Abstract
Genetic testing for personalizing pharmacotherapy is bound to become an important part of clinical routine. To address associated issues with data management and quality, we are creating a semantic knowledge base for clinical pharmacogenetics. The knowledge base is made up of three components: an expressive ontology formalized in the Web Ontology Language (OWL 2 DL), a Resource Description Framework (RDF) model for capturing detailed results of manual annotation of pharmacogenomic information in drug product labels, and an RDF conversion of relevant biomedical datasets. Our work goes beyond the state of the art in that it makes both automated reasoning as well as query answering as simple as possible, and the reasoning capabilities go beyond the capabilities of previously described ontologies.
Keywords: pharmacogenetics, pharmacogenomics, ontology, medical informatics, clinical decision support systems
Introduction
The discipline of pharmacogenetics is concerned with studying how genetic variation between individuals influences drug efficacy and safety. The clinical application of pharmacogenetics for drug selection and dosing holds great promise of improving the quality of care. However, the growing amount of data and knowledge in this area also makes it necessary to create information technologies for handling the sometimes complex and large sets of data, definitions and clinical guidelines. We created a semantic knowledge base for clinical pharmacogenetics to address these issues. The knowledge base is made up of three components: an expressive ontology formalized in the Web Ontology Language (OWL 2 DL), a Resource Description Framework (RDF) model for capturing detailed results of manual annotation of pharmacogenetic information in drug product labels, and an RDF conversion of relevant biomedical datasets. In this paper we describe the methodologies we used for assembling the knowledge base and preliminary results of applying it to problems from the pharmacogenetics domain.
Materials and Methods
The knowledge base is made up of three components that were first developed independently and were then mapped to each other. The components are based on the Resource Description Framework (RDF) and Web Ontology Language (OWL) standards of the World Wide Web Consortium (W3C).
Creation of OWL 2 reasoning component
We created an expressive OWL 2 ontology by automatically extracting and manually curating data from the Single Nucleotide Polymorphism Database (dbSNP), clinically relevant polymorphisms and allele definitions from the Pharmacogenomics Knowledge Base (PharmGKB) (1), clinically relevant polymorphisms from the Online Mendelian Inheritance in Man Database (OMIM), the Human Cytochrome P450 nomenclature database, guidelines issued by the Clinical Pharmacogenetics Working Group (CPIC) (2) and the Royal Dutch Pharmacogenetics Working Group (DPWG), based on snapshots of these data sources taken on February 2012. 385 polymorphic loci associated with 58 pharmacogenes were captured this way. Some design patterns used in the ontology are exemplified in Listing 1.For example, OWL property restrictions with qualified cardinality restrictions are used to infer haplotypes from combinations of SNPs, and to infer matching clinical recommendations from combinations of SNPs and haplotypes.
Listing 1. Examples of OWL axioms used in the OWL 2 ontology, formalized in Manchester OWL syntax. The first example shows axioms for inferring an allele from Single Nucleotide Polymorphisms. The second example show axioms for inferring an adequate clinical decision support message for warfarin dosing based on genetic data.
We used TrOWL (3), a highly scalable OWL 2 reasoner, for analysing the aggregated data and for testing the classification of individual genetic profiles.
Creation of RDF component for detailed capturing of structured product label text annotations
The Food and Drug Administration (FDA) provides a table of pharmacogenomics biomarkers present in the product labels of FDA-approved drug products (4). However, the information in this table is not sufficient for making informed decisions for either clinical or translational research applications. To address this issue, we created a semantic model of the pharmacogenomics information found in drug product labels. The model’s development was driven by a series of use cases that developed in collaboration with pharmacists and pharmacy doctoral students to demonstrate how structured pharmacogenomics information could be more effectively used to support clinical and translational efforts. Three clinical pharmacists and two fifth-year pharmacy doctoral students participated in the model’s development. Co-author RDB implemented an initial version of the model using the Knowtator plug-in for the Protégé modeling tool (5).
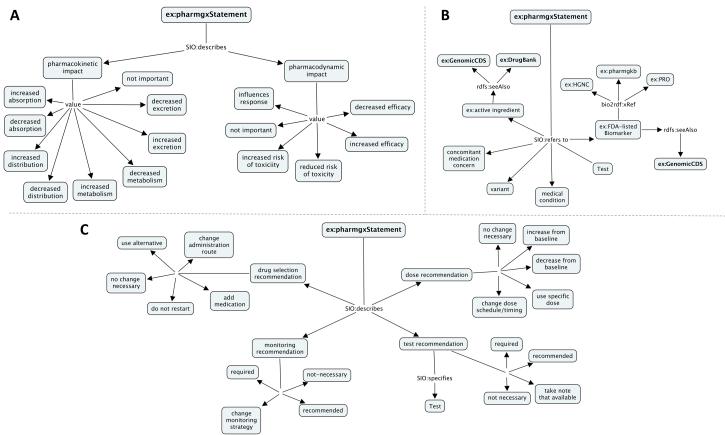
Using an iterative process, the semantic model was field-tested by five pharmacists who work at the University of Pittsburgh Medical Center until the model appeared to require no further revisions. A part of the structure of the model is shown in Figure 1. The pharmacists used the model to manually annotate a subset of the drug labels listed in the table provided by the FDA and considered high priority for pharmacogenomics decisions support. The pharmacists identified a total of 213 pharmacogenomic statements in the 29 sections during September and October of 2012. This initial round of annotation was analyzed to determine inter-rater reliability and make any necessary modifications to the annotation guidelines (available upon request).
Figure 1. Portions of the descriptive RDF model for representing pharmacogenomic statements in Structured Product Labels.
The annotation work is ongoing at the time of this writing with the goal of using the model to annotate all of the drug/biomarker combinations indexed in the FDA’s table. Each product label section is first annotated by two pharmacists and then reviewed by a different pair of pharmacists. Concerns and disagreements are resolved by discussion between all pharmacists at team meetings.
Creation of RDF conversion of relevant pharmacogenomic datasets
We created an RDF representation of relevant biomedical datasets and integrated them into the Bio2RDF infrastructure (6). Bio2RDF is an open source project that aims to provide linked data for the life sciences. PHP scripts were developed to download and convert PharmGKB, dbSNP and OMIM datasets from their source format into RDF. Bio2RDF follows a particular convention in the naming of entities. Provider identified data items are named with http://bio2rdf.org/prefix:identifier, where the dataset prefix (e.g. pharmgkb, dbsnp or omim) is obtained from a global registry of datasets. Following good practices of RDF data publishing, all identifiers can be resolved through the web to yield descriptions of the underlying resources. All scripts used for converting the datasets are available from https://github.com/bio2rdf/bio2rdf-scripts.
Dataset integration
Entities from the three dataset components were mapped to each other through rdfs:seeAlso relations. A part of this mapping was done automatically with simple scripts (e.g., mappings between SNP variants in the OWL 2 ontology and in the Bio2RDF representation of dbSNP), and another part was done manually (e.g., mappings between entities in the OWL 2 ontologies and entities in the RDF model of information from structured product labels).
Results
The OWL 2 ontology component can be downloaded from http://www.genomic-cds.org/. We tested the reasoning capabilities enabled by the ontology with a test dataset containing a hundred SNPs per patient, and verified that individual patients could be matched to clinical decision support messages through OWL reasoning.
The 5 pharmacists identified a total of 213 pharmacogenomic statements in the 29 structured product label sections they annotated. At least two pharmacists agreed that 11 sections (5 drug product labels) contained dosage recommendations, 5 sections (3 drug product labels) contained recommendations for alternative therapy based on genetic test results, 7 sections (3 drug product labels) provided recommendations for genetic testing, and 3 sections (in 2 drug product labels) provided specific monitoring recommendations. Additionally, pharmacological context was added regarding whether impact of the pharmacogenomic information involved altered pharmacokinetics (14 sections in 6 drug product labels) and/or pharmacodynamics (22 sections in 9 drug product labels). Interestingly, some sections listed in the FDA biomarker table as containing pharmacogenomic recommendations received no annotations by any of the 5 pharmacists (e.g., the citalopram Drug Interactions section for CYP2C19 and CYP2D6). The model showed potential to make the unstructured pharmacogenomic information currently written in product labeling more accessible and actionable through structured annotations of pharmacogenomics effects and clinical recommendations. For example, the model enables queries for information in the Warfarin structured product label on the pharmacokinetic/pharmacodynamic effects and dose selection recommendations related to the CYP2C9 biomarker. A simple example query is shown in Listing 2. The query is for any warfarin monitoring recommendations associated with CYP2C9. As the figure highlights, mappings from the semantic model (RDF) to the OWL data set for warfarin and CYP2C9 enable this cross-resource query.
Listing 2. Example of a SPARQL query for monitoring recommendations associated with CYP2C9 and warfarin.
Discussion
The integration of the three dataset components into an inter-linked knowledge base creates new opportunities for queries, and a flexible means of capturing pharmacogenetic knowledge in a range of granularities and degrees of expressivity. The unification of models with different expressivity (OWL 2 vs. light-weight RDF) makes it possible to reap the benefits of both in different scenarios (automated reasoning vs. simple creation and querying of triples with SPARQL). For example, the OWL 2 ontology component can first be used to match individual patients to certain treatment recommendations derived from product labels, and then the RDF component can be queried for detailed information about these recommendations and the corresponding product label.
The model of pharmacogenomics statements enables queries that can be used to create a more usable presentation of the potential impact of genetic variants on drug response. Using warfarin as an example, the structured product label has several pharmacogenomic statements regarding two biomarkers – variations in the genes Cytochrome P450 2C9 (CYP2C9) and Vitamin K epoxide reductase complex subunit 1 (VKORC1) – distributed within three product label sections (Dosage and Administration, Precautions, Clinical Pharmacology sections). Clinicians and translational researchers seeking pharmacogenomics information would have to integrate the text contained within the sections to understand that variants of CYP2C9 impact pharmacokinetics (decrease metabolism) and consequently drug response, while VKORC1 only impacts pharmacodynamics. Using the semantic model, the information in these sections can be automatically merged for both biomarkers, or queried for CYP2C9 or VKORC1 specific recommendations.
During our work we also identified potential difficulties with definitions of clinical phenotypes such “intermediate metabolizer”, “poor metabolizer” or “extensive metabolizer”. These phenotypes are often defined only vaguely, and each term can have quite different meanings when applied to different enzymes and drugs. We hope that such ambiguities can be better resolved through the use of well-defined ontologies and semantic data model such as those outlined in this paper.
The use of RDF/OWL in the domain of pharmacogenetics has been explored in some previous work. For example, the Suggested Ontology for Pharmacogenomics (SO-Pharm) (7) was one of the earliest projects that aimed to demonstrate the use of ontologies in this domain. Our work goes beyond the state of the art in that it a) integrates several components that make both automated reasoning as well as query answering as simple as possible, b) the reasoning capabilities offered by the ontology component and its expressive OWL 2 DL axioms go beyond the capabilities of previously described ontologies and c) considerable parts of the integrated knowledge base are created through automated or semi-automated processes, increasing the likelihood of successful long-term maintenance and growth of the knowledge base.
Conclusion
We invite stakeholders in clinical genetics to participate in the further development and application of the formalism and system we developed, with the potential goal of establishing it as an open standard for formalizing data and rules in clinical pharmacogenetics.
Acknowledgments
This work was supported in part by Austrian Science Fund (FWF): [PP 25608-N15] and the Agency for Healthcare Research and Quality (K12HS019461).
References
- 1.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002 Jan 1;30(1):163–5. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011 März;89(3):464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas E, Pan JZ, Ren Y. TrOWL: Tractable OWL 2 Reasoning Infrastructure; Proc. of the Extended Semantic Web Conference (ESWC2010); 2010. [Google Scholar]
- 4. [cited 2011 Feb 2];Genomics > Table of Pharmacogenomic Biomarkers in Drug Labels (FDA) [Internet] Available from: http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- 5.Ogren PV. Knowtator: A Protégé plug-in for annotated corpus construction; Proceedings of the 2006 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology: companion volume: demonstrations. Association for Computational Linguistics; 2006.pp. 273–5. [Google Scholar]
- 6.Belleau F, Nolin M-A, Tourigny N, Rigault P, Morissette J. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J Biomed Inform. 2008 Oct;41(5):706–16. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Coulet A, Smaïl-Tabbone M, Napoli A, Devignes M-D. Suggested Ontology for Pharmacogenomics (SO-Pharm): Modular Construction and Preliminary Testing [Internet] [cited 2011 Jan 12];HAL - CCSD. 2006 Available from: http://hal.inria.fr/inria-00089824/en/ [Google Scholar]