Abstract
The proinflammatory cytokine interleukin-17 (IL-17) is the signature cytokine of the T helper 17 (TH17) subset of CD4+ T cells, and antibodies targeting IL-17 or the IL-17 receptor (IL-17R) show clinical efficacy in several autoimmune diseases. Although important for protective immunity against microorganisms, IL-17 causes collateral damage in inflammatory settings. TNFAIP3 encodes the deubiquitinase A20 and is genetically linked to numerous autoimmune syndromes. A20, a potent inhibitor of tumor necrosis factor–α signaling, removes ubiquitin from signaling intermediates upstream of nuclear factor κB (NF-κB), thereby dampening NF-κB–mediated inflammation. We demonstrated that IL-17 stimulates TNFAIP3 expression. Enhanced IL-17–mediated induction of genes encoding proinflammatory factors, including IL-6 and various chemokines, occurred upon knockdown of A20 with short inhibitory RNA or in A20−/− cells. A20 associated with the E3 ubiquitin ligase TRAF6 (tumor necrosis factor receptor–associated factor 6) in an IL-17–dependent manner and restricted the IL-17–dependent activation of NF-κB and mitogen-activated protein kinases. A20 interacted directly with the distal domain of IL-17RA, a previously defined inhibitory domain. Together, these data describe a mechanism of restraining IL-17 signaling and reveal an aspect of A20 activity that may help to explain its role in autoimmunity in humans.
INTRODUCTION
Inflammatory cytokines, such as tumor necrosis factor–α (TNF-α), have long been recognized to promote the pathogenesis of devastating autoimmune diseases, such as rheumatoid arthritis (RA), among many others (1). It is not an exaggeration to say that biologic therapies targeting TNF-α and other inflammatory cytokines revolutionized the clinical management of many of these diseases. Despite these advances, many patients fail to respond to TNF-blocking drugs. In the last several years, interleukin-17 (IL-17; also known as IL-17A) emerged as a key player in autoimmune inflammation, and clinical trial data indicate exciting promise for anti–IL-17 drugs in treating psoriasis and other autoimmune conditions (2–5). IL-17 is produced by a subset of CD4+ T cells termed T helper 17 (TH17) cells, and a fast-moving body of literature has described the many mechanisms by which TH17 cells are generated and regulated (6, 7). In addition, it is increasingly apparent that IL-17 is produced by many innate cell types that bear marked similarities to classic TH17 cells and participate in mediating autoimmune inflammation (8).
In contrast to the efforts focused on investigating the immunology of TH17 cells, there has been far less emphasis on understanding how IL-17 activates downstream signaling pathways. IL-17 is the founding member of a distinct subclass of cytokines and receptors that exhibit distinct signaling properties compared to those of the better-defined cytokine receptors, such as the TNF receptor (TNFR) superfamily or the IL-1 receptor (IL-1R) and Toll-like receptor (TLR) families (9). However, IL-17 shares similar signaling end points with those of other inflammatory cytokines, particularly in terms of activation of nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling, as well as induced expression of genes encoding proinflammatory cytokines [such as IL-6 and G-CSF (granulocyte colony-stimulating factor)], antimicrobial peptides [including lipocalin 2 (also known as 24p3), S100A proteins, and β-defensins], and chemokines (including CXCL1, CXCL5, and CCL20) (10). The net effect of IL-17 signaling is effective host defense against bacterial and, especially, fungal infections. Indeed, in humans, mutations in the gene encoding IL-17R or in genes whose products control TH17 development, such as signal transducer and activator of transcription 3 (STAT3) or STAT1, cause increased susceptibility to infections by the commensal fungus Candida albicans (11, 12). Conversely, excess IL-17 is associated with numerous autoimmune diseases, and many genes identified as risk loci for autoimmunity in genome-wide association studies (GWAS) (for example, IL23R and STAT3) are associated with regulation of IL-17 or the TH17 differentiation pathway (13, 14).
IL-17 mediates signaling through a heterodimeric receptor composed of the IL-17RA and IL-17RC subunits (15). Both subunits contain a signaling motif unique to the IL-17R family known as a SEFIR (for SEF/IL-17R) domain (16). The SEFIR domain provides a platform for the binding of Act1 (also known as CIKS), a SEFIR-containing adaptor protein and E3 ubiquitin ligase (16–20). IL-17 engagement of the IL-17R recruits Act1 to the receptor complex. In turn, Act1 recruits and activates TNF receptor–associated factor 6 (TRAF6), ultimately leading to the activation of transcription factors, such as NF-κB, C/EBPβ, and C/EBPδ, as well as of MAPKs (21, 22). The C-terminal domain of IL-17RA, in contrast, is required for activation of C/EBPβ and is linked to inhibitory signaling through glycogen synthase kinase 3β (GSK-3β) and TRAF3 (23–26).
Ubiquitination is a posttranslational modification that is essential for modulating proinflammatory pathways (27). Ubiquitination involves covalent linking of ubiquitin moieties to target proteins through specific lysine residues, and it is regulated by a cascade of E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin-ligase) enzymes. Of the seven lysines in ubiquitin, Lys48 (K48) andLys63 (K63) are most commonly used to form polyubiquitin chains. K48-linked ubiquitin generally targets proteins for proteasomal degradation, whereas K63-linked ubiquitin triggers non-degradative functions, such as protein-protein interactions and cell signaling events (28). Notably, many of the currently identified IL-17R–proximal proteins are E3 ubiquitin ligases. For example, the E3 ligase activity of Act1 is required for the K63 ubiquitination of TRAF6, which is also an E3 ubiquitin ligase (17, 29, 30). Ubiquitination of TRAF6 is a key event in the downstream activation of the NF-κB and MAPK signaling pathways (15, 17, 31).
The reversal of ubiquitination is equally important in regulating inflammation, particularly to keep potentially damaging signals in check (32–34). A20 is a deubiquitinase (DUB) and tumor suppressor encoded by TNFAIP3 (TNF-α–induced protein 3) and was first identified as an inhibitor of the TNFR signaling pathway (35, 36). Subsequent studies identified roles for A20 in inhibiting the TLR, IL-1R, and Nod-like receptor (NLR) pathways (32, 37, 38). In the TNFR pathway, the E3 ubiquitin ligase and adaptor protein TRAF2 and the ribosome interacting protein kinase 1 (RIP1) are targets of A20, whereas in IL-1R and TLR signaling, TRAF6 is a key A20 target. Deubiquitination of these adaptors restricts the activation of the NF-κB and MAPK pathways (28, 32, 39). Confirming its essential role in restraining inflammation, A20-deficient (A20−/−) mice develop spontaneous multiorgan inflammation and die shortly after birth (39), and mice with cell type–specific knockout of A20 are prone to multiple autoimmune diseases (38, 40–42). Moreover, polymorphisms in the TNFAIP3 locus are associated with increased susceptibility to RA, lupus, systemic sclerosis, Crohn’s disease, and psoriasis (41, 43). Here, we show that A20 is a feedback inhibitor of the IL-17 signaling pathway. IL-17 increased the abundance of TNFAIP3 mRNA and the subsequent production of A20 protein, which inhibited IL-17–mediated activation of TRAF6, NF-κB, and MAPKs, as well as downstream gene expression. Furthermore, A20 bound to a domain of IL-17RA that is associated with inhibiting receptor signaling (23). Thus, these findings expand the known regulatory role of A20 in regulating inflammatory signaling and lend new insight into how IL-17–dependent inflammation is controlled.
RESULTS
A20 inhibits IL-17 signaling
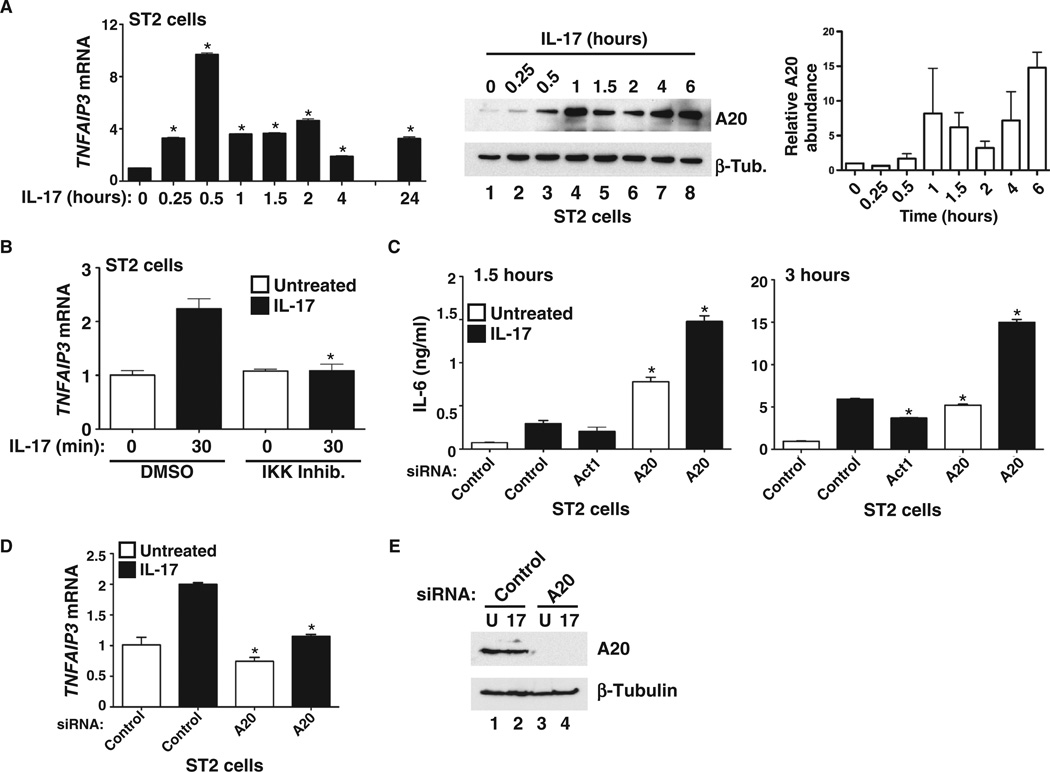
As its name implies, TNFAIP3 was first identified as a TNF-α–induced gene (44), and its gene product A20 serves as a feedback inhibitor of NF-κB (39). On the basis of similarities in the downstream pathways regulated by IL-17 and TNF-α, we hypothesized that A20 might also participate in inhibition of the IL-17 signaling pathway. Although produced primarily by T cells, IL-17 exerts its biological effects primarily in the nonhematopoietic compartment, particularly on mesenchymal cell types such as fibroblasts and stromal cells (45). Accordingly, we stimulated ST2 cells (a murine stromal cell line) with IL-17 over a time course of 24 hours and assessed endogenous TNFAIP3 mRNA abundance by real-time reverse transcription polymerase chain reaction (PCR) [hereafter termed quantitative PCR (qPCR)]. IL-17–induced expression of TNFAIP3 occurred within 15 min of stimulation (Fig. 1A). Expression peaked at 30 min after treatment and remained increased at four- to fivefold above baseline for at least 24 hours. These kinetics are similar to those reported for TNF-α–induced expression of TNFAIP3 (39). We verified that the production of endogenous A20 protein was also stimulated by IL-17 with slightly delayed kinetics compared to that of mRNA expression (Fig. 1A). The expression of TNFAIP3 is regulated at the promoter by various transcription factors, including NF-κB (44). Consistently, we found that an inhibitor targeting the inhibitor of κB (IκB) kinase (IKK) blocked the IL-17–mediated increase in TNFAIP3 mRNA in ST2 cells, implicating NF-κB in this pathway (Fig. 1B).
Fig. 1. IL-17 induces TNFAIP3 expression and production of A20, which suppresses IL-17–induced gene expression.
(A) Left panel: Rapid induction of endogenous TNFAIP3 mRNA expression by IL-17. ST2 cells were treated with IL-17 (200 ng/ml) for the indicated times, and TNFAIP3 mRNA abundance was assessed by qPCR. Data are presented as the fold induction in TNFAIP3 (A20) mRNA abundance in IL-17–treated cells compared to untreated cells. *P < 0.05 by analysis of variance (ANOVA) with post hoc Tukey’s test compared to untreated (at 0 min); n = 3 experiments. Middle panel: Endogenous A20 and β-tubulin (β-Tub.) proteins were assessed by Western blotting analysis of whole-cell lysates of ST2 cells treated with IL-17 for the indicated times. Right panel: Relative band intensities from pooled independent experiments were determined by densitometry and are presented as means ± SEM (n = 3 experiments). (B) IL-17 induction of endogenous A20 is mediated by the NF-κB pathway. ST2 cells were treated with IL-17 for 30 min in the presence of dimethyl sulfoxide (DMSO) or of an IKK inhibitor (IKK Inhib.) (0.1 µM), and TNFAIP3 mRNA abundance was assessed by qPCR analysis. *P < 0.05, ANOVA with post hoc Tukey’s test compared to untreated (at 30 min); n = 3 experiments. (C) Knockdown of A20 enhances the production of IL-6 protein. ST2 cells were transfected with siRNA specific for Act1 or A20 or with a scrambled siRNA control, and the amount of IL-6 in the culture medium was assessed at 1.5 hours (left panel) and 3 hours (right panel) after treatment with IL-17 (200 ng/ml). Black bars indicate IL-17–treated samples, and white bars indicate untreated samples. *P < 0.05, ANOVA with post hoc Tukey’s test compared to cells transfected with control siRNA and treated with IL-17; n = 3 experiments. (D) Successful knockdown of TNFAIP3 mRNA. TNFAIP3 mRNA abundance in cells treated with siRNA was assessed by qPCR. *P < 0.05, ANOVA with post hoc Tukey’s test compared to control samples treated with IL-17. (E) Successful knockdown of A20 protein. After siRNA-mediated knockdown of TNFAIP3 expression, the indicated cell lysates were analyzed by Western blotting with antibodies specific for A20 protein (top) or β-tubulin (bottom) as a loading control. n = 2 experiments.
Because inhibitory molecules are often induced by the cytokines that they suppress, we hypothesized that A20 would inhibit IL-17–mediated signaling. We transfected ST2 cells with short interfering RNA (siRNA) specific for A20 or with a nontargeting scrambled siRNA as a negative control. As a positive control, we used siRNA specific for Act1, an adaptor protein required for IL-17R signaling (18, 19). Forty-eight hours after transfection, we stimulated the cells with IL-17 for 1.5 or 3 hours and then measured IL-6 in culture supernatants by enzyme-linked immunosorbent assay (ELISA) and Il6 mRNA transcripts by qPCR (Fig. 1C and fig. S1). As expected, IL-6 protein and mRNA production increased even in untreated cells after transfection with A20-specific siRNA, indicating that A20 controls the tonic expression of genes encoding proinflammatory cytokines (36). Stimulation with IL-17 in combination with A20 knockdown led to a statistically significant (P < 0.001) increase in IL-6 production compared to that by stimulation with IL-17 alone, revealing an inhibitory role for A20 in the IL-17 pathway (Fig. 1C). Il6 mRNA was similarly affected (fig. S1). We also verified that A20 was efficiently knocked down in these experiments (Fig. 1, D and E). To determine whether the stimulation of A20 production by IL-17 led to termination of IL-6 generation, we evaluated the kinetics of Il6 mRNA induction after treatment with IL-17. We found that there was a slight reduction in Il6 mRNA abundance at the 1- and 1.5-hour time points, which coincided with the time when A20 protein was increased in abundance, but generally, Il6 mRNA was maintained at steady state for at least 4 hours (fig. S2). This finding supports a model in which A20 serves to keep IL-17 signaling in check but does not cause complete signal termination.
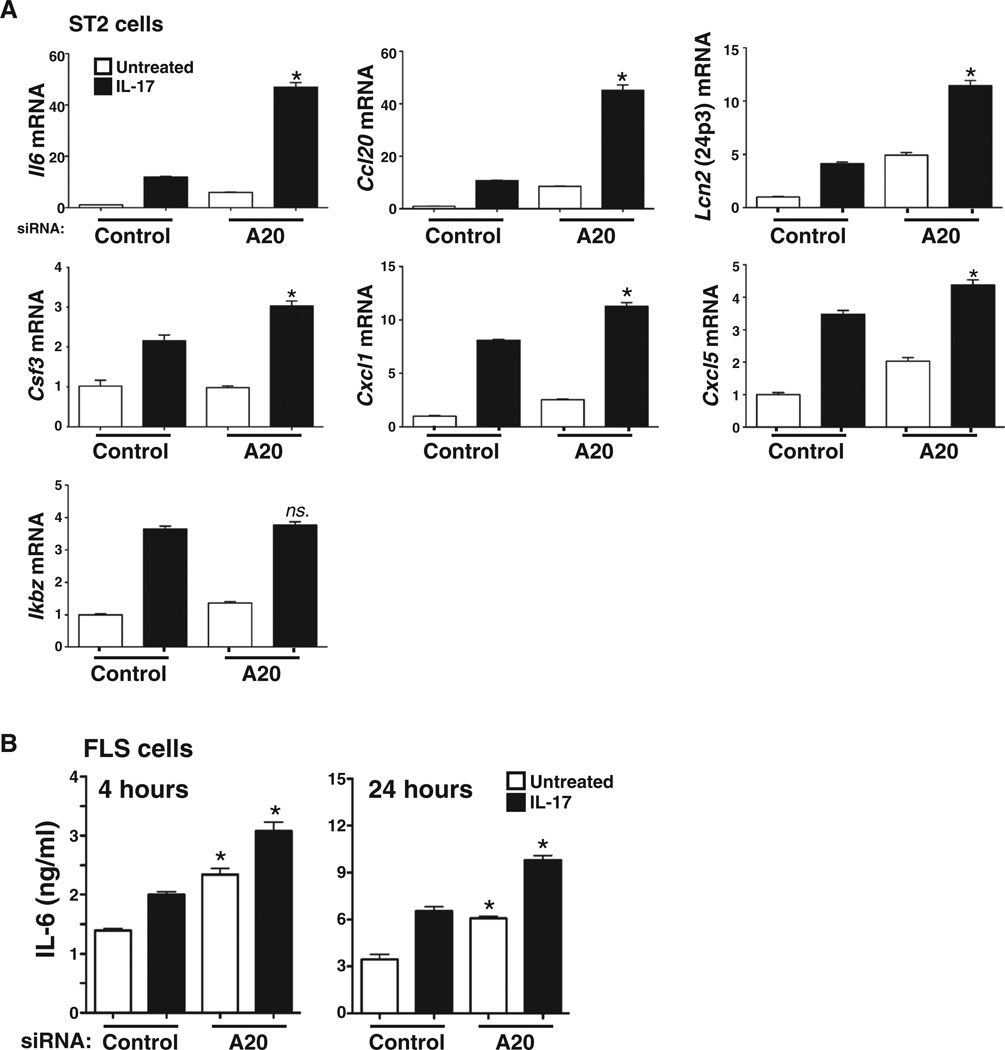
To further evaluate the role of A20 in IL-17 signaling, we analyzed the expression of a panel of additional well-defined IL-17 target genes in the context of A20 silencing (46–48). Consistent with an increase in IL-6 protein abundance, transfection with A20-specific siRNA enhanced IL-17–dependent induction of Il6 expression (Fig. 2A and fig. S1). Similarly, knockdown of A20 enhanced IL-17–induced expression of Ccl20, Lcn2 (which encodes lipocalin 2), Csf3 (which encodes G-CSF), Cxcl1, and Cxcl5 (Fig. 2A and fig. S1). Although the magnitude by which A20-specific siRNA enhanced the expression of IL-17 target genes was variable, the extent of expression of all of these genes was statistically significantly and reproducibly increased. However, there was no change in the expression of Ikbz in response to A20 knockdown (Fig. 2A and fig. S1), suggesting that there may be distinct modes of regulation for some IL-17–induced genes.
Fig. 2. Knockdown of A20 suppresses IL-17–mediated gene expression.
(A) Knockdown of A20 enhances the expression of most, but not all, IL-17 target genes. ST2 cells transfected with the indicated siRNAs and left untreated or treated with IL-17 for 3 hours were analyzed for the expression of the indicated genes by qPCR. *P < 0.05, ANOVA and post hoc Tukey’s test compared to cells transfected with control siRNA and treated with IL-17; n = 3 experiments. (B) Knockdown of A20 enhances IL-17R signaling in human FLS cells. FLS cells were transfected with the indicated siRNAs, and IL-6 secretion after 4 or 24 hours of IL-17 stimulation was assessed by ELISA. *P < 0.05, ANOVA and post hoc Tukey’s test compared to cells transfected with control siRNA and treated with IL-17; n = 2 experiments.
Increased amounts of IL-17 and an increase in the extent of expression of IL-17–induced genes are associated with pathology in RA (49). Accordingly, we tested the effect of A20 knockdown in human primary fibroblast-like synoviocytes (FLSs), a cell type that mediates pathogenesis in the inflamed joint. Transfection of FLS cells with A20-specific siRNA led to enhanced IL-17–dependent IL-6 production compared to that in FLS cells transfected with control siRNA (Fig. 2B). Therefore, we conclude that A20 mediates feedback inhibition of IL-17 signaling and that this regulation has the potential to be relevant in a disease in which IL-17 plays a well-documented role.
Overexpression of A20 inhibits activation of IL-17 target promoters
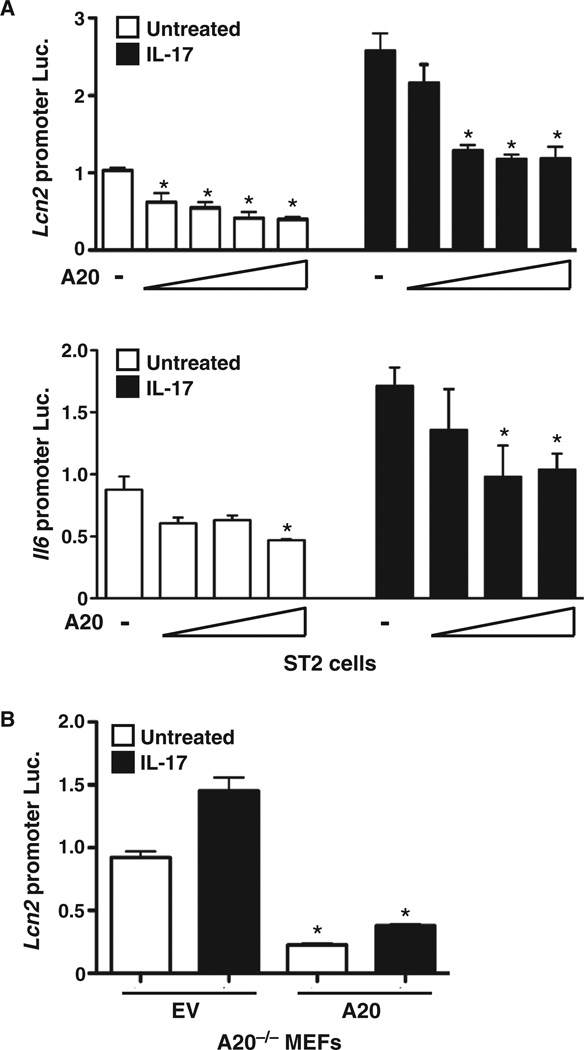
NF-κB is required for the activation of many IL-17 target genes, including those encoding lipocalin 2 (also known as 24p3) and IL-6 (46, 50). To determine whether A20 affected NF-κB–dependent promoters regulated by IL-17, we cotransfected ST2 cells with luciferase (Luc) constructs driven by the Lcn2 or Il6 promoters together with increasing amounts of plasmid encoding A20 (46, 50). IL-17 triggered a ~2.5-fold increase in Lcn2-Luc activity over baseline in the absence of A20 (Fig. 3A). Cotransfection with plasmid encoding A20 resulted in a dose-dependent reduction in Lcn2-Luc activity, supporting a model in which A20 inhibits IL-17–dependent activation of this gene at the promoter level. A mild suppression in luciferase activity was also seen in cells that were not treated with IL-17, consistent with the effect of A20 on tonic signaling (noted in Fig. 1). We obtained similar results in parallel experiments with the Il6 promoter construct (Fig. 3A). Further evidence of a role for A20 in suppressing target promoter activation came from the observation that IL-17–mediated induction of the Lcn2 promoter was more strongly enhanced in A20-deficient (A20−/−) cells than in cells reconstituted with A20 (Fig. 3B). Together, these data support the concept that A20 inhibits IL-17 signaling by modulating NF-κB–regulated gene expression.
Fig. 3. A20 suppresses IL-17–mediated activation of NF-κB–dependent gene promoters.
(A) Ectopic expression of A20 suppresses IL-17–dependent activation of the promoters of the genes encoding Lcn2 and IL-6. ST2 cells were transfected with luciferase (Luc.) constructs encoding the Lcn2 promoter (48) or the Il6 promoter (46) together with increasing concentrations of plasmid encoding A20. After 8 hours of treatment with IL-17, luciferase activity was assessed and was normalized to that of samples that were transfected with control DNA and were not treated with cytokines. *P < 0.05, ANOVA with post hoc Tukey’s test compared to untreated samples transfected with control DNA; n = 3 experiments. (B) Deficiency in A20 leads to enhanced IL-17–dependent activation of the Lcn2 promoter. A20−/− MEFs were transfected with the Lcn2-Luc construct and either an empty vector (EV) or a plasmid encoding A20. After 8 hours of treatment with IL-17, luciferase activity was assessed and was normalized to that of samples that were transfected with control DNA and were not treated with cytokines. *P < 0.05, ANOVA and post hoc Tukey’s test compared to unstimulated EV sample; n = 2 experiments.
Reconstitution of A20−/− murine embryonic fibroblasts with A20 reverses IL-17–dependent target gene expression
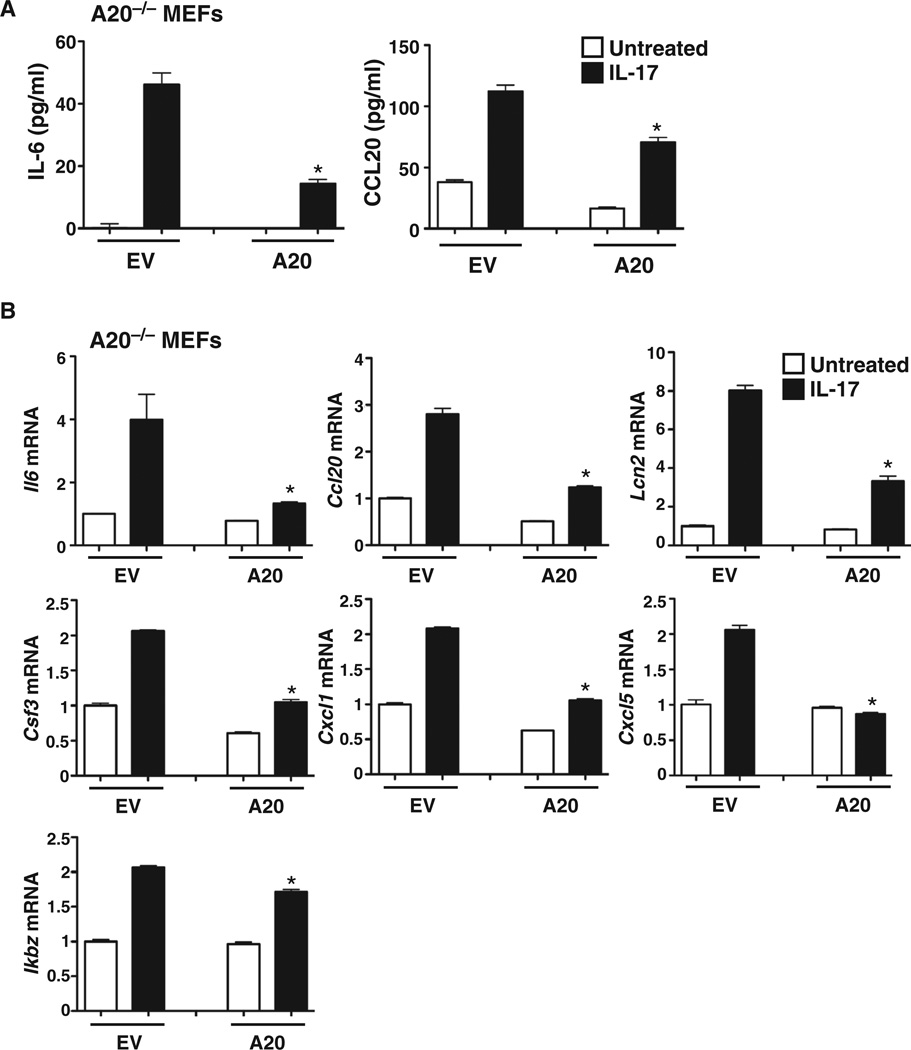
To independently verify that A20 inhibited the IL-17 pathway and to rule out possible nonspecific or off-target effects of siRNAs, we transfected A20−/− murine embryonic fibroblasts (MEFs) with either an EV or a plasmid encoding murine A20. Cells were stimulated with IL-17 for 4 hours, and IL-6 in culture supernatants was assessed by ELISA. A20−/− MEFs transfected with EV showed an IL-17–dependent increase in IL-6 production, which was reduced upon reconstitution with A20 (Fig. 4A). Production of CCL20 was similarly affected by reconstitution of the A20−/− MEFs with A20 (Fig. 4A). We next determined the effect of reconstituting A20−/− MEFs with A20 on the expression of a panel of IL-17 target genes. We found that IL-17–induced expression of Il6, Ccl20, Cxcl1, Cxcl5, Csf3, and lcn2 in A20−/− cells was repressed upon reconstitution with A20 (Fig. 4B). Similar to the findings in our siRNA-based experiments (Fig. 2A), we found that there was only a mild effect of A20 reconstitution on Ikbz expression (Fig. 4B), indicating that A20 represses the expression of most, but not all, IL-17 target genes.
Fig. 4. Reconstitution of A20−/− cells with A20 suppresses IL-17–dependent signaling.
(A and B) A20−/− MEFs were transfected with EV or with plasmid encoding murine A20 and were treated with IL-17 for 4 hours. (A) Culture supernatants were evaluated for IL-6 or CCL20 by ELISA. (B) Cell lysates were evaluated for the indicated mRNAs by qPCR. *P < 0.05, ANOVA and post hoc Tukey’s test compared to IL-17–treated samples transfected with EV; n = 3 experiments.
A20 inhibits activation of the IL-17–mediated TRAF6–NF-κB pathway
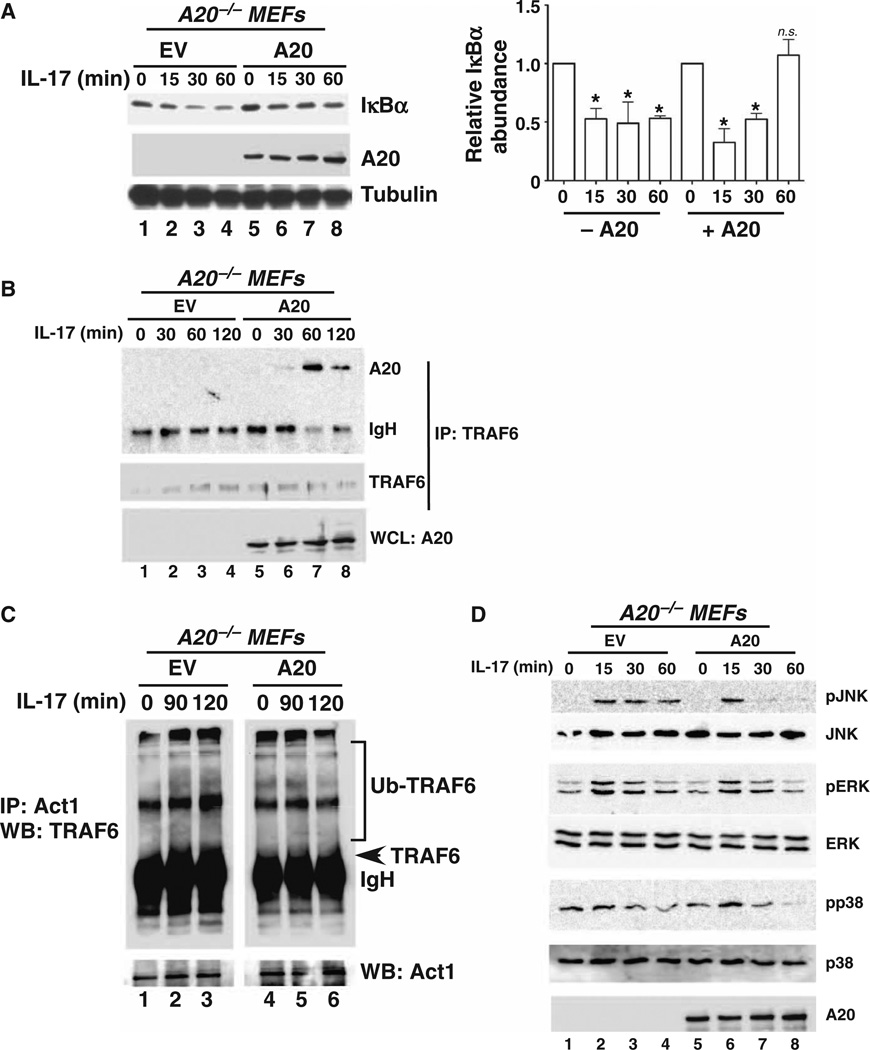
NF-κB is activated when IκBα is phosphorylated, ubiquitinated, and degraded, which results in the unmasking of the nuclear localization signal of NF-κB and its subsequent nuclear translocation. Expression of the gene encoding IκBα is, in turn, dependent on NF-κB in a feedback loop. To determine whether A20 inhibited IL-17–mediated activation of NF-κB, we monitored IL-17–dependent IκBα degradation in A20−/− MEFs that were reconstituted with EV or with A20. Lack of A20 resulted in continued IL-17–dependent degradation of IκBα, suggesting prolonged NF-κB activation. However, in cells reconstituted with A20, this degradation was inhibited very rapidly, suggesting inhibition of NF-κB activity (Fig. 5A). These data support a model in which A20 blocks the IL-17–dependent NF-κB pathway.
Fig. 5. A20 targets IL-17–stimulated TRAF6–NF-κB and MAPK pathways.
(A) A20 promotes IκBα degradation. A20−/− cells were transfected with EV (lanes 1 to 4) or plasmid encoding A20 (lanes 5 to 8), treated with IL-17 for the indicated times, and then assessed by Western blotting to determine IκBα abundance (top blot). Loading of A20 (middle blot) and β-tubulin (bottom blot) is shown. Bar graph: Relative band intensities from independent experiments were determined by densitometry and are shown as means ± SEM. *P < 0.05, ANOVA and post hoc Tukey’s test compared to the unstimulated time point for each condition; n = 3 experiments. n.s., not significant. (B) A20 associates with TRAF6 in an IL-17–dependent manner. A20−/− cells were transfected with EV (lanes 1 to 4) or with plasmid encoding A20 (lanes 5 to 8), and cell lysates were subjected to immunoprecipitation (IP) with antibody against TRAF6. Immunoprecipitated samples were analyzed by Western blotting (WB) with antibodies against A20 (top blot) and TRAF6 (middle blot). Samples of whole-cell lysates (WCLs) were analyzed by Western blotting with antibody against A20 (bottom blot) to verify transfection efficiency. n = 3 experiments. (C) A20 mediates the deubiquitination of TRAF6 in response to IL-17 signaling. A20−/− cells were transfected with EV (lanes 1 to 3) or plasmid encodingA20 (lanes 4 to 6), and cell lysates were subjected to immunoprecipitation with antibody against Act1. Lysates were analyzed by Western blotting with antibody against TRAF6. The arrowhead indicates the approximate migration of TRAF6 (band is obscured by immunoglobulin heavy chain). Note that lanes 1 to 3 and 4 to 6 are from the same gel, but the TRAF6 gel was subjected to different exposure times to optimize visualization of the larger, ubiquitinated TRAF6 bands. n = 3 experiments. (D) A20 suppresses IL-17–dependent activation of MAPK. A20−/− cells were transfected with EV (lanes 1 to 4) or plasmid encoding A20 (lanes 5 to 8), stimulated with IL-17 for the indicated times, and the cell lysates were analyzed by Western blotting for the indicated MAPK family members or A20. n = 2 experiments.
Because TRAF6 is essential for IL-17–induced NF-κB activation (31), we asked whether IL-17 stimulated an association between A20 and TRAF6. We stimulated A20−/− MEFs that were or were not reconstituted with A20 with IL-17 over a 2-hour time course. Cell lysates were subjected to immunoprecipitation with antibody against TRAF6 and analyzed by Western blotting to detect coimmunoprecipitated A20. We could not detect an association between A20 and TRAF6 under basal conditions (Fig. 5B, lane 5); however, A20 associated with TRAF6 60 min after treatment with IL-17. These kinetics are very similar to those of the association of A20 with TRAF6 and TRAF2, which is stimulated by IL-1β and TNF-α, respectively (Fig. 5B) (32, 51, 52). To determine whether the activation of A20 was associated with the deubiquitination of TRAF6, we treated reconstituted A20−/− cells with IL-17, immunoprecipitated Act1 from cell lysates, and assessed the ubiquitination status of TRAF6 by Western blotting analysis. Consistent with an association between A20 and TRAF6, TRAF6 exhibited sustained ubiquitination after stimulation of the A20−/− cells with IL-17 for 90 to 120 min, as evidenced by the increased intensity of the larger migrating forms reactive to the anti-TRAF6 antibodies (Fig. 5C, lanes 1 to 3). In contrast, reconstitution of A20−/− cells with A20 was associated with the reduced appearance of the larger migrating forms of TRAF6 (Fig. 5C, lanes 4 to 6). Although the signals were weak, they were reproducible and are consistent with findings from another study (53). These data probably also reflect the fact that IL-17–mediated activation of NF-κB is typically less substantial than that by classical inflammatory cytokines, such as IL-1β and TNF-α (46, 54).
A20 inhibits the IL-17–dependent activation of MAPK signaling, particularly the c-Jun N-terminal kinase pathway
In the IL-17 signaling pathway, TRAF6 is required for the activation of MAPKs (15). Although there is some variation among cell types, all three major MAPK signaling pathways, those activated by extracellular signal–regulated kinase (ERK), p38, and c-Jun N-terminal kinase (JNK), are activated in response to IL-17. To determine whether A20 suppressed MAPK activation, we transfected A20−/− cells with empty plasmid or with plasmid encoding A20 and assessed the IL-17–dependent phosphorylation of JNK, ERK, and p38 by Western blotting. A20 prevented the prolonged phosphorylation of JNK, as indicated by the absence of detectable phosphorylated JNK (pJNK) at the 30- and 60-min time points in A20-transfected cells compared to cells transfected with EV (Fig. 5D, compare lanes 3 and 4 with lanes 7 and 8). A similar, although less pronounced, pattern was observed for phosphorylated ERK (pERK) and phosphorylated p38 (pp38) (Fig. 5D). Therefore, we conclude that A20 suppresses the activation of MAPK signaling, particularly that of JNK.
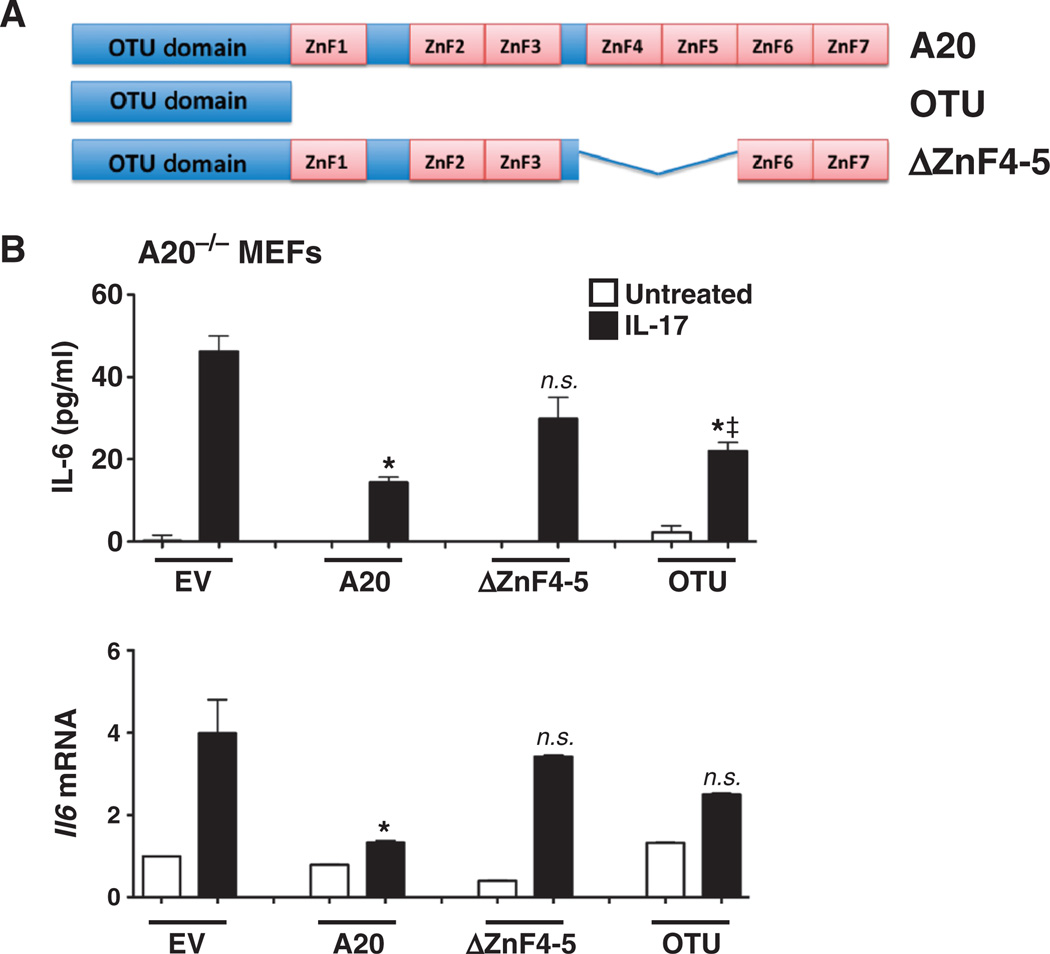
Inhibition of IL-17 signaling is mediated through the OTU and zinc finger domains of A20
Multiple subdomains of A20 contribute to its inhibitory capacity (Fig. 6A and fig. S3) (32), the best characterized of which is the N-terminal OTU (ovarian tumor) domain, which encodes DUB activity and is proposed to be important for the association of A20 with TRAF6 (55, 56). In addition, the seven C-terminal zinc finger (ZnF) domains are important for A20 activity (32, 55, 57, 58). In particular, ZnF4 and ZnF5 exhibit ubiquitin binding activity, which facilitates adaptor recruitment and substrate recognition (59, 60). To determine the roles of these domains in IL-17 reconstituted A20−/− MEFs with wild-type A20 or its OTU domain or ΔZnF4–5 mutants (Fig. 6A). Twenty-four hours later, cells were stimulated with IL-17, and supernatants were evaluated for IL-6 by ELISA. Neither mutant A20 protein inhibited IL-17–dependent IL-6 secretion as effectively as did wild-type A20 (Fig. 6B). Similarly, neither mutant detectably suppressed Il6 expression (Fig. 6B). Although in some experiments the OTU mutant mediated a mild suppressive effect on IL-17–induced IL-6 production (Fig. 6), this was not always reproducible (fig. S3) and may have been a result of the higher abundance of the mutant proteins compared to that of wild-type A20 (fig. S3A). To further confirm the roles of the OTU and ZnF domains, we reconstituted A20−/− MEFs with a construct encoding only the terminal four ZnF domains (fig. S3B). This mutant also failed to inhibit IL-17–induced IL-6 production. We conclude that A20-mediated inhibition of IL-17 signaling involves its catalytic OTU domain and its ZnF domains.
Fig. 6. Inhibition of IL-17 signaling is mediated through the OTU and zinc finger domains of A20.
(A) Schematic diagram of A20 subdomains. OTU, DUB domain. The locations of the ZnF are shown. (B) ZnF mutants of A20 impair its ability to regulate IL-17 signaling. A20−/− cells were transfected in triplicate with EV or with plasmids encoding wild-type A20 or the indicated mutants and were treated with IL-17 for 4 hours. IL-6 in the culture medium was assessed by ELISA measurements performed in triplicate (top panel), and the abundance of Il6 mRNA was assessed by qPCR (bottom panel). *P < 0.05, ANOVA and post hoc Tukey’s test compared to samples transfected with EV and treated with IL-17. ‡P < 0.05, ANOVA and post hoc Tukey’s test compared to samples transfected with A20 and treated with IL-17; n = 3 experiments.
A20 associates with IL-17RA at the C-terminal inhibitory domain of the receptor
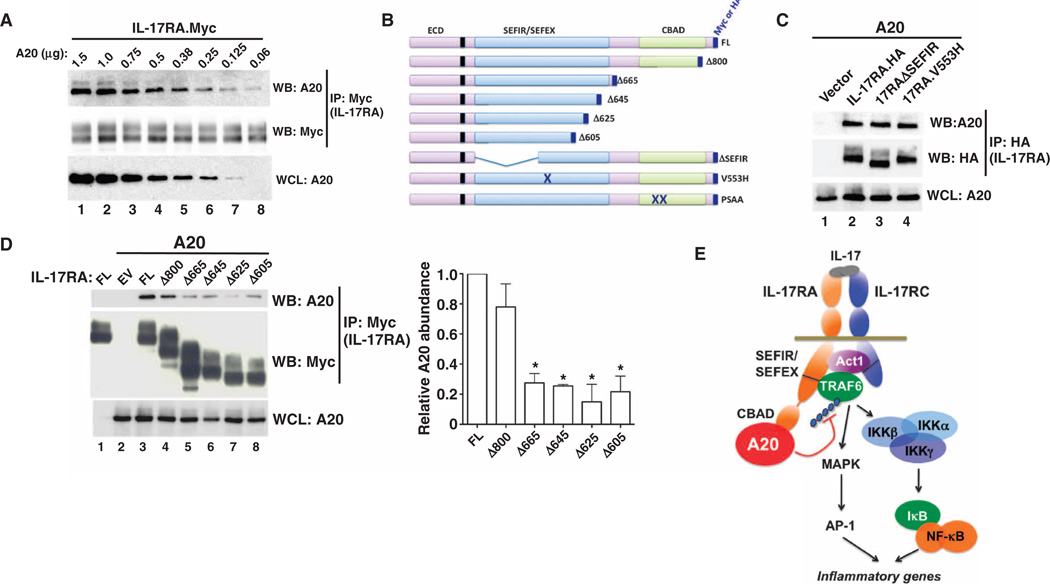
To determine whether IL-17R and A20 interacted directly, we cotransfected human embryonic kidney (HEK) 293T cells with plasmids encoding A20 and murine IL-17RA (tagged at the C terminus with Myc). Lysates were subjected to immuno-precipitation with anti-Myc antibodies to pull down the IL-17R, and its physical association with A20 was determined by Western blotting. We found that A20 associated with IL-17RA in response to IL-17 in a dose-dependent manner (Fig. 7A). Although A20 restricts the TNFR, IL-1R, and NLR pathways, it is not known whether A20 binds directly to any of these receptors.
Fig. 7. A20 binds to IL-17RA through the CBAD.
(A) IL-17RA binds to A20 in a dose-dependent manner. HEK 293T cells were transfected with plasmid encoding IL-17RA (tagged at the C terminus with Myc) together with decreasing concentrations of plasmid encoding A20 (1.5 to 0.06 µg). Anti-Myc antibody was used to immunoprecipitate IL-17RA, and the immunoprecipitates were analyzed by Western blotting with antibodies against A20 (top blot) or Myc (middle blot) to detect IL-17RA. WCLs from samples taken before immunoprecipitation were analyzed by Western blotting with antibody against A20 (bottom blot). n = 2 experiments. (B) Schematic diagram of IL-17RA mutants. ECD, extracellular domain. The locations of the SEFIR and SEFEX regions, the point mutants (V553H, PSAA), and the CBAD are indicated. (C) A20 binds to IL-17RA in a SEFIR- and SEFEX-independent manner. HEK 293T cells were cotransfected with plasmids encoding wild-type IL-17RA or the indicated mutants [tagged at the C terminus with hemagglutinin (HA)] together with plasmid encoding A20. Anti-HA antibody was used to pull down IL-17RA, and immunoprecipitates were analyzed by Western blotting with antibodies against A20 (top blot) or HA (middle blot) to detect IL-17RA. The presence of A20 in WCLs was determined (bottom blot). n = 3 experiments. (D) A20 binds to IL-17RA through the CBAD. HEK 293T cells were transfected with plasmids encoding full-length (FL) IL-17RA or the indicated IL-17RA constructs (tagged at the C terminus with Myc) as well as plasmid encoding A20. Anti-Myc antibodies were used to pull down IL-17RA, and immunoprecipitates were analyzed by Western blotting with antibodies against A20 (top blot) or Myc (middle blot) to detect IL-17RA. The presence of A20 in WCLs was also determined (bottom blot). *P < 0.05, ANOVA and post hoc Tukey’s test compared to FL; n = 3 experiments. (E) Schematic diagram of IL-17RA–mediated signaling and of the role of A20 in restricting this process.
IL-17RA contains several functional subdomains (Fig. 7B). The SEFIR domain is conserved among IL-17R family members and serves as a platform for its association with Act1, which in turn interacts with TRAF6 (19). A large, nonconserved extension of the SEFIR, termed the SEFEX, is also required for signaling in response to IL-17 (23, 24, 61). A point mutation within the SEFEX region (V553H) renders IL-17RA nonfunctional and impairs activation of the NF-κB and MAPK pathways (23, 25). Because A20 restricts the activation of both NF-κB and MAPK, we initially predicted that the SEFIR or SEFEX region would be the site of interaction with A20. Unexpectedly, we found that A20 associated with the IL-17RAΔSEFIR and IL-17RA.V553H mutants (Fig. 7C), indicating that A20 binds to IL-17RA in a SEFIR- and SEFEX-independent manner. Moreover, A20 coimmunoprecipitated with IL-17RA in TRAF6-deficient cells (fig. S4A), consistent with the lack of a requirement for the SEFIR or SEFEX regions in the recruitment of A20 to IL-17RA.
In addition to the SEFIR and SEFEX regions, IL-17RA contains a C-terminal domain located downstream of residue 665 that does not overlap with the SEFIR or SEFEX regions. This region is required for the IL-17–dependent activation of C/EBPβ and, hence, is termed the C/EBPβ activation domain (CBAD) (23). To determine whether A20 interacted with the CBAD of IL-17RA, we cotransfected HEK 293T cells with plasmid encoding A20 and with plasmids encoding a range of C-terminal truncations of IL-17RA. Binding of A20 to IL-17RAΔ800 was not significantly reduced compared to that of full-length IL-17RA. IL-17RAΔ665 and all of the larger truncation mutants were impaired in their ability to coimmunoprecipitate with IL-17RA (Fig. 7D). The CBAD contains a TRAF binding motif that is required for its interaction with TRAF3 (26); thus, we tested the association of A20 with an IL-17RA mutant lacking this site (IL-17RA.PSAA). We found that A20 coimmunoprecipitated with the IL-17RA.PSAA mutant similarly to wild-type IL-17RA (fig. S4B). We and others showed that the CBAD is an inhibitory domain because IL-17RA truncation mutants lacking this region exhibit enhanced IL-17–dependent signaling (23–26). The association of this region with A20 may explain the underlying basis for how the CBAD inhibits IL-17–dependent signaling.
DISCUSSION
The IL-17 family is the most recently discovered and least understood of the cytokine subclasses (15). Consisting of the ligands IL-17A to IL-17F and binding to the receptors IL-17RA to IL-17RE, the IL-17 family of cytokines has many unique structural and functional features. Since the discovery of the TH17 subset of helper T cells in 2005, considerable attention has been paid to how TH17 and other IL-17–producing cells are generated and maintained but far less to how IL-17 mediates downstream signaling (6, 62).
Although IL-17 is vital for host defense against certain pathogens, it has high potential for inducing pathological damage to inflamed tissue. Hence, it is not surprising that there are numerous mechanisms in place to constrain the activities of IL-17 and TH17 cells (63, 64). To list a few examples, the TH1- and TH2-specific cytokines interferon-γ (IFN-γ) and IL-4 and IL-2 block the differentiation of TH17 cells (65–67). TH17 cells often convert to regulatory T cells (Tregs) or TH1-like cells in vivo, tempering their inflammatory activity (68–70). Immunoregulatory cytokines, such as IL-25, IL-27, and IL-10, limit TH17-mediated pathology (71–74). At the level of the IL-17R signaling pathway, TRAF3 inhibits IL-17 by binding to IL-17RA and displacing Act1 (26). A report also implicates the microRNA miR-23b in limiting IL-17 activity (75). IL-17–inducible degradation of Act1 follows engagement of the IL-17R,mitigating the signaling response (76). In studying the activation of the C/EBP transcription factors, we found that IL-17–dependent phosphorylation of C/EBPβ by GSK-3β inhibits the expression of IL-17 target genes (25). Additionally, IL-17A exists either as a homodimer or as a heterodimer with IL-17F, and in the latter state, it has a reduced signaling capacity that probably moderates its activity in vivo (77–79). A report identified the DUB ubiquitin-specific protease 25 (USP25) in targeting TRAF5 and TRAF6 for deubiquitination and limiting IL-17 signaling (53).
Similarly to other inflammatory effectors, IL-17 activates the canonical NF-κB pathway, albeit far more modestly than does TNF-α or TLR ligands (15). The NF-κB pathway is intricately regulated by ubiquitination and deubiquitination reactions (28). A20 dampens TNFR-induced signaling (39) and is the gene product of a well-known susceptibility locus for autoimmunity. The primary enzymatic function of A20 is to serve as a DUB, although it can also act as a ubiquitin ligase and inhibit signaling independently of its catalytic activity (32, 36,55, 80). In the IL-17 pathway, both Act1 and TRAF6 are E3 ubiquitin ligases that target transforming growth factor β–activated kinase 1 (TAK1) and the IKK complex, ultimately causing degradation of IκBα and nuclear import of NF-κB (17, 22). In IL-1 and TLR signaling, A20 limits MAPK activation by targeting TRAF6 for degradation (39). Our work reveals that A20 is both a gene target and a potent feedback inhibitor of IL-17 signaling, acting through TRAF6, NF-κB, and MAPK. Therefore, regulation of ubiquitination is emerging as a central feature of IL-17 signaling (Fig. 7E).
Using RNA-silencing and A20−/− cells, we showed that the expression of various IL-17 target genes was inhibited by A20 (Figs. 1 to 3). As expected, nearly all of these genes are regulated by NF-κB, including genes encoding IL-6 and Lcn2, whose proximal promoters require an intact NF-κB element for their induction by IL-17 (46, 50). There were, however, some exceptions. For example, Ikbz expression was largely unaffected by A20 (Figs. 1E and 4B). The basis for this difference is not clear; however, IL-17 also regulates the stability of many target mRNAs (81). Best studied in the context of the gene Cxcl1, which encodes the chemokine KC (also known as Groα), and this event is mediated in a noncanonical manner by activation of splicing factor 2 (SF2) through TRAF2 and TRAF5 (82). A20 does not appear to influence mRNA stability, which perhaps explains its modest effect on some genes but not others (Figs. 1 and 2). Similar to the TNF pathway, A20 also inhibits IL-17–mediated activation of MAPK pathways, particularly that of JNK (Fig. 5D). MAPK activation leads to AP-1 activation, and AP-1 binding sites are statistically overrepresented in the promoters of IL-17 target genes (50); however, at least for the Il6 promoter, the AP-1 binding site is dispensable for IL-17–induced activation (46). In addition, MAPK signaling may participate in the regulation of mRNA stability, although it is not clear to what extent this is the case for IL-17 target genes.
We found that A20 bound directly to IL-17RA, which supports a direct model of inhibition of IL-17 (Fig. 7). Like most receptors, IL-17RA contains discrete functional subdomains (16, 23). Unexpectedly, A20 bound to the distal domain of IL-17RA, and not the SEFIR or SEFEX regions that are the sites of engagement for Act1 and TRAF6 (19, 24). This distal domain (the CBAD) was initially identified and defined by its ability to regulate the alternative translation and phosphorylation of C/EBPβ (23, 25). Phosphorylation of C/EBPβ by GSK-3β is mediated through the CBAD and is associated with dampened IL-17R signaling (25). ATRAF consensus site within the CBAD is an interaction site for TRAF3, but not TRAF6, which is thought to inhibit IL-17 signaling by competing with Act1. Here, we showed that A20 also bound to the CBAD, although not through the TRAF consensus site (Fig. 7 and fig. S3B), indicating that the CBAD may serve as a platform for the binding of multiple inhibitory proteins.
There are still many open questions regarding the details of how A20 restrains IL-17 signaling. In other systems, A20 cooperates with the Itchy E3 ubiquitin protein ligase (Itch), human T cell leukemia virus type I binding protein 1 (Tax1BP1), and Ring finger protein 11 (RNF11) to form a functional ubiquitin-editing complex (35), but it is not known whether any or all of these factors are required to inhibit IL-17R signaling. It is also not known whether A20 blocks other IL-17 family members that signal through IL-17RA, such as IL-17E (IL-25) or IL-17C. Because IL-17 has repeatedly been shown to use noncanonical pathways and signaling intermediates, it is likely that surprises will emerge regarding how this pathway is controlled.
The A20 gene locus, TNFAIP3, is strongly associated with many autoimmune diseases. Clearly, this is related in part through the inhibition of TNF-dependent signaling by A20 (83); however, A20 also constrains TLR and NLR signaling, which are increasingly recognized as important in mediating autoimmunity (84, 85). We now add IL-17 signaling to the list of proinflammatory pathways governed by A20. This finding dovetails well with the role for IL-17 in mediating at least some forms of autoimmune disease, as illustrated by the clinical success of biologic therapies targeting this cytokine (86). Consistently, silencing of A20 led to enhanced IL-17 signaling in human RA FLS cells (Fig. 1G), supporting its potential relevance in human cells. Nonetheless, more work is certainly needed to determine how much of the effect of A20 on IL-17 contributes to disease in vivo compared to the effect of A20 on other inflammatory stimuli. Defining molecular signaling intermediates, especially enzymes such as A20, has the potential to reveal strategies for developing small-molecule therapeutics that target IL-17 signaling (87, 88). Drugs that enhance A20 function could theoretically help to restrain IL-17 and other inflammatory cytokines that promote autoimmunity. Alternatively, blocking A20 might be useful in stimulating the host defense response in settings in which IL-17 activity is beneficial, such as during fungal or bacterial infections (10).
MATERIALS AND METHODS
Cell cultures, reagents, and luciferase assays
A20−/− and TRAF6−/− MEFs were cultured in α-MEM (minimum essential medium, Sigma) containing 10% fetal bovine serum (FBS) supplemented with l-glutamine and antibiotics (Invitrogen) (39). ST2 stromal cells, HEK 293T cells, and human FLS cells were cultured in α-MEM with 15% FBS, l-glutamine, and antibiotics. HEK 293T cells were transfected by the calcium phosphate method, whereas ST2 cells and A20−/− MEFs were transfected with FuGENE 6 (Roche) or FuGENE HD (Promega). Within each transfection experiment, three replicate samples were transfected and assayed separately. Luciferase assays were performed as previously described (50) with the Dual-Luciferase Reporter Assay System (Promega). Recombinant murine and human IL-17 proteins were purchased from PeproTech and were used at a final concentration of 200 ng/ml. The IKK Inhibitor VII (EMD Millipore) was used at a final concentration of 0.1 µM. Human subject research was performed in accordance with protocols approved by the Institutional Review Board (IRB) of the University of Pittsburgh.
siRNA and DNA plasmids
ON-TARGETplus SMARTpool siRNA targeting A20 (tnfaip3) and Act1 (traf3ip2) and scrambled controls were obtained from Dharmacon/Thermo Scientific. ST2 cells were seeded overnight in antibiotic-free medium and transfected the next day with 50 nM siRNA with DharmaFECT Reagent 1 (Dharmacon/Thermo Scientific). Twenty-four hours later, the culture medium was changed, and after a further 24 hours, cells were stimulated with IL-17 for the times indicated for each experiment. For each experiment, three replicate samples were analyzed separately. Plasmids encoding murine IL-17RA, its point mutants, and truncation mutants were constructed as described previously (23, 24, 26). Plasmids encoding murine A20 and its mutants were obtained from the plasmid repository at BCCM/LMBP (Belgian Coordinated Collections of Micro-organisms/Laboratory of Molecular Biology–Plasmid collection) (Belgium) (60).
RNA isolation and qPCR analysis
Total RNA was isolated from cells with an RNeasy Mini Kit (Qiagen). Complementary DNA synthesis was performed with SuperScript III First-Strand (Invitrogen). The extent of expression of Il6, Ccl20, 24p3, Csf3, Cxcl1, Cxcl5, IκBζ, and Tnfaip3 was determined by qPCR analysis with PerfeCTa SYBR Green FastMix ROX (Quanta BioSciences). The PCRs were performed on a 7300 Real-Time PCR System (Applied Biosystems). The abundances of the mRNAs of interest were normalized to that of Gapdh. Primers were purchased from Super Array Biosciences (Qiagen). For each experiment, three replicates were analyzed separately.
ELISAs and immunoprecipitations
Western blotting analysis and immunoprecipitations were performed as described previously (23, 61), and bands on blots corresponding to proteins of interest were analyzed by ImageJ software or on a ProteinSimple FluorChem E instrument. Anti-A20, anti-pJNK, anti-JNK, anti-pERK, anti-ERK, anti-pp38, anti-p38, and anti-Myc antibodies were from Cell Signaling Technology; anti-IκBα, anti-TRAF6, and anti-Act1 antibodies were from Santa Cruz Biotechnology; anti-tubulin antibody was obtained from Invitrogen; and anti-HA antibody was from Sigma. Western blots were developed with a FluorChem E imager (ProteinSimple). Murine and human IL-6 ELISA kits were from eBioscience, and the CCL20 ELISA kit was from R&D Systems. For each experiment, each sample was analyzed in duplicate or triplicate, and a minimum of three replicate samples were included per experiment.
Statistics
To assess statistical significance, we used Student’s t test (for pairwise comparisons) or ANOVA with post hoc Tukey’s analysis (for more than two comparisons in an experiment). P < 0.05 was considered statistically significant. Error bars reflect the means ± SEM of biological replicates within individual experiments. All experiments were repeated a minimum of two times to ensure reproducibility.
Supplementary Material
Acknowledgments
We thank E. C. Childs for valuable assistance and L. Kane and P. Biswas for critical reading of the manuscript.
Funding: This work was supported by NIH grant AR054389 to S.L.G. and a grant from the Nancy Taylor Foundation for Chronic Diseases to A.N.V.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/6/278/ra44/DC1
Fig. S1. Knockdown of A20 enhances the expression of most IL-17 target genes.
Fig. S2. Kinetics of IL-17–dependent IL-6 production.
Fig. S3. ZnF4 to ZnF7 of A20 are insufficient to suppress IL-17–mediated signaling.
Fig. S4. TRAF6 and TRAF3 are not required for the association between A20 and IL-17RA.
Author contributions: A.V.G. and M.A. performed the experiments; A.V.G., A.N.V., A.M., and S.L.G. contributed to experimental design and data interpretation; and A.V.G. and S.L.G. wrote the manuscript.
Competing interests: S.L.G. received travel reimbursement, honoraria, and research grants from Amgen and Novartis.
Data and Materials Availability: Use of the human FLS cells is covered by an IRB protocol.
REFERENCES AND NOTES
- 1.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol. 2007;7:429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 2.Genovese MC, Van den Bosch F, Roberson SA, Bojin S, Biagini IM, Ryan P, Sloan-Lancaster J. LY2439821, a humanized anti–interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebo-controlled, proof-of-concept study. Arthritis Rheum. 2010;62:929–939. doi: 10.1002/art.27334. [DOI] [PubMed] [Google Scholar]
- 3.Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, Kricorian G, Aras G, Li J, Russell CB, Thompson EH, Baumgartner S. Brodalumab, an anti–interleukin-17–receptor antibody for psoriasis. N. Engl. J. Med. 2012;366:1181–1189. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 4.Hueber W, Patel DD, Dryja T, Wright AM, Koroleva I, Bruin G, Antoni C, Draelos Z, Gold MH, Psoriasis Study Group. Durez P, Tak PP, Gomez-Reino JJ, Rheumatoid Arthritis Study Group. Foster CS, Kim RY, Samson CM, Falk NS, Chu DS, Callanan D, Nguyen QD, Uveitis Study Group. Rose K, Haider A, Di Padova F. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci. Transl. Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- 5.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, Edson-Heredia E, Braun D, Banerjee S. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N. Engl. J. Med. 2012;366:1190–1199. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 6.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds JM, Angkasekwinai P, Dong C. IL-17 family member cytokines: Regulation and function in innate immunity. Cytokine Growth Factor Rev. 2010;21:413–423. doi: 10.1016/j.cytogfr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cua DJ, Tato CM. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 9.Gaffen SL. Recent advances in the IL-17 cytokine family. Curr. Opin. Immunol. 2011;23:613–619. doi: 10.1016/j.coi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onishi RM, Gaffen SL. IL-17 and its target genes: Mechanisms of IL-17 function in disease. Immunology. 2010;129:311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppler AR, Bishu S, Gaffen SL. Mucocutaneous candidiasis: The IL-17 pathway and implications for targeted immunotherapy. Arthritis Res. Ther. 2012;14:217. doi: 10.1186/ar3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova JL. Inborn errors of mucocutaneous immunity to Candida albicans in humans: A role for IL-17 cytokines? Curr. Opin. Immunol. 2010;22:467–474. doi: 10.1016/j.coi.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenewicz LA, Abraham C, Flavell RA, Cho JH. Unraveling the genetics of autoimmunity. Cell. 2010;140:791–797. doi: 10.1016/j.cell.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novatchkova M, Leibbrandt A, Werzowa J, Neubüser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci. Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J. Biol. Chem. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 19.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17–dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 20.Leonardi A, Chariot A, Claudio E, Cunningham K, Siebenlist U. CIKS, a connection to IκB kinase and stress-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10494–10499. doi: 10.1073/pnas.190245697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zepp J, Wu L, Li X. IL-17 receptor signaling and T helper 17-mediated autoimmune demyelinating disease. Trends Immunol. 2011;32:232–239. doi: 10.1016/j.it.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Maitra A, Shen F, Hanel W, Mossman K, Tocker J, Swart D, Gaffen SL. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7506–7511. doi: 10.1073/pnas.0611589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onishi RM, Park SJ, Hanel W, Ho AW, Maitra A, Gaffen SL. SEF/IL-17R (SEFIR) is not enough: An extended SEFIR domain is required for IL-17RA-mediated signal transduction. J. Biol. Chem. 2010;285:32751–32759. doi: 10.1074/jbc.M110.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen F, Li N, Gade P, Kalvakolanu DV, Weibley T, Doble B, Woodgett JR, Wood TD, Gaffen SL. IL-17 receptor signaling inhibits C/EBPβ by sequential phosphorylation of the regulatory 2 domain. Sci. Signal. 2009;2:ra8. doi: 10.1126/scisignal.2000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S, Pan W, Shi P, Gao H, Zhao F, Song X, Liu Y, Zhao L, Li X, Shi Y, Qian Y. Modulation of experimental autoimmune encephalomyelitis through TRAF3-mediated suppression of interleukin 17 receptor signaling. J. Exp. Med. 2010;207:2647–2662. doi: 10.1084/jem.20100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malynn BA, Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X. Act1 modulates autoimmunity through its dual functions in CD40L/BAFF and IL-17 signaling. Cytokine. 2008;41:105–113. doi: 10.1016/j.cyto.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Lindén A. A role for the cytoplasmic adaptor proteins Act1 in mediating IL-17 signaling. Sci. STKE. 2007;2007:re4. doi: 10.1126/stke.3982007re4. [DOI] [PubMed] [Google Scholar]
- 31.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor–associated factor (TRAF)6 in interleukin 17 signal transduction. J. Exp. Med. 2000;191:1233–1240. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruland J. Return to homeostasis: Downregulation of NF-κB responses. Nat. Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 34.Sun SC. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shembade N, Harhaj EW. Regulation of NF-κB signaling by the A20 deubiquitinase. Cell. Mol. Immunol. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verstrepen L, Verhelst K, van Loo G, Carpentier I, Ley SC, Beyaert R. Expression, biological activities and mechanisms of action of A20 (TNFAIP3) Biochem. Pharmacol. 2010;80:2009–2020. doi: 10.1016/j.bcp.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 37.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 38.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maelfait J, Roose K, Bogaert P, Sze M, Saelens X, Pasparakis M, Carpentier I, van Loo G, Beyaert R. A20 (Tnfaip3) deficiency in myeloid cells protects against influenza A virus infection. PLoS Pathog. 2012;8:e1002570. doi: 10.1371/journal.ppat.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Mc Guire C, Vereecke L, Chu Y, Boon L, Staelens S, Matthys P, Lambrecht BN, Schmidt-Supprian M, Pasparakis M, Elewaut D, Beyaert R, van Loo G. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 42.Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, Reizis B, DeFranco A, Criswell LA, Nakamura MC, Ma A. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat. Immunol. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, Seldin MF, Gregersen PK, Behrens TW, Ma A, Kwok PY, Criswell LA. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J. Biol. Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- 45.Miossec P. Interleukin-17 in rheumatoid arthritis: If T cells were to contribute to inflammation and destruction through synergy. Arthritis Rheum. 2003;48:594–601. doi: 10.1002/art.10816. [DOI] [PubMed] [Google Scholar]
- 46.Ruddy MJ, Wong GC, Liu XK, Yamamoto H, Kasayama S, Kirkwood KL, Gaffen SL. Functional cooperation between interleukin-17 and tumor necrosis factor-a is mediated by CCAAT/enhancer binding protein family members. J. Biol. Chem. 2004;279:2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 47.Sønder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, Siebenlist U. IL-17-induced NF-κB activation via CIKS/Act1: Physiologic significance and signaling mechanisms. J. Biol. Chem. 2011;286:12881–12890. doi: 10.1074/jbc.M110.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen F, Ruddy MJ, Plamondon P, Gaffen SL. Cytokines link osteoblasts and inflammation: Microarray analysis of interleukin-17- and TNF-α-induced genes in bone cells. J. Leukoc. Biol. 2005;77:388–399. doi: 10.1189/jlb.0904490. [DOI] [PubMed] [Google Scholar]
- 49.Gaffen SL. The role of interleukin-17 in the pathogenesis of rheumatoid arthritis. Curr. Rheumatol. Rep. 2009;11:365–370. doi: 10.1007/s11926-009-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J. Biol. Chem. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 51.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heyninck K, Beyaert R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 1999;442:147–150. doi: 10.1016/s0014-5793(98)01645-7. [DOI] [PubMed] [Google Scholar]
- 53.Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, Reynolds JM, Dong C. Negative regulation of IL-17-mediated signalling and inflammation by the ubiquitin-specific protease USP25. Nat. Immunol. 2012;13:1110–1117. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 55.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 56.Ma A, Malynn BA. A20: Linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, Kirkpatrick DS, Dixit VM, Hymowitz SG. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 60.Klinkenberg M, Van Huffel S, Heyninck K, Beyaert R. Functional redundancy of the zinc fingers of A20 for inhibition of NF-κB activation and protein–protein interactions. FEBS Lett. 2001;498:93–97. doi: 10.1016/s0014-5793(01)02504-2. [DOI] [PubMed] [Google Scholar]
- 61.Ho AW, Shen F, Conti HR, Patel N, Childs EE, Peterson A, Hernández-Santos N, Kolls JK, Kane LP, Ouyang W, Gaffen SL. IL-17RC is required for immune signaling via an extended SEFIR domain in the cytoplasmic tail. J. Immunol. 2010;185:1063–1070. doi: 10.4049/jimmunol.0903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 63.Stumhofer JS, Silver J, Hunter CA. Negative regulation of Th17 responses. Semin. Immunol. 2007;19:394–399. doi: 10.1016/j.smim.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diveu C, McGeachy MJ, Cua DJ. Cytokines that regulate autoimmunity. Curr. Opin. Immunol. 2008;20:663–668. doi: 10.1016/j.coi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17–producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 68.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bluestone JA, Mackay CR, O’Shea JJ, Stockinger B. The functional plasticity of T cell subsets. Nat. Rev. Immunol. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, Feng XH, Jetten AM, Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stumhofer JS, Hunter CA. Advances in understanding the anti-inflammatory properties of IL-27. Immunol. Lett. 2008;117:123–130. doi: 10.1016/j.imlet.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, Villarino AV, Huang Q, Yoshimura A, Sehy D, Saris CJ, O’Shea JJ, Hennighausen L, Ernst M, Hunter CA. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol. 2006;7:937–945. doi: 10.1038/ni1376. [DOI] [PubMed] [Google Scholar]
- 73.Kleinschek MA, Owyang AM, Joyce-Shaikh B, Langrish CL, Chen Y, Gorman DM, Blumenschein WM, McClanahan T, Brombacher F, Hurst SD, Kastelein RA, Cua DJ. IL-25 regulates Th17 function in autoimmune inflammation. J. Exp. Med. 2007;204:161–170. doi: 10.1084/jem.20061738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen F, Gaffen SL. Structure–function relationships in the IL-17 receptor: Implications for signal transduction and therapy. Cytokine. 2008;41:92–104. doi: 10.1016/j.cyto.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, Shi Y, Harley JB, Shen N, Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med. 2012;18:1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 76.Shi P, Zhu S, Lin Y, Liu Y, Liu Y, Chen Z, Shi Y, Qian Y. Persistent stimulation with interleukin-17 desensitizes cells through SCFβ-TrCP-mediated degradation of Act1. Sci. Signal. 2011;4:ra73. doi: 10.1126/scisignal.2001653. [DOI] [PubMed] [Google Scholar]
- 77.Wright JF, Guo Y, Quazi A, Luxenberg DP, Bennett F, Ross JF, Qiu Y, Whitters MJ, Tomkinson KN, Dunussi-Joannopoulos K, Carreno BM, Collins M, Wolfman NM. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J. Biol. Chem. 2007;282:13447–13455. doi: 10.1074/jbc.M700499200. [DOI] [PubMed] [Google Scholar]
- 78.Wright JF, Bennett F, Li B, Brooks J, Luxenberg DP, Whitters MJ, Tomkinson KN, Fitz LJ, Wolfman NM, Collins M, Dunussi-Joannopoulos K, Chatterjee-Kishore M, Carreno BM. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J. Immunol. 2008;181:2799–2805. doi: 10.4049/jimmunol.181.4.2799. [DOI] [PubMed] [Google Scholar]
- 79.Chang SH, Dong C. A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 2007;17:435–440. doi: 10.1038/cr.2007.35. [DOI] [PubMed] [Google Scholar]
- 80.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamilton T, Novotny M, Pavicic PJ, Jr, Herjan T, Hartupee J, Sun D, Zhao C, Datta S. Diversity in post-transcriptional control of neutrophil chemoattractant cytokine gene expression. Cytokine. 2010;52:116–122. doi: 10.1016/j.cyto.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Datta S, Novotny M, Pavicic PG, Jr, Zhao C, Herjan T, Hartupee J, Hamilton T. IL-17 regulates CXCL1 mRNA stability via an AUUUA/tristetraprolin-independent sequence. J. Immunol. 2010;184:1484–1491. doi: 10.4049/jimmunol.0902423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tran TM, Temkin V, Shi B, Pagliari L, Daniel S, Ferran C, Pope RM. TNFα-induced macrophage death via caspase-dependent and independent pathways. Apoptosis. 2009;14:320–332. doi: 10.1007/s10495-009-0311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Q, Pope RM. Toll-like receptor signaling: A potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J. Leukoc. Biol. 2010;88:253–262. doi: 10.1189/jlb.0310126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abdollahi-Roodsaz S, van de Loo FA, Koenders MI, Helsen MM, Walgreen B, van den Bersselaar LA, Arntz OJ, Takahashi N, Joosten LA, van den Berg WB. Destructive role of myeloid differentiation factor 88 and protective role of TRIF in interleukin-17–dependent arthritis in mice. Arthritis Rheum. 2012;64:1838–1847. doi: 10.1002/art.34328. [DOI] [PubMed] [Google Scholar]
- 86.Maddur MS, Miossec P, Kaveri SV, Bayry J. Th17 cells: Biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am. J. Pathol. 2012;181:8–18. doi: 10.1016/j.ajpath.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 87.Lopez-Castejon G, Luheshi NM, Compan V, High S, Whitehead RC, Flitsch S, Kirov A, Prudovsky I, Swanton E, Brough D. Deubiquitinases regulate the activity of caspase-1 and interleukin-1β secretion via assembly of the inflammasome. J. Biol. Chem. 2012;288:2721–2733. doi: 10.1074/jbc.M112.422238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, Vorobyov A, Garg P, Beatty L, Chan PK, Juang YC, Landry MC, Yeh C, Zeqiraj E, Karamboulas K, Allali-Hassani A, Vedadi M, Tyers M, Moffat J, Sicheri F, Pelletier L, Durocher D, Raught B, Rotin D, Yang J, Moran MF, Dhe-Paganon S, Sidhu SS. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.