Abstract
Hepatocellular carcinoma (HCC) is the fifth most common form of cancer globally and is rarely curable once detected. The 5 year survival rate of patients diagnosed with late stage HCC may be as low as 27 %. HCC is a cancer largely driven by epigenetic changes which arise from exposure to exogenous environmental factors, rather than coding sequence mutations. The liver is susceptible to effects from; Hepatitis C and Hepatitis B viruses, exposure to aflatoxin and continuous excessive consumption of alcohol. The liver is a highly metabolic organ balancing many vital biochemical processes; exposure to any of the above environmental factors is associated with loss of liver function and is a major risk factor for the development of HCC. Emerging studies aim to examine the underlying metabolic processes which are abrogated in cancer and lead to the altered flux and availability of key metabolites important for epigenetic processes. Metabolites have been shown to act as substrates for many canonical epigenetic regulators. These enzymes are responsible for regulating histone modification, DNA methylation and micro RNA expression. By studying the impact of altered liver metabolism we may better understand the long term epigenetic processes which lead to the development and progression of HCC.
Introduction
Hepatocellular carcinoma (HCC) is a major cause of cancer related deaths globally. The incidence of HCC is rising in westernized countries, and has been rising over the past few decades.1, 2 The 5 year survival rate for HCC patients is low, and as of 2012 was estimated to be 27% (with the caveat of intrahepatic biliary cancer).2 This high mortality is due to detection of late stage HCC and the recurrence of tumors following resection.1 Hence, there is an urgent need to develop rapid and accurate diagnostic tools for early detection and treatment. The causes of HCC are heterogeneous, with both genetic and epigenetic alterations associated with tumor progression.3–10 Infection with hepatitis C (HCV) and hepatitis B (HBV ) are major risk factors, as are exposure to aflatoxin (found in stored peanut products) and chronic alcohol abuse.2 The liver produces metabolites essential for epigenetic processes. De-regulation of both the hepatic one carbon and oxidative phosphorylation (TCA) cycles in the liver have major roles in driving aberrant epigenetic changes during tumor progression.11–14 In a progressive model of HCC we may assume that infection of HCV, HBV or chronic alcohol abuse are initiators that cause functional changes in the liver which lead to HCC. Changes in both the TCA and hepatic one carbon cycles may provide good models to determine HCC progression.
Over 80 years ago Otto Warburg first reported that cancer cells preferentially convert glucose to lactate, even in the presence of sufficient oxygen for aerobic metabolism.13 Subsequent studies have identified links between tumor cell function and the switching of ATP production from oxidative phosphorylation to anaerobic glycolysis.15–17 Utilizing glycolysis as the primary source of energy production has several advantages for tumor cells. For example, by not producing mitochondrial reactive oxygen species (ROS), tumor cells have diminished capacity to undergo apoptosis.17, 18 Also, increasing glucose uptake allows tumor cells to grow rapidly and out-compete healthy tissue for available glucose.19, 20 Tumor cell metabolism can be linked to epigenetic changes during tumor progression, recent research has shifted focus from pure epigenetic studies to studies linking epigenetics with metabolic pathways. 18, 21, 22
The purpose of this review is to summarize the current literature with particular focus on the epigenetic study of HCC. We also highlight here, some of the emerging-novel investigations that combine epigenetics with cancer metabolism. By identifying metabolic deficiencies in HCC we may develop new models by which we can study HCC.
Hepatocellular carcinoma as a CIMP+ cancer
So far, tissue testing has revealed no candidate genetic markers that can be associated with the majority of HCC cases. The frequency of TP53 mutation in HCC is highly variable between studies ranging from 0 to 63%, depending on the cohort analyzed.3 Hepatocellular adenomas, from which HCC can arise, have been linked to β-catenin mutations.23 In approximately 20–40% of cases β-catenin coding sequence mutations are associated with HCC.24 With no common mutations of coding genes that can account for all cases of HCC it is likely epigenetic changes are the main driving mechanisms of HCC development. Mutations of coding genes which regulate epigenomic modifications may initiate HCC progression via changes to the epigenome, however these mutations are rarely observed.17 During the course of HCC development alterations to global DNA methylation have been observed. For example, global hypomethylation leads to aberrant over-expression of oncogenes and genome wide chromosomal instability, which increases the risk of chromosomal translocations. 25, 26 Alongside global DNA hypomethylation, localized gene promoter hypermethylation frequently occurs.25, 26 These two phenomena seem to be at odds and further study is required to elucidate the mechanisms driving these distinct processes.
HCC is characterised as a CpG Island Methylator Phenotype positive (CIMP+) cancer.7,27, 28 CIMP+ is a term used to describe cancers exhibiting DNA hypermethylation at CpG Islands, where the number of promoters that become hypermethylated may increase throughout tumor development.27, 28 Studies of gene promoter methylation have identified increasing DNA methylation in liver tissues from patients with HCC. The number of methylated gene promoters increases between cirrhotic and primary HCC tumor samples and between primary HCC and metastatic tumor samples.7, 27 This is important as it implies that worsening DNA hypermethylation can drive tumor progression.
CIMP markers have been shown to have predictive value in the detection of HCC recurrence in HBV-associated HCC following liver transplantation.28 Wu and colleagues (2010) studied the methylation profiles of 7 genes in DNA collected from tumor tissue from HCC patients and from normal healthy liver tissue. Patients were identified as having CIMP+ HCC by gain of methylation in a subset of genes. These patients had significantly lower recurrence free survival.28 However, it is to be noted that HBV associated HCC is more readily studied by DNA methylation, as the hepatitis B virus protein X (HBx) indirectly increases expression of the de novo methyltransferase DNMT3a.29
Much research has been undertaken to identify hypermethylated CpG Island (CGI) promoters. CGIs are usually unmethylated and the presence of CpG methylation at gene promoters is associated with gene silencing. Cumulative promoter hypermethylation is an indicator of tumor stage, as more genes involved in tumor suppression or growth regulation become silenced over time. Epigenetic biomarkers based on promoter hypermethylation have been developed as diagnostic tools to detect liver cirrhosis and HCC (Table 1).4–10 Many studies have shown that progressive promoter methylation accumulates throughout HCC development and is associated with tumor recurrence and prognosis.27, 28, 30–32
Table 1. Aberrant DNA methylation markers for hepatocellular carcinoma.
Synonyms of gene promoters are provided and presented alongside their genomic location. The frequency at which gene promoters are found to be hypermethylated are given for HCC tissues (HCC), cirrhotic tissues (CIR) and adjacent normal tissues (N).
| Gene | Location | Function | Methylation frequency %
|
Refs | ||
|---|---|---|---|---|---|---|
| HCC | CIR | N | ||||
| APC | 5q22.2 | Prostaglandin Synthesis | 53–81 | 13–44 | 0–14 | 7–9, 12 |
| BLU | 3q21.3 | Unknown Zinc Finger | 20 | - | - | 11 |
| COX2 | 1q31.1 | Prostaglandin Synthesis | 35–50 | 0 | 0 | 8, 9, 12 |
| CPS1 | 2q34 | Urea cycling | 80 | 0 | 0 | 13 |
| DAPK1 | 9q21.33 | Apoptosis | 10 | 10 | 0 | 8 |
| E2F-1 | 20q11.22 | Transcription factor | 70 | 24 | 0 | 10 |
| E-Cadherin | 16q22.1 | Cell Adhesion | 33–67 | 6–20 | 7 | 7–9 |
| GSTP1 | 11q13 | Glutathione Synthesis | 41–76 | 13–31 | 0–7 | 7–9 |
| hMLH1 | 3p21.3 | Mismatch repair | 0 | 0 | 0 | 8 |
| P14 | 11q13.1 | CDK Inhibitor | 6 | 0 | 0 | 7 |
| P15 | 18q12.2 | CDK Inhibitor | 42–47 | 0–19 | 0 | 7,10 |
| P16INK4a | 9q21.3 | CDK Inhibitor | 16–83 | 0–23 | 0–10 | 7–10, 12 |
| P21 | 6p21.2 | CDK Inhibitor | 63 | 53 | 10 | 10 |
| P27 | 12p13.1 | CDK Inhibitor | 48 | 36 | 0 | 10 |
| P300 | 22q13.2 | Growth/Cell division | 65 | 54 | 0 | 10 |
| P53 | 17p13.1 | Tumor suppressor | 14 | 8 | 0 | 10 |
| P73 | 1p36.32 | Tumor suppressor | 6 | 0 | 0 | 7 |
| RaR-Beta | 3p24.2 | Retinoic acid signalling | 12 | 13 | 7 | 7 |
| RASSF1A | 3p21.3 | Apoptosis | 59–75 | 0–55 | 0 | 8,9,11,12 |
| RB | 13q14.2 | Chromatin Structure | 32 | 25 | 0 | 10 |
| SEMA3B | 3p21.3 | Apoptosis | 83 | - | - | 11 |
| SOCS-1 | 16p13.13 | Cytokine inhibitor | 43–65 | 28–7 | 0–7 | 7,12 |
| TIMP3 | 22q12.3 | Cell adhesion | 13 | 0 | 0 | 8 |
| WT1 | 11p13 | Urogenital Devpt | 54 | 18 | 0 | 10 |
Genes often found to be hypermethylated in HCC, include those involved in gene expression (transcription factors), growth regulation, cell cycle progression and apoptosis (Table 1). 4–10 Many of the DNA methylation biomarkers identified distinguish between normal healthy tissue and cirrhotic tissue, however the distinction between cirrhotic tissue and HCC is less clear. Many gene promoters which are methylated in HCC are also methylated in the surrounding cirrhotic tissue (Table 1).4–10 The incidence of gene promoter methylation for any given gene is also highly variable and differs from study to study, many of these markers are not essential for HCC and are only generally indicative of the mechanisms regulating tumor cell progression.
Given the paucity of genetic mutations and the length of time liver cancer takes to develop, one likely route by which HCC may develop is by abrogation of metabolic pathways. Reduced turnover of metabolites or changes to metabolite synthesis may influence epigenetic modifications overtime. One of the primary examples of these epigenetic changes has been studied in conjunction with the activity of the histone deacetylase SIRT1. SIRT1 represses glycolysis in the liver by interacting with and deacetylating histones at the promoters of several genes including FOXO1, CRTC2 and STAT3, in an NAD-dependent manner.33 During glycolysis, energy overproduction reduces the availability of NAD+ which is reduced to NADH, as cancer progresses a lack of NAD+ reduces the activity of SIRT1.33 Studies of SIRT1 and NAD+ indicate that metabolite availability influences epigenetic modifications such as histone deacetylation enabling the expression of FOXO1, CRTC2 and STAT3. The paucity of NAD+ directly down regulates the activity of SIRT1 and is one mechanism by which the lack of a specific metabolite may affect the expression of a number of genes by directing epigenetic changes.
Epithelial to Mesenchymal transition, metastasis and microRNAs
Epithelial to mesenchymal transition (EMT) is an attractive model to study HCC as it involves the analysis of functional changes to gene expression which occur in epithelial tumor cells (ETCs). Although relatively confined, as ETCs transform into mesenchymal cells they become motile and invasive, and may enter the circulation.34–36 Our laboratory has developed a novel cell line LH86 derived from a well differentiated model of HCC without the associated hepatitis or cirrhosis.37 This LH86 cell line has been used in a novel mouse xenograft model of HCC/EMT to show that HGF and TGF-β-1 drive EMT via COX2, PGE2 and AKT gene overexpression.36 However, there remains much work to be done on the characterisation of EMT. Another prominent area of research is metastasis which is also a primary cause of death. Even after successful surgical removal of primary tumors, metastatic cancer recurrence does not reduce mortality with less than 30–40% of patients surviving 5 years post resection.38 This emphasizes the importance of studying changes in tumor cells before they enter the circulation (becoming circulating tumor cells, CTCs), undergo metastasis and spread to other parts of the body. EMT is required prior to tumor cell invasion and is characterised by key molecular changes including loss of E cadherin, gain of expression of Vimentin, collagen I and fibronectin. There is also an associated increase in the expression of several transcription factors including Twist, SLUG, SNAIL family members and Zinc finger E box containing transcription factors, ZEB1 and ZEB2.36, 40, 41 Furthermore, EMT has also been shown to be characterized by aberrant microRNA (miRNAs/miRs) expression. miRs are small highly conserved RNA molecules which regulate gene expression by binding to the 3′-untranslated regions of target mRNAs.42–44 Consequently, miRs silence gene expression and can also act on multiple gene targets.45, 46 Over 20 miRs have been shown to be aberrantly expressed in HCC and EMT phenotypes, miRs have also been studied to identify markers of HCC and other cancers, and may be overexpressed or down regulated causing aberrant expression of tumor suppressors or oncogenes.42–50 Both miR-200c and miR-141 expression are lost in many cancer types, DNA methylation of a putative CpG island upstream of the 200c/141 cluster has been shown to repress the expression of these miRs.42,43 By directly targeting ZEB1 and ZEB2 miR-200c and mIR-141 alter the expression of E-cadherin and other genes involved in cell polarity. miR-200c has been shown to regulate EMT, in cancer the loss of miR-200c due to gain of DNA methylation also prevents repression of the neurotropic receptor tyrosine kinase 2 (NTRK2). 51 NTRK2 expression confers anoikis resistance and enables cells to survive after they detach from the extracellular matrix and invade the circulation.51 miR-200c restoration could be used as a therapeutic intervention to induce anoikis in circulating tumor cells preventing the spread of cancer.
Several studies also provide evidence to support the hypothesis that aberrant miRNA expression may confer chemoresistance in certain cancer types. DNA methylation of miR-193-3p confers 5-fluorouracil (5-FU) resistance in HCC. In normal healthy cells SRSF2 expression regulates the pro-apoptotic splicing form of Caspase 2 (CASPL). The loss of DNA methylation in HCC allows the expression of miR-193-3p which in turn suppresses SRSF2 and subsequent up regulation of CASP2L, making the cells resistant to apoptosis induced by 5-FU. 50 The de-regulated expression of miRs is a feature common to many cancer types, and the study of microRNA expression provides another route whereby diagnostic and prognostic tools could be developed to understand cancer stage and risk. One caveat of miRs however, is that due to their small length (~22–24 nucleotides) they may target hundreds of coding genes making their direct effects difficult to study.
The mechanisms causing aberrant gene expression of genes driving EMT have not been fully elucidated. However, by incorporating studies of EMT with metabolomics and epigenetics, models for changes in gene expression could be developed to more fully understand the processes involved.
The continuing study of miRs may yet elucidate powerful predictors of tumor stage and aggression; and provide targets to induce tumor-sensitivity to chemotherapy. These markers may also be combined with associated DNA methylation markers for further diagnostic accuracy or as a putative target for miRNA restoration. A recent study of HCC caused by HCV identified a number of mIRs with altered expression which may indicate that different subsets of HCC have different miR profiles. 81
Evidence from mouse studies has shown that the expression of miRs in primary hepatocytes may be affected by the availability of certain adenine analogs.52 5-Aminoimidazole-4-carboxamide 1-β-D-ribofuranoside (AICAR) is an analogue of adenine and treatment of murine models of HCC, with exogenous AICAR has been shown to alter the expression of 41 miRs in primary hepatocytes indicating a relationship between miR expression and AMP kinase (AMPK) mediated energy sensing. However, the mechanism by which miR expression modulates AMPK remains to be determined. 52
Role of the hepatic one-carbon cycle in epigenetic regulation
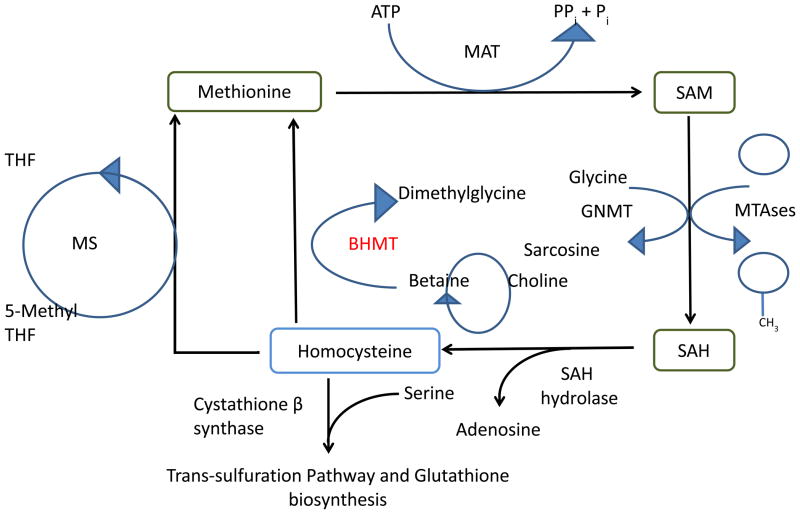
Another mechanism whereby metabolic and epigenetic processes intersect is the methionine synthesis pathway (or hepatic one-carbon cycle) and its components, which are central to liver function. More than half of the dietary methionine is converted into S-Adenosyl methionine (SAM) where over 85% of all methylation reactions take place.53 This makes the liver a unique driver of epigenetic processes with the labile methyl pool and the ratio of SAM and adenosyl-homocysteine (AdoHCY) regulating essential to liver function. SAM as a methyl donor has a diverse range of functions and is used a substrate for reactions including: methylation of DNA, histones, phospholipids and small molecules, as well as the synthesis of polyamines.54 Key enzymes involved in the hepatic one-carbon cycle include; methionine adenosyl transferase (MAT), S-adenosyl hydrolase (SAH), betaine homocysteine methyltransferase (BHMT), Glycine-N methyltransferase (GNMT) and cystathione β synthase (CBS) (Figure 1).
Figure 1. Schematic showing the hepatic carbon-one cycle.
SAM biosynthesis is catalyzed by methionine adenosyltransferase MAT1A in the liver. SAM donates methyl groups to methyltransferases and also Glycine N-methyltransferase for methylation of Glycine to become S-adenosyl homocysteine, which is hydrolyzed to form Hcy. Hcy is then re-methylated to form methionine by one of two enzymes, methionine synthase (MS), the canonical enzyme found in all cells, or betaine-homocysteine-methyltransferase BHMT which is found mainly in the liver and kidney. Low abundance of SAM can lead to a lack of methyl reactions in the liver, oxidative stress (via deregulation of the transulfuration pathway) and arrest of DNA synthesis (via the folate metabolic pathway). Maintenance of the SAM: Hcy ratio and the labile methyl pool are essential to liver function.
Many studies utilizing cell culture and mouse knockout models have helped to clarify the function of many one-carbon cycle genes, with the abrogation or knockout of these genes leading to liver disease and HCC.55–57 Avila et al identified that a number of key one-carbon cycle enzymes are under-expressed in HCC.58 In cirrhotic samples a reduced abundance of mRNA was identified in the following genes; GNMT, methionine synthase (MS), BHMT and CBS. Loss of expression was also identified in BHMT, CBS, GNMT and MS in tissues taken after resection from HCC patients and compared to normal liver tissue.58 These data indicate that abrogation of the TCA cycle may be a predisposing factor to developing HCC in a cirrhotic background.
Methionine adenosyltransferase (MAT1A) converts methionine into SAM and is also expressed exclusively in the liver. MAT1A is epigenetically regulated with studies of MAT1A identifying 2 CpG sites in the first exon, the methylation of which, correlates with reduced MAT1A expression.17, 58 De-regulation of MAT1A is associated with HCC development and is in turn linked to the overexpression of hepatocyte growth factor HGF. The primary function of HGF is to expand the liver during development and to drive regenerative growth following injury or insult. HGF is present in the circulation with concentrations increasing after partial hepatectomy, liver injury or fulminant hepatic failure.59 Chronic hepatic injury, as caused by hepatitis and cirrhosis, allows HGF to be constitutively expressed, this in turn drives hepatocyte expansion, placing a strong selective pressure towards tumorigenesis and the development of HCC.54 Interestingly, hepatic HGF overexpression correlates well with the switch in expression of MAT1A to MAT2A in HCC.54 HGF also drives growth of cells at low density but inhibits growth at high cell densities. However, the precise mechanism by which HGF and MAT2A interact has not been fully described.60, 51
Studies of the hepatic one-carbon cycle provide evidence for the global hypomethylation component of HCC. Down-regulation of one or more of the cycle’s constituent enzymes may reduce the availability of the hepatic labile methyl pool, but further studies are necessary. De-regulation of the hepatic one-carbon cycle has not been linked with promoter hypermethylation; therefore this process must be controlled by other mechanisms during HCC development.
Warburg effect on epigenetic processes
Under the partially hypoxic conditions found in many types of solid tumor, cells switch to glycolysis as the primary source of energy production.62 This change in metabolism which may also occur in normoxia has been linked to stabilization of HIF1α. The HIF1 heterodimer is a transcription factor which controls a suite of genes involved in the up regulation of glycolysis. The switch that takes place in non-hypoxic conditions is linked to rare mutations which alter the Hif Prolyl hydroxylase (PHD or EgIN) proteins; these in turn modify HIF1α for ubiquitination by Von-Hippel Lindau protein (pVHL). This change may also be linked to mutations of the pVHL protein which recognizes hydroxylated HIF1α and marks it for ubiquitination.17, 63 Either of these mechanisms permit the stabilization of HIF1α in the cytoplasm allowing it to form a heterodimer with HIF1β. This complex can then translocate to the nucleus where it acts as a transcription factor up-regulating the down-regulate the TCA cycle.14, 17,20,63, 64 One of the major consequences of depressing the TCA cycle is the reduced availability of α-ketoglutarate. This decreases the activity of α-ketoglutarate dependent proteins which are responsible for hydroxylating many substrates in the cell important in epigenomic control. Of primary interest are the Ten Eleven Translocation (TET) family of FeII-dependent, α-ketoglutarate-dependent methyl dioxygenases which convert methylated cytosine to 5 hydroxy methylcytosine (5hmC).
We hypothesize that loss of TET activity may explain gene promoter hypermethylation in HCC and other CIMP+ cancers. TET proteins confer the epigenetic modification of a hydroxyl group to 5 methylcytosine (5mC) to form 5hmC. 5hmC is a demethylation modification and its loss has been implicated in cancer development.65–67 First identified in 1953 in the bacteriophage T2.I, 5hmC functions in the bacteriophage genome in defence against excision by host enzymes.68 Recently this modification was identified in murine brain and ES cells, Purkinje neurons and in human embryonic kidney cells with induced overexpression of TET1.65, 66 The discovery of the TET family and its role in conferring the epigenetic mark 5hmC represents a major breakthrough in epigenetics and cancer research. Previously the regulation of DNA methylation remained enigmatic as no de-methylation enzymes had been identified that could account for the genome wide modification of DNA methylation. These discoveries have significant implications for previous investigations using bisulfite conversion to study DNA methylation since bisulfite sequencing and the use of methylation sensitive restriction enzymes, are unable to distinguish 5mC from 5hmC. There is, however, a new technique which is able to differentiate 5hmC from 5mC.69
TET proteins have been suggested to modify genomic DNA methylation in two ways; either by binding to unmethylated CpGs via a CxxC binding domain thereby preventing DNA methyltransferase activity, or by hydroxylation of 5mC marking it for subsequent demethylation.70 TET2 does not contain a CxxC binding domain but does possess a role in cancer development and may have multiple binding partners allowing it to modify alternative sites in the genome.70 Several mechanisms are postulated whereby methylated DNA can be demethylated. One is base excision repair (BER), while the other is modification by TET members followed by BER, although the addition of the hydroxyl group to 5mC alone may be sufficient for passive demethylation.71 Genomic distribution of 5hmC was first studied using an immunoprecipitation technique, 5hmC-IP-seq, which combines an antibody-specific for the enrichment of 5 hydroxyl-methylcytidine, and subsequent massive parallel sequencing of 5hmC-enriched DNA.72 Since these important initial studies, investigations have detected 5hmC in human tissues.72, 73 Loss of TET expression (which confers the 5hmC modification) leads to global genomic reduction of 5hmC and has been detected in several cancers including HCC.74, 75 5hmC also has a protective role in gene bodies, enabling transcription by preventing localization of methyl binding proteins,61, 62 thereby preventing transcriptional silencing associated with DNA methylation.
Thus far, detection of 5hmC in cancer has been performed by histological staining and other global methods which measure total 5hmC. 5hmC genomic distribution has been studied in human stem cells and mouse studies of 5hMC and TET distribution have also been conducted.76, 77
Future Directions
Since the observations made by Warburg almost 90 years ago,13 significant strides have been made to understand the molecular mechanisms whereby altered tumor cell metabolism may drive tumor progression. The field of cancer epigenetics has sought to identify how gene expression is modified during tumor development and studies of DNA methylation have offered insights into the genome wide epigenetic processes which occur. Many studies have sought to identify DNA methylation biomarkers of HCC, however there are few that could be applied clinically as methylation of specific gene targets is highly variable (Table 1). Better understanding of the methionine synthesis pathway, may be able to more accurately determine which genes are likely to become hypomethylated and drive HCC development.
Another epigenetic mechanism that may yield promising results is the study of microRNAs. miRs are often aberrantly expressed in HCC and have been shown to play a role in EMT. Future studies may seek to identify specific miRs required for HCC development and the subset of genes targeted by mIRs for post translational regulation. Further metabolic studies linking miR expression with nucleic acid metabolism may provide insights into restorative mechanisms of miR expression.
Studying the enzymes down-regulated in glycolytic tumor metabolism could also be vital in understanding cancer. The TET family genome distribution could unveil the mystery of gene promoter hypermethylation in CIMP+ cancers. Currently, we do not yet understand whether individual TET members target different genes exclusively or if they have a synergistic function (the answer may be both, and dependent on the presence of binding sites). Loss of TET expression could result in the aberrant hypermethylation observed in CIMP+ cancers, as TET may function to block the action of DNA methyltransferases at gene promoters. Another likely scenario is that TET expression dynamically regulates methylation marks which are laid down during cell division and may otherwise accumulate. Future studies may ascribe a role for TET expression and regulation of DNA methylation during EMT. By following methylation and hydroxymethylation during EMT we may be able to more fully understand how aberrant TET expression could drive tumor cell invasion and the development of circulating tumor cells.
Seeking therapeutic agents to restore oxidative phosphorylation could be a viable treatment option. By reinstating components of the TCA cycle such as α-ketoglutarate, the function of epigenetic modifiers such as EgINs, LOX and histone demethylases may also be restored.17 In this regard, studies undertaken to assess the effect of Dichloroacetate (DCA) showed positive results in vitro.78 However, due to the impermeability of DCA to cells the dosages required to treat HCC caused peripheral neuropathy in rats.79 Conversely, the cell-permeable α-ketoglutarate showed promise in reversing competitive inhibition of EgIN proteins by fumarate and succinate in cancer cells, illustrating the potential for the development of pharmaceuticals that could restore TCA cycle function.80 More in vivo experiments are required to test the efficacy of TCA metabolites in murine xenograft models of HCC, to identify the putative benefits in the treatment of HCC.
Comparative in vitro studies of HCC cell lines with different TET expression could clarify the epigenetic vivo studies of HCC cell lines in mice may also identify to what extent the loss of TET proteins play a role in cancer tumorigenicity and their response to anti-cancer chemotherapy. By studying the distribution of TET proteins in both hepatocyte and tumor genomes it may be possible to readily identify the genes effected by loss of TET expression and to use the data as candidate markers for tumor development, malignancy and recurrence. By approaching HCC in this way we may develop models for the study of all CIMP+ gastrointestinal cancers.
With the advent of genome wide platforms we are now able to study the multivariate changes occurring during cancer. By understanding how metabolic changes drive epigenetic changes during HCC progression, we can study the impact of altered metabolite availability. New investigations combining both tumor metabolism and epigenomics will be pivotal in developing future models by which we can identify the causes of HCC and other CIMP+ cancers.
Acknowledgments
WP and CL are funded by R01 AA019976 Liu, Robertson (MPI) 09/20/11-06/30/2016 We thank Dr Jonathan Whittamore for his careful reading of the manuscript
References
- 1.Cucchetti A, Piscaglia F, Cescon M, et al. Conditional Survival after Hepatic Resection for Hepatocellular Carcinoma in Cirrhotic Patients. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-2663. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55(3):724–725. doi: 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163(3):1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163(4):1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katoh H, Shibata T, Kokubu A, et al. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol. 2006;168(4):1375–1384. doi: 10.2353/ajpath.2006.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Guo X, Jiang G, et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123(5):998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 8.Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A, et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer. 2005;115(5):684–9. doi: 10.1002/ijc.20944. [DOI] [PubMed] [Google Scholar]
- 9.Um TH, Kim H, Oh BK, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54(5):939–947. doi: 10.1016/j.jhep.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Dong H, Robertson K, Liu C. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. Am J Pathol. 2011;178(2):652–661. doi: 10.1016/j.ajpath.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomasi ML, Li TW, Li M, Mato JM, Lu SC. Inhibition of human methionine adenosyltransferase 1A transcription by coding region methylation. J Cell Physiol. 2012;227(4):1583–1591. doi: 10.1002/jcp.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier K, Hofmann U, Reuss M, Mauch K. Dynamics and control of the central carbon metabolism in hepatoma cells. BMC Syst Biol. 2010;4:54. doi: 10.1186/1752-0509-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O, Wind F, Neglers E. Metabolism of tumours. London: Constable & Co; 1930. pp. 254–270. [Google Scholar]
- 14.Stubbs M, Griffiths JR. The altered metabolism of tumors: HIF-1 and its role in the Warburg effect. Adv Enzyme Regul. 2010;50(1):44–55. doi: 10.1016/j.advenzreg.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 15.WARBURG O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 16.Garber K. Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst. 2004;96(24):1805–6. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- 17.Kaelin WG. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–45. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cyr AR, Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal. 2011;15(2):551–89. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeluri S, Madhok B, Prasad KR, Quirke P, Jayne DG. Cancer’s craving for sugar: an opportunity for clinical exploitation. J Cancer Res Clin Oncol. 2009;135(7):867–77. doi: 10.1007/s00432-009-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39(3):231–4. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- 21.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339(6116):211–4. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanbhogue AK, Prasad SR, Takahashi N, Vikram R, Sahani DV. Recent advances in cytogenetics and molecular biology of adult hepatocellular tumors: implications for imaging and management. Radiology. 2011;258(3):673–93. doi: 10.1148/radiol.10100376. [DOI] [PubMed] [Google Scholar]
- 24.van Malenstein H, van Pelt J, Verslype C. Molecular classification of hepatocellular carcinoma anno 2011. Eur J Cancer. 2011;47(12):1789–97. doi: 10.1016/j.ejca.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21(1 Pt 1):15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 25.Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117(9):2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Liu W, Wang L, Li M, Wang J, Huang L, et al. CpG island methylator phenotype associated with tumor recurrence in tumor-node-metastasis stage I hepatocellular carcinoma. Ann Surg Oncol. 2010;17(7):1917–26. doi: 10.1245/s10434-010-0921-7. [DOI] [PubMed] [Google Scholar]
- 28.Wu LM, Zhang F, Zhou L, Yang Z, Xie HY, Zheng SS. Predictive value of CpG island methylator phenotype for tumor recurrence in hepatitis B virus-associated hepatocellular carcinoma following liver transplantation. BMC Cancer. 2010;10:399. doi: 10.1186/1471-2407-10-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, et al. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25(2):439–46. doi: 10.1016/j.cellsig.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Liu JB, Zhang YX, Zhou SH, Shi MX, Cai J, Liu Y, et al. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol. 2011;17(42):4718–24. doi: 10.3748/wjg.v17.i42.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu WJ, Wang L, Wang JP, Li JQ, Zhang CQ, Zheng L, et al. Correlations of CpG island methylator phenotype and OPCML gene methylation to carcinogenesis of hepatocellular carcinoma. Ai Zheng. 2006;25(6):696–700. [PubMed] [Google Scholar]
- 32.Cheng Y, Zhang C, Zhao J, Wang C, Xu Y, Han Z, et al. Correlation of CpG island methylator phenotype with poor prognosis in hepatocellular carcinoma. Exp Mol Pathol. 2010;88(1):112–7. doi: 10.1016/j.yexmp.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31(5):212–20. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding W, You H, Dang H, et al. Epithelial-to-mesenchymal transition of murine liver tumor cells promotes invasion. Hepatology. 2010;52(3):945–953. doi: 10.1002/hep.23748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Zijl F, Mall S, Machat G, et al. A human model of epithelial to mesenchymal transition to monitor drug efficacy in hepatocellular carcinoma progression. Mol Cancer Ther. 2011;10(5):850–860. doi: 10.1158/1535-7163.MCT-10-0917. [DOI] [PubMed] [Google Scholar]
- 36.Ogunwobi OO, Liu C. Hepatocyte growth factor upregulation promotes carcinogenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via Akt and COX-2 pathways. Clin Exp Metastasis. 2011;28(8):721–731. doi: 10.1007/s10585-011-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Dong H, Eksioglu E, et al. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133(5):1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7(4):445–454. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 41.Gregory PA, Bracken CP, Smith E, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22(10):1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neves R, Scheel C, Weinhold S, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vrba L, Jensen TJ, Garbe JC, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5(1):e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiklund ED, Bramsen JB, Hulf T, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128(6):1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 45.John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- 46.Schmeier S, Schaefer U, Essack M, Bajic VB. Network analysis of microRNAs and their regulation in human ovarian cancer. BMC Syst Biol. 2011;5:183. doi: 10.1186/1752-0509-5-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 48.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286(29):25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tellez CS, Juri DE, Do K, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma K, He Y, Zhang H, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287(8):5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13(2):R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Liu W, Ying H, Zhao W, Zhang H. Analysis of microRNA expression profile induced by AICAR in mouse hepatocytes. Gene. 2013;512(2):364–72. doi: 10.1016/j.gene.2012.09.118. [DOI] [PubMed] [Google Scholar]
- 53.Mudd SH, Poole JR. Labile methyl balances for normal humans on various dietary regimens. Metabolism. 1975;24(6):721–735. doi: 10.1016/0026-0495(75)90040-2. [DOI] [PubMed] [Google Scholar]
- 54.Martínez-López N, Varela-Rey M, Ariz U, et al. S-adenosylmethionine and proliferation: new pathways, new targets. Biochem Soc Trans. 2008;36(Pt 5):848–852. doi: 10.1042/BST0360848. [DOI] [PubMed] [Google Scholar]
- 55.Santamaría E, Muñoz J, Fernandez-Irigoyen J, et al. Molecular profiling of hepatocellular in human liver diseases. J Proteome Res. 2006;5(4):944–953. doi: 10.1021/pr050429v. [DOI] [PubMed] [Google Scholar]
- 56.Wang YC, Tang FY, Chen SY, Chen YM, Chiang EP. Glycine-N methyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation kinetics and DNA methylation. J Nutr. 2011;141(5):777–782. doi: 10.3945/jn.110.135954. [DOI] [PubMed] [Google Scholar]
- 57.Teng YW, Mehedint MG, Garrow TA, Zeisel SH. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J Biol Chem. 2011;286(42):36258–36267. doi: 10.1074/jbc.M111.265348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avila MA, Berasain C, Torres L, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33(6):907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 59.Fausto N. Growth factors in liver development, regeneration and carcinogenesis. Prog Growth Factor Res. 1991;3(3):219–234. doi: 10.1016/0955-2235(91)90008-r. [DOI] [PubMed] [Google Scholar]
- 60.Liu F, Schaphorst KL, Verin AD, et al. Hepatocyte growth factor enhances endothelial cell barrier function and cortical cytoskeletal rearrangement: potential role of glycogen synthase kinase-3beta. FASEB J. 2002;16(9):950–962. doi: 10.1096/fj.01-0870com. [DOI] [PubMed] [Google Scholar]
- 61.Yang H, Magilnick N, Noureddin M, Mato JM, Lu SC. Effect of hepatocyte growth factor on methionine adenosyltransferase genes and growth is cell density-dependent in HepG2 cells. J Cell Physiol. 2007;210(3):766–773. doi: 10.1002/jcp.20891. [DOI] [PubMed] [Google Scholar]
- 62.Simon LM, Robin ED, Theodore J. Differences in oxygen-dependent regulation of enzymes between tumor and normal cell systems in culture. J Cell Physiol. 1981;108(3):393–400. doi: 10.1002/jcp.1041080313. [DOI] [PubMed] [Google Scholar]
- 63.Kaelin WG. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8(11):865–73. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 64.Robey IF, Lien AD, Welsh SJ, Baggett BK, Gillies RJ. Hypoxia-inducible factor-1alpha and the glycolytic phenotype in tumors. Neoplasia. 2005;7(4):324–30. doi: 10.1593/neo.04430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hershey AD, Dixon J, Chase M. Nucleic acid economy in bacteria infected with bacteriophage T2. I. Purine and pyrimidine composition. J Gen Physiol. 1953;36(6):777–89. doi: 10.1085/jgp.36.6.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 70.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–64. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–52. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosinein the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012 Apr;103(4):670–6. doi: 10.1111/j.1349-7006.2012.02213.x. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012 doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen YC, Ou DL, Hsu C, Lin KL, Chang CY, Lin CY, et al. Activating oxidative phosphorylation by a pyruvate dehydrogenase kinase inhibitor overcomes sorafenib resistance of hepatocellular carcinoma. Br J Cancer. 2013;108(1):72–81. doi: 10.1038/bjc.2012.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calcutt NA, Lopez VL, Bautista AD, Mizisin LM, Torres BR, Shroads AL, et al. Peripheral neuropathy in rats exposed to dichloroacetate. J Neuropathol Exp Neurol. 2009;68(9):985–93. doi: 10.1097/NEN.0b013e3181b40217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27(9):3282–9. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133(4):816–24. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]