Abstract
Introduction
Small-cell lung cancer (SCLC) is the most aggressive subtype of lung cancer, with no early detection strategy or targeted therapy currently available. We hypothesized that difference gel electrophoresis (DIGE) may identify membrane-associated proteins (MAPs) specific to SCLC, advance our understanding of SCLC biology, and discover new biomarkers of SCLC.
Methods
MAP lysates were prepared from three SCLCs, three non–small-cell lung cancers, and three immortalized normal bronchial epithelial cell lines and coanalyzed by DIGE. Subsequent protein identification was performed by mass spectrometry. Proteins were submitted to Ingenuity Pathway Analysis. Candidate biomarkers were validated by Western blotting (WB) and immunohistochemistry (IHC).
Results
Principal component analysis on the global DIGE data set demonstrated that the four replicates derived from each of the nine cell lines clustered closely, as did samples within the same histological group. One hundred thirty-seven proteins were differentially expressed in SCLC compared with non–small-cell lung cancer and immortalized normal bronchial epithelial cells. These proteins were overrepresented in cellular/tissue morphology networks. Dihydropyrimidinase-related protein 2, guanine nucleotide–binding protein alpha-q, laminin receptor 1, pontin, and stathmin 1 were selected as candidate biomarkers among MAPs overexpressed in SCLC. Overexpression of all candidates but RSSA in SCLC was verified by WB and/or IHC on tissue microarrays. These proteins were significantly associated with SCLC histology and survival in univariables analyses.
Conclusion
DIGE analysis of a membrane-associated subproteome discovered overexpression of dihydropyrimidinase-related protein 2, guanine nucleotide–binding protein alpha-q, RUVB1, and stathmin 1 in SCLC. Results were verified by WB and/or IHC in primary tumors, suggesting that investigating their functional relevance in SCLC progression is warranted. Association with survival requires further validation in larger clinical data sets.
Keywords: Biomarker, Membrane-associated, Proteomics, Small-cell lung cancer
Small-cell lung cancer (SCLC) represents 10% to 15% of lung cancers and is clinically the most aggressive subtype, with a 5-year overall survival (OS) as low as 5%.1 Molecular steps leading to SCLC are still poorly understood, and this has translated into the absence of efficient early detection strategies or targeted therapies. In-depth proteomic analysis is therefore needed to improve our understanding of SCLC tumorigenesis and may have implications in the discovery of novel early detection and therapeutic strategies for this aggressive cancer.
Although (epi)genetic alterations are driving carcinogenesis and genomic studies have provided valuable information on cancer biology, the cellular phenotype is determined by proteins and cannot be predicted by genomics alone. Proteomic approaches are therefore powerful tools to study cancer biology.2,3 Among them, difference gel electrophoresis (DIGE) coupled with protein identification by mass spectrometry (MS) is widely used in comparative proteomics for the detection of protein differences with high reproducibility and reliability.4–6 However, these studies have not identified key underpinnings of tumor progression or specific molecular targets for SCLC diagnostics or therapeutics.
Membrane-associated proteins (MAPs) are excellent targets for diagnostic and therapeutic interventions. Although membrane proteins constitute only 20% to 30% of human proteome, they represent more than 60% of all drug targets.7 In this study, we hypothesized that the identification of SCLC-specific MAPs by DIGE may advance our understanding of SCLC biology and lead to the discovery of new candidate diagnostic or therapeutic biomarkers. We therefore analyzed MAPs from three SCLC, three non–small-cell lung cancer (NSCLC), and three immortalized normal bronchial epithelial (INBE) cell lines by DIGE.
MATERIALS AND METHODS
Cell Lines and Tissues
Nine cell lines were used for this study: National Cancer Institute (NCI)-H69, NCI-H82, NCI-H209, A549, NCI-H23, NCI-H520, BEAS-2B, HBEC-3KT, and 16-HBE. All were purchased from ATCC (Manassas, VA), except HBEC-3KT (Dr. Minna’s gift, University of Texas Southwestern) and 16-HBE (Dr. Gruenert’s gift, Children’s Hospital, Oakland Research Institute). They were cultured under recommended conditions.
Tissue microarrays (TMAs) made of SCLC and NSCLC specimens were prepared from formalin-fixed, paraffin-embedded tissue blocks8 retrieved from the Pathology Department’s archives at Vanderbilt University Medical Center, Nashville VA Medical Center, and St Thomas Hospital in Nashville, TN. They were obtained between 1996 and 2008 from 136 patients who had surgery or bronchoscopy. Samples were annotated with clinical data elements. The study was approved by the Institutional Review Board at each institution.
MAP Extraction
MAP extraction was performed four independent times for each cell line with the ProteoExtract Native Membrane Protein Extraction Kit (EMD Chemicals, San Diego, CA) per manufacturer’s protocol.
DIGE and MS
Minimal labeling was performed with N-hydroxysuccinimide–ester dyes Cy2/3/5 using the mixed internal standard methodology as described previously,9 with 150 μg of protein of each of the 36 experimental samples. DIGE-associated instrumentation was manufactured by GE Healthcare (Piscataway, NJ). Isoelectric focusing (pH 4–7, 24 cm) was performed after passive rehydration with sample and then focused using a manifold-equipped IPGphor per manufacturer’s protocol. Second-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis was performed using handcast gels for which one plate was pre-silanized using an Ettan DALT 12U per manufacturer’s protocol. Cy2/3/5-specific 16-bit data files were acquired at 100-μm resolution separately by dye-specific excitation and emission wavelengths using a Typhoon 9400 Variable Mode Imager and analyzed using DeCyder_6.5 software. Gels were counterstained for total protein content with SyproRuby (Molecular Probes/Invitrogen, Grand Island, NY), followed by robot excision of proteins of interest and subsequent processing for trypsin digestion and MS using an automated Spot Handling Workstation. Resulting peptides were subjected to C18 reverse-phase liquid chromatography coupled in-line with tandem MS using an LTQ Orbitrap tandem MS equipped with MicroAS autosampler and Surveyor HPLC pump, nanospray source, and Xcalibur_2.0 instrument control (Thermo Scientific, San Jose, CA). Tandem MS/MS data were searched against proteins extracted from UniProtKB database (www.uniprot.org), with taxonomy tag “Homo sapiens (human)” acquired on July 2009 (173,736 entries). This database was concatenated with protein sequences in reverse to enable false discovery rate calculations and also contained common laboratory contaminants. Searches were performed using Sequest and X! Tandem algorithms, allowing for cysteine carbamidomethylation and partial methionine oxidation. Results were validated and assembled into protein identifications using Scaffold_3.1.2 (Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at more than 95.0% probability as specified by Peptide Prophet algorithm.10 Protein identifications were accepted if they could be established at more than 99.0% probability and contained two or more identified peptides. Protein probabilities were assigned by Protein Prophet algorithm.11 Proteins containing similar peptides and not differentiated based on MS/MS alone were grouped to satisfy parsimony principles.
Ingenuity Pathway Analysis
Biological processes, molecular functions, and genetic/protein networks of identified proteins were analyzed using Ingenuity Pathway Analysis (IPA) tools (Ingenuity Systems, Mountain View, CA). The score associated with a particular network was the likehood of differentially expressed proteins being found together due to chance.
Western Blotting
Blots were probed with antibodies against human v-Akt murine thymoma viral oncogene homolog 1 (AKT) (1:1000; Cell Signaling, Danvers, MA), dihydropyrimidinase-related protein 2 (DPYL2) (1:1000; LSBio, Seattle, WA), phospho-DPYL2 (Thr514/Ser518) (1:1000; LSBio), E-cadherin (1:1000; BD Transduction Laboratories, San Jose, CA), endothelial growth factor receptor (EGFR) (1:750; Santa Cruz Biotechnology, Santa Cruz, CA), guanine nucleotide–binding protein alpha-q (GNAQ) (1:900; Abcam, Cambridge, MA), neural cell adhesion molecule (NCAM) (1:500; Sigma, St. Louis, MO), laminin receptor 1 (RSSA) (1:1000; Sigma), pontin (RUVB1) (1:1000; Atlas Antibodies, Stockholm, Sweden), stathmin 1 (STMN1) (1:1000; Cell Signaling), and phospho-STMN1 (Ser16) (1:1000; Cell Signaling). Protein expression was normalized to Actin (1:5000; Sigma).
Immunohistochemistry
Tissue sections were stained following previously reported protocol,12 using antibodies against human DPYL2 (1:500; LSBio), GNAQ (1:100; Abcam), RSSA (1:150; LSBio), RUVB1 (1:100; Atlas Antibodies), and STMN1 (1:50; Cell Signaling). Staining intensity of cases represented in triplicate was evaluated by two independent observers (B.W., S.O.) as follows: 0, no staining; 1, weak; 2, moderate; and 3, strong. Staining intensity was then multiplied by stained tumor cell percentage to obtain the final staining score (range, 0–300).
Statistics
DIGE
In DeCyder_6.5, the normalized volume ratio of each protein spot feature from a Cy3- or Cy5-labeled sample was quantified relative to the Cy2 signal from the pooled-sample internal standard corresponding to the same spot feature, and all ratios were normalized across all 18 DIGE gels using the individual signals from the Cy2-labeled standard. Univariable analyses (Student’s t test and analysis of variants) were used to establish confidence associated with expression changes. Multivariate principal components analysis and hierarchical clustering (using Euclidean correlation and average linkage) were used to assess global variation arising from individual samples.
iPA
One-side right-tailed Fisher’s exact test (α = 0.05) was used to calculate the significance of the values assigned to the analyzed functions of proteins.
Immunohistochemistry
Wilcoxon rank-sum or Kruskal-Wallis tests were used to correlate average staining scores with categorical clinical outcomes, and Spearman’s rank correlation was used for continuous outcomes. OS was calculated from the date of diagnosis to the date of death or last date of contact for those alive at the time of analysis. Descriptive statistics, including median and interquartile ranges for continuous and percentages or frequencies for categorical variables, were reported. Multivariable analyses were performed using Cox proportional hazards model to adjust for age, pack years, histology, and stage. All tests were two-sided, and p values less than 0.05 were considered statistically significant. Analyses were performed using R_2.15.1.
RESULTS
Identification by DIGE of 137 Proteins Differentially Expressed in SCLC
We extracted MAPs from three SCLC (NCI-H69, NCI-H82, NCI-H209), three NSCLC (A549, NCI-H23, NCI-H520), and three INBE (BEAS-2B, HBEC-3KT, 16-HBE) cell lines in quadriplicate, obtaining 36 extracts. By Western blotting (WB), membrane proteins E-cadherin, NCAM, and EGFR were mainly expressed in membrane-associated, while cytosolic protein AKT was mainly expressed in cytosolic extracts (Fig. 1). This confirmed enrichment for MAPs in MAP extracts.
FIGURE 1.
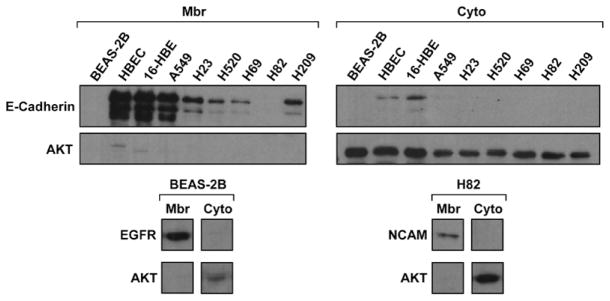
Isolation of membrane-associated proteins. Membrane-associated (mbr) and cytosolic (cyto) extracts were resolved with sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blots were incubated with anti-E-cadherin, NCAM, EGFR, AKT, or actin antibodies. E-cadherin, NCAM, EGFR, and AKT levels were normalized to actin. In each quadruplicate analysis, membrane proteins E-cadherin, NCAM, and EGFR were mainly expressed in membrane-associated extracts, whereas cytosolic protein AKT was mainly expressed in cytosolic extracts. Blots are representative of three independent experiments. AKT, v-Akt murine thymoma viral oncogene homolog 1; EGFR, epidermal growth factor receptor; NCAM, neural cell adhesion molecule.
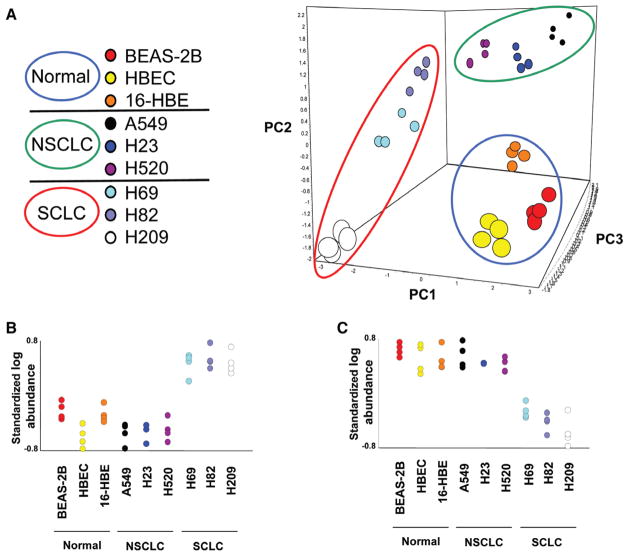
MAP extracts were analyzed by DIGE according to the experimental design summarized in Supplementary table 1 (Supplemental Digital Content 1, http://links.lww.com/JTO/A529). On each of the 18 gels used, many differentially expressed protein spots were observed (representative DIGE image in Supplementary figure 1, Supplemental Digital Content 2, http://links.lww.com/JTO/A530). Principal components analysis on the global data set demonstrated that the four replicates derived from each of the nine cell lines clustered very closely by global expression patterns, as did samples within each classification (12 samples per classification) using the first three principle components (cumulative variance = 62%; Fig. 2A).
FIGURE 2.
DIGE results. A, PCA of the DIGE global data set. The axes correspond to the first three principal components. The apparent groups yielded by PCA are enclosed in circles: blue circle for immortalized normal bronchial epithelial cell lines (normal), green circle for NSCLC cell lines, and red circle for SCLC cell lines. The four replicates from each of the nine cell lines clustered very closely, as did samples within each histological subgroup. B, C, Quantitative analysis of protein expression in the nine cell lines analyzed by DIGE. Representative graphs of protein features significantly overexpressed (B) and underexpressed (C) in SCLC compared with immortalized normal bronchial epithelial and NSCLC cell lines (p < 10−3). The Y axis represents the standardized log protein abundance generated by the DeCyder analysis. DIGE, difference gel electrophoresis; NSCLC, non small-cell lung cancer; SCLC, small-cell lung cancer; PC, principle component; PCA, principal component analysis.
For further analyses, we selected 135 DIGE protein spots significantly over/underexpressed in SCLC compared with NSCLC and INBE cells (p < 10−3) (Fig. 2B). These protein spots were digested into peptides for tandem MS analysis, and 137 unique proteins were identified. Among them, 83 were from overexpressed (Table 1) and 54 from underexpressed spots (Supplementary table 2, Supplemental Digital Content 1, http://links.lww.com/JTO/A529).
TABLE 1.
Proteins Overexpressed in Small-Cell Lung Cancer, Membrane-Associated, and with Functions Relevant to Cancer Progression and/or Neuroendocrine Phenotype and/or Part of Networks Related to Cell/Tissue Morphology
| Protein Symbol | Gene Symbol | Overexpressed in SCLC (N = 83)
|
Membrane-Associated (N = 32) | Interesting Functions for SCLC Progression (N = 17)
|
Final Candidate Biomarkers (N = 5) | |||
|---|---|---|---|---|---|---|---|---|
| Av. Ratio | t test | Relevant to Cancer Progression | Relevant to Neuroendocrine Phenotype | Part of Cell/Tissue Morphology Networks | ||||
| PRDX2 | PRDX2 | 7.23 | 2.90E–11 | x | x | x | ||
| STMN1 | STMN1 | 5.71 | 1.30E–12 | x | x | x | x | x |
| HNRPC | HNRNPC | 5.38 | 8.90E–10 | |||||
| FPPS | FDPS | 4.68 | 1.80E–09 | |||||
| STRAP | STRAP | 4.68 | 1.80E–09 | x | x | |||
| PSA1 | PSMA1 | 4.18 | 3.10E–12 | |||||
| ATD3A | ATAD3A | 3.92 | 4.90E–09 | x | ||||
| ERO1A | ERO1L | 3.92 | 4.90E–09 | x | ||||
| G3BP1 | G3BP1 | 3.92 | 4.90E–09 | x | ||||
| HSP71 | HSPA1A-1B | 3.92 | 4.90E–09 | |||||
| MIR7-1 | 3.92 | 4.90E–09 | ||||||
| HMGB1 | HMGB1 | 3.67 | 6.90E–07 | |||||
| CALD1 | CALD1 | 3.55 | 1.80E–09 | x | x | x | ||
| SPF27 | BCAS2 | 3.44 | 8.50E–13 | |||||
| ROAA | HNRNPAB | 3.31 | 1.70E–08 | |||||
| CAPG | CAPG | 3.06 | 2.40E–09 | x | x | |||
| RS10 | RPS10 | 3.06 | 5.90E–05 | |||||
| DPYL2 | DPYSL2 | 2.73 | 4.10E–08 | x | x | x | x | x |
| TCPG | CCT3 | 2.73 | 4.10E–08 | |||||
| AL9A1 | ALDH9 | 2.72 | 4.50E–10 | |||||
| EFR1 | ERVFRDE1 | 2.72 | 4.50E–10 | x | ||||
| IF2B | EIF2S2 | 2.72 | 4.50E–10 | |||||
| ODO2 | DLST | 2.72 | 4.50E–10 | x | ||||
| PTBP1 | PTBP1 | 2.72 | 4.50E–10 | |||||
| RUVB2 | RUVBL2 | 2.72 | 4.50E–10 | x | x | |||
| SPF45 | RBM17 | 2.72 | 4.50E–10 | |||||
| UAP56 | BAT1 | 2.72 | 4.50E–10 | |||||
| BUB3 | BUB3 | 2.71 | 2.50E–08 | |||||
| PCBP1 | PCBP1 | 2.71 | 2.50E–08 | |||||
| RBM4B | RBM4B | 2.71 | 2.50E–08 | |||||
| RSSA | RPSA | 2.71 | 2.50E–08 | x | x | x | x | x |
| EIF3H | EIF3H | 2.69 | 3.10E–10 | |||||
| AHNK | AHNAK | 2.64 | 2.40E–07 | |||||
| HSP7E | HSPA14 | 2.64 | 2.40E–07 | |||||
| IF5 | EIF5 | 2.64 | 2.40E–07 | |||||
| SFRS6 | SFRS6 | 2.64 | 2.40E–07 | |||||
| TCPQ | CCT8 | 2.64 | 2.40E–07 | |||||
| ARP2 | ACTR2 | 2.62 | 1.40E–06 | |||||
| PURA | PURA | 2.62 | 1.40E–06 | |||||
| AMPL | LAP3 | 2.61 | 4.20E–09 | |||||
| CSTF1 | CSTF1 | 2.61 | 4.20E–09 | |||||
| ODB2 | DBT | 2.61 | 4.20E–09 | |||||
| RUVB1 | RUVBL1 | 2.61 | 4.20E–09 | x | x | x | ||
| CT007 | C20orf7 | 2.56 | 6.00E–05 | x | ||||
| OCAD1 | OCIAD1 | 2.42 | 2.80E–05 | |||||
| SYYC | YARS | 2.39 | 2.80E–05 | |||||
| FUBP2 | KHSRP | 2.29 | 0.00059 | |||||
| DNJC3 | DNAJC3 | 2.14 | 6.20E–07 | |||||
| GLSK | GLS | 2.14 | 6.20E–07 | |||||
| PRDX4 | PRDX4 | 2.14 | 9.50E–06 | x | x | |||
| PRP19 | PRPF19 | 2.14 | 6.20E–07 | |||||
| ARSA1 | ASNA1 | 2.1 | 3.80E–08 | x | ||||
| FKBP8 | FKBP8 | 2.03 | 6.30E–08 | x | x | x | ||
| HNRH1 | HNRNPH1 | 2.03 | 6.30E–08 | |||||
| TBB5 | TUBB | 2.03 | 6.30E–08 | x | x | |||
| CH60 | HSPD1 | 1.97 | 2.60E–05 | x | x | x | ||
| NONO | NONO | 1.97 | 2.60E–05 | |||||
| SF3A3 | SF3A3 | 1.97 | 2.60E–05 | |||||
| TBL1R | TBL1XR1 | 1.97 | 2.60E–05 | |||||
| TPR | TPR | 1.92 | 0.0006 | x | x | |||
| ACTB | ACTB | 1.9 | 2.60E–06 | |||||
| RANG | RANBP1 | 1.9 | 2.60E–06 | |||||
| ACON | ACO2 | 1.83 | 3.40E–05 | |||||
| EFG2 | GFM2 | 1.83 | 3.40E–05 | |||||
| IMMT | IMMT | 1.83 | 3.40E–05 | x | ||||
| MIRO2 | RHOT2 | 1.83 | 3.40E–05 | x | ||||
| PLOD3 | PLOD3 | 1.83 | 3.40E–05 | x | x | |||
| XRCC5 | XRCC5 | 1.83 | 3.40E–05 | x | ||||
| CORO1A | CORO1A | 1.82 | 2.80E–05 | x | x | x | ||
| DLDH | DLD | 1.82 | 2.80E–05 | |||||
| SCOT1 | OXCT1 | 1.82 | 2.80E–05 | |||||
| EIF3G | EIF3G | 1.81 | 6.00E–07 | |||||
| SCMC1 | SLC25A24 | 1.81 | 6.00E–07 | x | ||||
| TADBP | TARDBP | 1.81 | 6.00E–07 | |||||
| GNAQ | GNAQ | 1.75 | 2.50E–05 | x | x | x | x | x |
| IDH3B | IDH3B | 1.75 | 2.50E–05 | |||||
| UBFD1 | UBFD1 | 1.75 | 2.50E–05 | |||||
| RBBP7 | RBBP7 | 1.73 | 0.0056 | |||||
| ATPB | ATP5B | 1.65 | 4.90E–05 | x | ||||
| TPD52 | TPD52 | 1.65 | 0.00054 | x | ||||
| KAP2 | PRKAR2A | 1.62 | 0.00062 | x | ||||
| PDIA6 | PDIA6 | 1.62 | 0.00062 | x | ||||
| PCBP2 | PCBP2 | 1.56 | 3.80E–06 | |||||
SCLC, small-cell lung cancer.
Bold indicates candidate biomarkers.
Overrepresentation in SCLC of Proteins Involved in Cellular/Tissue Morphology Networks
Using IPA, we found that the 137 proteins differentially expressed in SCLC were overrepresented in five specific networks: (1) gene expression, cellular response to therapeutics, cellular assembly, and organization (score: 60); (2) protein synthesis, RNA post-transcriptional modification, gene expression (score: 49); (3) cell death, cellular assembly and organization, cellular function, and maintenance (score: 47); (4) carbohydrate metabolism, small molecule biochemistry, DNA replication, recombination, and repair (score: 29); and (5) hematological system development/function, humoral immune response, tissue morphology (score: 23). Proteins related to cellular assembly, organization, and morphology and tissue morphology (Table 3) were overrepresented in three of these five networks, the most interesting one being the cell death, cellular assembly, organization, function, and maintenance network (Supplementary figure 2, Supplemental Digital Content 3, http://links.lww.com/JTO/A531). Many proteins represented in these three networks were underexpressed in SCLC (Table 4). Among proteins represented in these top five networks and overexpressed in SCLC, we found proteins related to the nervous system (Table 5) and involved in inhibition of apoptosis and induction of proliferation and motility (Supplementary table 3, Supplemental Digital Content 1, http://links.lww.com/JTO/A529).
TABLE 3.
Proteins Related to Cellular Assembly, Organization, Morphology, and Tissue Morphology (Including Proteins Related to Nervous System) in the Top Five Networks in Ingenuity Pathway Analysis
| Protein Symbol | Function | p | Number of Proteins |
|---|---|---|---|
| ACTN4, HMGB1, HSP71, K1C18, PRDX2, PSA, PTBP1, ROAA, STMN1, TMOD3, TPD52, TPR, VIME, VINC | Cell morphology | 0.000079 | 14 |
| ACTB, ACTN4, ARP2, CALD1, CH60, CORO1A, CRC8, HMGB1, NPM, STMN1, TBB5, TMOD3, VIME, VINC, SYYC | Cell motility | 0.0362 | 15 |
| CAPG, CORO1A, CRC8, DPYL2, GNAQ, PTBP1, VINC | Shape change of cells | 0.0439 | 7 |
| CRC8, DPYL2 | Cell blebbing | 0.0123 | 2 |
| CAPG, VINC | Biogenesis of plasma membrane projections | 0.0273 | 2 |
| CALD1 | Quantity of podosomes | 0.00633 | 2 |
| CALD1, CRC8 | Formation of podosomes | 0.00633 | 2 |
| PTBP1, VINC | Cell spreading | 0.0412 | 2 |
| CALD1, GNAQ, VIME | Contraction of cells | 0.0314 | 3 |
| STMN1, VIME | Quantity of microtubules | 0.0123 | 2 |
| DPYL2, STMN1, TBB5 | Assembly of microtubules | 0.00468 | 3 |
| STMN1, TBB5 | Polymerization of microtubules | 0.0199 | 2 |
| RRBP1, TBB5 | Organization of microtubules | 0.0159 | 2 |
| CORO1A, K1C18, RRBP1, TBB5 | Organization of filaments | 0.0299 | 4 |
| CRC8, K1C18, STMN1 | Stabilization of filaments | 0.0131 | 3 |
| ACTN4 | Elongation of actin filaments | 0.0397 | 1 |
| K1C18, VIME | Quantity of intermediate filaments | 0.0397 | 1 |
| VIME | Assembly of intermediate filaments | 0.0397 | 1 |
| K1C18 | Organization and stabilization of keratin filaments | 0.0161 | 1 |
| VINC | Organization of zonula adherens, junctions and complexes integrity | 0.0161 | 1 |
| CALD1, PTBP1 | Size of focal adhesions | 0.00175 | 2 |
| HSP71 | Reformation of focal adhesions | 0.00806 | 1 |
| CRC8 | Formation of cell-matrix contacts, (de)branching of actin filaments | 0.0161 | 1 |
| RSSA | Cell adhesion | 0.0474 | 1 |
| GNAQ | Remodeling of neurons | 0.0474 | 1 |
| DPYL2 | Induction of axons and formation of supernumerary axons | 0.00806 | 1 |
| VIME | Loss of synapse | 0.0474 | 1 |
| FKBP8 | Morphology of neural tube | 0.024 | 1 |
| VINC | Development of spinal nerve | 0.00806 | 1 |
| STIP1 | Neuritogenesis of hippocampal neurons | 0.0397 | 1 |
ACTB, β actin; ACTN4, actinin alpha 4; ARP2, actin-related protein 2; CALD1, caldesmon 1; CAPG, capping protein (actin filament), gelsolin-like; CH60, 60 kDa chaperonin; COR1A, coronin 1A; CRC8, cortactin; DPYL2, dihydropyrimidinase-related protein 2; FKBP8, FK506-binding protein 8; GNAQ, guanine nucleotide–binding protein alpha-q; HMGB1, high mobility group box 1; HSP71, heat shock 70 kDa protein 1A; K1C18, cytokeratin 18; NPM, nucleophosmin; PRDX2, peroxiredoxin 2; PSA, puromycin-sensitive aminopeptidase; PTBP1, polypyrimidine tract–binding protein 1; ROAA, heterogeneous nuclear ribonucleoprotein A/B; RRBP1, ribosome-binding protein 1; RSSA, laminin receptor 1; STIP1, stress-induced-phosphoprotein 1; STMN1, stathmin 1; SYYC, tyrosyl-tRNA synthetase; TBB5, tubulin β5; TMOD3, tropomodulin 3; TPD52, tumor protein D52; TPR, translocated promoter region—to activated MET oncogene; VIME, vimentin; VINC, vinculin.
Bold indicates candidate biomarkers.
TABLE 4.
Proteins Overrepresented in the Cellular Assembly, Organization, Morphology, and Tissue Morphology Networks and Downregulated in Small-Cell Lung Cancer Cell Lines
| Protein Symbol | Function |
|---|---|
| ACTN4 | Elongation of actin filaments Cellular adhesion |
| CRC8 | Debranching and branching of actin filaments Cell blebbing Formation of podosomes and of cell-matrix contacts |
| K1C18 | Organization and stabilization of keratin filaments |
| TMOD3 | Cell morphology |
| VIME | Contraction of cells Quantity of microtubules Quantity and assembly of intermediate filaments |
| VINC | Organization of zonula adherens and adherens junctions Integrity of junctional complexes Formation of membrane projections and cell spreading |
ACTN4, actinin alpha 4; CRC8, cortactin; K1C18, cytokeratin 18; TMOD3, tropomodulin 3; VIME, vimentin; VINC, vinculin.
TABLE 5.
Proteins Related to the Nervous System in the Top Five Networks in Ingenuity Pathway Analysis
| Protein Symbol | Function | p |
|---|---|---|
| GNAQ | Remodeling of neurons | 0.0474 |
| DPYL2 | Induction of axons and formation of supernumerary axons | 0.00806 |
| VIME | Loss of synapse | 0.0474 |
| FKBP8 | Morphology of neural tube | 0.024 |
| VINC | Development of spinal nerve | 0.00806 |
| STIP1 | Neuritogenesis of hippocampal neurons | 0.0397 |
DPYL2, dihydropyrimidinase-related protein 2; FKBP8, FK506-binding protein 8; GNAQ, guanine nucleotide–binding protein alpha-q; STIP1, stress-induced-phosphoprotein 1; VIME, vimentin; VINC, vinculin.
Bold indicates candidate biomarkers.
The top biological functions of the proteins differentially expressed in SCLC were also determined using IPA (Supplementary table 4, Supplemental Digital Content 1, http://links.lww.com/JTO/A529). Consistent with the network analysis, we found that the functions of these proteins were related to the cytoskeleton, morphology, and interactions with the environment and to the neuroendocrine system, cell cycle, growth, apoptosis, and metabolism.
Selection of DPYL2, GNAQ, RSSA, RUVB1, and STMN1 as SCLC Candidate Biomarkers
To select SCLC candidate biomarkers among the 137 proteins differentially expressed in SCLC, we used three criteria: (1) proteins overexpressed in SCLC (N = 81), (2) confirmed membrane-associated (N = 32) (including 10 plasma membrane, 12 endomembrane, and 10 MAPs), and (3) with functions relevant to cancer progression and/or neuroendocrine phenotype and/or part of networks related to cell/tissue morphology (Table 1). Five proteins met these criteria: DPYL2, GNAQ, RSSA, RUVB1, and STMN1 (main functions reported on Table 2).
TABLE 2.
Subcellular Location and Functions of the Final Candidate Biomarkers
| Protein Symbol | Subcellular Location | Interesting Functions for SCLC Progression |
|---|---|---|
| STMN1 | Cytosolic MAP | Role in cell cycle progression and migration through microtubule depolymerization Possible role in axon formation during neurogenesis and neuron polarization Overexpression plays a role in tumor progression in various cancers, and contributes to chemoresistance |
| DPYL2 | PMP and cytosolic MAP | Role in neuronal differentiation, axon guidance, neuronal growth, and cell migration in developing nervous system Associated with various neuropathologic or psychiatric disorders but not with cancer |
| RSSA | PMP (mainly), cytosolic and nuclear | Laminin receptor, role in cell adhesion to basal membrane and activation of signaling transduction pathways Overexpression correlated with invasive phenotype in various cancers |
| RUVB1 | Nuclear (mainly) and PMP | AAA+ ATPase interacting with several transcription factors involved in carcinogenesis Role in tumor invasion by binding plasminogen at cell surface and increasing its activation into plasmin Overexpression observed in many cancers |
| GNAQ | PMP and cytosolic MAP | Couples cell surface transmembrane receptors to intracellular signaling pathways, including the MAPK pathway Activating mutations described in melanoma |
MAP, membrane-associated protein; PMP, plasma membrane protein; SCLC, small-cell lung cancer.
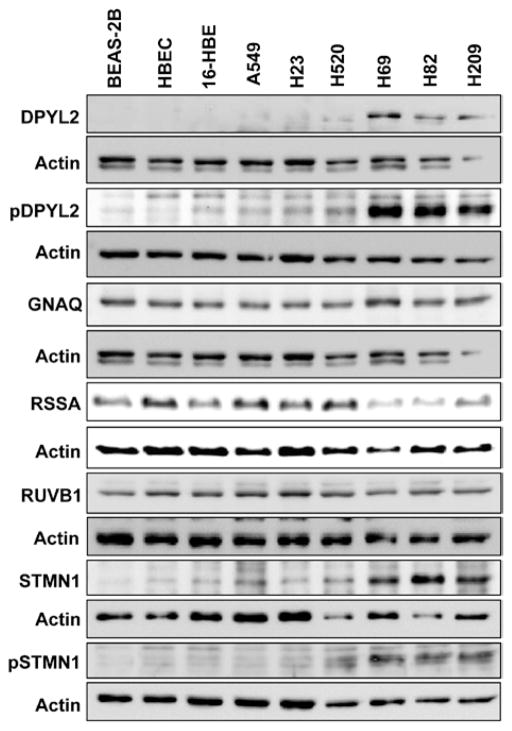
To verify the expression of our candidates, we analyzed the MAP lysates initially used for DIGE by WB. Although there was no expression difference among SCLC, NSCLC, and INBE cells for RSSA and RUVB1, we confirmed DPYL2, GNAQ, and STMN1 overexpression in SCLC (Fig. 3A). Moreover, we found phospho-DPYL2 (Thr514/Ser518) and phospho-STMN1 (Ser16) overexpression in SCLC, suggesting the activation of DPYL2 and STMN1 in SCLC (Fig. 3).
FIGURE 3.
Verification of the candidate biomarkers’ expression by Western blotting. Membrane-associated protein lysates were resolved with sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blots were incubated with an antibody against DPYL2, phospho-DPYL2 (Thr514/Ser518), GNAQ, RSSA, RUVB1, STMN1, or phospho-STMN1 (Ser16). Expression levels were normalized to actin. Overexpression of DPYL2, GNAQ, and STMN1 was observed in SCLC compared with immortalized normal bronchial epithelial (normal) and NSCLC cell lines, while RSSA and RUVB1 expression was similar in all the cell lines. Increased phosphorylation of DPYL2 and STMN1 was also observed in SCLC compared with normal and NSCLC cell lines, suggesting an activation of DPYL2 and STMN1 in SCLC. DPYL2, dihydropyrimidinase-related protein 2; GNAQ, guanine nucleotide binding protein alpha-q; NSCLC, non small-cell lung cancer; RSSA, laminin receptor 1; RUVB1, pontin; SCLC, small-cell lung cancer; STMN1, stathmin 1.
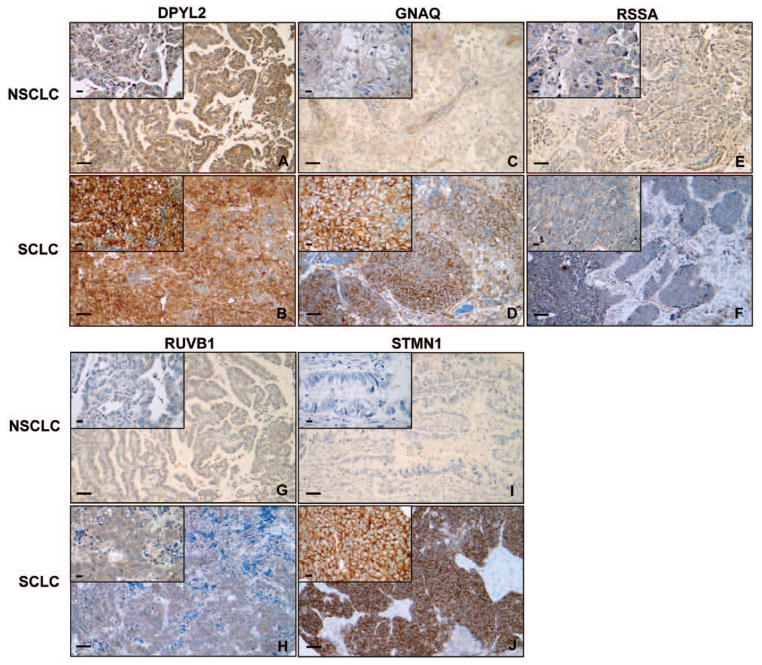
Next, we performed immunohistochemistry (IHC) on TMAs made of 100 NSCLCs and 36 SCLCs. Protein expression was membranous and cytosolic for DPYL2, GNAQ, and STMN1 and cytosolic for RSSA and RUVB1. Median staining scores in NSCLC and SCLC, respectively, were 180 (range, 50–300) and 300 (range, 60–300) for DPYL2 (p < 0.001), 80 (range, 0–300) and 250 (range, 30–300) for GNAQ (p < 0.001), 100 (range, 0–300) and 150 (range, 0–300) for RSSA (p = 0.051), 28 (range, 0–300) and 180 (range, 0–200) for RUVB1 (p < 0.001), and 0 (range, 0–240) and 240 (range, 0–300) for STMN1 (p < 0.001). Thus, IHC on TMAs verified overexpression in SCLC of all candidate biomarkers but RSSA. STMN1 was strongly expressed in SCLCs, whereas it was absent in most NSCLCs. Representative images of NSCLC and SCLC tissues with staining scores close to average staining scores for each biomarker in each histological subgroup are displayed in Figure 4.
FIGURE 4.
Verification of the candidate biomarkers’ expression by immunohistochemistry: representative cases of 130 studied NSCLC and 36 studied SCLC. Tissue sections were incubated with an antibody against DPYL2, GNAQ, RSSA, RUVB1, or STMN1. A J, Representative images of NSCLC and SCLC tissues with a staining score close to the average staining score for each biomarker in each histological subgroup. A, NSCLC tumor with a DPYL2 staining score of 200. B, SCLC tumor with a DPYL2 staining score of 300. C, NSCLC tumor with a GNAQ staining score of 100. D: SCLC tumor with a GNAQ staining score of 300. E, NSCLC tumor with an RSSA staining score of 100. F, SCLC tumor with an RSSA staining score of 100. G, NSCLC tumor with an RUVB1 staining score of 100. H, SCLC tumor with an RUVB1 staining score of 200. I, NSCLC tumor with an STMN1 staining score of 0. J, SCLC tumor with an STMN1 staining score of 300 (magnification: ×10 in the large windows and ×40 in the small windows; scale bars: 100 μm in the large windows and 20 μm in the small windows). DPYL2, dihydropyrimidinase-related protein 2; GNAQ, guanine nucleotide binding protein alpha-q; NSCLC, non small-cell lung cancer; RSSA, laminin receptor 1; RUVB1, pontin; SCLC, small-cell lung cancer; STMN1, stathmin 1.
Correlation of the Candidate Biomarkers’ Expression with Clinical Characteristics
Results of the correlations between the candidate biomarkers’ staining scores and various clinical characteristics in univariable analyses are presented in Table 6. In these univariable analyses, staining scores were significantly higher in SCLC than in NSCLC for DPYL2, GNAQ, RUVB1, and STMN1 (p < 0.001). OS was significantly shorter in the presence of higher staining scores for DPYL2 (p = 0.022), GNAQ (p = 0.026), RUVB1 (p = 0.024), and STMN1 (p = 0.001). However, these were not associated with OS after tumors were broken by histology.
TABLE 6.
Patient Characteristics and Statistical Significance of the Association with DPYL2, GNAQ, RSSA, RUVB1, and STMN1 Staining Scores in Univariables Analyses
| n (%) | IQR | 95% CI |
p
|
|||||
|---|---|---|---|---|---|---|---|---|
| DPYL2 | GNAQ | RSSA | RUVB1 | STMN1 | ||||
| Sex | ||||||||
| Male | 78 (57) | 0.583 | 0.236 | 0.22 | 0.211 | 0.258 | ||
| Female | 58 (43) | |||||||
| Race | ||||||||
| Caucasian | 126 (93) | 0.240 | 0.222 | 0.422 | 0.949 | 0.857 | ||
| African American | 10 (7) | |||||||
| Age at diagnosis | ||||||||
| Median, yr | 67 (58–73) | 0.358 | 0.710 | 0.278 | 0.646 | 0.897 | ||
| ≤60 | 45 (33) | 0.740 | 0.475 | 0.760 | 0.713 | 0.852 | ||
| >60 | 91 (67) | |||||||
| Smoking status | ||||||||
| Current smoker | 45 (33) | 0.689 | 0.408 | 0.633 | 0.059 | 0.071 | ||
| Ex smoker | 85 (63) | |||||||
| Never smoker | 5 (4) | |||||||
| Smoking history | ||||||||
| Median pack year | 50 (35–75) | 0.301 | 0.163 | 0.659 | 0.643 | 0.231 | ||
| Lung cancer histology | ||||||||
| NSCLC | 100 (74) | <0.001 | <0.001 | 0.051 | <0.001 | <0.001 | ||
| SCLC | 36 (26) | |||||||
| NSCLC histology | ||||||||
| ADC | 49 (36) | 0.006 | <0.001 | 0.541 | 0.947 | 0.184 | ||
| SqCC | 42 (31) | |||||||
| SCLC path stage | ||||||||
| Limited | 24 (67) | 0.262 | 0.687 | 0.378 | 0.309 | 0.240 | ||
| Extensive | 12 (33) | |||||||
| NSCLC path stage | ||||||||
| I | 65 (66) | 0.072 | 0.126 | 0.209 | 0.813 | 0.009 | ||
| II | 13 (13) | |||||||
| III | 17 (17) | |||||||
| IV | 3 (3) | |||||||
| Overall survival | ||||||||
| Median, yr | 2.7 (1.9–4.0) | 0.022 | 0.026 | 0.651 | 0.024 | 0.001 | ||
| Alive at 3 yr | 66 (49) | 0.594 | 0.072 | 0.392 | 0.189 | 0.017 | ||
| SCLC overall survival | ||||||||
| Median, yr | 1.1 (0.7–2.3) | 0.466 | 0.427 | 0.643 | 0.778 | 0.705 | ||
| Alive at 3 yr | 9 (25) | 0.779 | 0.102 | 0.955 | 0.576 | 0.415 | ||
| NSCLC overall survival | ||||||||
| Median, yrs | 4.0 (2.6–5.6) | 0.134 | 0.763 | 0.312 | 0.947 | 0.801 | ||
| Alive at 3 yrs | 57 (57) | 0.139 | 0.64 | 0.103 | 0.624 | 0.863 | ||
| ADC overall survival | ||||||||
| Median, yr | 4.4 (2.5–5.8) | 0.295 | 0.926 | 0.678 | 0.543 | 0.642 | ||
| Alive at 3 yr | 31 (63) | 0.502 | 0.610 | 0.848 | 0.922 | 0.749 | ||
| SqCC overall survival | ||||||||
| Median, yr | 3.9 (2.4–5.7) | 0.147 | 0.855 | 0.146 | 0.504 | 0.862 | ||
| Alive at 3 yr | 23 (55) | 0.082 | 0.345 | 0.017 | 0.176 | 0.946 | ||
ADC, adenocarcinoma; CI, confidence interval; DPYL2, dihydropyrimidinase-related protein 2; GNAQ, guanine-nucleotide-binding protein alpha-q; IQR, interquartile range; NSCLC, non–small-cell lung carcinoma; RSSA, laminin receptor 1; RUVB1, pontin; SCLC, smallcell lung carcinoma; SqCC, squamous cell carcinoma; STMN1, stathmin 1.
Bold indicates statistically significant P values.
In multivariable analyses including each biomarker’s average staining score, lung cancer histology, disease stage, age, sex, and smoking status, we did not confirm the association between staining scores and OS (Table 7). However, age was an independent predictor of OS (nonlinear trend) for all biomarkers. For the subset of 36 SCLCs, we did not find a significant association between staining scores and OS in multivariable analyses including each biomarker’s average staining score, disease stage, and age (Table 8).
TABLE 7.
Multivariable Analyses for the Association of Candidate Biomarkers’ Staining Scores with Overall Survival for the Whole Data Set, Including Non Small-Cell and Small-Cell Lung Cancers (n = 136)
|
p
|
|||||
|---|---|---|---|---|---|
| DPYL2 | GNAQ | RSSA | RUVB1 | STMN1 | |
| Average staining score | 0.0881 | 0.5177 | 0.1500 | 0.7983 | 0.6491 |
| Lung cancer histology | 0.0047 | 0.1720 | 0.0046 | 0.0540 | 0.1711 |
| Disease stage | 0.0962 | 0.1038 | 0.0943 | 0.0647 | 0.0760 |
| Age | 0.0375 | 0.0328 | 0.0175 | 0.0380 | 0.0280 |
| Sex (male vs. female) | 0.0604 | 0.2517 | 0.0515 | 0.0984 | 0.1114 |
| Smoking status (ex vs. current) | 0.8785 | 0.9432 | 0.7353 | 0.9380 | 0.8702 |
DPYL2, dihydropyrimidinase-related protein 2; GNAQ, guanine nucleotide–binding protein alpha-q; RSSA, laminin receptor 1; RUVB1, pontin; STMN1, stathmin 1.
Bold indicates statistically significant P values.
TABLE 8.
Multivariable Analyses for the Association of Candidate Biomarkers’ Staining Scores with Overall Survival for the Subset of Small-Cell Lung Cancers (n = 36)
|
p
|
|||||
|---|---|---|---|---|---|
| DPYL2 | GNAQ | RSSA | RUVB1 | STMN1 | |
| Average staining score | 0.3468 | 0.0795 | 0.9800 | 0.8892 | 0.3109 |
| Disease stage | 0.4081 | 0.2269 | 0.2836 | 0.2329 | 0.1399 |
| Age | 0.0251 | 0.0111 | 0.0349 | 0.0926 | 0.0459 |
DPYL2, dihydropyrimidinase-related protein 2; GNAQ, guanine nucleotide–binding protein alpha-q; RSSA, laminin receptor 1; RUVB1, pontin; STMN1, stathmin 1.
Bold indicates statistically significant P values.
DISCUSSION
The main objective of this study was to test the hypothesis that the identification of MAPs specific to SCLC by DIGE may advance our understanding of SCLC biology and lead to the discovery of new candidate biomarkers of SCLC. To do so, we analyzed a membrane-associated subproteome in three SCLC, three NSCLC, and three INBE cells by DIGE and validated these results in primary SCLCs.
DIGE is a proteomic tool widely used to study cancer biology. In a previous study, DIGE analysis of 30 lung cancer cell lines including SCLCs, SqCCs, and ADCs led to the discovery of a 32-protein spots signature able to distinguish between the three histological types, further validated in tissues (29 NSCLCs and 1 SCLC).6 Other DIGE studies found proteins overexpressed in SCLC when compared with control cells13 or normal bronchial epithelium tissues.14 Only one DIGE study specifically analyzed MAPs in the SCLC cell line H69 and its doxorubicin-resistant subcell line H69AR and found, in H69AR, upregulation of Serca2, a calcium pump located in the endoplasmic reticulum membrane.15
In our DIGE analysis, replicates from each cell line clustered very closely, demonstrating low technical noise and high reproducibility in membrane fractionation. Samples within each histological group also clustered very closely, indicating that many variations were descriptive of differences between classifications. Further analyses revealed 135 distinct protein features differentially expressed in SCLC when compared with NSCLC and INBE cells, including 137 proteins. These were overrepresented in the cellular assembly, organization, morphology, and tissue morphology networks. Among proteins represented in these networks and underexpressed in SCLC, many were involved in cell–cell and cell–extracellular matrix interactions. This was consistent with the nonadherent phenotype that many SCLC cells have in vitro (including these in this study). Among proteins part of these networks and overexpressed in SCLC, there were proteins involved in nerve/brain development and endocrine functions, according to the neuroendocrine nature of SCLC cells and previous proteomic studies.13 Many proteins were involved in cell cycle, growth, apoptosis, and metabolism, which were also consistent with SCLC aggressive behavior. DPYL2, GNAQ, RSSA, RUVB1, and STMN1 were selected as candidate biomarkers among MAPs overexpressed in SCLC and with cellular functions relevant to cancer progression (all of them) and/or the neuroendocrine phenotype (GNAQ, DPYL2, and STMN1), and/or part of networks related to cell/tissue morphology (all of them but RUVB1). Our ultimate goal was to find molecular tools to move our discoveries toward SCLC molecular imaging or targeted therapies. Interestingly, overexpression of DPYL2, GNAQ, RUVB1, and STMN1 in SCLC was verified by WB and/or IHC on TMAs, supporting their potential as targets in SCLC.
In our DIGE analysis, STMN1 was the most significantly overexpressed protein in SCLC cell lines. By IHC, STMN1 was also strongly expressed in most SCLC tissues, while the expression was absent in most NSCLCs and very low in the remaining minority. Moreover, in IPA, STMN1 was part of molecular networks related to cellular/tissue morphology. STMN1 is a membrane-associated phosphoprotein playing a role in cell cycle progression and migration through microtubule depolymerization.16,17 STMN1 phosphorylation at Ser16 may also be required for axon formation during neurogenesis and neuron polarization, which is relevant for cells with neuroendocrine differentiation. STMN1 overexpression plays a role in tumor progression and metastasis in various cancers and contributes to chemoresistance.18 Therefore, STMN1 appears as an attractive therapeutic target candidate. Recent integrative genome analyses showed high STMN1 gene expression levels in 15 SCLC tumors,19 confirming our data adequately. Although STMN1 protein expression was absent in most of the NSCLC tissues we tested, previous studies demonstrated STMN1 overexpression in lung ADC and SqCC.20,21 However, consistent with our results, other studies described higher STMN1 expression in SCLC than NSCLC.22,23 Moreover, by WB, we showed higher STMN1 phosphorylation in SCLC than NSCLC and INBE cells, suggesting the activation of STMN1 in SCLC and a potential role in SCLC progression. STMN1 expression in NSCLC and SCLC was not found to be an independent predictor of OS. This is in contrast with observations made in other cancers18 and may be explained by the relative small number of SCLCs studied here (N = 36).
DPYL2 plays a role in neuronal differentiation, axon guidance, neuronal growth, and cell migration in the developing nervous system,24 which is once again relevant for SCLC. DPYL2 has been associated with various neuropathologic or psychiatric disorders24 but never with cancer, even though the gene is located in 8p, an oncogene- and tumor suppressor gene-rich region.25 This is therefore the first report of its overexpression in SCLC. Moreover, we demonstrated higher DPYL2 phosphorylation in SCLC than NSCLC and INBE cells, suggesting DPYL2 activation in SCLC and its possible role in SCLC progression.
GNAQ is a membrane and membrane-bound protein from the family of guanine nucleotide–binding proteins that couple cell surface transmembrane receptors to intracellular signaling pathways, including the mitogen activated protein kinase (MAPK) pathway. Activating mutations of GNAQ have been described in melanoma, rendering them potentially susceptible to mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase (a kinase in the MAPK pathway) inhibition26,27 but have not been found in other tumors and particularly not in lung cancer.28 GNAQ overexpression in SCLC has never been reported before and deserves further investigation.
Finally, RUVB1 is an AAA+ ATPase interacting with several transcription factors involved in carcinogenesis, such as c-MYC.29 RUVB1 localized at cell surface may also be able to bind plasminogen and increase its activation into plasmin,30 which plays a role in tumor invasion. RUVB1 overexpression has been observed in many cancers, including NSCLC.31 However, this is the first report of RUVB1 overexpression in SCLC, especially when compared with NSCLC. Although RUVB1 expression has been correlated with poor outcome in colorectal carcinoma,32 we did not confirm this correlation with OS in our study. Small molecule inhibitors of RUVB1 have recently been developed, are commercially available,33 and will facilitate testing the functional implications of RUVB1.
Among the five SCLC candidate biomarkers we selected by DIGE, the only one for which we did not verify the overexpression in SCLC when compared with NSCLC was RSSA, a laminin receptor mainly localized at the surface of the plasma membrane. RSSA plays a role in cell adhesion to the basal membrane and the consequent activation of signaling transduction pathways. Its overexpression in many cancers,34–36 including NSCLC and SCLC,37 has already been described. A correlation between RSSA upregulation in cancer cells and their invasive phenotype has also been reported.38 Consistently, the expression of RSSA has been found to be higher in SCLC than in NSCLC37 and to be a predictor of poor prognosis in many cancers, including NSCLC.39 However, the higher expression of RSSA in SCLC than in NSCLC and the prognostic value of RSSA were not verified in our study. These discordant results may be related to differences in methodologies and antibodies used to evaluate RSSA expression or to sample size issues.
We specifically focused on MAPs because of their potential as diagnostic or therapeutic target. Yet, cell surface proteins are often poorly represented in two-dimensional gels due to their hydrophobic nature and high molecular weight.40 Therefore, this study probably underestimated the number of MAPs important for SCLC progression. By restricting our study to MAPs, we omitted proteins potentially relevant to SCLC biology. Moreover, potentially relevant proteins with high expression level did not fulfil our three selection criteria, leaving aside other potential candidate biomarkers (available in Table 1). Another limitation of the study is the small number of cell lines used for the DIGE analysis, although the results were validated in a larger group of tumors from patients with SCLC and NSCLC. Of note, the mutation status of the cell lines may have limited the generalizability of the findings. We therefore searched the Sanger Center database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) for the mutation status of well-known oncogenes or tumor suppressor genes in the cell lines we used. We found that none of them had AKT1, EGFR, v-Raf Murine sarcoma viral oncogene homolog B, MAP2K4, MET, Myc, or NRAS mutation, while KRAS mutation was found in NCI-H23, PIK3CA mutation in NCI-H69, PTEN mutation in NCI-H23, retinoblastoma 1 mutation in all the three SCLC cell lines, and TP53 mutation in NCI-H69, NCI-H209, NCI-H23, and NCI-H520. To the exception of retinoblastoma 1 mutation found in all the SCLC cell lines (reported in >90% of SCLCs), no other mutation was found in every cell line of the same histological subgroup.
In summary, using DIGE coupled with tandem MS to analyze a membrane-associated subproteome, we identified 137 proteins differentially expressed in SCLC compared with NSCLC and INBE cells. These proteins were enriched for cellular/tissue morphology networks. DPYL2, GNAQ, RUVB1, and STMN1 overexpression in SCLC was verified by WB and/or IHC, suggesting that these results need to be tested for functional implication in SCLC progression. The association with survival requires further validation in larger clinical data sets.
Supplementary Material
Acknowledgments
The authors thank Kathy Taylor, Director of the Research Institute at St. Thomas Health Services, Nashville, TN, for sharing archived SCLC tissue blocks. This work was supported by a Merit Review Grant from the Department of Veterans Affairs and a SPORE in Lung Cancer (CA90949), USA (Pierre P. Massion), and a Télévie Grant from the Fonds National de la Recherche Scientifique (FNRS), Belgium (Sebahat Ocak).
Dr. Ocak was supported by a Fondation Mont-Godinne Grant, Belgium, and a Clinician-Researcher Mandate from Secteur des Sciences de la Santé, Université Catholique de Louvain (UCL), Belgium.
Footnotes
Disclosure: The authors declare no conflict of interest.
References
- 1.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 2.Ocak S, Chaurand P, Massion PP. Mass spectrometry-based proteomic profiling of lung cancer. Proc Am Thorac Soc. 2009;6:159–170. doi: 10.1513/pats.200809-108LC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ocak S, Sos ML, Thomas RK, Massion PP. High-throughput molecular analysis in lung cancer: insights into biology and potential clinical applications. Eur Respir J. 2009;34:489–506. doi: 10.1183/09031936.00042409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfonso P, Núñez A, Madoz-Gurpide J, Lombardia L, Sánchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602–2611. doi: 10.1002/pmic.200401196. [DOI] [PubMed] [Google Scholar]
- 5.Evans CA, Tonge R, Blinco D, et al. Comparative proteomics of primitive hematopoietic cell populations reveals differences in expression of proteins regulating motility. Blood. 2004;103:3751–3759. doi: 10.1182/blood-2003-09-3294. [DOI] [PubMed] [Google Scholar]
- 6.Seike M, Kondo T, Fujii K, et al. Proteomic signatures for histological types of lung cancer. Proteomics. 2005;5:2939–2948. doi: 10.1002/pmic.200401166. [DOI] [PubMed] [Google Scholar]
- 7.Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 8.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 9.Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multivariable difference gel electrophoresis and mass spectrometry: a case study on transforming growth factor-beta and ERBB2 signaling. Mol Cell Proteomics. 2007;6:150–169. doi: 10.1074/mcp.D600001-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 11.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 12.Massion PP, Taflan PM, Jamshedur Rahman SM, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63:7113–7121. [PubMed] [Google Scholar]
- 13.Ziv T, Barnea E, Segal H, et al. Comparative proteomics of small cell lung carcinoma. Cancer Biomark. 2006;2:219–234. doi: 10.3233/cbm-2006-2601. [DOI] [PubMed] [Google Scholar]
- 14.Jeong HC, Kim GI, Cho SH, et al. Proteomic analysis of human small cell lung cancer tissues: up-regulation of coactosin-like protein-1. J Proteome Res. 2011;10:269–276. doi: 10.1021/pr100714b. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson H, Lengqvist J, Hedlund J, et al. Quantitative membrane proteomics applying narrow range peptide isoelectric focusing for studies of small cell lung cancer resistance mechanisms. Proteomics. 2008;8:3008–3018. doi: 10.1002/pmic.200800174. [DOI] [PubMed] [Google Scholar]
- 16.Gavet O, Ozon S, Manceau V, Lawler S, Curmi P, Sobel A. The stathmin phosphoprotein family: intracellular localization and effects on the microtubule network. J Cell Sci. 1998;111 (Pt 22):3333–3346. doi: 10.1242/jcs.111.22.3333. [DOI] [PubMed] [Google Scholar]
- 17.Niethammer P, Bastiaens P, Karsenti E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–1866. doi: 10.1126/science.1094108. [DOI] [PubMed] [Google Scholar]
- 18.Nemunaitis J. Stathmin 1: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets. 2012;16:631–634. doi: 10.1517/14728222.2012.696101. [DOI] [PubMed] [Google Scholar]
- 19.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Wang H, Gharib TG, et al. Overexpression of oncoprotein 18 correlates with poor differentiation in lung adenocarcinomas. Mol Cell Proteomics. 2003;2:107–116. doi: 10.1074/mcp.M200055-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Singer S, Malz M, Herpel E, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res. 2009;69:2234–2243. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
- 22.Pisanu S, Ghisaura S, Pagnozzi D, et al. The sheep milk fat globule membrane proteome. J Proteomics. 2011;74:350–358. doi: 10.1016/j.jprot.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Tanca A, Addis MF, Pagnozzi D, et al. Proteomic analysis of formalin-fixed, paraffin-embedded lung neuroendocrine tumor samples from hospital archives. J Proteomics. 2011;74:359–370. doi: 10.1016/j.jprot.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Hensley K, Venkova K, Christov A, Gunning W, Park J. Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications. Mol Neurobiol. 2011;43:180–191. doi: 10.1007/s12035-011-8166-4. [DOI] [PubMed] [Google Scholar]
- 25.Tabarés-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Küsters-Vandevelde HV, Klaasen A, Küsters B, et al. Activating mutations of the GNAQ gene: a frequent event in primary melanocytic neoplasms of the central nervous system. Acta Neuropathol. 2010;119:317–323. doi: 10.1007/s00401-009-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano E, Schwartz GK, Chapman PB, Wolchock JD, Carvajal RD. Treatment implications of the emerging molecular classification system for melanoma. Lancet Oncol. 2011;12:913–922. doi: 10.1016/S1470-2045(10)70274-6. [DOI] [PubMed] [Google Scholar]
- 28.Lamba S, Felicioni L, Buttitta F, et al. Mutational profile of GNAQQ209 in human tumors. PLoS One. 2009;4:e6833. doi: 10.1371/journal.pone.0006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellosta P, Hulf T, Balla Diop S, et al. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci U S A. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawley SB, Tamura T, Miles LA. Purification, cloning, and characterization of a profibrinolytic plasminogen-binding protein, TIP49a. J Biol Chem. 2001;276:179–186. doi: 10.1074/jbc.M004919200. [DOI] [PubMed] [Google Scholar]
- 31.Dehan E, Ben-Dor A, Liao W, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175–184. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Lauscher JC, Elezkurtaj S, Dullat S, et al. Increased Pontin expression is a potential predictor for outcome in sporadic colorectal carcinoma. Oncol Rep. 2012;28:1619–1624. doi: 10.3892/or.2012.1968. [DOI] [PubMed] [Google Scholar]
- 33.Elkaim J, Castroviejo M, Bennani D, et al. First identification of small-molecule inhibitors of Pontin by combining virtual screening and enzymatic assay. Biochem J. 2012;443:549–559. doi: 10.1042/BJ20111779. [DOI] [PubMed] [Google Scholar]
- 34.Cioce V, Castronovo V, Shmookler BM, et al. Increased expression of the laminin receptor in human colon cancer. J Natl Cancer Inst. 1991;83:29–36. doi: 10.1093/jnci/83.1.29. [DOI] [PubMed] [Google Scholar]
- 35.Ménard S, Castronovo V, Tagliabue E, Sobel ME. New insights into the metastasis-associated 67 kD laminin receptor. J Cell Biochem. 1997;67:155–165. [PubMed] [Google Scholar]
- 36.Ménard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Res Treat. 1998;52:137–145. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- 37.Satoh K, Narumi K, Isemura M, et al. Increased expression of the 67kDa-laminin receptor gene in human small cell lung cancer. Biochem Biophys Res Commun. 1992;182:746–752. doi: 10.1016/0006-291x(92)91795-r. [DOI] [PubMed] [Google Scholar]
- 38.Wewer UM, Taraboletti G, Sobel ME, Albrechtsen R, Liotta LA. Role of laminin receptor in tumor cell migration. Cancer Res. 1987;47:5691–5698. [PubMed] [Google Scholar]
- 39.Fontanini G, Vignati S, Chiné S, et al. 67-Kilodalton laminin receptor expression correlates with worse prognostic indicators in non-small cell lung carcinomas. Clin Cancer Res. 1997;3:227–231. [PubMed] [Google Scholar]
- 40.Shin BK, Wang H, Yim AM, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.