of radiographic abnormalities, age, and BMI [46]. Moreover, it is also plausible that some of the biomarkers discovered in our study may be associated with baseline factors predicting conversion, rather than the outcome itself. Additional discovery efforts using larger sample sets will address these issues, as it will allow for analyses adjusted for potential confounders as well as permit using lower false discovery rate thresholds for reporting. Despite these limitations our data contribute to further understanding of the complexity of changes occurring during anti-TB treatment and provide us with biomarkers that warrant further investigation for the prediction of treatment response.
In conclusion, our study has identified biomarkers predictive of the currently established surrogate endpoint for phase 2 TB trials, and has also highlighted the complexity of proteomic changes accompanying TB treatment. Studies that follow patients long term to capture clinical endpoints of interest, namely treatment failure and relapse, along with additional serial time points for serum collection, will be needed to fully scrutinize the predictive capabilities of the biomarkers discovered.
Acknowledgments
We thank the TB patients enrolled in Study 29 and the study coordinators and staff at the CDC TBTC and in Uganda for their contributions. We thank Drs. Susan Dorman, Richard Chaisson and Neil Schluger, Chair and Co-Chairs of Study 29, as well as the Study 29 Protocol Team. We thank the CDC TBTC Biomarker Working Group for their critical input into the development of the TBTC serum storage facility. We also thank our SomaLogic colleagues that contributed to this study (in particular S. Williams, R. Ostroff, and M. Messenbaugh), and the SomaLogic scientists who performed the SOMAscan assay and developed the tools for this technology.
Financial Support. Funding for recruitment, enrollment, and clinical and laboratory follow-up of TBTC Study 29 participants was provided by the United States Centers for Disease Control and Prevention. The work of P.N. is supported by the National Institutes of Health through National Institute of Allergy and Infectious Diseases funding (1R01AI104589), and by the Centers for Disease Control and Prevention TB Trials Consortium. The entire sample analysis was funded by SomaLogic, Inc. No additional external funding was received for this study.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Potential conflicts of interest. UAO, NJ, and DS are employees and shareholders of SomaLogic. MAD is a paid SomaLogic consultant.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov. 2013;12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 2.Dodd LE, Proschan MA. Innovative Trial Designs to Improving Tuberculosis Drug Development. J Infect Dis. 2013;207:544. doi: 10.1093/infdis/jis704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsberg AM. Tuberculosis drug development: progress, challenges, and the road ahead. Tuberculosis (Edinb) 2010;90:162–7. doi: 10.1016/j.tube.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Lienhardt C, Vernon A, Raviglione MC. New drugs and new regimens for the treatment of tuberculosis: review of the drug development pipeline and implications for national programmes. Curr Opin Pulm Med. 2010;16:186–93. doi: 10.1097/MCP.0b013e328337580c. [DOI] [PubMed] [Google Scholar]
- 5.Spigelman M, Woosley R, Gheuens J. New initiative speeds tuberculosis drug development: novel drug regimens become possible in years, not decades. Int J Tuberc Lung Dis. 2010;14:663–4. [PubMed] [Google Scholar]
- 6.Wallis RS. Sustainable tuberculosis drug development. Clin Infect Dis. 2013;56:106–13. doi: 10.1093/cid/cis849. [DOI] [PubMed] [Google Scholar]
- 7.Burman WJ. The hunt for the elusive surrogate marker of sterilizing activity in tuberculosis treatment. Am J Respir Crit Care Med. 2003;167:1299–301. doi: 10.1164/rccm.2302003. [DOI] [PubMed] [Google Scholar]
- 8.Chakera A, Lucas A, Lucas M. Surrogate markers of infection: interrogation of the immune system. Biomark Med. 2011;5:131–48. doi: 10.2217/bmm.11.17. [DOI] [PubMed] [Google Scholar]
- 9.Nahid P, Saukkonen J, Mac Kenzie WR, et al. CDC/NIH Workshop. Tuberculosis biomarker and surrogate endpoint research roadmap. Am J Respir Crit Care Med. 2011;184:972–9. doi: 10.1164/rccm.201105-0827WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemeth J, Winkler HM, Boeck L, et al. Specific cytokine patterns of pulmonary tuberculosis in Central Africa. Clin Immunol. 2011;138:50–9. doi: 10.1016/j.clim.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Wallis RS. Surrogate markers to assess new therapies for drug-resistant tuberculosis. Expert Rev Anti Infect Ther. 2007;5:163–8. doi: 10.1586/14787210.5.2.163. [DOI] [PubMed] [Google Scholar]
- 12.Wallis RS, Doherty TM, Onyebujoh P, et al. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis. 2009;9:162–72. doi: 10.1016/S1473-3099(09)70042-8. [DOI] [PubMed] [Google Scholar]
- 13.Wallis RS, Kim P, Cole S, et al. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013;13:362–72. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim PS, Makhene M, Sizemore C, Hafner R. Viewpoint: Challenges and opportunities in tuberculosis research. J Infect Dis. 2012;205:S347–352. doi: 10.1093/infdis/jis190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeGroote MA, Nahid P, Jarlsberg L, et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. Plos One. 2013;8:e61002. doi: 10.1371/journal.pone.0061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaught JD, Bock C, Carter J, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–51. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 17.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer S, Vaught JD, Bock C, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehan MR, Ostroff R, Wilcox SK, et al. Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Adv Exp Med Biol. 2013;734:283–300. doi: 10.1007/978-1-4614-4118-2_20. [DOI] [PubMed] [Google Scholar]
- 20.Dorman SE, Goldberg S, Stout JE, et al. Substitution of rifapentine for rifampin during intensive phase treatment of pulmonary tuberculosis: study 29 of the tuberculosis trials consortium. J Infect Dis. 2012;206:1030–40. doi: 10.1093/infdis/jis461. [DOI] [PubMed] [Google Scholar]
- 21.Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol. 2012;29:543–9. doi: 10.1016/j.nbt.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Ostroff R, Foreman T, Keeney TR, Stratford S, Walker JJ, Zichi D. The stability of the circulating human proteome to variations in sample collection and handling procedures measured with an aptamer-based proteomics array. J Proteomics. 2010;73:649–66. doi: 10.1016/j.jprot.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Hollander M, Wolfe DA. Nonparametric Statistical Methods. 2. John Wiley; 1999. [Google Scholar]
- 24.Seber GAF, Lee AJ. Linear Regression Analysis. 2. John Wiley; 2003. [Google Scholar]
- 25.Storey JD. A direct approach to false discovery rates. J Royal Stat Soc, Series B. 2002;64:479–498. [Google Scholar]
- 26.Dabney A, Storey JD, Warnes GR. qvalue: Q-value estimation for false discovery rate control. R package version 1.26.0 2011. http://CRAN.R-project.org/package=qvalue.
- 27.Meinshausen N, Buhlmann P. High-dimensional graphs and variable selection with the lasso. The Annals of Statistics. 2006;34:1436–1462. [Google Scholar]
- 28.Kohda K, Ishibashi T, Shimbara N, Tanaka K, Matsuda Y, Kasahara M. Characterization of the mouse PA28 activator complex gene family: complete organizations of the three member genes and a physical map of the approximately 150-kb region containing the alpha- and beta-subunit genes. J Immunol. 1998;160:4923–35. [PubMed] [Google Scholar]
- 29.Yang YC, Yin T. Interleukin-11 and its receptor. Biofactors. 1992;4:15–21. [PubMed] [Google Scholar]
- 30.Jang YS, Kim JH, Seo GY, Kim PH. TGF-beta1 stimulates mouse macrophages to express APRIL through Smad and p38MAPK/CREB pathways. Mol Cells. 2011;32:251–5. doi: 10.1007/s10059-011-1040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert S, Nau R, Michel U. Role of activin in bacterial infections: a potential target for immunointervention? Immunotherapy. 2010;2:673–84. doi: 10.2217/imt.10.64. [DOI] [PubMed] [Google Scholar]
- 32.Allen SS, Cassone L, Lasco TM, McMurray DN. Effect of neutralizing transforming growth factor beta1 on the immune response against Mycobacterium tuberculosis in guinea pigs. Infect Immun. 2004;72:1358–63. doi: 10.1128/IAI.72.3.1358-1363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai Y, Uchida K, Nakayama H. Histopathological features and expression profiles of cytokines, chemokines and SOCS family proteins in trehalose 6,6′-dimycolate-induced granulomatous lesions. Inflamm Res. 2011;60:371–8. doi: 10.1007/s00011-010-0280-7. [DOI] [PubMed] [Google Scholar]
- 34.Yu G, Boone T, Delaney J, et al. APRIL and TALL-I and receptors BCMA and TACI: system for regulating humoral immunity. Nat Immunol. 2000;1:252–6. doi: 10.1038/79802. [DOI] [PubMed] [Google Scholar]
- 35.Elkington PT, Ugarte-Gil CA, Friedland JS. Matrix metalloproteinases in tuberculosis. Eur Respir J. 2011;38:456–64. doi: 10.1183/09031936.00015411. [DOI] [PubMed] [Google Scholar]
- 36.LaPan P, Brady J, Grierson C, et al. Optimization of total protein and activity assays for the detection of MMP-12 in induced human sputum. BMC Pulm Med. 2010;10:40. doi: 10.1186/1471-2466-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadhwa S, Embree MC, Bi Y, Young MF. Regulation, regulatory activities, and function of biglycan. Crit Rev Eukaryot Gene Expr. 2004;14:301–15. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.50. [DOI] [PubMed] [Google Scholar]
- 38.Cardona PJ. A spotlight on liquefaction: evidence from clinical settings and experimental models in tuberculosis. Clin Dev Immunol. 2011;2011:868246. doi: 10.1155/2011/868246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit van Dixhoorn MG, Munir R, Sussman G, et al. Gene expression profiling of suppressor mechanisms in tuberculosis. Mol Immunol. 2008;45:1573–86. doi: 10.1016/j.molimm.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Weijer S, Wieland CW, Florquin S, van der Poll T. A thrombomodulin mutation that impairs activated protein C generation results in uncontrolled lung inflammation during murine tuberculosis. Blood. 2005;106:2761–8. doi: 10.1182/blood-2004-12-4623. [DOI] [PubMed] [Google Scholar]
- 41.Hesseling AC, Walzl G, Enarson DA, et al. Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int J Tuberc Lung Dis. 2010;14:560–70. [PubMed] [Google Scholar]
- 42.Holtz TH, Sternberg M, Kammerer S, et al. Time to sputum culture conversion in multidrug-resistant tuberculosis: predictors and relationship to treatment outcome. Ann Intern Med. 2006;144:650–9. doi: 10.7326/0003-4819-144-9-200605020-00008. [DOI] [PubMed] [Google Scholar]
- 43.Sahin F, Yildiz P. Serum CA-125: biomarker of pulmonary tuberculosis activity and evaluation of response to treatment. Clin Invest Med. 2012;35:E223–8. [PubMed] [Google Scholar]
- 44.Huang WC, Tseng CW, Chang KM, Hsu JY, Chen JH, Shen GH. Usefulness of tumor marker CA-125 serum levels for the follow-up of therapeutic responses in tuberculosis patients with and without serositis. Jpn J Infect Dis. 2011;64:367–72. [PubMed] [Google Scholar]
- 45.Agranoff D, Fernandez-Reyes D, Papadopoulos MC, et al. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet. 2006;368:1012–21. doi: 10.1016/S0140-6736(06)69342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillespie SH, Kennedy N. Weight as a surrogate marker of treatment response in tuberculosis. Int J Tuberc Lung Dis. 1998;2:522–3. [PubMed] [Google Scholar]
- 47.Desjardin LE, Perkins MD, Wolski K, et al. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med. 1999;160:203–10. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 48.Perrin FM, Lipman MC, McHugh TD, Gillespie SH. Biomarkers of treatment response in clinical trials of novel antituberculosis agents. Lancet Infect Dis. 2007;7:481–90. doi: 10.1016/S1473-3099(07)70112-3. [DOI] [PubMed] [Google Scholar]