Abstract
HOXB13 (G84E) was reported to significantly increase risk for prostate cancer. The goal of the current analysis was to assess the prevalence of G84E in ethnically-diverse high-risk men undergoing prostate cancer screening and place the carrier frequency within the context of prevalence estimates from reported studies to gain insight into the future role of this mutation in genetic counseling. PRAP is a prostate cancer screening program for unaffected men ages 35–69 with a family history of prostate cancer or African descent. HOXB13 G84E was genotyped by pyrosequencing in 649 PRAP participants with available DNA. Prevalence of the mutation was calculated for PRAP and for reported studies and exact binomial confidence intervals were generated. Prevalence of the G84E mutation in non-African PRAP men was 0.73 %. When placed in context of the literature, this was higher than reported controls. One G84E mutation carrier was notably of Hispanic background. While the HOXB13 G84E mutation may be rare, there may be a future role in genetic testing for this mutation after further studies of clinical utility in assessing prostate cancer risk.
Keywords: HOXB13, Family history, Genetic mutations, Prostate cancer
Introduction
Men with a family history of prostate cancer and African American men are at increased risk for developing prostate cancer (Cancer Facts and Figures 2013), with subsets at increased risk of aggressive disease and/or death from the disease (Bigler et al. 2011; Powell et al. 2010; Brandt et al. 2009; Hemminki et al. 2011). Screening studies in high-risk men (i.e. men with a family history of prostate cancer and African American men) have found that approximately 10 % of high-risk men will be diagnosed with prostate cancer at a mean age of 56.5 years and at an average interval of 2 years after starting screening (Giri et al. 2007) with cancer detection rates of 2–15 % (Giri et al. 2007; Catalona et al. 2002). While the risk for prostate cancer is increased in this group, many high-risk men undergo biopsies with no cancer detected based on prostate-specific antigen (PSA) criteria, incurring potential risks of biopsies. There is a need to better refine risk estimates for prostate cancer among high-risk men in order to provide individualized screening recommendations based on predicted risk.
Several genetic alterations have been reported to associate with prostate cancer risk, but the application to high-risk men or the clinical utility has been limited/uncertain. BRCA1 and BRCA2 mutations have been reported to raise the risk for prostate cancer in males from families with hereditary breast and ovarian cancer syndrome (HBOC) (Ford et al. 1994; BCLC 1999; Liede et al. 2004). However, mutations in these genes have not been shown to significantly account for prostate cancer in multi-case prostate cancer families (Zuhlke et al. 2004). Furthermore, there is very limited information of BRCA mutations and prostate cancer risk in African American men. Thus genetic testing for BRCA mutations for prostate cancer susceptibility is not clinically indicated for unaffected men from prostate cancer families or for African American men. Genomewide association studies (GWAS) have identified approximately 80 genetic variants associated with prostate cancer risk (Eeles et al. 2013). These variants identified from GWAS are relatively common in the population and modestly raise the risk for prostate cancer (Hindorff et al. 2009). However, due to their poor discriminative ability for risk for prostate cancer or for aggressive disease, panels of GWAS-identified markers have been determined to be of uncertain clinical utility at the present time (Pomerantz and Freedman (2013).
Recently, a recurrent mutation in HOXB13 (G84E) was identified to significantly increase the risk for prostate cancer particularly among families with prostate cancer (Ewing et al. 2012). A large confirmatory study from the International Consortium of Prostate Cancer Genetics which included 2, 443 prostate cancer families confirmed this finding (Xu et al. 2013). In particular, HOXB13 (G84E) was identified in ~5 % of prostate cancer families (meeting features of hereditary prostate cancer) of European ancestry and was associated with an approximate 4-fold increase in risk (Xu et al. 2013). Other studies have confirmed and further characterized this mutation for prostate cancer risk. One study reported findings that G84E appears to be restricted men primarily of Northern European descent (Chen et al. 2013). A recent pooled analysis including HOXB13 G84E results published in European Americans found an overall mutation frequency of 1.34 % among cases and 0.28 % among controls (Witte et al. 2013). The overall odds ratio for prostate cancer comparingHOXB13 G84E carriers vs. non-carriers was 4.86, and increased to an OR of 8.41 among men diagnosed with prostate cancer at age ≤55 years. HOXB13 G84E mutation carriers have been estimated to have a 33 % lifetime risk for prostate cancer in one study (Karlsson et al. 2012). Thus, this mutation may have a future role in providing high-risk men, particularly men with a family history of prostate cancer, with individualized risk for prostate cancer to inform prostate cancer screening recommendations in order to individualize prostate cancer screening strategies.
The goal of this descriptive analysis was to assess the prevalence of HOXB13 G84E in ethnically-diverse high-risk men undergoing prostate cancer screening and place the carrier frequency within the context of prevalence estimates reported from studies in the literature to gain insight into the potential future role of this mutation in genetic counseling and genetic testing for high-risk men. The cohort for this study was the Prostate Cancer Risk Assessment Program (PRAP)—a prostate cancer screening and research program for high-risk men with 60 % African American participation (Giri et al. 2007).
Methods
Prostate Cancer Risk Assessment Program (PRAP)
PRAP was established in 1996 to provide prostate cancer screening for men at high-risk for prostate cancer and to perform research into genetic susceptibility to the disease (Giri et al. 2007). Eligibility for PRAP include any man between ages 35–69 years without a previous diagnosis of prostate cancer with one first-degree relative with prostate cancer, 2 second-degree relatives with prostate cancer on the same side of the family, or any African American man regardless of family history. Accrual to PRAP is ongoing and participants are followed longitudinally for prostate cancer screening and cancer detection. This analysis includes participants consecutively accrued to PRAP from 1996 to 2009 with complete clinical and genotype data. The PRAP study is approved by the Institutional Review Board at FCCC.
Screening Approach in PRAP
Participants undergo annual prostate cancer screening, which includes the total PSA, digital rectal examination (DRE), and estimation of PSA velocity. Prostate cancer detection rates in PRAP and cancer characteristics have been described in detail previously (Giri et al. 2007). Current biopsy criteria include: (1) PSA greater than 2.0 ng/ml, (2) PSA 1.5 to 2.0 ng/ml with fPSA 25 % or less, (3) any abnormality on DRE or (4) PSAv 0.75 ng/ml/year. All biopsies are transrectal ultrasound guided 5-region biopsies with additional biopsies obtained at physician discretion.
Genotyping of HOXB13 G84E
DNA was isolated from blood samples collected from PRAP participants and stored per standard operating procedure in the Biosample Repository Facility at FCCC. Genotyping for the G84E mutation was performed using pyrosequencing. Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences are available on request. The forward primers were biotinylated to facilitate single-strand DNA template preparation. Reactions were prepared using Choice Taq Blue Mastermix (Denville Scientific Inc.), 10 µM of each primer, and 20 ng of genomic DNA according to manufacturer’s instructions. Thermal cycling conditions are available upon request. The biotinylated PCR product was immobilized on Streptavidin-coated Sepharose High Performance beads (GE Healthcare.) The beads were captured with the PyroMark Q96 Vacuum (Qiagen) generating single-stranded DNA suitable for pyrosequencing. The template DNA was deposited into the pyrosequencing reaction plate containing the sequencing primer, according to manufacturer’s instructions. The sequencing-by-synthesis reaction of the complementary strand was automatically performed using PyroMark Gold Q96 reagents (Qiagen) and the PSQ 96MA instrument (Qiagen) at room temperature. SNP assignment and quality assessment of the raw data was performed using PyroMark Q96 Software (Qiagen.)
Statistical Methods
The number of mutations reported within self-reported ethnic groups was first determined, and subsequently the main analysis was limited to participants with non-African American ancestry who had G84E genotype data available since no African American men tested positive for the G84E mutation. To summarize clinical and pathologic characteristics of the cohort, we calculated means, medians, ranges, and percentages. The prevalence of mutations was calculated as the percent known positive in the cohort, and because mutations were rare, we generated confidence intervals using the Clopper-Pearson exact binomial method (Clopper and Pearson 1934). For comparison, we also calculated prevalences and confidence intervals based on the number of mutations found in previously published studies, also using the exact binomial method. Statistical analyses were performed using STATA (version 12.1) and R (version 3.0.1).
Results
At the time of this study, 649 out of 777 PRAP participants (83.5 %) had DNA available for analysis. Genotype data was available for 248 White participants, 375 African American participants, 6 Hispanic men, and 20 with self-reported race as “other”. Consistent with previous studies reporting low carrier frequencies in African Americans (Ewing et al. 2012; Witte et al. 2013), none of the African American participants were found to carry the G84E mutation; therefore, we only describe characteristics and carrier frequency for G84E for non-African American PRAP participants. Table 1 shows the characteristics of the 274 non-African American participants included in this analysis. As seen in Table 1, the mean age at entry into PRAP for non-African American participants was 50 years, and 80 % of this group returned for follow-up. All of these men reported a family history of prostate cancer. Prostate cancer was diagnosed in 15 % of this group, with the mean age at diagnosis of 58.5 years. The median Gleason score was 6, with a range of 5–7.
Table 1.
Characteristics of 274 non-African American PRAP participants
| Characteristics | Non-African American participants with G84E data (n =274) |
|---|---|
| Age at entry (years) mean (range) | 50 (34–69) |
| Race, n(%) | |
| -White | 248 (91 %) |
| -Hispanic | 6 (2 %) |
| -Other | 20 (7 %) |
| PSA at entry, mean (range) ng/mLa | 1.66 (0.20, 22.50) |
| Digital rectal exam at entry, n(%)b | |
| -Normal/BPH | 259 (96 %) |
| -Abnormal | 10 (4 %) |
| Duration of Follow-up, months Mean (range) | 61.9 (0.6–189.0) |
| Follow up>= 1 visit, n(%) | 220 (80 %) |
| Prostate cancer diagnosis, n(%) | 40 (15 %) |
| Age at diagnosis (years), mean (range) | 58.5 (44–69) |
| PSA prior to diagnosis, mean (range) ng/mL | 3.99 (1.50–22.50) |
| Gleason score, median (range) | 6 (5–7) |
PSA missing on 1 participant
DRE data missing for 5 participants
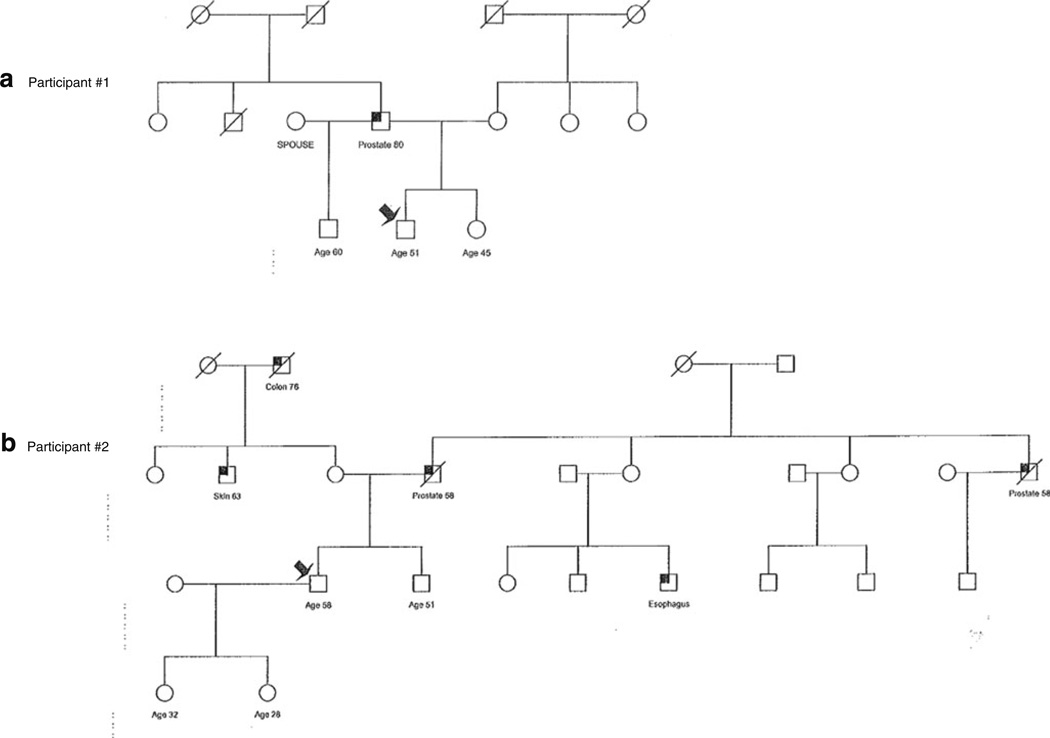
Two of the 274 non-African American participants were found to carry the G84E mutation in HOXB13, with a carrier frequency in this high-risk group of 0.73%. Of interest, one of the participants found to carry the G84E mutation was of self-reported Hispanic ethnicity. The pedigrees of these two PRAP participants found to carry the mutation are shown in Fig. 1. Of note, the first participant did not have a strong family history of prostate cancer. Participant #2 had a stronger family history of prostate cancer with his father and paternal uncle with prostate cancer reportedly diagnosed in their late 50’s.
Fig. 1.
Pedigrees of Two HOXB13 G84E+ PRAP Participants. a Pedigree of Participant #1 with G84E mutation shown by arrow who has father with prostate cancer at age 80. The proband is of self-reported Hispanic origin. b Pedigree of Participant #2 with G84E mutation showing father and paternal uncle with prostate cancer at younger ages of onset
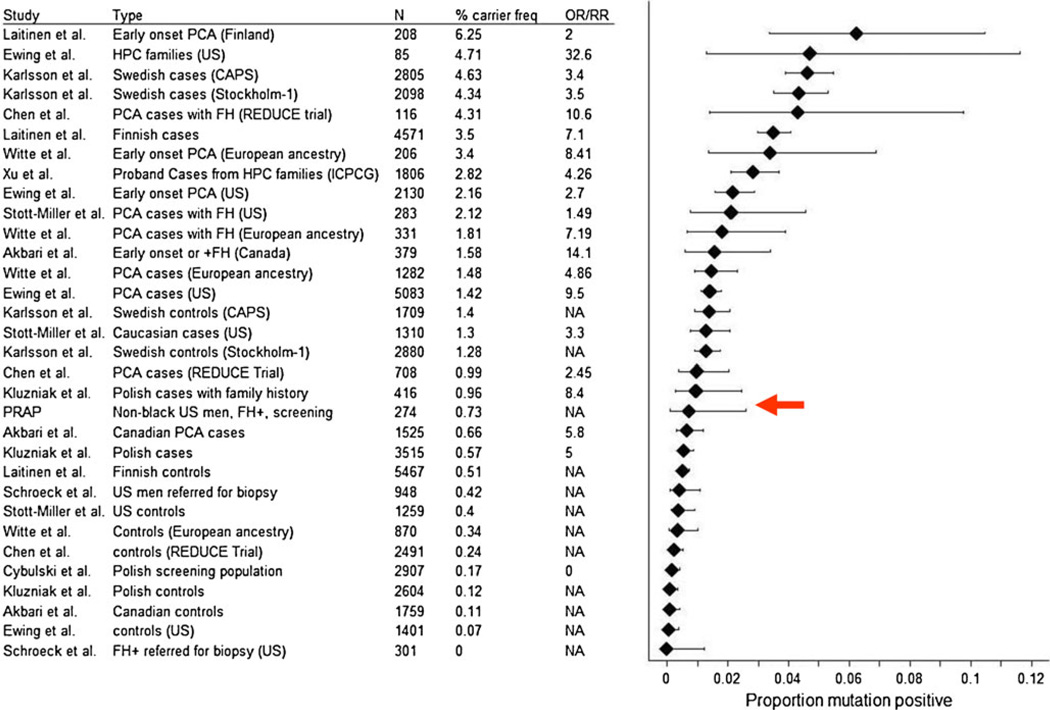
This carrier frequency of 0.73% from PRAP was placed in context of prevalence estimates from studies in the literature (Ewing et al. 2012; Chen et al. 2013; Witte et al. 2013; Karlsson et al. 2012; Laitinen et al. 2013; Xu et al. 2013; Stott-Miller et al. 2013; Akbari et al. 2012; Kluzniak et al. 2013; Schroeck et al. 2013; Cybulski et al. 2013). Figure 2 shows the forest plot of carrier frequencies of the G84E mutation from reported studies. As can be seen from Fig. 1, the highest carrier frequency is observed in prostate cancer cases from Scandinavia, with prevalence estimates of 3.5 %–6.25 %. Cases from the US/European ancestry rank second as a group in carrier frequency, with estimates in the range of .57 %–3.4 % depending on the study. The highest carrier frequency in controls has been reported from Sweden, with prevalence of 1.4 % (Karlsson et al. 2012). However, the majority of controls have been reported to have a carrier frequency of 0–0.51 % (Fig. 1). Based on this overview, non-African American men in PRAP with a family history of prostate cancer are intermediate in the carrier frequency spectrum between controls and Scandinavian cases. Thus, though the mutation is rare, these findings may support future genetic testing for the G84E mutation in HOXB13 to men with a family history of prostate cancer who are of non-African ancestry.
Fig. 2.
Carrier frequencies of HOXB13 G84E
Discussion
Men with a family history of prostate cancer have traditionally been considered to be at increased risk for the disease as a group, with estimates of risk for prostate cancer ranging from 1.7 to 5.5 fold above population level depending on number of affected relatives and age at diagnosis (Zeegers et al. 2003; Kicinski et al. 2011). However, at the individual level, family history lacks precision when informing prostate cancer screening decisions and strategies. Previous screening studies from high-risk cohorts have shown that approximately 10 % of high-risk men will be diagnosed with prostate cancer (Giri et al. 2007). Therefore, a substantial proportion of men with a family history of prostate cancer may undergo unnecessary intensive screening. Personalized approaches to predicting risk for prostate cancer has potential to inform prostate cancer screening and management at the individual level.
After several years of research, the G84E recurrent mutation in HOXB13 was identified as an alteration strongly predisposing to prostate cancer risk particularly in families with a prostate cancer history and for families with early-onset disease (Ewing et al. 2012). This finding was confirmed in a large international study including 2,443 hereditary prostate cancer families (Xu et al. 2013). Since the initial publication, several studies have been reported from various cohorts to better characterize this mutation and the risk associated with prostate cancer development. In particular, G84E appears to increase the risk for prostate cancer in men with a family history of prostate cancer and of Northern European descent (Xu et al. 2013; Chen et al. 2013; Witte et al. 2013; Karlsson et al. 2012; Laitinen et al. 2013; Stott-Miller et al. 2013; Akbari et al. 2012; Kluzniak et al. 2013; Schroeck et al. 2013; Cybulski et al. 2013). Since there is a spectrum of prevalence of the G84E mutation based primarily on strength of family cancer history and ancestry, our study was performed among high-risk men in a US cohort to describe the prevalence of G84E and place the findings in the context of reported studies of G84E in order to gain insight into the potential future role of G84E in prostate cancer risk assessment particularly for high-risk men.
The overall carrier frequency among non-African American PRAP participants with a family history of prostate cancer was 0.73 %. Of interest, one participant with the mutation was of self-reported Hispanic origin, the first to our knowledge. Our study did not find this mutation among African American participants, which is consistent with prior studies (Ewing et al. 2012; Witte et al. 2013). Overall, the carrier frequency of 0.73 % in men with a family history in PRAP was higher than published controls (Ewing et al. 2012; Chen et al. 2013;Witte et al. 2013; Laitinen et al. 2013; Stott-Miller et al. 2013; Akbari et al. 2012; Kluzniak et al. 2013; Schroeck et al. 2013; Cybulski et al. 2013). While PRAP is not fully representative of the US male population, our findings might indicate that a certain percentage of unaffected men with a family history of prostate cancer will carry the G84E mutation. Further population-based studies describing penetrance as well as the impact on screening for prostate cancer based on G84E carrier status are needed to gain insight into how this mutation will play a role in prostate cancer risk assessment for high-risk men. One limitation to note is that the two PRAP participants with the G84E mutation have not returned for follow-up, and therefore their cancer status is unknown.
The information gained from the review of carrier frequencies lends insight into the probability of carrying the G84E mutation to inform future genetic counseling approaches for men interested in prostate cancer risk assessment. While the clinical utility of G84E needs to be further defined, future counseling will likely need to factor in the following information in order for men to make an informed decision regarding whether to proceed with genetic testing for HOXB13 G84E: (1) approximately 1–6 % of men studied have been found to carry G84E, with the highest carrier frequency of 6 % in men of Scandinavian origin, (2) the lifetime risk for prostate cancer conferred by the mutation has been reported to be approximately 35 %, (3) controls from prior studies have also been found to carry the mutation, so understanding of the penetrance of the mutation is still evolving, and (4) studies focused on prostate cancer detection and outcomes among male HOXB13 G84E mutation carriers are needed to inform optimal prostate cancer screening strategies in this group.
Conclusion
Approximately 1 % of unaffected Caucasian men with a family history of prostate cancer in our cohort were found to carry HOXB13 G84E. While this mutation is associated with prostate cancer risk particularly among men with a family history of prostate cancer and with early-onset disease, the role in decision-making for prostate cancer risk assessment remains to be determined. Unaffected male carriers stand to potentially benefit from further research focused on the clinical utility of G84E.
Acknowledgements
Funding Grant support for this project was provided by 1R03CA150079-01 (VNG), with support from the P30 CA006927 grant from the National Cancer Institute (Cancer Center Support Grant). VNG is also supported by the Department of Defense Physician Research Training Award (DOD W81XWH-09-1-0302);
Participants We are also grateful to the participants of the Prostate Cancer Risk Assessment Program.
Footnotes
Conflict of Interest Authors Elizabeth Handorf, Nicole Crumpler, Laura Gross, and Veda N. Giri declare they have no conflict of interest.
Contributor Information
Elizabeth Handorf, Cancer Prevention and Control Program, Fox Chase Cancer Center, Philadelphia, PA, USA.
Nicole Crumpler, Cancer Prevention and Control Program, Fox Chase Cancer Center, Philadelphia, PA, USA.
Laura Gross, Department of Clinical Genetics, Fox Chase Cancer Center, 4th Floor, Robert C. Young Pavilion 333 Cottman Avenue, Philadelphia, PA 19111, USA.
Veda N. Giri, Email: Veda.Giri@fccc.edu, Cancer Prevention and Control Program, Fox Chase Cancer Center, Philadelphia, PA, USA; Department of Clinical Genetics, Fox Chase Cancer Center, 4th Floor, Robert C. Young Pavilion 333 Cottman Avenue, Philadelphia, PA 19111, USA.
References
- Akbari MR, Trachtenberg J, Lee J, Tam S, Bristow R, Loblaw A, et al. Association between germline HOXB13 G84E mutation and risk of prostate cancer. Journal of the National Cancer Institute. 2012;104(16):1260–1262. doi: 10.1093/jnci/djs288. [DOI] [PubMed] [Google Scholar]
- Bigler SA, Pound CR, Zhou X. A retrospective study on pathologic features and racial disparities in prostate cancer. Prostate Cancer. 2011 doi: 10.1155/2011/239460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A, Bermejo JL, Sundquist J, Hemminki K. Age at diagnosis and age at death in familial prostate cancer. The Oncologist. 2009;14(12):1209–1217. doi: 10.1634/theoncologist.2009-0132. [DOI] [PubMed] [Google Scholar]
- Breast Cancer Linkage Consortium (BCLC. Cancer risks in BRCA2 mutation carriers. Journal of the National Cancer Institute. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- Cancer Facts & Figures. Atlanta, GA: American Cancer Society; 2013. [cited 2013 May 29]. 2013; Available from: http://www.cancer.org/research/cancerfactsfigures/index. [Google Scholar]
- Catalona WJ, Antenor JA, Roehl KA, Moul JW. Screening for prostate cancer in high risk populations. The Journal of Urology. 2002;168(5):1980–1983. doi: 10.1016/S0022-5347(05)64276-0. discussion 3–4. [DOI] [PubMed] [Google Scholar]
- Chen Z, Greenwood C, Isaacs WB, Foulkes WD, Sun J, Zheng SL, et al. The G84E mutation of HOXB13 is associated with increased risk for prostate cancer: results from the REDUCE trial. Carcinogenesis. 2013;34(6):1260–1264. doi: 10.1093/carcin/bgt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- Cybulski C, Wokolorczyk D, Kluzniak W, Kashyap A, Golab A, Slojewski M, et al. A personalised approach to prostate cancer screening based on genotyping of risk founder alleles. British Journal of Cancer. 2013;108(12):2601–2609. doi: 10.1038/bjc.2013.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature Genetics. 2013;45(4):385–391. doi: 10.1038/ng.2560. 91e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. The New England Journal of Medicine. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast cancer linkage consortium. Lancet. 1994;343(8899):692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- Giri VN, Beebe-Dimmer J, Buyyounouski M, Konski A, Feigenberg SJ, Uzzo RG, et al. Prostate cancer risk assessment program: a 10-year update of cancer detection. The Journal of Urology. 2007;178(5):1920–1924. doi: 10.1016/j.juro.2007.07.010. discussion 4. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Sundquist J, Brandt A. Familial mortality and familial incidence in cancer. Journal of Clinical Oncology. 2011;29(6):712–718. doi: 10.1200/JCO.2010.30.5664. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson R, Aly M, Clements M, Zheng L, Adolfsson J, Xu J, et al. A Population-based Assessment of Germline HOXB13 G84E Mutation and Prostate Cancer Risk. European Urology. 2012 doi: 10.1016/j.eururo.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Kicinski M, Vangronsveld J, Nawrot TS. An epidemiological reappraisal of the familial aggregation of prostate cancer: a meta-analysis. PloS One. 2011;6(10):e27130. doi: 10.1371/journal.pone.0027130. PMCID: 3205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluzniak W, Wokolorczyk D, Kashyap A, Jakubowska A, Gronwald J, Huzarski T, et al. The G84E mutation in the HOXB13 gene is associated with an increased risk of prostate cancer in Poland. The Prostate. 2013;73(5):542–548. doi: 10.1002/pros.22594. [DOI] [PubMed] [Google Scholar]
- Laitinen VH, Wahlfors T, Saaristo L, Rantapero T, Pelttari LM, Kilpivaara O, et al. HOXB13 G84E mutation in Finland: population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiology, Biomarkers and Prevention. 2013;22(3):452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- Liede A, Karlan BY, Narod SA. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. Journal of Clinical Oncology. 2004;22(4):735–742. doi: 10.1200/JCO.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Pomerantz M, Freedman ML. Clinical uncertainty of prostate cancer genetic risk panels. Science Translational Medicine. 2013;5(182):182ed6. doi: 10.1126/scitranslmed.3004696. [DOI] [PubMed] [Google Scholar]
- Powell IJ, Bock CH, Ruterbusch JJ, Sakr W. Evidence supports a faster growth rate and/or earlier transformation to clinically significant prostate cancer in black than in white American men, and influences racial progression and mortality disparity. The Journal of Urology. 2010;183(5):1792–1796. doi: 10.1016/j.juro.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeck FR, Zuhlke KA, Siddiqui J, Siddiqui R, Cooney KA, Wei JT. Testing for the recurrent HOXB13 G84E germline mutation in men with clinical indications for prostate biopsy. The Journal of Urology. 2013;189(3):849–853. doi: 10.1016/j.juro.2012.09.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott-Miller M, Karyadi DM, Smith T, Kwon EM, Kolb S, Stanford JL, et al. HOXB13 mutations in a population-based, case-control study of prostate cancer. The Prostate. 2013;73(6):634–641. doi: 10.1002/pros.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS, Mefford J, Plummer SJ, Liu J, Cheng I, Klein EA, et al. HOXB13 mutation and prostate cancer: studies of siblings and aggressive disease. Cancer Epidemiology, Biomarkers and Prevention. 2013;22(4):675–680. doi: 10.1158/1055-9965.EPI-12-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lange EM, Lu L, Zheng SL, Wang Z, et al. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium for Prostate Cancer Genetics (ICPCG) Human Genetics. 2013;132(1):5–14. doi: 10.1007/s00439-012-1229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeegers MP, Jellema A, Ostrer H. Empiric risk of prostate carcinoma for relatives of patients with prostate carcinoma: a meta-analysis. Cancer. 2003;97(8):1894–1903. doi: 10.1002/cncr.11262. [DOI] [PubMed] [Google Scholar]
- Zuhlke KA, Madeoy JJ, Beebe-Dimmer J, White KA, Griffin A, Lange EM, et al. Truncating BRCA1 mutations are uncommon in a cohort of hereditary prostate cancer families with evidence of linkage to 17q markers. Clinical Cancer Research. 2004;10(18 Pt 1):5975–5980. doi: 10.1158/1078-0432.CCR-04-0554. [DOI] [PubMed] [Google Scholar]