Highlights
-
•
In 2012 a novel coronavirus emerged in the Middle East region.
-
•
MERS-CoV causes a severe lower respiratory tract infection in humans.
-
•
Dromedary camels were found to be positive for MERS-CoV.
-
•
MERS-CoV chains of transmission in humans do not seem to be self-sustaining.
-
•
Isolation of MERS patients combined with limiting the zoonotic events may be crucial in controlling the outbreak.
Abstract
A novel coronavirus (CoV) that causes a severe lower respiratory tract infection in humans, emerged in the Middle East region in 2012. This virus, named Middle East respiratory syndrome (MERS)-CoV, is phylogenetically related to bat CoVs, but other animal species like dromedary camels may potentially act as intermediate hosts by spreading the virus to humans. Although human to human transmission has been demonstrated, analysis of human MERS clusters indicated that chains of transmission were not self-sustaining, especially when infection control was implemented. Thus, timely identification of new MERS cases followed by their quarantine, combined with measures to limit spread of the virus from the (intermediate) host to humans, may be crucial in controlling the outbreak of this emerging CoV.
Current Opinion in Virology 2014, 5:58–62
This review comes from a themed issue on Emerging viruses
Edited by Christopher F Basler and Patrick CY Woo
For a complete overview see the Issue and the Editorial
Available online 28th February 2014
1879-6257/$ – see front matter, © 2014 Elsevier B.V. All rights reserved.
Introduction
Human coronaviruses (CoVs) such as NL63, 229E, OC43, and HKU1 are distributed worldwide and cause a significant percentage of all colds. Most of these viruses emerged through zoonotic transmission from bats. Ten years ago, a more pathogenic CoV named severe acute respiratory syndrome (SARS)-CoV, crossed the species barrier [1, 2]. SARS-CoV originated from horseshoe bats, but palm civets acted as intermediate host allowing animal-to-human transmission [3, 4]. Current knowledge indicates that human CoVs emerged from animal ancestors and that various animal CoVs also crossed the species barrier [5, 6, 7, 8, 9, 10, 11, 12]. The high frequency of genetic recombination and mutation make CoVs well capable of adapting to new hosts [13, 14, 15, 16, 17, 18]. More recently a novel human coronavirus emerged, the Middle East respiratory syndrome (MERS)-CoV.
Human lower respiratory tract infection with unknown cause in the Middle East region
Although a cluster of 11 patients including 10 health care workers with severe lower respiratory tract infection with unknown aetiology was reported in an intensive care unit in Zarga, Jordan in March/April 2012 [19], no causative agent could be identified at that time. Only half a year later, this outbreak was linked to a patient in Jeddah, Saudi Arabia; a 60-year-old man who died as a consequence of acute respiratory infection and renal failure [20••]. A 49-year old Qatari who was treated in the United Kingdom represented another case identified around the same time [21]. As of 27 December 2013, the World Health Organization announced a total of 170 confirmed cases of human infection, including 72 fatalities [22]. All cases were linked directly or indirectly to the Middle East region including Saudi Arabia, Jordan, Qatar, Oman, Kuwait and the United Arab Emirates [23••, 24]. The largest number of MERS-CoV cases has been reported from Saudi Arabia. Patients diagnosed in the United Kingdom, France, Germany, Italy, Spain, and Tunisia, were also linked to the Middle East [22]. In addition to severe lower respiratory tract infection, gastrointestinal symptoms including diarrhoea, vomiting, and abdominal pain are also infrequently observed. Approximately 75 percent of patients was reported to have at least one underlying medical condition.
Identification and characterization of MERS-CoV
Clinical samples collected from the patient in Jeddah that were used to inoculate monkey kidney cells, were instrumental in the initial identification and characterization of MERS-CoV. Using a pan-CoV reverse transcription-PCR (RT-PCR), a short fragment from a highly conserved region of the RNA-dependent RNA polymerase (RdRp) gene was amplified and the identity of this novel human coronavirus was thus revealed [20••]. Initial phylogenetic analysis of the sequence fragment along with those of known coronaviruses showed that MERS-CoV clustered together with bat CoVs HKU4 and HKU5 that belong to subgroup 2c of the linage Betacoronavirus [20••, 25]. It is the first lineage 2c virus to infect humans. The virus, tentatively named HCoV-EMC/2012 (GenBank accession number: JX869059) [25], was renamed Middle East respiratory syndrome (MERS)-CoV upon consultation with the coronavirus study group of the International Committee on the Taxonomy of Viruses [26].
The MERS-CoV genome is 30119 nucleotides (nt) in length and contains 10 predicted open reading frames (ORFs). The single-stranded, positive sense polyadenylated RNA genome has 5′ and 3′ untranslated regions (UTR) of 278 and 300 nt in length, respectively [25]. The 5′ end of the genome is translated to yield a large polyprotein that is cotranslationally cleaved in cis by two viral proteases into 16 functional nonstructural proteins that cooperatively form the complex machinery for viral RNA synthesis and RNA recombination. The region downstream of ORF1b is characterized by containing a variable number of structural proteins, including the spike, envelope, membrane, and nucleocapsid protein. MERS-CoV accessory proteins (ORF3, ORF4a, ORF4b, ORF5 and ORF8b) share no homology with any known host or virus protein, other than those of the closely related HKU4 and HKU5 strains of bat CoV.
Zoonotic transmission of MERS-CoV
Because most human CoVs originally emerged upon transmission from bats to other animal species and given the phylogenetic relation of MERS-CoV with bat CoVs like HKU4 and HKU5, MERS-CoV most likely originated from bats. Partial genome sequences from viruses closely related to MERS-CoV have been detected in bats from Africa and Europe [27, 28•]. Insectivore bats like Pipistrellus Pipistrellus are most likely a major reservoir of these group 2c bat CoVs. The identification of a relative small and conserved RdRp fragment from an Egyptian cave bat shown to be identical to the human MERS-CoV EMC isolate [29•], however, needs further investigation.
Evidence that bats may have served as the original MERS-CoV host species also comes from studies on the receptor usage by MERS-CoV. Dipeptidyl peptidase 4 (DPP4; also known as CD26) — expressed in the lower respiratory tract of humans — acts as a functional receptor for MERS-CoV [30•]. Importantly, MERS-CoV can also use the evolutionarily conserved DPP4 protein of Pipistrellus Pipistrellus bats to infect cells [30•]. It remains unclear whether MERS-CoV-like viruses in bats are able to use the DPP4 receptor, although recent investigations revealed that bat DPP4 genes have been subject to significant adaptive evolution, suggesting that the evolutionary lineage leading to MERS-CoV may have circulated in bats for a substantial time period [31]. Three positively selected residues in DPP4 were identified that directly interact with the viral spike protein. Interestingly, recent investigations on the origin of SARS-CoV revealed that closely related SARS-CoV-like viruses in horseshoe bats are able to infect human cells by using the human ACE2 receptor [32]. Therefore, direct transmission of bat CoVs to humans, or indirect transmission without requirement of virus adaptation in an intermediate host, is now considered a likely scenario to explain the emergence of novel human CoVs.
Considering that direct contact of humans with bats or their secreta may be rare, intermediate hosts that are susceptible to MERS-CoV may be involved in transmitting this virus to humans. In case of SARS-CoV, civet cats are thought to have been responsible for the transmission of this virus to humans, although other animal species present at the wet markets in southern China such as ferret badgers were also found to carry a SARS-CoV like virus. As a consequence, upon detection of MERS-CoV emergence, different animal species commonly found in the Middle East, such as camels and goats, are considered as potential intermediate hosts in the MERS-CoV outbreak (Figure 1 ). Characterization of crucial amino acid residues in DPP4 that are involved in binding the MERS-CoV spike protein revealed that these animal species are more likely able to use DPP4 as a functional receptor for MERS-CoV entry as compared to other animal species such as mice, cats, dogs, hamsters, and ferrets [33]. Cell lines originating from goats and camels were shown to be permissive to efficient replication of MERS-CoV [34].
Figure 1.
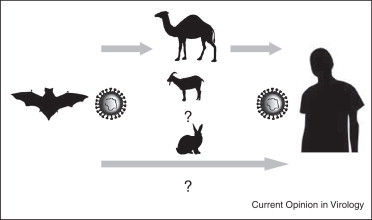
Schematic diagram of the potential zoonotic transmission of MERS-CoV.
Further evidence for the involvement of a specific animal species as an (intermediate) host comes from studies analyzing the host antibody response to MERS-CoV or closely related viruses. Initially, serological studies using samples from different animal species in Oman, Egypt, and the Canary Islands indeed provided clues for the presence of MERS-CoV neutralizing antibodies in dromedary camels [35, 36]. Subsequently, studies in camels in other affected regions, in Jordan and Saudi Arabia, confirmed these findings. Whereas a very high percentage of dromedary camels turned out to have circulating MERS-CoV neutralizing antibodies, other animal species such as sheep, goats, and cows were found negative for virus neutralizing antibodies [37, 38]. These observations have now been supported by more recent studies showing that dromedary camels from a farm in Qatar proved to be positive for MERS-CoV and virus neutralizing antibodies [39••]. In addition, the viral sequences obtained from these dromedary camels were almost identical to sequences from two human MERS-CoV cases linked to this farm. Therefore, dromedary camels most likely acquired the virus some time ago from bats and the virus has subsequently spread efficiently between animals in the Middle East region. Dromedary camels are used for racing and beauty contests and are kept in large groups at these festivities, likely promoting subsequent circulation of this virus. Although MERS-CoV infection in humans is mainly observed in the lower respiratory tract, in camels nose swabs were found virus positive. Conclusive evidence for the route of transmission from animals to humans, however, is still lacking. In addition, it is not clear whether the virus is introduced multiple times through zoonotic transmission or that human to human transmission is the main driver of the spread of the virus.
Human to human transmission
Human to human transmission of MERS-CoV has been reported in several clusters of cases in France, the United Kingdom, Italy, Jordan, Tunisia, Saudi Arabia, the United Arab Emirates, and Qatar, including among family members and health care workers [23••, 40•, 41, 42, 43, 44, 45, 46•, 42, 47, 48]. These include a cluster of cases in Saudi Arabia involving 3 family members living within the same house [43] and a family cluster of 3 brothers in Riyadh [46•]. A large cluster of 23 confirmed and 2 probable cases has been reported in a hospital in Al-Hasa, Saudi Arabia [23••]. The majority of patients experienced severe respiratory diseases and some had acute renal failure, whereas most common symptoms were fever, fever with chills or rigours, cough, shortness of breath, and myalgia. The patterns of spread of MERS-CoV among family or hospital clusters suggest that transmission occurs through droplets or contacts. Differences in receptor expression in the upper and lower respiratory tract of humans could potentially explain limited human to human transmission. Transmission appears to occur more readily if the recipient is immunocompromised or has comorbidities, such as diabetes. Since most identified patients had underlying diseases, it is possible that MERS-CoV is a more common infection, at least in Saudi Arabia, and that patients without significant comorbidities develop a mild respiratory disease or remain asymptomatic. However, spread of MERS-CoV is considered to be relatively inefficient, as two studies indicated that this viral infection does not seem to occur frequently in the normal human population in the Middle East region. Among 130 blood donors sampled in Jeddah in 2012 and 226 abattoir workers sampled in Jeddah and Makkah in October 2012, only 8 reactive sera were seen upon immunofluoresence testing that were all found to be specific for established human CoVs, but not for MERS-CoV [49]. In addition, Gierer et al. did not detect MERS-CoV neutralizing antibodies in any of the 268 samples tested that were obtained from persons from the Eastern province of Saudi Arabia [50]. Using independent data sources, different investigators demonstrated that R, the basic reproduction number representing the number of secondary cases per index case in a fully susceptible population, cannot be much above 1, with an upper bound of 1.2–1.5 [42, 51•]. In the absence of a clear picture of how the virus spreads, intervention strategies may be ineffective. In the case of predominant human to human transmission and absence of approved medication such as vaccines and antivirals, timely identification of new MERS cases followed by their isolation and quarantine may be crucial in controlling the outbreak of this emerging CoV. These measures may need to be combined with actions to limit spread and emergence of MERS-CoV from the (intermediate) host.
Conclusions
Until now, 170 confirmed MERS cases have been reported. The clinical symptoms caused by MERS-CoV infection mainly relate to the acute respiratory disease that is induced by the virus. By contrast to the SARS epidemic that rapidly was controlled, MERS-CoV still spreads more than 1.5 year after it was first detected. Therefore there is an urgent need to clarify whether the virus is introduced multiple times through zoonotic transmission or that human to human transmission is the main driver of the spread of the virus. A further understanding of the emergence and spread of this novel human CoV may halt its emergence and establishment in the human population.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The study was financed by the European Union FP7 projects EMPERIE (contract number 223498) and ANTIGONE (contract number 278976) and NIAID/NIH contract HHSN266200700010C.
References
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 4.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekseev K.P., Vlasova A.N., Jung K., Hasoksuz M., Zhang X., Halpin R., Wang S., Ghedin E., Spiro D., Saif L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol. 2008;82:12422–12431. doi: 10.1128/JVI.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin L., Cebra C.K., Baker R.J., Mattson D.E., Cohen S.A., Alvarado D.E., Rohrmann G.F. Analysis of the genome sequence of an alpaca coronavirus. Virology. 2007;365:198–203. doi: 10.1016/j.virol.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorusso A., Decaro N., Schellen P., Rottier P.J., Buonavoglia C., Haijema B.J., de Groot Gain R.J. Preservation, and loss of a group 1a coronavirus accessory glycoprotein. J Virol. 2008;82:10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorusso A., Desario C., Mari V., Campolo M., Lorusso E., Elia G., Martella V., Buonavoglia C., Decaro N. Molecular characterization of a canine respiratory coronavirus strain detected in Italy. Virus Res. 2009;141:96–100. doi: 10.1016/j.virusres.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Muller M.A., Annan A., Vallo P., Adu-Sarkodie Y., Kruppa T.F., Drosten C. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 14.Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau S.K., Lee P., Tsang A.K., Yip C.C., Tse H., Lee R.A., So L.Y., Lau Y.L., Chan K.H., Woo P.C., Yuen K.Y. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin–esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hijawi B., Abdallat M., Sayaydeh A., Alqasrawi S., Haddadin A., Jaarour N., Alsheikh S., Alsanouri T. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19:S12–S18. [PubMed] [Google Scholar]
- 20••.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]; First report on the identification of MERS-CoV.
- 21.Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 22.World Health Organization. Middle East syndrom coronavirus (MERS-CoV) – update – 27 December 2013. http://www.who.int/csr/don/2013_12_27/en/index.html
- 23••.Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H. Hospital outbreak of middle east respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive study that describes a large MERS-CoV hospital outbreak.
- 24.Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molercular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3 doi: 10.1128/mBio.00473-12. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS–CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–460. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C., Drosten C., Drexler J.F., Preiser W. Close relative of human middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]; This investigation provided evidence for the existence of group 2C betacoronaviruses in bats.
- 29•.Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; Preliminary evidence for the possible involvement of bats in the emergence of MERS-CoV.
- 30•.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of the receptor for MERS-CoV and showing it's localization in the human lower respiratory tract.
- 31.Cui J., Eden J.S., Holmes E.C., Wang L.F. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol J. 2013;10:304. doi: 10.1186/1743-422X-10-304.10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raj V.S., Smits S.L., Provacia L.B., van den Brand J.M., Wiersma L., Ouwendijk W.J., Bestebroer T.M., Spronken M.I., van Amerongen G., Rottier P.J. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4 mediated entry of the middle east respiratory syndrome coronavirus. J Virol. 2014;88:1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckerle I., Corman V.M., Müller M.A., Lenk M., Ulrich R.G., Drosten C. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg Infect Dis. 2014 doi: 10.3201/eid2002.131182. http://dx.doi.org/10.3201/eid2002.131182, Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Vries L.S., Corman V.M. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera R., Wang P., Gomaa M., El-Shesheny R., Kandeil A., Bagato O., Siu L., Shehata M., Kayed A., Moatasim Y. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 37.Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M., Poon L.L., Saif L., Alnaeem A., Peiris M. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.50.20659. Available online: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20659. [DOI] [PubMed] [Google Scholar]
- 38.Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S., Godeke G.J., Bestebroer T.M., Zutt I., Müller M.A. Middle East Respiratory Syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.50.20662. Available online: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20662. [DOI] [PubMed] [Google Scholar]
- 39••.Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70690-X. Available online 17 December 2013, ISSN 1473-3099, http://dx.doi.org/10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detection of MERS-CoV in dromedary camels revealing their possible role in the zoonotic transmission to humans.
- 40•.Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A., Al-Tawfiq J.A., Alhakeem R.F., Madani H., Alrabiah F.A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013 doi: 10.1016/S0140-6736(13)61887-5. pii: S0140-6736(13)61887-5. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Characterization of a large set of complete MERS-CoV genomes.
- 41.Guery B., Poissy J., el Mansouf L., Séjourné C., Ettahar N., Lemaire X., Vuotto F., Goffard A., Behillil S., Enouf V. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Health Protection Agency (HPA) UK Novel Coronavirus Investigation team Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013;18:20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- 43.Memish Z.A., Zumla A.I., Al-Hakeem R.F., Al-Rabeeah A.A., Stephens G.M. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 44.World Health Organization. MERS-CoV summary and literature update – 31 May 2013. http://www.who.int/csr/disease/coronavirus_infections/update_20130531/en/index.html.
- 45.Omrani A.S., Matin M.A., Haddad Q., Al-Nakhli D., Memish Z.A., Albarrak A.M. A family cluster of Middle East Respiratory Syndrome Coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int J Infect Dis. 2013;17:e668–e672. doi: 10.1016/j.ijid.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Breban R., Riou J., Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; A modelling study that estimated the risk of a MERS pandemic.
- 47.Mailles A., Blanckaert K., Chaud P., van der Werf S., Lina B., Caro V., Campese C., Guéry B., Prouvost H., Lemaire X. First cases of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill. 2013;18:20502. [PubMed] [Google Scholar]
- 48.Puzelli S., Azzi A., Santini M., Di Martino A., Facchini M., Castrucci M., Meola M., Arvia R., Corcioli F., Pierucci F. Investigation of an imported case of Middle East Respiratory Syndrome Coronavirus (MERS–CoV) infection in Florence, Italy, May to June 2013. Euro Surveill. 2013;18:20564. doi: 10.2807/1560-7917.es2013.18.34.20564. [DOI] [PubMed] [Google Scholar]
- 49.Aburizaiza A.S., Mattes F.M., Azhar E.I., Hassan A.M., Memish Z.A., Muth D., Meyer B., Lattwein E., Müller M.A., Drosten C. Investigation of anti-middle east respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia. J Infect Dis. 2012;Fall doi: 10.1093/infdis/jit589. 2013 Dec 4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gierer S., Hofmann-Winkler H., Albuali W.H., Bertram S., Al-Rubaish A.M., Yousef A.A., Al-Nafaie A.N., Al-Ali A.K., Obeid O.E., Alkharsah K.R., Pöhlmann S. Lack of MERS coronavirus neutralizing antibodies in humans, eastern province, Saudi Arabia. Emerg Infect Dis. 2013;19:2034–2036. doi: 10.3201/eid1912.130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Cauchemez S., Fraser C., Van Kerkhove M.D., Donnelly C.A., Riley S., Rambaut A., Enouf V., van der Werf S., Ferguson N.M. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70304-9. Nov 12. pii: S1473-3099(13)70304-9. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; An in depth modelling analysis of the MERS outbreak.


