Abstract
Darwin proposed two seemingly contradictory hypotheses for a better understanding of biological invasions. Strong relatedness of invaders to native communities as an indication of niche overlap could promote naturalization because of appropriate niche adaptation, but could also hamper naturalization because of negative interactions with native species (‘Darwin’s naturalization hypothesis’). Although these hypotheses provide clear and opposing predictions for expected patterns of species relatedness in invaded communities, so far no study has been able to clearly disentangle the underlying mechanisms. We hypothesize that conflicting past results are mainly due to the neglected role of spatial resolution of the community sampling. In this study, we corroborate both of Darwin’s expectations by using phylogenetic relatedness as a measure of niche overlap and by testing the effects of sampling resolution in highly invaded coastal plant communities. At spatial resolutions fine enough to detect signatures of biotic interactions, we find that most invaders are less related to their nearest relative in invaded plant communities than expected by chance (phylogenetic overdispersion). Yet at coarser spatial resolutions, native assemblages become more invasible for closely-related species as a consequence of habitat filtering (phylogenetic clustering). Recognition of the importance of the spatial resolution at which communities are studied allows apparently contrasting theoretical and empirical results to be reconciled. Our study opens new perspectives on how to better detect, differentiate and understand the impact of negative biotic interactions and habitat filtering on the ability of invaders to establish in native communities.
Species transported far from their original range that spread and maintain viable populations (i.e. naturalized non-native species sensu Richardson and Pysek 2006) often pose significant challenges to conserving native biodiversity. Predicting which species can invade which communities is essential if control measures are to be successfully implemented (Marco et al. 2010). The composition of local native assemblages and the phylogenetic relatedness of an invader to these communities can influence invasion success and thus provide a predictive tool. Closely related species are more likely to be ecologically similar, provided that traits determining responses of species to environment and co-existence show a signal along the phylogeny (sensu Blomberg and Garland 2002; i.e. similar trait values between closely-related species). Under these conditions, species’ phylogenetic distances can be used as a proxy for ecological similarity and have the advantage of combining multiple functional trait information.
There are two opposing hypotheses originally proposed by Darwin to link the phylogenetic relatedness between potential invaders and native communities with probabilities of successful invasion (Darwin 1859). On the one hand, close relatedness is predicted to hamper local naturalization due to niche overlap and competition with native species (i.e. ecologically similar species compete more than dissimilar species; ‘Darwin’s naturalization hypothesis’) (Elton 1958). The resulting pattern is commonly referred to as phylogenetic overdispersion in studies of community assembly rules. On the other hand, appropriate niche adaptation may instead favor the naturalization of closely-related introduced species due to habitat filtering, which leads to a spatial pattern of phylogenetic clustering or underdispersion of niches (Duncan and Williams 2002). Previous studies have found support for both of these hypotheses, leading to a fierce controversy in recent literature (Daehler 2001, Lambdon and Hulme 2006, Diez et al. 2009, Ricotta et al. 2010, Schaefer et al. 2011). Two aspects are likely to play a major role in explaining the discrepancy between theoretical predictions and among empirical studies: methodological differences and most importantly differences in the scale considered (Thuiller et al. 2010).
A standard methodological framework to address Darwin’s naturalization hypothesis was outlined in a recent review paper (Thuiller et al. 2010). The authors suggest that the niche overlap between the invader and the members of the recipient community can be explored through a series of metrics based on functional or phylogenetic distances among species (when traits show a phylogenetic signal) and then tested with an appropriate null hypothesis and associated algorithm (Hardy 2008, Thuiller et al. 2010). As different metrics and methods have specific assumptions and may lead to different conclusions, adopting a combination of approaches both for the quantification of niche overlap and for the statistical test is a good method of corroborating results.
Ecological patterns are inherently scale-dependent and the resolution at which communities are sampled may have a major impact on the conclusions that can be drawn from data (Huston 1999, Willis and Whittaker 2002, Hanan and Ross 2010, Qian and Kissling 2010, Rocchini et al. 2010). Theoretically, we anticipate more niche dissimilarity among species (overdispersion) at finer resolutions where biotic interactions take place because of the effect of interspecific competition/facilitation or shared natural enemies. On the other hand, we cannot conclusively predict direct biotic interaction between co-occurring species at a coarser resolution, as species can segregate along environmental gradients encompassed within large sampling units. Therefore we rather anticipate greater similarity (clustering/underdispersion) among species because of shared resource requirements in this case. In fact, community assembly studies have shown that both phylogenetic clustering and overdispersion of native species within communities can appear within the same system, but at different spatial resolutions (Cavender-Bares et al. 2006, Swenson et al. 2006). In a similar way, spatial resolution may be the key to reconciling apparently contrasting hypotheses and empirical results in the field of invasion ecology (Stohlgren et al. 1997, 2002, Catford and Downes 2010, Jones et al. 2010).
Although the issue of spatial scale is clearly important in this context and has been addressed theoretically (Proches et al. 2008, Thuiller et al. 2010), there are few studies that have investigated the effect of scale on invaders’ relatedness patterns using nested resolutions in the field (Cadotte et al. 2009, Davies et al. 2011, Schaefer et al. 2011). While promising evidence comes from an analysis conducted by Diez et al. (2008), which however did not include progressively finer sampling resolutions, to our knowledge no empirical study has been able to demonstrate the theoretically predicted spatial turning point from phylogenetic clustering to phylogenetic overdispersion of invaders. For example, Davies et al. (2011) found that native and non-native species were more distantly related than expected by chance not only at a fine resolution (plot scale), but also at a coarse one (hectare scale). Conversely, Cadotte et al. (2009) demonstrated phylogenetic clustering of invader success at the continental scale, but only found a random pattern at the smallest scale of analysis they considered (landscape). No study has yet included a comprehensive enough set of sampling resolutions to empirically reconcile the expected divergent patterns across sampling resolutions. Consequently, there has been a call for cross-scale field-based approaches to tackle the issue of spatial scale in the context of phylogenetic patterns of biological invasions (Proches et al. 2008, Thuiller et al. 2010).
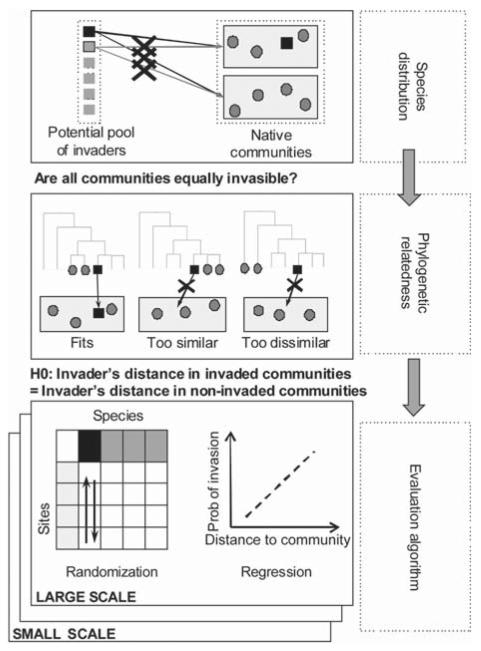
In this paper we explore the effect of sampling resolution on patterns of plant invasions with field data using a comprehensive set of metrics and statistical tests (Fig. 1). Focusing on Mediterranean coastal sand dunes, we test patterns of naturalized non-native species (‘invaders’ hereafter) in local communities at three spatial resolutions. We built a phylogenetic supertree to derive two complementary community scale measurements of the phylogenetic distance of invaders: the Mean Distance of the invader relative to Native Species (MDNS) and the Distance of the invader to its Nearest Native Species in the native community (DNNS). Finally, we assess the results by testing the hypothesis that relatedness of invaders is different in invaded communities from what it would be in non-invaded communities. To do so we rely both on randomization tests using an algorithm simulating ‘random invasions’ and on phylogenetic mixed effects models in a Bayesian framework. Specifically, we address the following crucial questions: 1) how phylogenetically distant should species be to successfully invade a native community? 2) Are the observed patterns different from random expectations? 3) Do the observed patterns change with increasing sampling resolution?
Figure 1.
Conceptual diagram depicting the hypotheses relating naturalizations/invasions and phylogenetic relatedness/distance to the community and the testing procedure adopted in this paper. Non-native species are represented as squares and successful invaders in at least one community in the study area are in black.
Methods
The system under study is located in Mediterranean coastal sand dunes known to be prone to invasions (Chytrý et al. 2009). The vegetation of sandy shores in central Italy has been extensively sampled in the past few years at nested sampling resolutions and (contrary to what is often the case with phytosociological surveys) with no bias towards native species (Acosta et al. 2008, 2009). The conditions were therefore ideal for testing phylogenetic structure in relation to invasion patterns at different sampling resolutions with a relevant number of invaders to generalize the findings.
Study area and sampling resolutions
We specifically focused on recent coastal dunes (Holocene) extending over 400 km on the central Italian coasts: 250 km along the Tyrrhenian Sea (Lazio Region, from 42°23′N, 11°39′E to 41°11′N, 13°20′E) and 150 km along the Adriatic Sea (Molise and Abruzzi Regions). Holocene dunes represent ca 80% of the overall extent examined. Here we examined three different sampling resolutions: one very coarse and two progressively finer resolutions. For the coarse resolution, we relied on a survey of the vascular flora of central Italian coastal dunes carried out from 2004 to 2007 in 3′ by 5′ grid-cells (about 35 km2) which was limited to the geologic class of Holocenic dunes (Acosta et al. 2008, Carboni et al. 2010). Within each grid cell all vascular plant species (natives and introduced) were recorded wherever they occurred on recent dunes. 91 grid cells fall within the limits of the studied regions, however only 71 contained holocenic dunes. For the finer resolutions we used presence–absence data from a long-term (2002–2009) random sampling campaign of coastal dune vegetation in several study sites, comprising most of the best conserved remnant dune systems of the region (about 80 km on the western coast and 22 km on the eastern coast) (for more details see Acosta et al. 2009, Carboni et al. 2011). In this work we specifically consider two nested plot dimensions: 2 × 2 m (4 m2) as the finest resolution and an expansion of the plots to 8 × 8 m (64 m2) as the intermediate resolution of analysis. We considered 690 plots for each of these two dimensions. In these environments the finest plot size is compatible with the identification of homogenous plant communities, some habitat heterogeneity already occurs within the intermediate sized plots, and many different habitats occur within the grid cells at the coarsest resolution (sorted along a strong sea-inland environmental gradient; Carboni et al. 2011). Our sampling design is therefore arranged as to include decreasing environmental heterogeneity within sampling units from the coarse to the intermediate resolution, while we assume that environmental conditions are relatively homogenous within the fine resolution plots.
We classified only species introduced after the 15th century as non-natives in this study, but we included both invasive and naturalized species (Pyšek et al. 2004) according to the classification by Celesti-Grapow et al. (2009). From this list, we additionally excluded all species that were clearly not naturalized on coastal dunes based on expert knowledge. See Supplementary material Appendix 1 for a list of all the non-native species (invasive and naturalized) sampled at each resolution. We refer to these as ‘invaders’ in a general sense, although we focus on all naturalized species, not only on those with high spread potential or with documented negative effects. Irrespective of the term used, we only make inferences on the naturalization process, not on the level of spread and further impact of introduced species.
Supertree construction
We created a supertree of all the taxa in the communities sampled by combining a backbone tree based on the APG III phylogeny (<www.mobot.org/MOBOT/research/APweb/>), which was generated by Phylomatic (<www.phylodiversity.net/phylomatic/phylomatic.html>), along with subtrees that were created using other literature sources to include e.g. gymnosperms and ferns (Chaw et al. 1997, Frohlich and Chase 2007). We assigned branch lengths to the phylogenetic tree using the branch length adjustment algorithm (BLADJ) in Phylocom (Webb et al. 2008), based on the minimum age of nodes estimated from the fossil record (Wikstrom et al. 2001). To produce phylogenetic distance matrices and calculate distance-based metrics, we used the sum of branch lengths separating pairs of species. In the absence of more precise species phylogenies obtained by sequencing proper DNA regions these matrices provide a useful measurement expressing the phylogenetic relatedness for community analyses and have proved to be effective in other studies (Pillar and Duarte 2010, Ricotta et al. 2010, Davies et al. 2011). As our working phylogeny still contained many polytomies, we ran sensitivity analyses to test whether our limited tree resolution was substantially distorting our results (see Results section and Supplementary material Appendix 3).
We also checked for a phylogenetic signal (sensu Abouheif 1999, Blomberg and Garland 2002) in a set of functional traits known to relate to species strategies or resource use (Westoby 1998, Supplementary material Appendix 4). Tests of phylogenetic signal showed that most traits examined were more similar for closely-related species than under random expectations, corroborating our assumption that closely-related species shared more similar ecological characteristics than two species taken at random in the phylogeny. All details on traits, methods and results for these preliminary tests can be found in Supplementary material Appendix 4.
Spatial structure of invaders
In order to better calibrate the subsequent randomization algorithms and regression analyses we performed a series of preliminary tests to check whether invaders were spatially clustered as a subgroup. To verify how invaders were spatially arranged at the three scales we measured co-occurrence patterns (species.dist function in the R package picante, R Development Core Team, Kembel et al. 2010). We calculated pairwise values of co-occurrence using Schoener’s index (Cij), which is based on proportional similarity (Schoener 1970): Cij = 1 – 0.5 × Σ∣pih – pjh∣, where Cij is the co-occurrence of species i and j and p is the proportion of occurrences of the ith species in the hth plot. With presence/absence data, pjh is zero if the species is absent from site h, otherwise it is the inverse of the number of sites where species i occurs. Mean observed Cij at the three scales was compared with randomized indices. Co-occurrence patterns were assessed for the two separate subgroups of invaders and natives by using a null-model which maintains the overall frequency of each species in the study region, i.e. shuffling sites within each species (Gotelli 2000). This allowed us to see if there is clustering of invaders as a subgroup (Table 1, column 1) and then compare with patterns of natives (Table 1, column 2). Finally, to verify if there are differences between the two, clustering of invaders is assessed with respect to natives by comparing the observed ratio of ‘Cij values for invaders/Cij values for all species’ with randomized values obtained by randomly selecting invaders from among the species in the species pool (Table 1, column 3).
Table 1.
Spatial co-occurrence patterns of invaders and natives. Cij co-occurrence Schoener’s index at the three resolutions (by rows) compared with randomized indices to address (by columns): co-occurrence patterns assessed separately for invaders (1) and for natives (2), and (3) for invaders in comparison to natives.
| Hypothesis: Data: Randomization: | (1) Some invaders tend to co-occur invaders (shuffle w/in sp) |
(2) Some natives tend to co-occur native species (shuffle w/in sp) |
(3) Invaders co-occur more often than natives all species (shuffle invaders w/random sp) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| obs | mean rand | p | obs | mean rand | p | obs | mean rand | p | |
| 4 m2 | 0.016 | 0.014 | 0.279 | 0.022 | 0.014 | <0.001 | 0.716 | 1.019 | 0.402 |
| 64 m2 | 0.014 | 0.007 | <0.001 | 0.018 | 0.009 | <0.001 | 0.784 | 1.016 | 0.692 |
| 35 km2 | 0.09 | 0.040 | <0.001 | 0.09 | 0.043 | <0.001 | 1.043 | 0.998 | 0.349 |
Testing Darwin’s naturalization hypothesis
We considered two complementary distance-based metrics to quantify invaders’ relatedness to the community: the Mean Distance of the invader relative to Native Species (MDNS) and the Distance of the invader to its Nearest Native Species in the native community (DNNS) (Thuiller et al. 2010). It has been shown that phylogenetic distance-based metrics can be confounded with species richness (Pavoine and Bonsall 2011) and that there can be associations between species richness and the presence of invaders (Stohlgren et al. 2002, Stachowicz and Tilman 2005). In order to partial out the effect of species richness and focus only on the effect of phylogenetic relatedness, we used residuals of MDNS/DNNS regressed against total plot richness rather than the observed MDNS/DNNS values in the following analyses (Davies and Buckley 2011). These residuals are measures of phylogenetic distance independent of species richness (for simplicity just MDNSresid and DNNSresid hereafter). Examples of regression plots of MDNS and DNNS vs species richness are reported in Supplementary material Appendix 2.
To test whether patterns of invasion measured with MDNSresid and DNNSresid were different from random expectations, we adopted two complementary approaches.
First, we defined an ad-hoc randomization scheme (Fig. 1), testing whether there are differences between invaded and non-invaded communities. We simulated ‘random invasions’ by manipulating the invaders in the species by site matrix and permuting, independently for each invader, local presences/absences among sites. At each examined resolution we thus generated null distributions of MDNSresid and DNNSresid averaged across sites for each invader recorded, to which the observed values could be compared. We concluded that the test was significant if the actual values were greater than 97.5% (overdispersion) of the generated values or lower than 97.5% of the values (underdispersion), i.e. if the overall two-tailed p-value was <0.05. In other words, for each scale by metric combination (3 × 2 = 6 combinations) there are two one-tailed tests at α = 0.025 distinguishing phylogenetic clustering and overdispersion of each invader. To assess overall significance of patterns for each spatial scale but across species, we performed a Fisher’s test that combined the p-values for each hypothesis (function ‘combine.test’ in package ‘survcomp’; Haibe-Kains et al. 2008).
Second, we implemented mixed effects models to assess whether the probability of community invasion was related to the phylogenetic distance between invasive species and native communities. We independently modeled the effect of MDNSresid and DNNSresid on the binary response variable ‘invaded/non-invaded’ assuming a binomial distribution of the response. To account for the fact that more than one alien species could invade one plot and to allow for different intercepts for each invader, we included two random factors in each model: plot identity and invader identity. Additionally we accounted for non-independence among individual invaders with a matrix of phylogenetic relatedness among species. We took a Bayesian approach using the R package MCMCglmm, which enables both random factors and a correlation structure depending on species phylogenetic relationships to be included (Hadfield 2010). Each model was run for 250 000 Markov chain Monte Carlo (MCMC) steps, with a burn-in of 50 000 iterations. Uninformative prior distributions were used for parameters, with mean of 0 and residual variance–covariance matrices set to 1. We checked for convergence in the parameter estimation by inspecting trace plots of the MCMC iterations. We chose a thinning interval of 200 iterations, which resulted in posteriori distributions with 1000 samples. From these posteriori distributions we calculated mean parameter estimates, and 95% Highest Posterior Density (HPD) or Credible Intervals (CI). Significance of model parameters was estimated by examining CIs: parameters with CIs overlapping with zero were considered not to be significant.
Results
Phylogeny
We obtained a phylogenetic supertree for our study system comprising a total of 798 species, of which 51 species were invaders (Supplementary material Appendix 1). A supertree constructed with Phylomatic is typically not fully resolved, with many species as polytomies within genera and some genera as polytomies within families. To test the influence of the polytomies on MDNSresid and DNNSresid, we performed a sensitivity analysis on the basis of randomly resolved trees (using the ‘polytomy resolver’ phylogenetic tool, Supplementary material Appendix 3). We found that the metrics from the unresolved tree were unbiased estimates and that the uncertainty was consistently moderate (Supplementary material Appendix 3). Hence, in a pragmatic way we decided to perform all analyses with the unresolved trees to avoid working with hundreds of randomly resolved trees. Visual inspection of the tree showed that invaders tended to be grouped in several independent clusters with likely different evolutionary histories and ecological niches.
Spatial structure of invader distribution
In order to fine-tune the randomization approach we analyzed species’ spatial co-occurrence patterns. When a community is invaded by more than one species it is not obvious how all invaders of the community should be treated in the randomization tests. It is indeed unknown whether a native species has been excluded by an invader, in which case this invader would reflect characteristics of the lost native. The spatial differences in co-occurrence patterns of invaders with respect to natives can be informative in this sense. We found that invaders as a subgroup did not tend to co-occur at fine resolutions, though they appeared to be somewhat spatially clustered at coarser resolutions (Table 1, column 1). However, they were never more spatially clustered than the native species in the species pool (compare columns 1 and 2 in Table 1; column 3). Given these results on spatial occurrences, we adopted the more conservative approach whereby other invaders occurring in the community were not excluded in permutations for generating ‘random invasions’.
Testing Darwin’s naturalization hypothesis
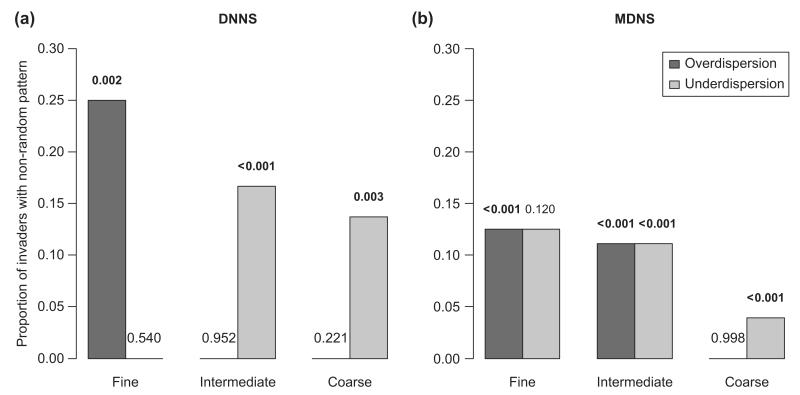
When considering only the most related native species (DNNSresid – Fig. 2a), results from randomization tests corroborated Darwin’s naturalization hypothesis (phylogenetic overdispersion) at the finest sampling resolution (2 × 2 m). For this resolution, a high proportion of invaders (ca 25%; Fisher’s p-value = 0.002) were more distantly related to the closest relative in the invaded local community than under random expectations. However, none of the invaders showed a DNNSresid greater than expected by chance at the intermediate and coarse sampling resolutions. The trend was even inverted at the coarsest resolutions, with ca 10–15% of the invaders having smaller DNNSresid values than expected by chance (Fisher’s p-value < 0.001 at intermediate and 0.003 at coarse resolution). In other words, at coarser resolutions, the invaders tended to preferentially invade communities where at least one close relative already occurred (i.e. a species likely to share similar niche requirements).
Figure 2.
Proportion of invaders for which phylogenetic distance of the invader to the native community (after partialling out the effect of species richness) deviated from random patterns generated through randomizations. Significantly greater distances than expected by chance indicate overdispersion, whereas smaller distances indicate underdispersion. P-values on top of the bars are obtained by combining the p-values of randomization tests for each single invader through a Fisher’s test. Bold type indicates overall significant deviations from random expectations (i.e. across invaders). In each panel sampling resolutions from left to right are 4 m2, 64 m2 and ca 35 km2.
Patterns were similar, but less clear when considering all the species in the community (MDNSresid). Fisher’s test showed that invaders were significantly more distant from natives than expected by chance at the finest resolution (p<0.001) and closer than expected at the coarsest resolution (p<0.001). However at all scales examined, a relatively small proportion of invaders had MDNSresid values that differed from random expectations, with approximately equal proportions of overdispersion and underdispersion at the fine and intermediate scales (Fig. 2b).
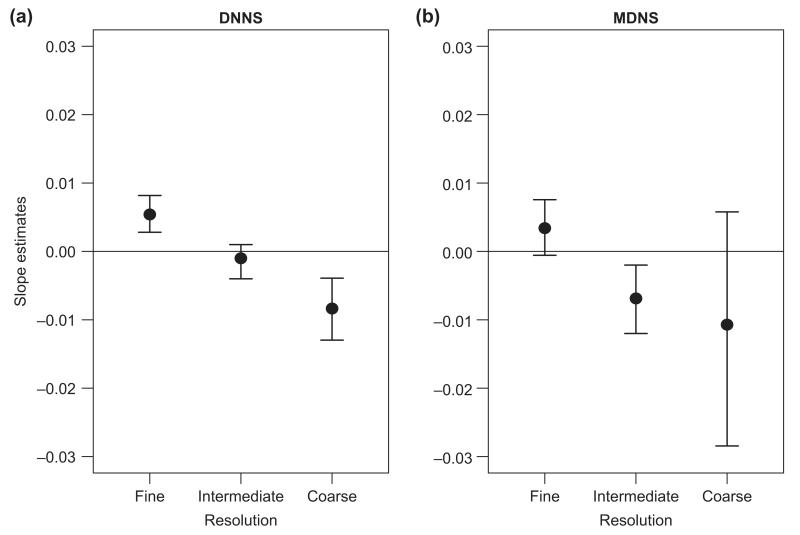
Mixed effect models supported the results obtained by comparing observed patterns with simulated ‘random invasions’ (Fig. 3a, b). Specifically, at the finest resolution we found a significant positive relation of the probability of invasion with the phylogenetic distance of the invader to the closest relative (DNNSresid). This relationship was inversed at the coarsest resolution, where the slope of the relationship was negative. The models with MDNSresid as the explanatory variable showed the same trends at both fine and coarse resolutions, although the slope was not significantly different from zero in either case.
Figure 3.
Results of MCMC mixed effects models for the probability of community invasion as a function of the phylogenetic distance of the invader to the community (after partialling out the effect of richness). Plots depict the mean posterior distributions for the slope parameter (with 95% credible intervals plotted as bars) for the effect of DNNSresid and MDNSresid, at fine, intermediate and coarse spatial resolutions (i.e. 4 m2, 64 m2 and ca 35 km2).
Discussion
Recognition that community invasibility depends on the match between the characteristics of the invader and those of members of the recipient native community (Richardson and Pysek 2006) has been a major shift in the field of invasion ecology. This has generated a growing interest in assessing the role that functional similarity and phylogenetic relationships play in biological invasions (Daehler 2001, Duncan and Williams 2002, Lambdon and Hulme 2006, Strauss et al. 2006, Winter et al. 2009). However, most studies have provided only partial or even diverging conclusions. Our study empirically investigated one of the main conceptual reasons put forward to explain conflicting results (Proches et al. 2008, Thuiller et al. 2010). Here we demonstrated using empirical data the crucial importance of spatial resolution for detecting phylogenetic patterns of invasion and we explored the implications of the choice of metrics and statistical tests.
When comparing DNNSresid of potential invaders in invaded and non-invaded communities we found support for Darwin’s naturalization hypothesis (phylogenetic overdispersion) at the fine sampling resolution at which competitive interactions take place on Mediterranean coastal dunes. In other words, invaders were more likely to be present in plots when they were more phylogenetically distant from their native relatives occurring in that plot (e.g. in the case of C. acinaciformis, one of the most invasive species in these environments. See Supplementary material Appendix 5 for a discussion on patterns of single invaders and their likely interactions with native species). This finding was reported at relatively small spatial scales in other contexts (Ricciardi and Atkinson 2004, Jiang et al. 2010). As we found that several traits tended to show a phylogenetic signal (Supplementary material Appendix 4), it is likely that relatedness of the invader indeed reflected high functional similarity and thus niche overlap with the native species. Consequently, our results are in line with Darwin’s and Elton’s theoretical expectations on the biotic resistance of the native community to invasion. At coarser resolutions we instead found an opposite pattern suggesting a more dominant effect of habitat filtering (phylogenetic clustering). At the coarsest resolution (ca 35 km2) a high proportion of invaders was more related to the invaded communities than expected by chance. This trend mirrors patterns previously observed at regional and continental scales, for example in the floras of New Zealand and Australia (Duncan and Williams 2002, Diez et al. 2009). In fact, when considering a coarser resolution, species can co-occur while avoiding direct biotic interactions. The main reason is presumably that greater environmental variation is encompassed within larger sites, providing opportunities for species to sort across environmental gradients (Willis et al. 2010). It is therefore possible to reconcile apparently contrasting hypotheses and results for the patterns of relatedness of invading plants through the explicit consideration of the scale or resolution at which communities are sampled and defined. This is true in our system even though we restricted our analyses to coastal dunes, so that our coarse scale species pool is already quite filtered compared to earlier studies (Duncan and Williams 2002, Cadotte et al. 2009, Diez et al. 2009). However, given the strong sea-inland environmental gradient and strong zonation of the vegetation the species pool is still broad enough to detect the underdispersion in a high proportion of invaders.
Determining the spatial resolution at which the effects of biotic resistance are outbalanced by environmental filtering so that closely-related introduced species are no longer excluded has proven a difficult task. This is because competition is most plausible only at fairly fine spatial resolutions and among fairly related species (the ‘Darwin– Hutchinson zone’ according to Vamosi et al. 2009). The few studies explicitly searching for a turning point may have failed because they may have considered a range of scales inappropriate for the specific study system/taxon or because they may have missed the Darwin–Hutchinson zone (Vamosi et al. 2009). In contrast, our study picked up within a single area (central Italy) and ecosystem (coastal dunes) both phylogenetic overdispersion and phylogenetic clustering of invaders, effectively establishing the spatial turning point from one pattern to the other. Interestingly, in recent work on serpentine ecosystems, Davies et al. (2011) found that native and non-native species were more distantly related than expected by chance at a fine sampling resolution roughly comparable to the one used in this study. However, contrary to their expectations, they also found overdispersion at the rather coarse resolution of one hectare blocks. In contrast, we were able to detect the shift to phylogenetic clustering, presumably on two main grounds. First, our chosen system is characterized by marked habitat heterogeneity within relatively small extensions, as the strong sea–inland environmental gradient determines a compressed vegetation zonation across the dune profile. Second, we included a very broad range of sampling resolutions so that the grid cells used for our coarse scale analysis are much larger (35 km2) than the ones used by Davies et al. (2011). Intriguingly, Cadotte et al. (2009) were able to show phylogenetic clustering of invader success as a result of habitat filtering only at an extremely large continental scale. Our results, forming a bridge between these two studies, seem to imply that the spatial turning point at which invaders become more similar to native species must be searched for at a resolution coarse enough to encompass a variety of different habitats (e.g. from annual beach communities to backdune Mediterranean macchia in our case). The exact plot size depends on the study system and on the amount of heterogeneity characteristic for the specific ecosystem examined, as well as on the taxa under consideration. Our study focusing on a single type of environment (coastal dunes) allows highlighting phylogenetic clustering at relatively finer scales than previous state wide or continental assessments (Strauss et al. 2006, Cadotte et al. 2009).
Quite surprisingly, in a recent study addressing ‘Darwin’s naturalization hypothesis’ at different scales, Schaefer et al. (2011) found that introduced plant species were more likely to become invasive in the absence of close relatives in the overall native flora of the Azores, but could not confirm this trend with a fine sampling resolution. The authors argue that on these islands the exclusion of similar invaders seemed to be mostly driven by the clustering of common enemies, such as herbivores and pathogens rather than by competition. They argue that enemy release should act at all spatial scales in a system like the Azores and thus produce a signal of over-dispersion not only at small but also at large scales, explaining why their outcome partially contradicts theoretical expectations. A similar result is reported by van Wilgen and Richardson (2011) for reptiles in North America.
By using a combination of two metrics of community relatedness we were able to investigate whether biotic resistance to invaders was best predicted by a single closely-related species or by the community structure overall. Using the DNNSresid metric we could highlight clear patterns in accordance with theoretical expectations at all scales, whereas the MDNSresid measure of community relatedness gave less clear or non-significant results. In fact, within small homogenous plots we found a strong proportion of invaders, which tended to avoid the single most closely-related species (DNNSresid metric). It has previously been hypothesized that the biotic resistance of a given community would be mainly driven by the closest native species because one strong competitor is sufficient for competitive exclusion (Kraft et al. 2007). Furthermore, we found stronger support for the influence of habitat filtering at coarse resolutions when focusing specifically on the most related taxon (DNNSresid). This is in contrast with the common belief that total community relatedness tests should perform better with habitat filtering (Kraft et al. 2007). In large grained grid cells the non-limiting availability of resources and the included habitat heterogeneity may support a number of different environmental conditions and thus a high native diversity of phylogenetic lineages. The niche of the invader may need to be close to one of these suitable environmental conditions, but the mean distance to all species may be uninformative and obscure any pattern. Focusing only on closely-related taxa may instead provide the necessary precision to reveal the availability of favorable environmental conditions for the invader within the heterogeneous site. In addition, the use of a not fully resolved phylogenetic tree (affecting ‘phylogenetic scale’) may also more strongly limit the analytical power of MDNS, given that randomly resolving the tree resulted in greater variation in MDNS than in DNNS (Thuiller et al. 2010; Supplementary material Appendix 3). The identified patterns were consistent among statistical tests (randomizations and regressions) and generally indicated that the closest relative was most informative for both the habitat preference of the invader and for the biotic resistance of the community. In contrast MDNSresid was a less effective proxy with few invaders that had significant patterns in randomization tests and large widely overlapping credible intervals for slope estimates in regressions. However, considering that these results may strongly depend on system-specific habitat heterogeneity or on the phylogenetic resolution available and that a collection of related species may have more impact than a single closely-related one (Strauss et al. 2006), it is generally advisable to check both metrics when analyzing invasion patterns.
As we have seen, non-random phylogenetic patterns of plant invasions may be detected either by a comparison with random expectations generated through various algorithms or through regression models. Using a combination of approaches enabled an assessment to be made as to whether our results were not dependent on methodological assumptions. Randomization techniques have been widely used in community ecology (Gotelli 2000) and phylogenetics (Cavender-Bares et al. 2006) as they are easy to implement and make relatively few a priori assumptions about expected relationships. Nevertheless, particular caution must be applied when formulating randomization schemes in order to avoid permutations that alter more patterns than the ones that are being specifically tested (Hardy 2008). We specifically chose a randomization procedure that breaks down the phylogenetic relationship between introduced and native species, but not the phylogenetic relationships between native species of recipient communities. However, restrictions in the randomization procedure reduce the statistical power. Alternatively, the regression approach has also often been adopted when studying phylogenetic patterns of invasions (Duncan and Williams 2002, Lambdon and Hulme 2006). While caution when formulating precise hypotheses and choosing adequate species pools should also be applied to regression models, these methods generally have higher testing power in comparison with randomizations. Moreover, in this paper we fit regression models including a random component that accommodates different intercepts for each invader. We can therefore evaluate patterns of more than one species in a single analysis, rather than only inferring trends from the proportion of invaders with significantly non-random patterns. Besides, we can take species phylogenetic relationships into account in the modeling process, thus compensating for statistical non-independence among invaders due to shared ancestry. In summary, although both approaches we employed, i.e. randomization and regression based, have shortcomings and specific assumptions, the convergence in the obtained results suggests that the patterns we highlight in this study are not dependent on methodological choices.
In conclusion, we found that the relationship between phylogenetic distance and probability of occurrence of an invader changes with spatial resolution and that we can confirm Darwin’s naturalization hypothesis for fine resolutions where biotic interactions are also expected to be most important. Our results appear robust as both statistical tests applied supported the same conclusions. This paper therefore offers a new methodological framework for using the composition of local native species assemblages as a predictive tool for the new establishment of invaders. The specific resolution at which a community is no longer driven by biotic interactions, but rather by habitat filtering depends on habitat heterogeneity, and should therefore vary depending on the system being studied. In general, our results are promising for the perspective of incorporating information on the phylogenetic identity of resident native species into fine-grained predictive models for species invasions.
Supplementary Material
Acknowledgements
The research leading to these results received funding from the European Research Council under the European Community’s Seven Framework Programme FP7/2007-2013 Grant Agreement no. 281422 (TEEMBIO). TM acknowledges support from the EraNet BiodivERsA project ANR-11-EBID-002 CONNECT. WT and SL acknowledge support from the funded by the French ‘Agence Nationale de la Recherche’ with the project SCION (ANR-08-PEXT-03). We thank Cristina Roquet for her help with the sensitivity test on polytomies. We are grateful to Jens-Christian Svenning for his insightful comments.
Footnotes
Supplementary material (Appendix E7479 at <www.oikosoffi ce.lu.se/appendix>). Appendix 1 – 5.
References
- Abouheif E. A method for testing the assumption of phylogenetic independence in comparative data. Evol. Ecol. Res. 1999;1:895–909. [Google Scholar]
- Acosta A, et al. Patterns of native and alien plant species occurrence on coastal dunes in central Italy. In: Tokarska-Guzik B, et al., editors. Plant invasions: human perception, ecological impacts and management. Backhuys Publishers; 2008. pp. 235–248. [Google Scholar]
- Acosta A, et al. Are there habitats that contribute best to plant species diversity in coastal dunes? Biodivers. Conserv. 2009;18:1087–1098. [Google Scholar]
- Blomberg SP, Garland T. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 2002;15:899–910. [Google Scholar]
- Cadotte MW, et al. Phylogenetic relatedness and plant invader success across two spatial scales. Divers. Distrib. 2009;15:481–488. [Google Scholar]
- Carboni M, et al. Disentangling the relative effects of environmental versus human factors on the abundance of native and alien plant species in Mediterranean sandy shores. Divers. Distrib. 2010;16:537–546. [Google Scholar]
- Carboni M, et al. Dealing with scarce data to understand how environmental gradients and propagule pressure shape fine-scale alien distribution patterns on coastal dunes. J. Veg. Sci. 2011;22:751–765. [Google Scholar]
- Catford JA, Downes BJ. Using multi-scale species distribution data to infer drivers of biological invasion in riparian wetlands. Divers. Distrib. 2010;16:20–32. [Google Scholar]
- Cavender-Bares J, et al. Phylogenetic structure of floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:S109–S122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Celesti-Grapow L, et al. Inventory of the non-native flora of Italy. Plant Biosystems. 2009;143:386–430. [Google Scholar]
- Chaw SM, et al. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Mol. Biol. Evol. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- Chytrý M, et al. European map of alien plant invasions based on the quantitative assessment across habitats. Divers. Distrib. 2009;15:98–107. [Google Scholar]
- Daehler CC. Darwin’s naturalization hypothesis revisited. Am. Nat. 2001;158:324–330. doi: 10.1086/321316. [DOI] [PubMed] [Google Scholar]
- Darwin C. The origin of species. John Murray. 1859 [Google Scholar]
- Davies KF, et al. Native communities determine the identity of exotic invaders even at scales at which communities are unsaturated. Divers. Distrib. 2011;17:35–42. [Google Scholar]
- Davies TJ, Buckley LB. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Phil. Trans. R. Soc. B. 2011;366:2414–2425. doi: 10.1098/rstb.2011.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez JM, et al. Darwin’s naturalization conundrum: dissecting taxonomic patterns of species invasions. Ecol. Lett. 2008;11:674–681. doi: 10.1111/j.1461-0248.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- Diez JM, et al. Learning from failures: testing broad taxonomic hypotheses about plant naturalization. Ecol. Lett. 2009;12:1174–1183. doi: 10.1111/j.1461-0248.2009.01376.x. [DOI] [PubMed] [Google Scholar]
- Duncan RP, Williams PA. Ecology – Darwin’s naturalization hypothesis challenged. Nature. 2002;417:608–609. doi: 10.1038/417608a. [DOI] [PubMed] [Google Scholar]
- Elton CS. The ecology of invasions by plants and animals. Methuen. 1958 [Google Scholar]
- Frohlich MW, Chase MW. After a dozen years of progress the origin of angiosperms is still a great mystery. Nature. 2007;450:1184–1189. doi: 10.1038/nature06393. [DOI] [PubMed] [Google Scholar]
- Gotelli NJ. Null model analysis of species co-occurrence patterns. Ecology. 2000;81:2606–2621. [Google Scholar]
- Hadfield JD. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 2010;33:1–22. [Google Scholar]
- Haibe-Kains B, et al. A comparative study of survival models for breast cancer prognostication on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanan EJ, Ross MS. Across-scale patterning of plant–soil–water interactions surrounding tree islands in southern Everglades landscapes. Landscape Ecol. 2010;25:463–476. [Google Scholar]
- Hardy OJ. Testing the spatial phylogenetic structure of local communities: statistical performances of different null models and test statistics on a locally neutral community. J. Ecol. 2008;96:914–926. [Google Scholar]
- Huston MA. Local processes and regional patterns: appropriate scales for understanding variation in the diversity of plants and animals. Oikos. 1999;86:393–401. [Google Scholar]
- Jiang L, et al. An experimental test of Darwin’s naturalization hypothesis. Am. Nat. 2010;175:415–423. doi: 10.1086/650720. [DOI] [PubMed] [Google Scholar]
- Jones CC, et al. Combining local- and large-scale models to predict the distributions of invasive plant species. Ecol. Appl. 2010;20:311–326. doi: 10.1890/08-2261.1. [DOI] [PubMed] [Google Scholar]
- Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Kraft NJB, et al. Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 2007;170:271–283. doi: 10.1086/519400. [DOI] [PubMed] [Google Scholar]
- Lambdon PW, Hulme PE. How strongly do interactions with closely-related native species influence plant invasions? Darwin’s naturalization hypothesis assessed on Mediterranean islands. J. Biogeogr. 2006;33:1116–1125. [Google Scholar]
- Marco A, et al. From the backyard to the backcountry: how ecological and biological traits explain the escape of garden plants into Mediterranean old fields. Biol. Invasions. 2010;12:761–779. [Google Scholar]
- Pavoine S, Bonsall MB. Measuring biodiversity to explain community assembly: a unified approach. Biol. Rev. 2011;86:792–812. doi: 10.1111/j.1469-185X.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- Pillar VD, Duarte LDS. A framework for meta-community analysis of phylogenetic structure. Ecol. Lett. 2010;13:587–596. doi: 10.1111/j.1461-0248.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- Proches S, et al. Searching for phylogenetic pattern in biological invasions. Global Ecol. Biogeogr. 2008;17:5–10. [Google Scholar]
- Pyšek P, et al. Alien plants in checklists and floras: towards better communication between taxonomists and ecologists. Taxon. 2004;53:131–143. [Google Scholar]
- Qian H, Kissling WD. Spatial scale and cross-taxon congruence of terrestrial vertebrate and vascular plant species richness in China. Ecology. 2010;91:1172–1183. doi: 10.1890/09-0620.1. [DOI] [PubMed] [Google Scholar]
- Ricciardi A, Atkinson SK. Distinctiveness magnifies the impact of biological invaders in aquatic ecosystems. Ecol. Lett. 2004;7:781–784. [Google Scholar]
- Richardson DM, Pysek P. Plant invasions: merging the concepts of species invasiveness and community invasibility. Progr. Phys. Geogr. 2006;30:409–431. [Google Scholar]
- Ricotta C, et al. Invasiveness of alien plants in Brussels is related to their phylogenetic similarity to native species. Divers. Distrib. 2010;16:655–662. [Google Scholar]
- Rocchini D, et al. Spectral variation versus species beta-diversity at different spatial scales: a test in African highland savannas. J. Environ. Monit. 2010;12:825–831. doi: 10.1039/b921835a. [DOI] [PubMed] [Google Scholar]
- Schaefer H, et al. Testing Darwin’s naturalization hypothesis in the Azores. Ecol. Lett. 2011;14:389–396. doi: 10.1111/j.1461-0248.2011.01600.x. [DOI] [PubMed] [Google Scholar]
- Schoener TW. Nonsynchronous spatial overlap of lizards in patchy habitats. Ecology. 1970;51:408–418. [Google Scholar]
- Stachowicz JJ, Tilman D. Species invasions and the relationships between species diversity, community saturation, and ecosystem functioning. In: Sax DF, et al., editors. Species invasions: insights into ecology, evolution, and biogeography. Sinauer; 2005. pp. 41–64. [Google Scholar]
- Stohlgren TJ, et al. Multiscale sampling of plant diversity: effects of minimum mapping unit size. Ecol. Appl. 1997;7:1064–1074. [Google Scholar]
- Stohlgren TJ, et al. Assessing vulnerability to invasion by nonnative plant species at multiple spatial scales. Environ. Manage. 2002;29:566–577. doi: 10.1007/s00267-001-0006-2. [DOI] [PubMed] [Google Scholar]
- Strauss SY, et al. Exotic taxa less related to native species are more invasive. Proc. Natl Acad. Sci. USA. 2006;103:5841–5845. doi: 10.1073/pnas.0508073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson NG, et al. The problem and promise of scale dependency in community phylogenetics. Ecology. 2006;87:2418–2424. doi: 10.1890/0012-9658(2006)87[2418:tpapos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Thuiller W, et al. Resolving Darwin’s naturalization conundrum: a quest for evidence. Divers. Distrib. 2010;16:461–475. [Google Scholar]
- Vamosi SM, et al. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 2009;18:572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- van Wilgen NJ, Richardson DM. Is phylogenetic relatedness to native species important for the establishment of reptiles introduced to California and Florida? Divers. Distrib. 2011;17:172–181. [Google Scholar]
- Webb CO, et al. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- Westoby M. A leaf–height–seed (LHS) plant ecology strategy scheme. Plant Soil. 1998;199:213–227. [Google Scholar]
- Wikstrom N, et al. Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis CG, et al. Phylogenetic community structure in Minnesota oak savanna is influenced by spatial extent and environmental variation. Ecography. 2010;33:565–577. [Google Scholar]
- Willis KJ, Whittaker RJ. Ecology – species diversity – scale matters. Science. 2002;295:1245–1248. doi: 10.1126/science.1067335. [DOI] [PubMed] [Google Scholar]
- Winter M, et al. Plant extinctions and introductions lead to phylogenetic and taxonomic homogenization of the European flora. Proc. Natl Acad. Sci. USA. 2009;106:21721–21725. doi: 10.1073/pnas.0907088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.