Abstract
A quartet of attachment proteins and a trio of fusion protein subunits play the cell entry concert of parainfluenza viruses. While many of these viruses bind sialic acid to enter cells, wild type measles binds exclusively two tissue-specific proteins, the lymphatic receptor signaling lymphocytic activation molecule (SLAM), and the epithelial receptor nectin-4. SLAM binds near the stalk-head junction of the hemagglutinin. Nectin-4 binds a hydrophobic groove located between blades 4 and 5 of the hemagglutinin β-propeller head. The mutated vaccine strain hemagglutinin binds in addition the ubiquitous protein CD46, which explains attenuation. The measles virus entry concert has four movements. Andante misterioso: the virus takes over the immune system. Allegro con brio: it rapidly spreads in the upper airway’s epithelia. “Targeting” fugue: the versatile orchestra takes off. Presto furioso: the virus exits the host with thunder. Be careful: music is contagious.
Theme: cell entry of Paramyxoviridae
Measles virus (MeV) [1] belongs to the Morbillivirus genus of the Paramyxoviridae family of negative strand RNA viruses; a family that also includes deadly emerging zoonotic viruses like Hendra and Nipah, as well as other prevalent human pathogens such as mumps, parainfluenza and respiratory syncytial viruses [2]. Most Paramyxoviruses enter cells by fusion at the plasma membrane at neutral pH [2-4]. This is unlike most other enveloped viruses, which use either low pH or proteases in endosomal compartments to trigger fusion [2,5].
Virus entry is mediated by the concerted action of a tetrameric attachment protein (the “quartet”) and a fusion protein trimer (the “trio”). The attachment proteins are known as hemagglutinin (H) for MeV and the other Morbilliviruses [6], glycoprotein (G) for the Henipaviruses [3], and hemagglutinin-neuraminidase (HN) for those paramyxoviruses with both hemagglutination and neuraminidase activities [2]. H and G bind protein receptors while HN binds sialic acid [3,7]. All are type II transmembrane glycoproteins with a short cytoplasmic tail, a membrane-spanning region, a membrane-proximal stalk and a membrane-distal head domain [2]. All head domains fold into six-bladed β-propellers [8-15] (Fig. 1). Most stalks form disulfide-linked dimers [16] which then further associate [17,18]. The only stalk structure available to date is that of HN, a four-helix bundle [14,19,20]. Receptors bind the heads, located above the F-trimers, triggering their refolding and membrane fusion [21-24].
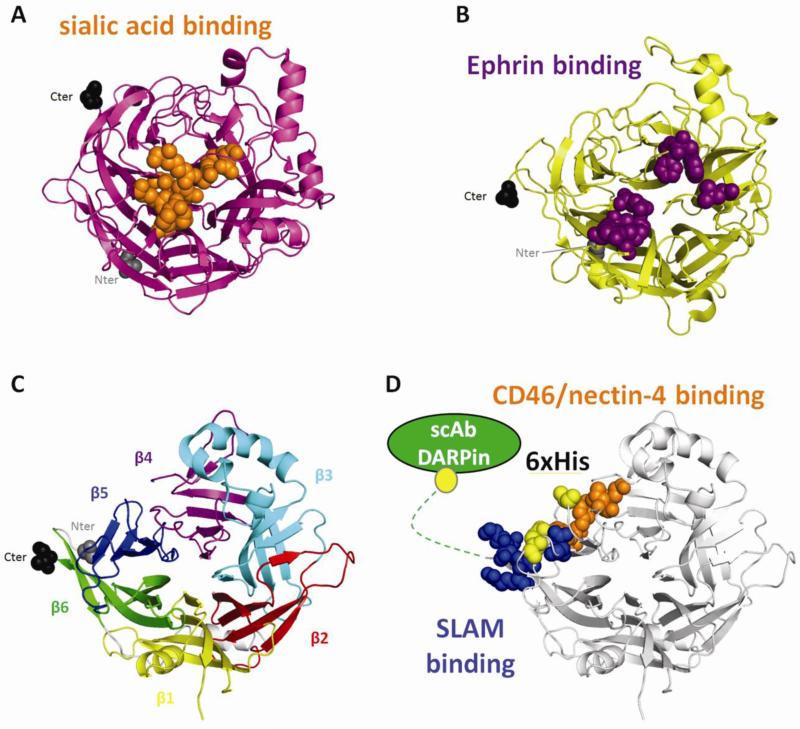
Figure 1.
Structures of paramyxovirus attachement protein heads and modes of receptor binding. (A) Structure of PIV5 HN. Residues contacting the sialic acid are shown in orange. (B) Structure of Nipah G. Residues contacting ephrin B2 are colored purple. (C) Structure of MeV H. Each blade in the 6-bladed β-propeller head is represented in a different color. (D) MeV H showing the most important residues binding CD46/nectin-4 (orange), those most important for binding SLAM (blue), the positions of hexahistidine tags supporting fusion (yellow), and C-terminal addition of specificity domains (green). In A, B and C, the N-terminal residue and the C-terminal residues are in spherical representation grey and black, respectively. The structures were aligned using PyMOL (www.pymol.org).
HN-heads bind sialic acid in a central pocket of the β-propeller (Fig. 1A) [10]. The active sites in the HN-head dimers are tilted by 90° to each other. Henipavirus G-heads (Fig. 1B) are structurally similar to HN but do not bind sialic acid and hence have neither hemagglutination nor neuraminidase activity [25,26]. Instead, G binds different ephrins, membrane anchored proteins involved in embryonic development [27-29]. While the binding occurs on the same face of the molecule as does sialic acid, ephrins bind closer to the top of the β-propeller [13,15,30] (Fig. 1B). Similar to HN, the G-protein’s receptor-binding faces are tilted at 90° [26].
As Henipavirus G-, the MeV H-protein does not bind sialic acid and lacks neuraminidase activity. The H-head is cuboidal (Fig. 1C-D) [11], in contrast to the more globular structures of HN and G. The H-monomers are also tilted and somewhat twisted. N-linked oligosaccharides cover the central pocket of the H-monomer, thus occluding the potential sialic acid binding site [11]. Instead, H binds protein receptors on the side of the β-propeller (Fig. 1D).
Andante misterioso: measles virus takes over the immune system
MeV biological niche is defined by the receptors. Wild-type MeV uses two proteins to enter cells, the signaling lymphocyte activation molecule SLAM/CD150 [31] and nectin-4, a cell junction organizer also named PVRL4 for poliovirus receptor-like 4 [32,33]. SLAM is expressed on activated immune cells [34] while nectin-4 is expressed at the basolateral surface of airway epithelial cells [35]. SLAM is the primary receptor and accounts for the immunosuppressive activity of MeV. It supports infection of alveolar macrophages patrolling the airway lumen and of dendritic cells [36,37]. These cells transfer the infection to lymphocytes including memory T-cells, hence causing temporary loss of immunity to other pathogens [38-40]. Circulating lymphatic cells then transmit the infection to the basolateral side of nectin-4 expressing cells in the upper respiratory epithelium [41,42]. After epithelial spread MeV is aerosolized by coughing and sneezing, promoting efficient contagion [43]. Both SLAM and nectin-4 are type I transmembrane proteins from the immunoglobulin superfamily that bind H with their membrane-distal V domains [32,44]. A ubiquitous regulator of complement activation, named membrane cofactor protein or CD46, is used as receptor only by the vaccine strain [45,46].
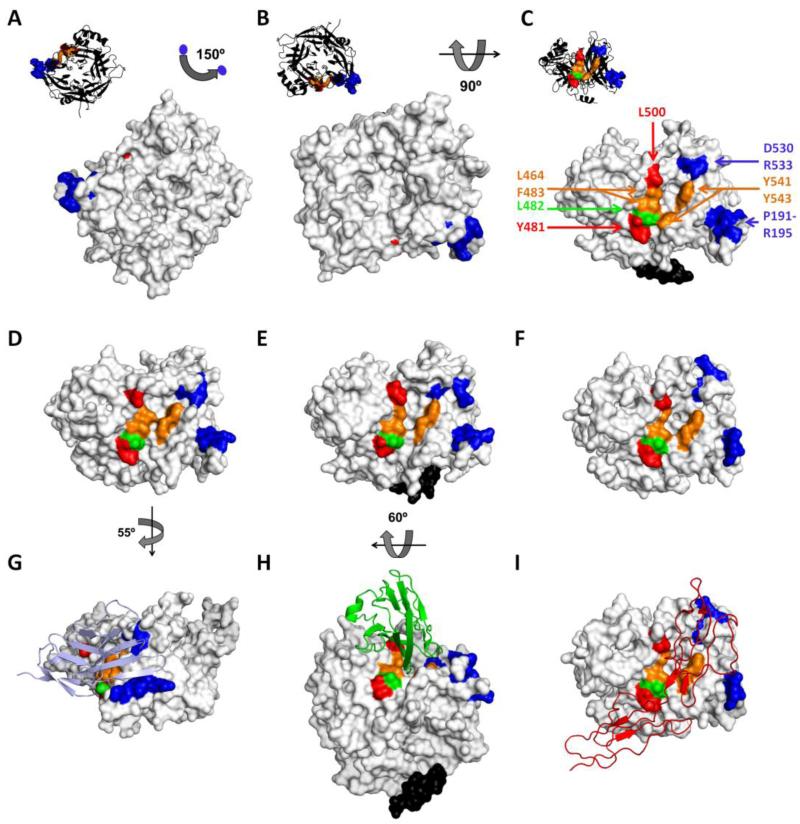
SLAM binding defines the first movement of the MeV entry concert. While avoiding recognition, the virus takes over the immune system. Figure 2 shows the MeV H protein alone (panels A-F, surface representation) or in complex with the three receptors (panels G-I, ribbon representation). Residues important for SLAM-dependent entry are shown in blue [47-49]. Residues D530 and R533 in the β5-blade (Fig. 2C, top right) are necessary for cell entry through SLAM [49] and the residue I194 in the β6-blade (Fig. 2C, bottom right) is necessary for SLAM binding as demonstrated by surface plasmon resonance [48]. In the H/SLAM co-crystal, the orientation of R533 is slightly modified compared to the unbound form in order to accommodate one loop of SLAM (compare panels G and D to panel C). In addition, the P191-R195 β-strand gets closer to R533 in the SLAM bound form to lock-on to the receptor. Whereas the SLAM-H co-structure indicates that residues Y541 and Y543 can form a hydrophobic interaction with SLAM [50], mutating these residues does not block SLAM usage in functional assays [51-53].
Figure 2.
Surface representation of the MeV H head unbound (A-C) [11], after SLAM binding (D) [50], after nectin-4 binding (E) [54] or after CD46 binding (F) [58]. With the exception of the H-nectin-4 co-crystal obtained with a wild-type H protein, all other structures were based on vaccine-lineage H proteins. In panel A, MV H has the same orientation as in Figure 1C, as shown also in the upper left black ribbon backbone representation, and is rotated according to the indicated angles in B and C. D, E and F panels are oriented as in C. G, H and I show also the receptors in ribbon representation: SLAM (light blue), nectin-4 (green) and CD46 (red), respectively. The MeV H residues important for function through CD46, nectin-4 and SLAM are colored red, green and blue, respectively. Residues shared by the CD46 and nectin-4 binding sites are colored orange. Stalk-connecting residues are colored black. Certain parts of the H protein structures were not resolved, also accounting for part of the visible differences between the structures. For example, the upper stalk residues 156 to 166 are only visible in the H/nectin-4 and unbound structures (panels C, E and H, black residues). Minor differences in structure noted upon binding to different receptors are mainly due to the orientation of the amino acid side chains (see main text).
Allegro con brio: the virus rapidly spreads in the upper airway’s epithelia
Brisk multiplication in the upper airways defines the second movement. Infected lymphatic cells spread the virus in the whole body, but the infection is transmitted only to nectin-4 expressing cells [41,42]. In Figure 2 residues important for nectin-4 dependent entry are shown in green and orange; orange indicates that the residue is also functionally relevant for entry through CD46 binding. About 20 residues in the β3, β4 and β5 blades are in close contact with MeV H in the complex [54]. Functionally important residues include L464, L482, F483, Y541 and Y543 (Fig. 2C). Interestingly Y541 and Y543 in the β5-blade delimit a hydrophobic groove along with residues L464 and F483 in the β4 blade (Fig. 2E, center). This groove is wider in the unbound (Fig. 2C) and SLAM-bound (Fig. 2G) forms of H. In the structure of the epithelial receptor bound complex, a nectin-4 loop protrudes deep into the groove (Fig. 2H), which may contribute to the high affinity interaction with H (about 20 nM versus 80-100 nM for SLAM and CD46) [32]. In addition, residue L482 located at the bottom edge of the groove (Fig. 2, green residue) gives some specificity for nectin-4 as its mutation modulates nectin-4-specific cell-to-cell fusion [52].
For illustrative purposes, this variation ends with a morendo (dying): binding CD46 causes entry into non-target cells that alert the immune system, which tries to eliminate the infection. In truth, only the vaccine strain H protein binds CD46 [48]. Hemagglutination of simian erythrocytes, which express CD46 [45,46], is restricted to the vaccine strain. Thus, “H” is a misnomer for the MeV attachment protein because wild type strains do not hemagglutinate. Moreover, CD46 has no discernible role in wild type infections [55], and its interactions with the live-attenuated vaccine strain may contribute to its outstanding safety record [1]. CD46 can be considered a “decoy” receptor that successfully misleads the vaccine strain, but not wild type MeV.
Like the nectin-4 interaction, one CD46 loop inserts in the β4-β5 hydrophobic groove (Fig. 2I). Substitution N481Y is the main determinant of CD46 adaptation [56,57], but at least 10 other residues interact in the co-crystal [58]. These residues include four forming the nectin-4 binding hydrophobic groove (Fig. 2C and 2F, orange residues L464, F483, Y541 and Y543). Y481 is located at the bottom edge of the groove (Fig. 2I, red residue) and does not move after receptor binding (compare Fig. 2C and 2F). At the top of the groove, L500 (red) also contributes to CD46 docking [52] (Fig. 2C, 2F, 2I). The CD46 binding surface may have evolved from the nectin-4 binding surface based on minimal changes.
“Targeting” fugue: the versatile orchestra takes off
The “targeting” fugue was recently added to the entry concert. The orchestra now plays on new stages, operating mainly with cancer marker proteins as designated receptors. We explain how this became possible. All receptors bind between H β-propeller blades 3 and 6. Nectin-4 and CD46 bind a β4-β5 hydrophobic groove, while residues important for SLAM binding map mainly to the β5-β6 blades. Thus, receptor binding to adjacent H-head locations can trigger fusion. But can binding anywhere on the H-head do it? This question was addressed by adding hexahistidine “handles” to surface-exposed loops of the H-head. These hexahistidines are recognized by a membrane-linked anti-His pseudoreceptor. The pseudoreceptor triggered fusion only when the “handles” were located close to the natural receptor binding sites or at the H carboxyl-terminus (Fig. 1D, yellow residues) [59].
Thus, targeted cell entry can be achieved simply by adding specificity domains to the H carboxyl-terminus (Fig. 1D). Indeed, membrane fusion through designated receptors was shown first with small domains [60], then with single chain antibodies that can be engineered to have any desired specificity [61-64]. Smaller and stackable specificity domains named DARPins were recently displayed on H to achieve double targeting [65,66]. Re-targeted H proteins and F-trimers inserted in the membrane of lentiviruses in place of their own envelope protein do re-target cell entry of these gene delivery vectors [67,68]. Thus, the MeV glycoprotein complex has become a widely used tool for targeted cell entry [67,69,70]: it can accept many input signals and convert these into the same output that triggers membrane fusion.
Presto furioso: the virus exits the host with thunder
The final movement illustrates MeV exit from the host, resulting in efficient contagion. Before coming to it, we note that two processes can trigger membrane fusion. First, binding the nectin-4 site while inserting a loop in the β4-β5 hydrophobic groove, as done by nectin-4 and CD46. Second, binding the SLAM site, which maps near the head-stalk junction: since specificity domains added to the H carboxyl-terminus are located close to this site, they may operate through the latter process.
But what activates membrane fusion? We know that natural, decoy, and designated receptors can trigger it. These are all membrane-bound proteins that may simply “pull” on H-tetramers. It is possible that the receptor’s cytoplasmic tails are connected with the cytoskeleton; in this case, binding to multiple receptor molecules would be followed by “pulling” the viral envelope in different directions, destabilizing ordered viral glycoprotein arrays [71] and triggering membrane fusion.
And how do H-tetramers integrate these triggering signals and activate F-trimers? Through the central segment of the H-stalk that changes conformation and hits the F-trimer, possibly at its base, separating the subunits [23,24,72,73]. This process is conserved among different genera of the Paramyxoviridae family [74]: provided that the H-stalk to F-trimer interaction is maintained, chimeric attachment proteins are functional [75-78]. Moreover, under certain conditions attachment protein stalks alone can trigger F-trimer refolding and membrane fusion [79,80]. On the other hand, the H-dependent triggering process differs from the HN-process in at least two aspects. First, while the HN-and F-oligomers interact only at the cell surface [2], the H- and F-oligomers associate early in their biogenesis [81]. Second, the HN-dimer interface [82], but not the H-dimer interface [59] must open at triggering.
In Figure 3 we consider two poses of the H-tetramer, in analogy with poses observed in HN-ectodomain crystals [14,19,20]. The first pose (Fig. 3A) locates the H-head dimers parallel to the sides of the tetrameric stalk. The second (Fig. 3B) puts them above the stalk. Our analyses of the H-stalk tetramer-forming propensity [73] suggest that in the pre-triggering conformation the Cys139-Cys154 segment bifurcates into two dimers, as in Figure 3A. Upon receptor binding, an extended tetrameric stalk may form where the four Cys154 converge at the top of the stalk [50], as in Figure 3B. An intermediate one-head-dimer-up and one-head-dimer-down pose [20] (not shown in Figure 3) may allow association of H-tetramers with F-trimers in the endoplasmic reticulum and co-transport of the glycoprotein complex to the cell surface [81]. Receptors would trigger fusion by pulling on this intermediate pose, rather than on that shown in Figure 3A.
Figure 3.
Two possible poses of the MV H-head dimers on the stalk. A) The H-head dimers are depicted in a 4-heads-down orientation similar to that seen for HN [14]. One monomer in each H-dimer is colored such that each β-propeller blade is shaded differently, as in Fig. 1A. The other monomer is shaded grey. Cysteine (Cys) 154 at the top of the stalk is shown in a space-filling representation and shaded yellow. A large gap in the crystal structure which corresponds to a flexible loop in the head-connecting segment is shown as a purple wavy line. Each horizontal line represents a residue in the stalk which was systematically substituted with Cys [73]. Numbering between the panels refers to MV H-stalk residues. Length of the horizontal line is proportional to the covalent tetramer trapping propensity of the Cys substitution at that position. The color of the line indicates the fusion support efficiency of the Cys substitutions: green, wild type levels of fusion; blue, intermediate fusion; orange, greatly reduced fusion; and red, no fusion. B) The H-head dimers are depicted in the form I arrangement [50]. Since a mutated H-head construct lacking the head-connecting segment was used to crystallize the form I tetramer, the head connecting segment of the original H-head dimer structure [11] was modeled into the form I structure. While modeling revealed some steric clash issues, all four Cys154 residues come together at the top of the stalk in this H-head conformation.
The MeV entry concert comes to its final movement, host exit. About two weeks after contagion the sick host coughs and sneezes, spreading the virus. Be careful: music is highly contagious. Vaccinate!
Highlights.
The β-propeller head of the wild type measles virus hemagglutinin binds exclusively two tissue-specific receptors
The signaling lymphocytic activation molecule (SLAM) and the epithelial receptor nectin-4 bind adjacent surfaces of the hemagglutinin head
A mutated hydrophobic groove of the vaccine strain hemagglutinin binds the ubiquitous protein CD46, a decoy receptor
Specificity domains added to the hemagglutinin can target viral entry to designated receptors expressed on cancer cells
Acknowledgements
This work was funded by National Institutes of Health grants R01CA90636, R01AI063476 and R01CA139389. MM is a Merck fellow of the Life Sciences Research Foundation. CKN was a Kendall fellow of the Mayo Foundation. We thank Denis Gerlier, Iris Kemler, Yoshi Kawaoka, Thilo Stehle, and Steve Harrison for insightful comments on the manuscript, Martin Billeter for making a genetic approach to the study of MeV biology possible, and Jacques Ogg (Lyra Baroque Orchestra) for inspiration. Christian Pfaller and Grazia Isaya suggested La Rubeola (with apologies to Giuseppe Verdi) as title of a planned opera.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Griffin D. Measles. In: Knipe D, Howley P, editors. Fields’ Virology. Sixth Edition Vol. 1. Lippincott, Williams and Wilkins; 2013. pp. 1042–1069. [Google Scholar]
- 2.Lamb RA, Parks G. Paramyxoviridae. In: Knipe D, Howley P, editors. Fields’ Virology. Sixth edition Vol. 1. Lippincott, Williams & Wilkins; 2013. pp. 957–995. [Google Scholar]
- 3.Lee B, Ataman ZA. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol. 2011;19:389–399. doi: 10.1016/j.tim.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plattet P, Plemper RK. Envelope protein dynamics in paramyxovirus entry. MBio. 2013;4:e00413–13. doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modis Y. Relating structure to evolution in class II viral membrane fusion proteins. Curr Opin Virol. 2014 doi: 10.1016/j.coviro.2014.01.009. COVIRO-S-13-00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ader N, Brindley M, Avila M, Orvell C, Horvat B, Hiltensperger G, Schneider-Schaulies J, Vandevelde M, Zurbriggen A, Plemper RK, et al. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J Virol. 2013;87:314–326. doi: 10.1128/JVI.01826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Mol Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 11*.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. First structural analysis of the measles virus hemagglutinin dimer, allowing to interpret previous genetic analyses in a structural context.
- 12.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 13.Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 14**.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. First structure of attachment protein heads with the stalk, revealing the “heads down” arrangement.
- 15.Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plemper RK, Hammond AL, Cattaneo R. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J Virol. 2000;74:6485–6493. doi: 10.1128/jvi.74.14.6485-6493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brindley M, Plemper R. Blue Native-PAGE and biomolecular complementation reveal tetrameric or higher order oligomer organization of the physiological measles virus attachment (H) protein. J Virol. 2010;84:12174–12184. doi: 10.1128/JVI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maar D, Harmon B, Chu D, Schulz B, Aguilar HC, Lee B, Negrete OA. Cysteines in the stalk of the Nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol. 2012;86:6632–6642. doi: 10.1128/JVI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011;85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 2013;9:e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Paal T, Brindley MA, St. Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83:10480–10493. doi: 10.1128/JVI.01195-09. Demonstration that the length of the upper half of the measles virus H-stalk can be altered without compromising signal transmission.
- 23.Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W, Cattaneo R. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J Biol Chem. 2012;287:33026–33035. doi: 10.1074/jbc.M112.373308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apte-Sengupta S, Navaratnarajah CK, Cattaneo R. Hydrophobic and charged residues in the central segment of the measles virus hemagglutinin stalk mediate transmission of the fusion-triggering signal. J Virol. 2013;87:10401–10404. doi: 10.1128/JVI.01547-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu M, Hansson E, Langedijk JP, Eaton BT, Wang LF. The attachment protein of Hendra virus has high structural similarity but limited primary sequence homology compared with viruses in the genus Paramyxovirus. Virology. 1998;251:227–233. doi: 10.1006/viro.1998.9302. [DOI] [PubMed] [Google Scholar]
- 26.Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 29.Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Muhlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, Lee B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu K, Chan YP, Rajashankar KR, Khetawat D, Yan L, Kolev MV, Broder CC, Nikolov DB. New insights into the Hendra virus attachment and entry process from structures of the virus G glycoprotein and its complex with ephrin-B2. PLoS One. 2012;7:e48742. doi: 10.1371/journal.pone.0048742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. Discovery of measles virus primary receptor, accounting for lymphatic cell tropism and immunosuppression.
- 32**.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. Discovery of the measles virus epithelial receptor, accounting for upper airways tropism and efficient contagion.
- 33**.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. Discovery of the measles virus epithelial receptor, accounting for upper airways tropism and efficient contagion.
- 34.Sintes J, Engel P. SLAM (CD150) is a multitasking immunoreceptor: from cosignalling to bacterial recognition. Immunol Cell Biol. 2011;89:161–163. doi: 10.1038/icb.2010.145. [DOI] [PubMed] [Google Scholar]
- 35.Reymond N, Fabre S, Lecocq E, Adelaide J, Dubreuil P, Lopez M. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J Biol Chem. 2001;276:43205–43215. doi: 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 36.Ferreira CS, Frenzke M, Leonard VH, Welstead GG, Richardson CD, Cattaneo R. Measles virus infection of alveolar macrophages and dendritic cells precedes spread to lymphatic organs in transgenic mice expressing human signaling lymphocytic activation molecule (SLAM, CD150) J Virol. 2010;84:3033–3042. doi: 10.1128/JVI.01559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemon K, de Vries RD, Mesman AW, McQuaid S, van Amerongen G, Yuksel S, Ludlow M, Rennick LJ, Kuiken T, Rima BK, et al. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 2011;7:e1001263. doi: 10.1371/journal.ppat.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Condack C, Grivel JC, Devaux P, Margolis L, Cattaneo R. Measles virus vaccine attenuation: suboptimal infection of lymphatic tissue and tropism alteration. J Infect Dis. 2007;196:541–549. doi: 10.1086/519689. [DOI] [PubMed] [Google Scholar]
- 39.de Swart RL, Ludlow M, de Witte L, Yanagi Y, van Amerongen G, McQuaid S, Yuksel S, Geijtenbeek TB, Duprex WP, Osterhaus AD. Predominant infection of CD150+ lymphocytes and dendritic cells during measles virus infection of macaques. PLoS Pathog. 2007;3:e178. doi: 10.1371/journal.ppat.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries RD, McQuaid S, van Amerongen G, Yuksel S, Verburgh RJ, Osterhaus AD, Duprex WP, de Swart RL. Measles immune suppression: lessons from the macaque model. PLoS Pathog. 2012;8:e1002885. doi: 10.1371/journal.ppat.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frenzke M, Sawatsky B, Wong XX, Delpeut S, Mateo M, Cattaneo R, von Messling V. Nectin-4-dependent measles virus spread to the cynomolgus monkey tracheal epithelium: role of infected immune cells infiltrating the lamina propria. J Virol. 2013;87:2526–2534. doi: 10.1128/JVI.03037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludlow M, Lemon K, de Vries RD, McQuaid S, Millar EL, van Amerongen G, Yuksel S, Verburgh RJ, Osterhaus AD, de Swart RL, et al. Measles virus infection of epithelial cells in the macaque upper respiratory tract is mediated by sub-epithelial immune cells. J Virol. 2013;87:4033–4042. doi: 10.1128/JVI.03258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Racaniello V. Virology. An exit strategy for measles virus. Science. 2011;334:1650–1651. doi: 10.1126/science.1217378. [DOI] [PubMed] [Google Scholar]
- 44.Ono N, Tatsuo H, Tanaka K, Minagawa H, Yanagi Y. V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J Virol. 2001;75:1594–1600. doi: 10.1128/JVI.75.4.1594-1600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 46.Naniche D, Varior-Krishnan G, Cervoni F, Wild TF, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leonard VH, Hodge G, Reyes-Del Valle J, McChesney MB, Cattaneo R. Measles virus selectively blind to signaling lymphocytic activation molecule (SLAM; CD150) is attenuated and induces strong adaptive immune responses in rhesus monkeys. J Virol. 2010;84:3413–3420. doi: 10.1128/JVI.02304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navaratnarajah CK, Vongpunsawad S, Oezguen N, Stehle T, Braun W, Hashiguchi T, Maenaka K, Yanagi Y, Cattaneo R. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150) J Biol Chem. 2008;283:11763–11771. doi: 10.1074/jbc.M800896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: Identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J Virol. 2004;78:302–313. doi: 10.1128/JVI.78.1.302-313.2004. First study of the functional footprints of two receptors on the measles virus hemagglutinin.
- 50.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 51.Leonard VH, Sinn PL, Hodge G, Miest T, Devaux P, Oezguen N, Braun W, McCray PB, Jr., McChesney MB, Cattaneo R. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J Clin Invest. 2008;118:2448–2458. doi: 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.Mateo M, Navaratnarajah CK, Syed S, Cattaneo R. The measles virus hemagglutinin beta-propeller head β4-β5 hydrophobic groove governs functional interactions with nectin-4 and CD46 but not those with the signaling lymphocytic activation molecule. J Virol. 2013;87:9208–9216. doi: 10.1128/JVI.01210-13. Detailed characterization of the functional footprints of all three receptors on the measles virus hemagglutinin.
- 53.Tahara M, Takeda M, Shirogane Y, Hashiguchi T, Ohno S, Yanagi Y. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J Virol. 2008;82:4630–4637. doi: 10.1128/JVI.02691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol. 2013;20:67–72. doi: 10.1038/nsmb.2432. Co-structure of the measles virus hemagglutinin with nectin-4, including discussion of how the structural footprints of three receptors compare.
- 55.Takeuchi K, Nagata N, Kato SI, Ami Y, Suzaki Y, Suzuki T, Sato Y, Tsunetsugu-Yokota Y, Mori K, Van Nguyen N, et al. Wild-type measles virus with the hemagglutinin protein of the Edmonston vaccine strain retains wild-type tropism in macaques. J Virol. 2012;86:3027–3037. doi: 10.1128/JVI.06517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu EC, Sarangi F, Iorio C, Sidhu MS, Udem SA, Dillehay DL, Xu W, Rota PA, Bellini WJ, Richardson CD. A single amino acid change in the hemagglutinin protein of measles virus determines its ability to bind CD46 and reveals another receptor on marmoset B cells. J Virol. 1998;72:2905–2916. doi: 10.1128/jvi.72.4.2905-2916.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lecouturier V, Fayolle J, Caballero M, Carabana J, Celma ML, Fernandez-Munoz R, Wild TF, Buckland R. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70:4200–4204. doi: 10.1128/jvi.70.7.4200-4204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 59*.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VHJ, Braun W, Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. Structure-based functional analysis of the mechanism by which receptors trigger the measles virus fusion apparatus.
- 60.Schneider U, Bullough F, Vongpunsawad S, Russell SJ, Cattaneo R. Recombinant measles viruses efficiently entering cells through targeted receptors. J Virol. 2000;74:9928–9936. doi: 10.1128/jvi.74.21.9928-9936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammond AL, Plemper RK, Zhang J, Schneider U, Russell SJ, Cattaneo R. Single-chain antibody displayed on a recombinant measles virus confers entry through the tumor-associated carcinoembryonic antigen. J Virol. 2001;75:2087–2096. doi: 10.1128/JVI.75.5.2087-2096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura T, Peng KW, Harvey M, Greiner S, Lorimer IA, James CD, Russell SJ. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat Biotechnol. 2005;23:209–214. doi: 10.1038/nbt1060. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura T, Peng KW, Vongpunsawad S, Harvey M, Mizuguchi H, Hayakawa T, Cattaneo R, Russell SJ. Antibody-targeted cell fusion. Nat Biotechnol. 2004;22:331–336. doi: 10.1038/nbt942. [DOI] [PubMed] [Google Scholar]
- 64.Navaratnarajah CK, Miest TS, Carfi A, Cattaneo R. Targeted entry of enveloped viruses: measles and herpes simplex virus I. Curr Opin Virol. 2012;2:43–49. doi: 10.1016/j.coviro.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedrich K, Hanauer JR, Prufer S, Munch RC, Volker I, Filippis C, Jost C, Hanschmann KM, Cattaneo R, Peng KW, et al. DARPin-targeting of measles virus: unique bispecificity, effective oncolysis, and enhanced safety. Mol Ther. 2013;21:849–859. doi: 10.1038/mt.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rasbach A, Abel T, Munch RC, Boller K, Schneider-Schaulies J, Buchholz CJ. The receptor attachment function of measles virus hemagglutinin can be replaced with an autonomous protein that binds Her2/neu while maintaining its fusion-helper function. J Virol. 2013;87:6246–6256. doi: 10.1128/JVI.03298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cattaneo R. Paramyxovirus entry and targeted vectors for cancer therapy. PLoS Pathog. 2010;6:e1000973. doi: 10.1371/journal.ppat.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Funke S, Maisner A, Muhlebach MD, Koehl U, Grez M, Cattaneo R, Cichutek K, Buchholz CJ. Targeted cell entry of lentiviral vectors. Mol Ther. 2008;16:1427–1436. doi: 10.1038/mt.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Russell SJ, Peng KW. Measles virus for cancer therapy. Curr Top Microbiol Immunol. 2009;330:213–241. doi: 10.1007/978-3-540-70617-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miest TS, Cattaneo R. New viruses for cancer therapy: meeting clinical needs. Nat Rev Microbiol. 2014 doi: 10.1038/nrmicro3140. doi: 10:1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci U S A. 2012;109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JPM, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem. 2012;287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73*.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J Biol Chem. 2012;287:38543–38551. doi: 10.1074/jbc.M112.410563. Characterization of the tetramer-forming propensity of the measles virus hemagglutinin stalk, revealing segments with different structure and function.
- 74.Jardetzky TS, Lamb RA. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol. 2014 doi: 10.1016/j.coviro.2014.01.005. COVIRO-S-13-00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mirza AM, Aguilar HC, Zhu Q, Mahon PJ, Rota PA, Lee B, Iorio RM. Triggering of the Newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J Biol Chem. 2011;286:17851–17860. doi: 10.1074/jbc.M111.233965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porotto M, Salah Z, DeVito I, Talekar A, Palmer SG, Xu R, Wilson IA, Moscona A. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol. 2012;86:5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talekar A, Devito I, Salah Z, Palmer SG, Chattopadhyay A, Rose JK, Xu R, Wilson IA, Moscona A, Porotto M. Identification of a region in the stalk domain of the nipah virus receptor binding protein that is critical for fusion activation. J Virol. 2013;87:10980–10996. doi: 10.1128/JVI.01646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Talekar A, Moscona A, Porotto M. Measles fusion machinery activated by sialic acid binding globular domain. J Virol. 2013 doi: 10.1128/JVI.02256-13. doi: 10.1128/JVI.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A. 2012;109:E2625–2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. A Stabilized Headless Measles Virus Attachment Protein Stalk Efficiently Triggers Membrane Fusion. J Virol. 2013;87:11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 82.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J Virol. 2008;82:10386–10396. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]