Abstract
Five large multigene families encoding innate-type immune receptors that are comprised of immunoglobulin domains have been identified in bony fish, of which four do not possess definable mammalian orthologs. The members of some of the multigene families exhibit unusually extensive patterns of divergence and the individual family members demonstrate marked variation in interspecific comparisons. As a group, the gene families reveal striking differences in domain type and content, mechanisms of intracellular signaling, basic structural features, haplotype and allelic variation and ligand binding. The potential functional roles of these innate immune receptors, their relationships to immune genes in higher vertebrate species and the basis for their adaptive evolution are of broad interest. Ongoing investigations are expected to provide new insight into alternative mechanisms of immunity.
1. Introduction
Although many effectors of innate immunity have experienced relatively minimal change over extended periods of evolutionary time, others, many of which are encoded in large multigene families, exhibit striking degrees of variation in both form and function. In mammals, natural killer (NK) cell function is mediated through the killer-cell immunoglobulin (Ig)-like receptors (KIRs), which are encoded in a diversified immunoglobulin gene family in man; however, in mouse, equivalent function is effected through lectins encoded in the Ly49 multigene family. Additional molecules encoded in multigene families mediate immune effector function in mammals and include: CD300s and CD300-like proteins, triggering receptors expressed on myeloid cells (TREMs and TREM-like proteins), sialic acid binding proteins, Ig-like lectins (SIGLECs) and the natural cytotoxicity triggering receptors (NCRs). The gene families encoding these molecules have been well defined genetically; however, their functions and particularly the scope of their interactions with other components of the immune system are less well understood. As a group, these molecules exhibit significant interspecific differences as evidenced, in some cases, by the lack of recognizable orthologs between mouse and man. Although it is recognized that multiple gene birth and death events account for the rapid evolution and functional variation of this large and diverse group of molecules, the nature of the genomic structure and selective pressure events that drive the process are not at all clear.
Several years ago as part of an investigation of alternative forms of immune function we initiated studies to examine the broad question of innate immune receptor diversity in the members of taxonomic classes outside of the mammalia. These efforts principally have been focused on the osteichthyes, specifically teleost fish, which represent the largest subclass of animals and possess particularly remarkable morphological variation as well as physiological diversity that reflect the extensive variation in their habitats and responses to selective pressures. The studies, which are based primarily on the recognition of genetic homologies, were initiated with a search for diversified receptors that contain variable-like (V)-type immunoglobulin domains and later were extended to include multigene families encoding molecules possessing other types of domains. In the course of these investigations, some of the gene families that have been identified have been shown to represent orthologs of immune effector genes in mammals, whereas others are unique to bony fish. Taken together with studies from other laboratories as well as extensive work in mammals, a picture has emerged that reveals particularly complex patterns of dynamic genetic change that likely are driven by selective pressures and host adaptations that are unique to major animal groups and overall reflect the enormous complexity and extensive system of interactions of the mediators of immunity.
2. Immunoglobulin (Ig) domains
An individual Ig domain is approximately 100 amino acids in length and forms an “Ig-fold” structure consisting of two anti-parallel β-sheets packed face to face. Four sets of Ig domains have been described: variable-like domains (V), constant-like domains (C1 and C2) and intermediate domains (I) (Harpaz and Chothia 1994). Although Ig domains vary in the number and size of strands in the β-sheets and in the size and conformation of the links between the strands, one β-sheet is anchored by strands A, B and E while strands G, F and C form the core of the second β-sheet (Barclay 2003;Harpaz and Chothia 1994). For example, most members of the V set possess ten strands forming the two β-sheets with strands A, B, E and D forming one β-sheet and strands A', G, F, C, C', and C” forming the other. In contrast, most members of the C1 set are smaller with one β-sheet formed by strands A, B, E and D and the other by strands G, F, C and C' (Harpaz and Chothia 1994).
Two highly conserved cysteines, C23 and C104 (numbering based on the IMGT system; Lefranc et al. 2009), form an intrachain disulfide bond between the B and F strands stabilizing Ig-fold conformation. The inter-cysteine distance in V domains ranges from 65 to 75 residues and is appreciably shorter in C1 and C2 domains (55 to 60 residues) (Williams and Barclay 1988). Intermediate (I-type) Ig domains possess structural features of V domains but exhibit shorter intercysteine distances (Harpaz and Chothia 1994). As discussed below, additional cysteines within Ig domains can also contribute to intrachain disulfide bridges.
3. Inhibitory and activating receptors
Many mammalian innate immune receptor families include receptors with opposing functions. Engagement of ligands by some receptors activates the immune cell, whereas other receptors inhibit this process. The KIR, leukocyte immunoglobulin-like receptor (LILR), CD300, TREM and SIGLEC multigene families encode Ig domain-containing transmembrane proteins that include both activating and inhibitory receptors.
Some activating receptors possess a short cytoplasmic tail and a charged residue within the transmembrane domain which permits partnering with and signaling through an adaptor protein, such as DAP12, DAP10, CD3ζ and FcRγ. These adaptor proteins possess a cytoplasmic activation motif such as an immunoreceptor tyrosine-based activation motif [ITAM; (D/E)X2YX2(L/I)X6–8YX2(L/I)]. Other activating receptors possess a cytoplasmic tail that includes an ITAM (e.g. select FcR-Like Molecules; Ehrhardt and Cooper 2011).
Inhibitory receptors typically possess one or more immunoreceptor tyrosine-based inhibition motif [ITIM; (I/V/L/S)XYX2L)] within their cytoplasmic tail and do not utilize an adaptor protein (Ravetch and Lanier 2000;Vely and Vivier 1997). For example, ligand engagement by activating KIR-DAP12 complexes on natural killer cells results in the phosphorylation of the DAP12 ITAM by a Src family kinase and subsequent phosphorylation and activation of the SYK/ZAP70 – PI3K – MAPK/ERK signaling pathway which leads to the production of cytokines and chemokines and the release of cytolytic granules. Ligand engagement by inhibitory KIRs results in the phosphorylation of the ITIM(s) and recruitment of SHP family phosphatases to inhibit this signaling cascade. Shifts in the balance of these opposing signals determine if a natural killer cell will attack or ignore a cell or pathogen.
Some receptors can function both in an activating or inhibitory manner depending on the cellular context. A cytoplasmic immunoreceptor tyrosine-based switch motif [ITSM; TXYX2(V/I)] is found in some of these receptors (Ostrakhovitch and Li 2006;Shlapatska et al. 2001). Multigene families that encode activating and inhibitory receptors and their associated adaptor proteins have been identified from the genomes of various teleost species including zebrafish (Danio rerio) (Haire et al. 2012;Montgomery et al. 2011;Stafford et al. 2006;Stet et al. 2005;Yoder et al. 2007;Yoder 2009).
4. Families of Ig domain-containing innate immune receptors in zebrafish
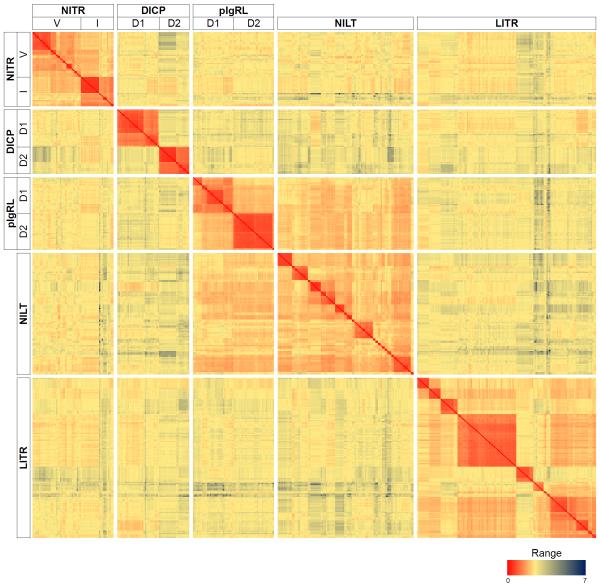
The genomes of teleost species, including zebrafish, encode hundreds to thousands of Ig domains. Many of these domains belong to the immunoglobulin (e.g. IgM, IgA, IgZ/T) and T cell receptor genes that undergo V(D)J recombination as part of the adaptive immune system (Hikima et al. 2011), whereas other Ig domains belong to genes encoding cell adhesion molecules, growth factor receptors (e.g. VEGFRs and FGFRs) and innate immune receptors. The best described Ig domain-containing innate immune receptor families in teleosts are the novel immune-type receptors (NITRs), leukocyte immune-type receptors (LITRs), novel immunoglobulin-like transcripts (NILTs), diverse immunoglobulin domain containing proteins (DICPs), and polymeric immunoglobulin receptor-like proteins (PIGRLs); we have described the zebrafish NITRs, DICPs and PIGRLs (Haire et al. 2012;Kortum et al. 2014;Yoder et al. 2001;Yoder et al. 2004;Yoder et al. 2008;Yoder et al. 2010). NILTs and LITRs have been characterized in other fish species, but can be identified in the zebrafish genome (Stafford et al. 2006;Stet et al. 2005) and it is likely that additional, related gene families remain to be described. Current estimates predict the zebrafish genome encodes a minimum of 39 NITR genes, 29 DICP genes and 29 PIGRL genes. In addition, at least 104 NILT and 137 LITR Ig domains can be identified in the zebrafish reference genome (Zv9; unpublished observations). A pairwise sequence comparison of these Ig domains demonstrates that the NITR, DICP and PIGRL families each possess two distinct types of Ig domains. The NILT and LITR families include multiple different types of Ig domains (discussed in detail below) (Figure 1).
Figure 1. Heat map of pairwise genetic distances between zebrafish NITR, DICP, PIGRL, NILT and LITR Ig domains.
The entire dataset (418 Ig domains) as well as two pIgR Ig domains (included with the PIGRLs) were aligned using default settings of Clustal Omega (Sievers et al. 2011). Poisson corrected distances were calculated using Meta-PIGA (Helaers and Milinkovitch 2010). The heat map was generated using Microsoft Excel 2013. Minimum and maximum distances were 0.4423 and 7.072 respectively.
As the NITR, DICP, PIGRL, NILT and LITR gene families lack easily recognizable genetic orthologs in mammals, predicting their function based on sequence alone is challenging. Multiple features can be considered when proposing functional roles for these protein families and one must consider the general function of each protein family and then the specific function of each family member. When considering the roles of individual proteins within a family, the simplest strategy is to catalog them into four groups based on protein architecture: 1) inhibitory defined as being membrane-bound and possessing cytoplasmic ITIMs, 2) activating defined as being membrane-bound and possessing cytoplasmic ITAMs or a charged residue within the transmembrane domain, 3) functionally ambiguous defined as possessing contradictory signaling motifs, ITSMs or simply lacking signaling motifs and 4) secreted. Each individual NITR, DICP, PIGRL, NILT and LITR protein can be assigned to one of these four groups (although mRNA splice variants from a single gene can produce multiple protein isoforms). Defining a receptor as inhibitory or activating is attractive; however, it is important to remember that multiple receptors that were originally described as inhibitory (via an ITIM) also can function to activate immune cells and that partially phosphorylated ITAMs can promote an inhibitory signal (Barrow and Trowsdale 2006;Waterman and Cambier 2010). The roles of secreted variants of these proteins remain to be elucidated; however, in mammals, secreted TREM-1 has been shown to compete with membrane bound TREM-1 for ligand binding thus dampening the immune response (Derive et al. 2010;Gibot et al. 2004).
4.1. Novel immune-type receptors (NITRs)
The NITR genes were identified initially as V domain-containing transmembrane receptors from the compact genome of the Southern pufferfish (Spheroides nephelus): 26 NITR genes were identified from a single genomic clone of ~100 kbp (Rast et al. 1995;Strong et al. 1999). NITRs subsequently were identified in two gene clusters on zebrafish chromosomes 7 and 14 (Yoder et al. 2001;Yoder et al. 2004;Yoder et al. 2008). To date, 36 NITR genes have been described on zebrafish chromosome 7 and three on chromosome 14. NITRs possess one or two Ig domains. For NITRs with two Ig domains, the membrane distal Ig domain is of the variable (V) type and the membrane proximal Ig domain is of the intermediate (I) type. Although all NITR genes encode a V domain, some NITR genes lack an I domain that results in proteins with a single V domain. NITR proteins that lack a V domain are generated by alternative mRNA splicing (Shah et al. 2012). Based on sequence similarities between V domains, NITRs form 14 groups, which are designated numerically (within an individual species) based on the order of their discovery, e.g. the NITR1, NITR2, NITR3 and NITR4 groups were the first to be described; the numbering does not reflect interspecific sequence homology. Individual gene names include a letter indicating the order of their description, e.g. nitr3b was the second gene described from group 3. A pairwise sequence comparison demonstrates a higher level of sequence variability between the NITR V domains as compared to the more conserved I domains (Figure 1). In addition, the 15-member NITR1 group displays very high levels of identity (observed as dark red regions of identity).
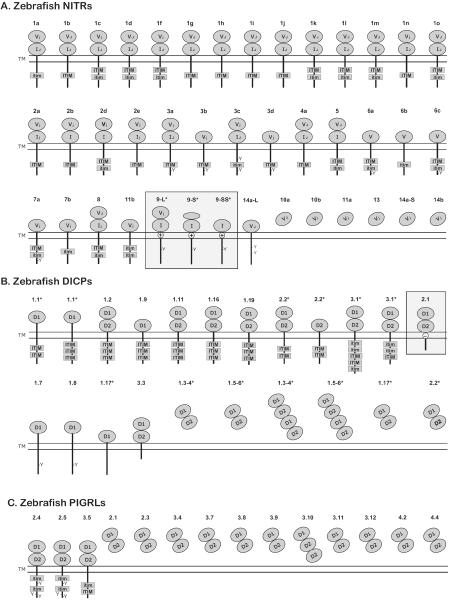
The zebrafish NITR family includes a single gene that encodes three activating receptor isoforms, multiple genes that encode inhibitory receptors possessing ITIM and/or ITIM-related (itim) sequences, and genes that encode receptors with no identifiable signaling motifs and secreted proteins (Figure 2; Yoder 2009). In the context of human NK cells, engagement of the ITIM-containing Nitr3a (previously Nitr3.1) has been shown to down regulate MAPK in an ITIM-dependent manner (Yoder et al. 2001). The single activating gene, nitr9, was described originally as encoding one V domain, one I domain and a positively charged residue within the transmembrane domain. After the discovery of mRNA splice variants that partially or completely lack the exon encoding the V domain, the original isoform was renamed Nitr9-long (or Nitr9L), whereas the proteins encoded by the splice variants were designated Nitr9-short (Nitr9S) and Nitr9-super short (Nitr9SS) (Wei et al. 2007). Within the context of mammalian cell culture, Nitr9L partners with and signals through Dap12 via pairing of reciprocally charged residues in their transmembrane domains (Wei et al. 2007;Yoder et al. 2004).
Figure 2. Predicted structures of zebrafish NITR, DICP, and PIGRL proteins.
Sequences have been reported (Haire et al. 2012;Kortum et al. 2014;Yoder et al. 2004;Yoder et al. 2008). Proteins are organized by inhibitory, activating, functionally ambiguous and secreted forms. Activating receptors are boxed. The Ig domains of (A) NITR (V and I), (B) DICP (D1 and D2) and (C) PIGRL (D1 and D2) proteins are indicated. Ig domains that include a joining (J) or J-like (j) sequence are labeled with a subscript J or j. Cytoplasmic ITIMs, ITIM-like (itim) sequences and tyrosines as well as charged residues (⊕ or ⊖) within transmembrane domains are labeled. Proteins encoded by splice variants are indicated with asterisks (*). This is not meant to be a complete catalog of proteins from these families as additional proteins sequences and structures will likely be identified in the future.
PCR-based expression studies reveal that transcripts from all NITR groups can be detected from zebrafish lymphocytes but not from myeloid cells (Yoder et al. 2010). Studies employing activating NITRs from channel catfish (Ictalurus punctatus) and NK-like catfish cell lines indicate that NITRs are involved in allorecognition (Cannon et al. 2008); however, specific molecular ligands have not been identified. These observations, along with the structural and signaling similarities to mammalian KIRs, led us to hypothesize that NITRs function as NK receptors in teleosts (Yoder and Litman 2011).
NITR genes or transcripts have been identified in most major lineages of teleosts including tetraodontiformes (pufferfish), perciformes (stickleback), ovalentariea (medaka), gadiformes (cod), salmoniformes (trout), siluriforms (catfish), cypriniformes (zebrafish), clupeiformes (lake whitefish), as well as in a Holostean fish (gar); however, NITRs have not been identified in cartilaginous fish or other vertebrate lineages (Yoder 2009). Comparisons of NITR Ig domain sequences between fish species indicates that NITR gene cluster expansions have occurred in a species-specific manner. Different numbers of NITR genes are found in different species and any given NITR within a species is more related to other NITRs within that species than to NITRs in a different species (Yoder 2009). Despite the species-specific differences in NITR sequences and gene number, NITRs likely are present in all teleosts.
4.2. Diverse immunoglobulin domain containing proteins (DICPs)
Efforts to identify NITRs from cartilaginous fish, resulted in the identification of the Ig domain-containing modular domain immune-type receptor (MDIR) gene family from clearnose skate (Raja eglanteria) (Cannon et al. 2006). Using the MDIRs as search queries, three gene clusters were identified in the zebrafish reference genome (Zv8) that are distinct from NITRs and named DICPs (Cannon et al. 2006;Haire et al. 2012). DICPs possess one or two types of Ig domains referred to as D1 and D2. All DICP genes encode at least one D1 domain which can be divided into three groups based on sequence similarity and chromosomal location. This relationship is reflected in their gene nomenclature. The DICP1, DICP2 and DICP3 groups are encoded in clusters on chromosomes 3, 14 and 16, respectively and individual genes are named in the order that their D1 domains were identified (e.g. dicp1.2 is encoded by the DICP1 group and encodes the second D1 domain identified in this cluster). Some DICP genes encode a single D1 domain, whereas others encode a D1 domain and membrane proximal D2 domain, although differential mRNA splicing can produce DICPs with a single D2 domain (Haire et al. 2012). As observed in Figure 1, the DICP D1 and D2 domains each are highly conserved in sequence and share little sequence similarity to Ig domains from other families.
Multiple DICP transcripts are predicted to encode a single putative activating receptor, multiple receptors with two or three cytoplasmic ITIM or itim sequences, membrane bound receptors with no identifiable signaling motifs and secreted forms (Figure 2), reminiscent in part of the structural variation observed in NITRs. The single activating receptor, Dicp2.1 possesses a negatively charged residue within the transmembrane domain and is predicted to partner with and signal through an adaptor protein as observed for mammalian CLM-5 (CD300LD) (Haire et al. 2012). Thus, the zebrafish DICP family is predicted to include both inhibitory and activating forms. Expression patterns of DICPs have not yet been reported.
Recombinant DICP Ig domains were shown to bind various phospholipids and lipid extracts from various bacteria in an ELISA-based assay (Haire et al. 2012). Of the seven DICP Ig domains examined, it was shown that the D1 domain of Dicp1.5 bound phospholipids with the broadest specificity while the D1 domain of Dicp1.14 displayed a narrow specificity for binding extracts from Mycobacterium. Although the ability to bind phospholipids is conserved between the DICP1 and DICP2 families (e.g. the D2 domain of Dicp2.1 binds with broad specificity), no lipid binding was detected for the D1 domain of Dicp3.1. The ability of DICPs to bind phospholipids is shared with the mammalian CD300 and TREM family of receptors and suggests they may share a common or at least related function (Cannon et al. 2012;Choi et al. 2011).
4.3. Polymeric immunoglobulin receptor-like proteins (PIGRLs)
In mammals, the polymeric immunoglobulin receptor (pIgR) is an integral transmembrane glycoprotein that transports soluble polymeric Igs such as pIgA across mucosal epithelial cells. For example, pIgA is expressed by plasma cells present in the lamina propria underlying the intestinal epithelium and is bound by pIgR on the basolateral surface of epithelial cells. pIgR transports pIgA through the epithelial cell by transcytosis (Asano and Komiyama 2011;Kaetzel 2005). The transport of pIgA by pIgR to the intestinal lumen is essential for protecting the host from invading pathogens and maintaining homeostasis (Johansen et al. 1999).
The mammalian pIgR is encoded by a single copy gene (PIGR) that encodes five extracellular Ig domains (D1–D5) (Asano and Komiyama 2011;Kaetzel 2005). Alternatively spliced PIGR transcripts lacking the D2 and D3 domains have been reported in rabbit and cow (Deitcher and Mostov 1986;Kulseth et al. 1995). Chicken and Xenopus pIgR possesses four Ig domains; the D2 domain found in mammals is lacking in these latter species (Braathen et al. 2007;Wieland et al. 2004). Full-length transcripts encoding a pIgR homolog, which possesses two Ig domains, have been identified in multiple fish species including zebrafish, fugu (Takifugu rubripes), grouper (Epinephelus coioides), common carp (Cyprinus carpio), Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) (Feng et al. 2009;Hamuro et al. 2007;Kortum et al. 2014;Rombout et al. 2008;Tadiso et al. 2011;Zhang et al. 2010). Teleost pIgR is expressed by lymphoid organs including mucosal tissues (intestine, skin and gill) and has been shown to bind both IgM and IgZ/IgT (Feng et al. 2009;Hamuro et al. 2007;Rombout et al. 2008;Tadiso et al. 2011;Zhang et al. 2010).
pIgR-like (PIGRL) transcripts were identified originally from Atlantic salmon and common carp (Cannon et al. 2006;Ribeiro et al. 2011;Tadiso et al. 2011). The carp PIGRL protein was shown to be expressed abundantly in macrophages and is secreted upon immune stimulation (referred to as a soluble immune-type receptor, SITR, in: Ribeiro et al. 2011). Recently a PIGRL multigene family was described adjacent to the single copy zebrafish pigr gene on chromosome 2 (Kortum et al. 2014). Twenty-nine distinct PIGRL genes were identified and transcripts corresponding to ten of these genes encode membrane-bound receptors with two cytoplasmic ITIM or itim sequences or secreted forms (Figure 2). Activating PIGRL receptors have not yet been identified. All PIGRL transcripts encode two Ig domains, D1 and D2. Although PIGRL D2 domains are highly similar in sequence, phylogenetic analyses of the PIGRL D1 domains define four groups of PIGRL proteins (PIGRL1 – PIGRL4). Individual gene names include the group number and a second number indicates the order in which they were identified (e.g. pigrl3.2 is the second gene identified in PIGRL group 3) (Kortum et al. 2014).
Zebrafish PIGRL genes are differentially expressed in lymphoid and myeloid cells. PIGRL1 and transcripts are predominantly detected in lymphoid cells whereas PIGRL2 transcripts are predominantly detected in myeloid cells. PIGRL3 and PIGRL4 transcripts are detected from both leukocyte lineages. An examination of PIGRL gene expression in adult tissues after bacterial or viral infection produced contradictory outcomes: bacterial infection led to a generalized increase in PIGRL transcript levels, whereas viral infection led to a generalized decrease in transcript levels of PIGRLs. Although there are a variety of mechanisms that may cause these different responses, one possibility is that PIGRL expression is down regulated through virally induced immune suppression (Kortum et al. 2014).
Recombinant PIGRL1 Ig domains have been shown to bind phospholipids in a manner similar to DICPs (see above). The Ig domains expressed from PIGRL2, PIGRL3 and PIGRL4 proteins did not show lipid binding and their ligands remain unknown (Kortum et al. 2014).
4.4. Novel immunoglobulin-like transcripts (NILTs)
NILTs have been described from carp, trout and salmon and are encoded on zebrafish chromosome 1 (Kock and Fischer 2008;Ostergaard et al. 2009;Ostergaard et al. 2010;Stet et al. 2005). Individual NILTs encode one or two Ig domains, which exhibit sequence and structural similarity to human TREM/NKp44, human CD300 and Xenopus pIgR Ig domains (Kock and Fischer 2008;Stet et al. 2005). NILTs encode type I transmembrane receptors that possess cytoplasmic ITIMs or ITAMs as well as secreted isoforms. NILTs are encoded by a multigene family as evidenced by Southern blot analyses in carp and sequencing of six NILT genes from a single salmon genomic (BAC) clone (Ostergaard et al. 2010;Stet et al. 2005). Although no reports have been published describing zebrafish NILT transcripts, nilt1 and nilt2 (GenBank: BN001234 and BN001235), respectively, are predicted to encode one and two Ig ectodomains and cytoplasmic itims (Ostergaard et al. 2009;Stet et al. 2005). This is likely an under-representation of the NILT genes encoded by zebrafish as 104 NILT-related Ig domains can be identified on chromosome 1 of the current (Zv9) zebrafish reference genome (Wcisel and Yoder, unpublished observations).
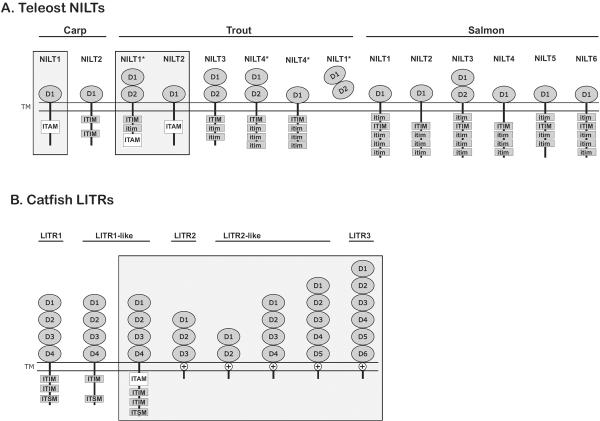
The two types of carp NILTs are: Cyca-NILT1 which encodes a single Ig domain and a cytoplasmic ITAM, and Cyca-NILT2 which encodes a single Ig domain and cytoplasmic ITIMs (Figure 3; Stet et al. 2005). Four types of NILTs have been described in trout. Onmy-NILT1 undergoes alternative splicing to generate a membrane bound isoform that possesses two Ig ectodomains, a cytoplasmic ITIM, a cytoplasmic itim and cytoplasmic ITAM as well as a secreted isoform. Onmy-NILT2 encodes a single Ig ectodomain and a cytoplasmic ITAM. Onmy-NILT3 encodes two Ig domains and three cytoplasmic ITIM/itim sequences. Onmy-NILT4 encodes a type I transmembrane protein that possesses four cytoplasmic ITIM/itim sequences and undergoes alternative splicing resulting in isoforms that possess one or two Ig ectodomains (Figure 3; Kock and Fischer 2008;Ostergaard et al. 2009). There is evidence for more than nine NILT genes in salmon in which the nomenclature has named new sequences sequentially (e.g. Sasa-NILT1 was the first described) and the structural variation of salmon NILTs reflects that observed in trout (Figure 3). As RT-PCR amplicons from a single carp reveal 53 different NILT sequences (Stet et al. 2005), it is highly likely that many more NILTs remain to be described from all of these species and not all structural variants have been recognized. The genomic organization and structural variation of zebrafish NILTs remains to be described and annotated.
Figure 3. Predicted structures of teleost NILT and LITR proteins.
Sequences have been reported (Montgomery et al. 2011;Ostergaard et al. 2009;Ostergaard et al. 2010;Stet et al. 2005). Proteins are organized numerically: activating receptors are boxed. The Ig domains of (A) NILT (D1 and D2) and (B) LITR (D1, D2, D3, D4, D5 and D6) proteins are indicated. Cytoplasmic ITAMs, ITIMs, ITIM-like (itim) sequences and tyrosines as well as charged residues (⊕) within transmembrane domains are labeled. Proteins encoded by splice variants are indicated with asterisks (*). This is not meant to be a complete catalog of proteins from these families as additional proteins sequences and structures will likely be identified in the future.
The expression of carp and trout NILTs has been described. The highest expression is seen in lymphoid tissues (kidney, spleen and thymus) and it is anticipated that zebrafish NILTs will display similar expression patterns (Kock and Fischer 2008;Ostergaard et al. 2009;Stet et al. 2005). Ligands for NILTs remain unknown.
4.5. Leukocyte immune-type receptors (LITRs)
LITRs are type I transmembrane proteins that possess two to six Ig ectodomains and have been best described from channel catfish (Figure 3; reviewed by Montgomery et al. 2011). Catfish LITRs are encoded by a multi-gene family located at multiple loci and include inhibitory and activating forms (Figure 3; Cortes et al. 2012;Montgomery et al. 2012;Stafford et al. 2006;Stafford et al. 2007). LITR Ig domains share similarity to mammalian FcR, FcR-like, KIR, LILR and NKp46 sequences (Stafford et al. 2006). Although zebrafish LITR transcripts have not been identified, putative LITR genes from Zv8 have been described (Stafford et al. 2006). Conservative estimates suggest at least 137 Ig domains from the current zebrafish reference genome (Zv9) belong to the LITR family which share 33–58% identity with catfish LITRs (Wcisel and Yoder, unpublished observations). This high level of sequence diversity between zebrafish and catfish LITRs confound efforts to predict the number and organization of individual zebrafish LITR genes based only on genomic Ig domain exons.
Expression analyses reveal catfish LITR transcripts to be the highest in kidney, gill, spleen and PBLs, as compared to liver, muscle and intestine. LITR transcripts were detected in macrophage, B cell, cytotoxic T cell, and NK-like cell lines (Stafford et al. 2006). In addition, multiple LITR transcripts are increased dramatically in PBLs and cytotoxic T cell lines after exposure to alloantigen (Stafford et al. 2007). Although ligands have not been identified for LITRs, some LITRs are predicted to bind MHCI (Stafford et al. 2007). Recombinant forms of ITIM-containing LITRs associate with SHP-1 and SHP-2 following treatment with the protein phosphatase inhibitor, pervanadate (Montgomery et al. 2009). In the context of transfected mammalian cells, surface expression of a recombinant activating LITR, which possesses a positively charged residue within the transmembrane domain, is increased by co-expression with the ITAM-containing adaptor proteins FcRγ and FcRγ-like. Activating LITRs associate with these adaptor proteins as well as with CD3ζ-like (Mewes et al. 2009). In addition, recombinant activating LITRs have been shown to form non-covalent homo- and heterodimers suggesting a complex mechanism for regulating LITR-mediated signaling.
The nomenclature of catfish LITRs has been based on the structures of the initial LITR transcripts designated, LITR1 (which encodes an inhibitory receptor) and LITR2 and LITR3 (both of which are activating receptors) (Figure 3; Stafford et al. 2006). Subsequent LITR transcripts have been named based on their similarity to the original transcripts (e.g. LITR1-like and LITR2-like) (Montgomery et al. 2011;Stafford et al. 2007). The complete genomic organization and structural variation of LITRs remains to be described and annotated in any fish species including zebrafish.
5. Defining the Ig domain differences
The zebrafish protein families discussed above can be distinguished from one another based on gene organization, sequence variation (see Figure 1) and Ig domain structure including the spacing of cysteine residues within Ig domains. The general characteristics of each family are summarized in Table 1. Whereas overall protein structures and signaling capabilities (via ITIMs/ITAMs or adaptor molecules) are conserved between protein families, many unique features distinguish each gene family, even in the absence of complete functional data. Given the remarkably large dataset now available, consisting of 418 different Ig domains, broad classifications can be attributed.
Table 1.
Sequence and structural characteristics of immunoglobulin domain-containing innate immune receptor families in zebrafish
| Feature | NITR | DICP | PIGRL | NILT | LITR |
|---|---|---|---|---|---|
| Min. estimate of genes | 39 | 29 | 29 | >52a | >23a |
| Ig domains per molecule | 1–2 | 1–2 | 2 | 1–2b | 1–6b |
| Encoded by zebrafish chromosomes | 7, 14 | 3, 14, 16 | 2 | 1 | 3, 7 |
| Members with cytoplasmic ITAM | No | No | No | Yesb | Nob |
| Members with charged residue in TM | Yes, + charge | Yes, − charge | Unknown | Nob | Yes, + chargeb |
| Adaptor protein | Dap12 | Unknown | Unknown | Unknown | FcRγ, FcRγLb |
| Members with cytoplasmic ITIM | Yes | Yes | Yes | Yesb | Yesb |
| Ig types | V, I | V-like | V | V-like | C2-like |
| C23 to C104 spacing | 64–73 | 59–67 | 59–66 | 56–63b | 36, 40–52b |
| Spacing of additional cysteine pairs | No internal C's in V domainc | 35–43 | 6, 7, 9d | 3, 6, 7b | No internal C'sb |
| Notable Features | J domains; mediates allogeneic recognitionb | Poly-serine in D2 | Conserved plgR motif | CX3C motifb | Putative MHC binding siteb |
| (I/V)(F/Y)X(L/V)X4LX5GXYXCX(I/V) motif | Yes | No | Yes | Yesb | Nob |
| Ligands | Unknown | Phospholipids | Phospholipids | Unknown | Unknown |
Approximate numbers of NILT and LITR genes were calculated by dividing the total number of Ig domains by two (NILTs) or six (LITRs)
Based on transcripts from catfish, carp, trout and/or salmon
NITR I domains contain six conserved cysteine residues and are excluded from this analysis
Not all Ig domains possess an extra cysteine pair
Within the Ig domains of each family, there is a large degree of amino acid sequence variation which is complicated by the observation that each gene family possesses multiple groups of similar Ig domains. In contrast to the NITR, DICP and PIGRL families which encode two very distinct types of Ig domains, the NILT and LITR families possess at least three and six different types of Ig domains, respectively (Haire et al. 2012;Kortum et al. 2014;Montgomery et al. 2011;Ostergaard et al. 2010;Yoder 2009). Pairwise sequence comparisons reveal that homology between Ig domains of the same type, for example NITR V domains, can be as low as 45% identical and 50% similar (Wcisel and Yoder, unpublished observations). Despite the relatively reduced level of sequence conservation, detailed comparisons reveal that Ig domains group together by Ig type within each gene family (Figure 1), revealing signatures within the predicted structures of the Ig domains that are unique to each receptor family.
The spacing between conserved pairs of cysteine residues is relatively unique among the gene families. As discussed above, the spacing between C23 and C104 can be employed to characterize an Ig domain as V, C1, C2, or I and are separated by 44–75 residues (Barclay 2003;Harpaz and Chothia 1994;Smith and Xue 1997). When available, x-ray crystallography or protein structure modeling confirmed the classifications for these protein families (Cannon et al. 2008;Ostrov et al. 2007;Stet et al. 2005). Additional pairs of cysteine residues also appear to be unique among each gene family (Table 1). For example, NITR I domains possess four additional highly conserved cysteines: three that fall between C23 and C104 and one at position C109, whereas NITR V domains do not possess additional conserved cysteines (Yoder 2009). In DICP D1 and D2 domains, a wide spacing between two additional conserved cysteines (CX35–43 C) present between C23 and C104 is observed. In the case of PIGRL D2 domains, two additional cysteines separated by six residues (CX6C) are seen, whereas only the PIGRL1, PIGRL3, and PIGRL4 D1 domains possess two additional cysteines (C42 and C49/C50/C52) spaced seven or nine residues apart (CX7C or CX9C). D1 domains of PIGRL2 proteins do not possess these additional cysteines (Kortum et al. 2014). NILT Ig domains can be divided into at least three subgroups based on the spacing between cysteines present in the C and C' strands: CX3C, CX6C and CX7C (Ostergaard et al. 2010). LITR Ig domains lack additional conserved cysteine pairs.
The Ig domains of NILTs and NITR I domains share a 21 residue motif (I/VF/YXL/VX4LX5GXYXCXI/V) that includes C104 as well as the residues that are conserved in classical V domains (Kock and Fischer 2008); many PIGRL proteins also contain this sequence. This motif spans the E and F strands in opposing β-sheets of the Ig fold and may be important for defining a ligand binding pocket (Kock and Fischer (2008).
Many of the gene families also contain unique sequences not shared across the other families. Many NITR V and I domains contain a sequence similar to joining (J) domains (FGXGTXLXV) present in immunoglobulins and T-cell receptors after V(D)J recombination (Litman et al. 2001;Yoder 2009). PIGRL proteins share a conserved IPCXY motif at the C23 position with pIgR molecules (Kortum et al. 2014). DICP D2 domains typically contain a poly-serine stretch in the amino-terminus which is caused by a triplet repeat expansion at the beginning of this exon (Haire et al. 2012).
6. Evidence for haplotypic variation
The human KIR gene cluster displays polymorphic and copy number variation between individuals resulting in over one hundred known KIR haplotypes (Middleton and Gonzelez 2010). Figure 4 demonstrates that the NITR, DICP, PIGRL, NILT and LITR gene clusters are located in chromosomal regions with high levels of copy number variation. In addition, there is growing evidence that the polymorphic NITR, DICP and PIGRL gene clusters display multiple haplotypes that encode different genes (Haire et al. 2012;Kortum et al. 2014;Yoder et al. 2004;Yoder et al. 2008). A comparison of the NITR, DICP and PIGRL loci to the Database of Genomic Variants archive (DGVa; Lappalainen et al. 2013) reveals that these gene clusters are indeed in chromosomal regions that display high levels of copy number variation, suggestive of gain and loss of haplotypes between individuals (Figure 5). Detailed analyses of the zebrafish NILT and LITR clusters remain to be examined, but are expected to display multiple haplotypes as well.
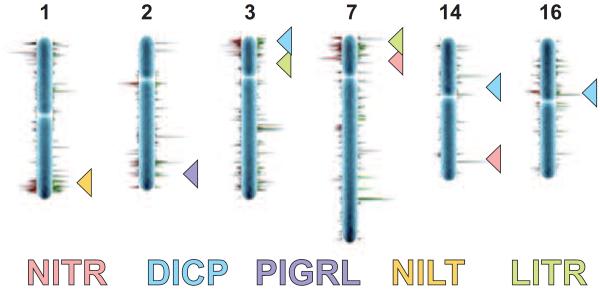
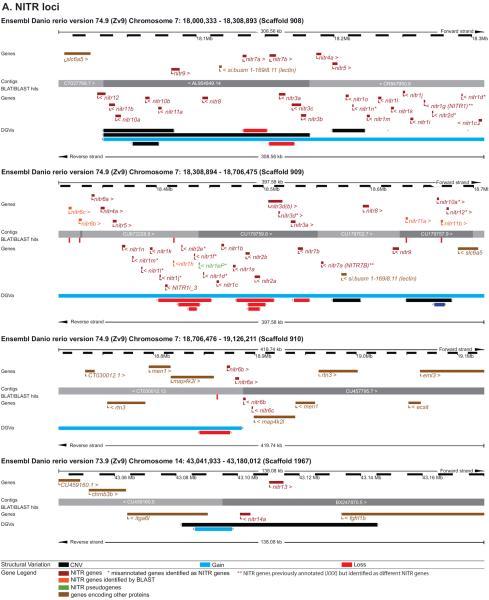
Figure 4. Genomic loci of zebrafish NITR, DICP, PIGRL, NILT and LITR gene clusters.
The location of each zebrafish NITR, DICP, PIGRL, NILT and LITR locus is shown in relation to a published chromosomal map of copy number variation (CNV). 31,749 CNVs found in 80 individual zebrafish were combined into a non-redundant dataset of 6,080 CNV elements (CNVEs) (Brown et al. 2012). Lengths of green (CNV gain) and red (CNV loss) horizontal lines reflect relative CNVE frequencies at respective chromosomal locations. CNV image courtesy of Charles Lee, The Jackson Laboratory Institute for Genomic Medicine.
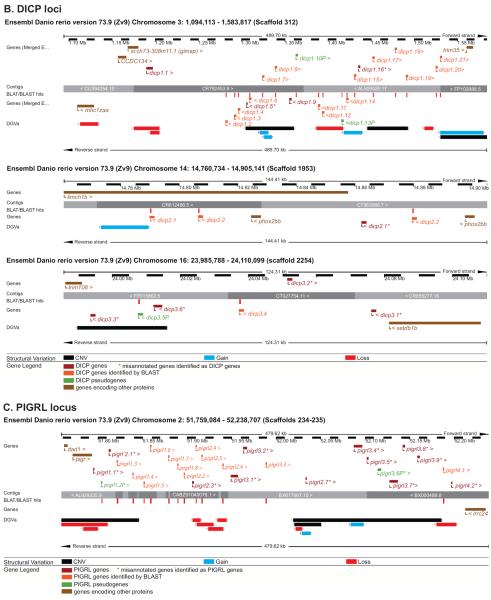
Figure 5. Evidence for haplotypic variation at the NITR, DICP and PIGRL loci.
The regions of (A) chromosomes 7 and 14 that encode the NITR genes, (B) chromosome 3, 14 and 16 that encode the DICP genes and (C) chromosome 2 that encodes the PIGRL genes are shown in detail. Graphics were adapted from the Ensembl Genome Browser, www.ensembl.org/Danio_rerio/. The NITR, DICP and PIGRL genes (red and orange) and relevant flanking genes (brown) are annotated above and below the genomic contigs. The Database of Genomic Variants archive (DGVa) provides the genomic structural variants: regions of copy number variation (CNV) are indicated in black, regions of gene loss are indicated in red, and regions of gene gain are indicated in blue.
Haplotypic variation may explain the difficulties experienced in assembling these regions of the reference genome as DNA from multiple individuals were merged into the reference genome. For example chromosome 7 genomic scaffolds 908 and 909 that encode numerous highly similar NITR genes (Figure 5A) show different organizations for these genes: in scaffold 908, the nitr3 genes are flanked by nitr4 and nitr7 genes while in scaffold 909 the nitr3 genes are flanked by nitr2 and nitr7 genes. Although this could be a result of recent gene duplication events, it may also be the result of misassembled scaffolds resulting from joining contigs derived from different haplotypes. In addition, chromosome 7 scaffold 910 joins two contigs that both encode the map4k2l, men1 and rtn3 genes adjacent to two nitr6 genes; in this case, it seems likely that each contig may represent a different haplotype. A similar scenario is observed for scaffold 1953 that joins two contigs that both encode dicp2.1, dicp2.2 and phox2bb (Figure 5B). It is likely that the haplotypic variation at these loci is far more complex than described here and may play a significant role in inter-individual immune health.
7. Conclusions
Investigations of large multigene families encoding receptor-like molecules that are predicted to effect innate immune function have been carried out in a number of different species of bony fish. In most cases, orthology with molecules identified in higher vertebrate forms cannot be recognized. In the case of the NITRs, the most extensively characterized of these gene families, the patterns of divergence in disparate groups are similar, although the genetic variants are extensively diverged both within and between multigene families. Taken together with the extensive differences in domain types and numbers, putative mechanisms of intracellular signaling and in some cases haplotypic and extensive allelic variation, it appears as if chromosomal mechanisms are in place that facilitate rapid divergence and functional specialization of different classes of immune-type receptors even though species are presumed to be subjected to similar selective processes. Further exploration of the functional variation in such molecules will rely heavily on the use of new methods of targeted gene disruption. Investigations into the mechanisms of genetic variation that occurs in these receptor gene families and how they are stabilized in the different species represent key focus areas for future avenues of research involving alternative pathways of innate immunity.
Highlights
Ig domain-containing innate immune receptor multifamilies are present in bony fish.
Orthologs for many of these receptors cannot be identified in higher vertebrates.
Receptors include inhibitory, activating, functionally ambiguous and secreted forms.
The genes encoding these receptors display polymorphic and haplotypic variation.
Acknowledgements
We are very grateful to Charles Lee (The Jackson Laboratory Institute for Genomic Medicine) for providing the chromosomal map of copy number variation and to Barb Pryor for editorial assistance. The authors are supported by the National Institutes of Health (R01 AI057559 to GWL and JAY and R01 AI23337 to GWL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Asano M, Komiyama K. Polymeric immunoglobulin receptor. J. Oral Sci. 2011;53:147–156. doi: 10.2334/josnusd.53.147. [DOI] [PubMed] [Google Scholar]
- Barclay AN. Membrane proteins with immunoglobulin-like domains—a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–223. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Barrow AD, Trowsdale J. You say ITAM and I say ITIM, let's call the whole thing off: the ambiguity of immunoreceptor signalling. Eur. J. Immunol. 2006;36:1646–1653. doi: 10.1002/eji.200636195. [DOI] [PubMed] [Google Scholar]
- Braathen R, Hohman VS, Brandtzaeg P, Johansen FE. Secretory antibody formation: conserved binding interactions between J chain and polymeric Ig receptor from humans and amphibians. J. Immunol. 2007;178:1589–1597. doi: 10.4049/jimmunol.178.3.1589. [DOI] [PubMed] [Google Scholar]
- Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, Chong WW, Chen JY, Yoo P, David S, Peterson SM, Raj T, Choy KW, Stranger BE, Williamson RE, Zon LI, Freeman JL, Lee C. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavez F, Young NT, Guethlein LA, Rajalingam R, Khakoo SI, Shum BP, Parham P. Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J Immunol. 2001;167:5786–5794. doi: 10.4049/jimmunol.167.10.5786. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA. A Bony Fish Immune Receptor of the NITR Multigene Family Mediates Allogeneic Recognition. Immunity. 2008;29:228–237. doi: 10.1016/j.immuni.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Mueller MG, Litman RT, Eason DD, Tinnemore D, Amemiya CT, Ota T, Litman GW. Ancient divergence of a complex family of immune-type receptor genes. Immunogenetics. 2006;58:362–373. doi: 10.1007/s00251-006-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, O'Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012;64:39–47. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
- Choi SC, Simhadri VR, Tian L, Gil-Krzewska A, Krzewski K, Borrego F, Coligan JE. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J. Immunol. 2011;187:3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes HD, Montgomery BC, Verheijen K, Garcia-Garcia E, Stafford JL. Examination of the stimulatory signaling potential of a channel catfish leukocyte immune-type receptor and associated adaptor. Dev. Comp Immunol. 2012;36:62–73. doi: 10.1016/j.dci.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Deitcher DL, Mostov KE. Alternate splicing of rabbit polymeric immunoglobulin receptor. Mol. Cell Biol. 1986;6:2712–2715. doi: 10.1128/mcb.6.7.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derive M, Massin F, Gibot S. Triggering receptor expressed on myeloid cells-1 as a new therapeutic target during inflammatory diseases. Self Nonself. 2010;1:225–230. doi: 10.4161/self.1.3.12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt GR, Cooper MD. Immunoregulatory roles for fc receptor-like molecules. Curr. Top. Microbiol. Immunol. 2011;350:89–104. doi: 10.1007/82_2010_88. [DOI] [PubMed] [Google Scholar]
- Feng LN, Lu DQ, Bei JX, Chen JL, Liu Y, Zhang Y, Liu XC, Meng ZN, Wang L, Lin HR. Molecular cloning and functional analysis of polymeric immunoglobulin receptor gene in orange-spotted grouper (Epinephelus coioides) Comp Biochem. Physiol B Biochem. Mol. Biol. 2009;154:282–289. doi: 10.1016/j.cbpb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr. Opin. Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibot S, Kolopp-Sarda MN, Bene MC, Bollaert PE, Lozniewski A, Mory F, Levy B, Faure GC. A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J Exp. Med. 2004;200:1419–1426. doi: 10.1084/jem.20040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire RN, Cannon JP, O'Driscoll ML, Ostrov DA, Mueller MG, Turner PM, Litman RT, Litman GW, Yoder JA. Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics. 2012;99:282–291. doi: 10.1016/j.ygeno.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamuro K, Suetake H, Saha NR, Kikuchi K, Suzuki Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J. Immunol. 2007;178:5682–5689. doi: 10.4049/jimmunol.178.9.5682. [DOI] [PubMed] [Google Scholar]
- Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- Helaers R, Milinkovitch MC. MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC. Bioinformatics. 2010;11:379. doi: 10.1186/1471-2105-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima J, Jung TS, Aoki T. Immunoglobulin genes and their transcriptional control in teleosts. Dev. Comp Immunol. 2011;35:924–936. doi: 10.1016/j.dci.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P, Betsholtz C, Brandtzaeg P. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 1999;190:915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- Kock H, Fischer U. A novel immunoglobulin-like transcript from rainbow trout with two Ig-like domains and two isoforms. Mol. Immunol. 2008;45:1612–1622. doi: 10.1016/j.molimm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Kortum AN, Rodriguez-Nunez I, Yang J, Shim A, Runft D, O'Driscoll M, Haire RN, Cannon JP, Turner PM, Litman RT, Kim CH, Neely MN, Litman GW, Yoder JA. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics. 2014 doi: 10.1007/s00251-014-0759-4. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulseth MA, Krajci P, Myklebost O, Rogne S. Cloning and characterization of two forms of bovine polymeric immunoglobulin receptor cDNA. DNA Cell Biol. 1995;14:251–256. doi: 10.1089/dna.1995.14.251. [DOI] [PubMed] [Google Scholar]
- Lappalainen I, Lopez J, Skipper L, Hefferon T, Spalding JD, Garner J, Chen C, Maguire M, Corbett M, Zhou G, Paschall J, Ananiev V, Flicek P, Church DM. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Hawke NA, Yoder JA. Novel immune-type receptor genes. Immunol Rev. 2001;181:250–259. doi: 10.1034/j.1600-065x.2001.1810121.x. [DOI] [PubMed] [Google Scholar]
- Mewes J, Verheijen K, Montgomery BC, Stafford JL. Stimulatory catfish leukocyte immune-type receptors (IpLITRs) demonstrate a unique ability to associate with adaptor signaling proteins and participate in the formation of homo- and heterodimers. Mol. Immunol. 2009;47:318–331. doi: 10.1016/j.molimm.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BC, Cortes HD, Burshtyn DN, Stafford JL. Channel catfish leukocyte immune-type receptor mediated inhibition of cellular cytotoxicity is facilitated by SHP-1-dependent and -independent mechanisms. Dev. Comp Immunol. 2012;37:151–163. doi: 10.1016/j.dci.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Montgomery BC, Cortes HD, Mewes-Ares J, Verheijen K, Stafford JL. Teleost IgSF immunoregulatory receptors. Dev. Comp Immunol. 2011;35:1223–1237. doi: 10.1016/j.dci.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Montgomery BC, Mewes J, Davidson C, Burshtyn DN, Stafford JL. Cell surface expression of channel catfish leukocyte immune-type receptors (IpLITRs) and recruitment of both Src homology 2 domain-containing protein tyrosine phosphatase (SHP)-1 and SHP-2. Dev. Comp Immunol. 2009;33:570–582. doi: 10.1016/j.dci.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Ostergaard AE, Lubieniecki KP, Martin SA, Stet RJ, Davidson WS, Secombes CJ. Genomic organisation analysis of novel immunoglobulin-like transcripts in Atlantic salmon (Salmo salar) reveals a tightly clustered and multigene family. Bmc Genomics. 2010;11:697. doi: 10.1186/1471-2164-11-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard AE, Martin SA, Wang T, Stet RJ, Secombes CJ. Rainbow trout (Oncorhynchus mykiss) possess multiple novel immunoglobulin-like transcripts containing either an ITAM or ITIMs. Dev. Comp Immunol. 2009;33:525–532. doi: 10.1016/j.dci.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Ostrakhovitch EA, Li SS. The role of SLAM family receptors in immune cell signaling. Biochem. Cell Biol. 2006;84:832–843. doi: 10.1139/o06-191. [DOI] [PubMed] [Google Scholar]
- Ostrov DA, Hernandez Prada JA, Haire RN, Cannon JP, Magis AT, Bailey K, Litman GW. Crystallization and X-ray diffraction analysis of a novel immune-type receptor from Ictalurus punctatus and phasing by selenium anomalous dispersion methods. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2007;63:1035–1037. doi: 10.1107/S1744309107054231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R. Soc. Lond B Biol. Sci. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patowary A, Purkanti R, Singh M, Chauhan R, Singh AR, Swarnkar M, Singh N, Pandey V, Torroja C, Clark MD, Kocher JP, Clark KJ, Stemple DL, Klee EW, Ekker SC, Scaria V, Sivasubbu S. A sequence-based variation map of zebrafish. Zebrafish. 2013;10:15–20. doi: 10.1089/zeb.2012.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast JP, Haire RN, Litman RT, Pross S, Litman GW. Identification and characterization of T-cell antigen receptor related genes in phylogenetically diverse vertebrate species. Immunogenetics. 1995;42:204–212. doi: 10.1007/BF00191226. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- Ribeiro CM, Bird S, Raes G, Ghassabeh GH, Schijns VE, Pontes MJ, Savelkoul HF, Wiegertjes GF. A novel soluble immune-type receptor (SITR) in teleost fish: carp SITR is involved in the nitric oxide-mediated response to a protozoan parasite. PLoS. One. 2011;6:e15986. doi: 10.1371/journal.pone.0015986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombout JH, van der Tuin SJ, Yang G, Schopman N, Mroczek A, Hermsen T, Taverne-Thiele JJ. Expression of the polymeric Immunoglobulin Receptor (pIgR) in mucosal tissues of common carp (Cyprinus carpio L.) Fish. Shellfish. Immunol. 2008;24:620–628. doi: 10.1016/j.fsi.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Shah RN, Rodriguez-Nunez I, Eason DD, Haire RN, Bertrand JY, Wittamer V, Traver D, Nordone SK, Litman GW, Yoder JA. Development and characterization of anti-nitr9 antibodies. Adv. Hematol. 2012;2012:596925. doi: 10.1155/2012/596925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlapatska LM, Mikhalap SV, Berdova AG, Zelensky OM, Yun TJ, Nichols KE, Clark EA, Sidorenko SP. CD150 association with either the SH2-containing inositol phosphatase or the SH2-containing protein tyrosine phosphatase is regulated by the adaptor protein SH2D1A. J. Immunol. 2001;166:5480–5487. doi: 10.4049/jimmunol.166.9.5480. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DK, Xue H. Sequence profiles of immunoglobulin and immunoglobulin-like domains. J Mol Biol. 1997;274:530–545. doi: 10.1006/jmbi.1997.1432. [DOI] [PubMed] [Google Scholar]
- Stafford JL, Bengten E, Du PL, McIntosh RD, Quiniou SM, Clem LW, Miller NW, Wilson M. A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics. 2006;58:758–773. doi: 10.1007/s00251-006-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Bengten E, Du PL, Miller NW, Wilson M. Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenetics. 2007;59:77–91. doi: 10.1007/s00251-006-0169-3. [DOI] [PubMed] [Google Scholar]
- Stet RJ, Hermsen T, Westphal AH, Jukes J, Engelsma M, Lidy Verburg-van Kemenade BM, Dortmans J, Aveiro J, Savelkoul HF. Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics. 2005;57:77–89. doi: 10.1007/s00251-005-0771-9. [DOI] [PubMed] [Google Scholar]
- Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, Rast JP, Amemiya CT, Litman GW. A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci U S A. 1999;96:15080–15085. doi: 10.1073/pnas.96.26.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiso TM, Sharma A, Hordvik I. Analysis of polymeric immunoglobulin receptor- and CD300-like molecules from Atlantic salmon. Mol. Immunol. 2011;49:462–473. doi: 10.1016/j.molimm.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Vely F, Vivier E. Conservation of structural features reveals the existence of a large family of inhibitory cell surface receptors and noninhibitory/activatory counterparts. J Immunol. 1997;159:2075–2077. [PubMed] [Google Scholar]
- Waterman PM, Cambier JC. The conundrum of inhibitory signaling by ITAM-containing immunoreceptors: potential molecular mechanisms. FEBS Lett. 2010;584:4878–4882. doi: 10.1016/j.febslet.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Zhuo J-M, Chen X, Shah R, Liu J, Orcutt TM, Traver D, Djeu JY, Litman GW, Yoder JA. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics. 2007;59:813–821. doi: 10.1007/s00251-007-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland WH, Orzaez D, Lammers A, Parmentier HK, Verstegen MW, Schots A. A functional polymeric immunoglobulin receptor in chicken (Gallus gallus) indicates ancient role of secretory IgA in mucosal immunity. Biochem. J. 2004;380:669–676. doi: 10.1042/BJ20040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Ann. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Yoder JA. Form, function and phylogenetics of NITRs in bony fish. Dev. Comp Immunol. 2009;33:135–144. doi: 10.1016/j.dci.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW. Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics. 2008;60:257–265. doi: 10.1007/s00251-008-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman GW. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics. 2011;63:123–141. doi: 10.1007/s00251-010-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Ota T, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW. Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A. 2004;101:15706–15711. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc. Natl. Acad. Sci. USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–164. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW. Developmental and tissue-specific expression of NITRs. Immunogenetics. 2010;62:117–122. doi: 10.1007/s00251-009-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]