Abstract
Phenotype-based screening of bacterial metagenomic libraries provides an avenue for the discovery of novel genes, enzymes and metabolites that have a variety of potential clinical and industrial uses. Here we report the identification of a functionally diverse collection of antibacterially active enzymes from the phenotypic screening of 700,000 cosmid clones prepared from Arizona soil DNA and hosted in Ralstonia metallidurans. Environmental DNA clones surrounded by zones of growth inhibition in a bacterial overlay assay were found, through bioinformatics and functional analyses, to encode enzymes with predicted peptidase, lipase and glycolytic activities conferring antibiosis. The antibacterial activities observed in our R. metallidurans-based assay could not be replicated with the same clones in screens using Escherichia coli as a heterologous host, suggesting that the large-scale screening of metagenomic libraries for antibiosis using phylogenetically diverse hosts should be a productive strategy for identifying enzymes with functionally diverse antibacterial activities.
Introduction
Analyses of bacterial culture broths have traditionally been a route for the discovery of novel small molecules and enzymes (Demain & Sanchez, 2009; Trincone, 2011). While productive, these studies are limited by our inability to culture the vast majority of bacteria from the environment. The metabolomes and proteomes of a more representative sample of environmental bacteria can be accessed using functional metagenomic approaches that involve the extraction of DNA directly from environmental samples (environmental DNA, eDNA), the cloning of this DNA into model cultured bacteria and, finally, the phenotypic screening of these clones in diverse assays (Handelsman et al., 1998; Iqbal et al., 2012).
To date, the majority of metagenomic library screens targeting antibiosis have relied on top agar overlay assays on Escherichia coli-based libraries to identify antibacterially active small molecules [Figure 1]. Large-scale functional screens using E. coli-based metagenomic libraries have seldom reported the discovery of heterologously expressed antibacterially active enzymes, despite the fact that cultured bacteria have been a prolific source of antimicrobial proteins (Veiga-Crespo et al., 2007; Thallinger et al., 2013). Phage endolysins, in particular, have recently garnered attention for their potential roles in enzyme-based antibiotic therapies (Fischetti, 2010; Thallinger et al., 2013).
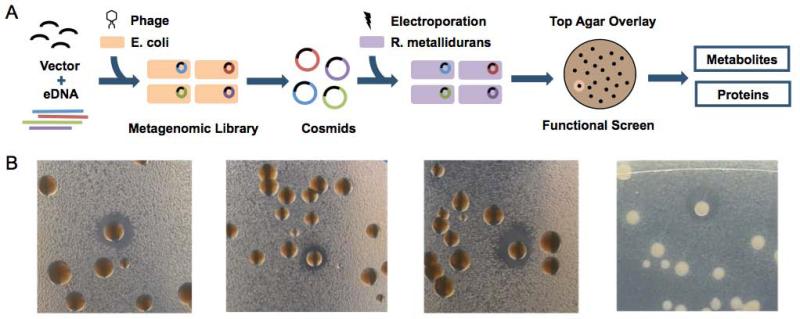
Figure 1.
(A) Overview of metagenomic library construction and screening methodology. In this study, eDNA extracted from soil samples was ligated to a shuttle cosmid vector and introduced into E. coli by phage transduction. DNA prepared from these libraries was electroporated into R. metallidurans and screened for antibiosis using the top agar overlay method. (B) Representative antibacterially active hits obtained by screening soil metagenomic libraries hosted in R. metallidurans using the top agar overlay method. R. metallidurans-based libraries were arrayed onto LB-agar plates and overlaid with a layer of top agar containing a B. subtilis assay strain; zones of growth inhibition around individual colonies indicate heterologous expression of the antibacterial phenotype.
While most metagenomic functional screening has used E. coli as a host, the utility of E. coli utility as a heterologous host in functional metagenomic screens is likely to be limited due to its limited heterologous expression capacity (Gabor et al., 2004). Since the success of functional metagenomic screening is contingent upon the ability of the library host to heterologously express genes found on foreign eDNA, the identification of transcriptionally diverse model hosts that can express these foreign genes will likely be critical to the overall success of metagenomic screening strategies. We hypothesized that by changing the host used in phenotypic metagenomic library screens it might be possible to begin to identify the diverse antibacterial enzymes that are undoubtedly encoded within soil microbiomes.
Here we show that enzymes conferring antibiosis can be found in R. metallidurans hosted soil DNA libraries. R. metallidurans is a gram negative beta proteobacteria that we have explored as an alternative host for small molecule-based functional metagenomic studies because of its previously described heterologous expression capabilities, its genetic tractability and the ease with which it can be grown in the laboratory (Craig et al., 2010). Based on recent interest in lytic enzyme-based antibiotic therapies, the large-scale screening of environmental DNA libraries hosted in R. metallidurans may be a productive strategy for identifying enzymes with diverse activities for potential use as novel therapeutics (Fischetti, 2010; Thallinger et al., 2013).
Materials and Methods
eDNA Library Construction
DNA extracted from soil collected in the Sonoran Desert of Arizona (USA) was used to create a 700,000-membered cosmid-based metagenomic library in E. coli (Brady, 2007). To obtain crude eDNA, 250 g of soil was passed through a 1/8 inch sieve to remove rocks and large debris, and then incubated in lysis buffer (100 mM Tris-HCl, 100 mM NaEDTA, 1.5 M NaCl, 1% (w/v) cetyl trimethyl ammonium bromide, 2% (w/v) SDS, pH 8.0) (1:1 wt:vol) at 70°C for 2 hours (Zhou et al., 1996). Heat lysed samples were centrifuged (4,000 g, 10 min) to remove soil particulates. Crude eDNA was precipitated from the supernatant by the addition of 0.7 volume isopropanol and collected by centrifugation (4,000 g, 10 min, 4°C). The pellet was washed with 70% ethanol and the eDNA resuspended in minimum volume of TE buffer. eDNA was separated from the remaining soil material by ethidium bromide-free agarose (1%) gel electrophoresis (1 hour at 100V, 5 hours at 20V). High molecular weight eDNA (>25 kb) was extracted from the gel by electroelution, concentrated by isopropanol precipitation and blunt-end repaired (End-It, Epicentre Biotechnologies). Blunt-ended eDNA was ligated with either the previously reported broad-host-range cosmid vector pJWC1 or pJSS, a pJWC1 derivative with a DNA linker containing a ScaI cloning site [TGGCCTGTCATGAGCAGGATC] replacing the sacB gene (Craig et al., 2009). Cosmids vectors were prepared for ligation by digestion with ScaI and dephosphorylation with calf-intestinal alkaline phosphatase. Ligation reactions were packaged with lambda phage packaging extracts (MaxPlax - Lambda Packaging Extracts, Epicentre) and transfected into E. coli EPI 300 (TransforMax, Epicentre Biotechnologies) grown to OD600 1.0 by shaking at 37°C for 1.5 hours. 1/1000 of the transfection reaction was plated on a LB-tetracycline plate (20 μg/mL) to estimate the size of the library. The remainder of the transfection reaction was selected overnight in LB broth containing 20 μg/mL tetracycline (37°C with shaking). The number of colonies on the titer plate was counted to estimate the number of clones in the library. In total, 5 x 105 and 2 x 105 unique cosmid clones were constructed using vectors pJWC1 (vector size 14 kb) and pJSS (vector size 12 kb) respectively, constituting 22.5 GB eDNA. The library was stored as 15% glycerol stocks.
To transform libraries into Ralstonia metallidurans, cosmid DNA was miniprepped (Qiagen) from overnight cultures inoculated from library glycerol stocks and pooled together the next day in equivalent volumes. 1 – 2 μg of DNA was transformed by electroporation (0.8 kV/1.0 mm cuvette) into 80 μl aliquots of electrocompetent R. metallidurans CH34 cells prepared as previously described (Taghavi et al., 1994). Following electroporation, cells were diluted in 1 mL SOC medium and incubated for 3 hours (30°C with shaking) before plating onto LB-tetracycline plates (20 μg/mL). The resulting R. metallidurans-based library comprising two times coverage of the original eDNA cosmid library was scraped from the selection plates after two days at 30°C and archived as glycerol stocks.
Functional Screening of Libraries in R. metallidurans
The R. metallidurans-based library was screened for clones with antibacterial activity in a top agar overlay assay against Bacillus subtilis. For this assay, the library was diluted directly from glycerol stocks and plated onto 150 mm LB-tetracycline (20 μg/mL) plates at a density of 1,000 – 1,500 clones per plate. Colonies were allowed to mature at 30°C overnight and then incubated at room temperature for 4 – 5 days to allow for heterologous expression. A thin layer of 0.7% top agar (10 – 12 mL) was overlaid onto the plates. The top agar was inoculated with a 1:200 dilution of tetracycline-resistant B. subtilis 1E9 (Bacillus Genome Stock Center, Ohio) grown to OD600 0.5. Plates were incubated at 30°C overnight. To cleanly obtain the naturally kanamycin-resistant R. metallidurans clones without residual B. subtilis assay strain contamination, colonies forming zones of growth inhibition in the B. subtilis lawn were picked from the assay plates and struck for single colonies on LB plates containing tetracycline (20 μg/mL) and kanamycin (30 μg/mL). DNA was miniprepped from overnight cultures of single colonies, retransformed into R. metallidurans and patched onto LB-tetracycline (20 μg/mL) plates. These plates were re-assayed using top agar overlays; clones that showed the antibiosis phenotype were considered true hits and archived as 15% glycerol stocks in R. metallidurans and E. coli.
Sequencing and Bioinformatics
High quality cosmid DNA was obtained from antibacterially active hits by miniprepping overnight E. coli EPI 300 cultures induced with CopyControl Induction Solution (Epicentre Biotechnologies). Cosmid DNA from each clone was pooled and sequenced using 454 GS-FLX Titanium pyrosequencing technologies (MSKCC Genomics Core Laboratory) and assembled on GS De Novo Assembler software (Roche) (GenBank Accession numbers KF835381-KF835386 for SZR1, SZR5, WZR9, WZR11, WZR18 and WZR21, respectively). Individual cosmid clones were also end sequenced by Sanger sequencing using primers designed to recognize vector sequences flanking the eDNA cloning site. End sequencing data was used to identify contigs assembled from the 454 sequencing data. Sequences were annotated using the online tool SoftBerry to predict open reading frames, and alignments to BLAST and PFAM databases were used to predict gene function (Altschul et al., 1990; Solovyev & Salamov, 2011; Punta et al., 2012). For phylogenetic analysis, predicted antibacterial protein found in clones SZR1, WZR21 and WZR9 were trimmed based on their alignment to PFAM families PF01464 (SLT Transglycosylase), PF01520 (Amidase) or PF07859 (Alpha/beta hydrolase), respectively. Phylogenetic trees were constructed from ClustalW alignments using the MEGA5 program with the Neighbor-Joining method and 1000 bootstrap replications (Tamura et al., 2011).
Subclone Library Construction
Cosmid DNA (2 μg) was sheared to 3 kb using Blue miniTubes in the Covaris S220 Focused-ultrasonicator. Sheared DNA was blunt-end repaired, ligated into ScaI-digested dephosphorylated pJWC1, and transformed into electrocompetent E. coli EPI 300 cells using the same methods described in the eDNA library construction section. Subclone libraries were transformed into electrocompetent R. metallidurans and assayed in a top agar overlay assay for antibacterial activity. Clones displaying zones of growth inhibition were struck onto LB-tetracycline-kanamycin plates. Single colony cultures were miniprepped and transformed into electrocompetent E. coli EPI 300 to obtain sufficient DNA for sequencing with vector specific primers.
Transposon Mutagenesis
E. coli-based random transposon mutagenesis libraries were created using the HyperMu transposon system (HyperMu <Kan-1>, Epicentre). DNA prepared from overnight cultures of the transposon mutagenesis library was transformed into electrocompetent R. metallidurans cells and selected on LB-tetracycline plates. The resulting R. metallidurans-based transposon mutant libraries were assayed using top agar overlays to identify clones that failed to display a zone of growth inhibition. DNA isolated from LB-tetracycline overnight cultures of these clones was transformed into E. coli EPI 300 cells, and DNA prepped from overnight cultures of transformants was Sanger sequenced using HyperMu specific primers.
Top Agar Overlays in E. coli
Cosmid DNA from antibacterial clones identified in our original functional screen in R. metallidurans were transformed into electrocompetent E. coli EPI 300 and assayed for the ability to confer the same antibiosis phenotype in E. coli. Each antibacterially active clone transformed into E. coli, and negative control (empty pJWC1 vector), were struck onto LB-tetracycline (20 μg/mL) plates to obtain single colonies. Plates were incubated at 30°C overnight, followed by 3 days at room temperature and then assayed using top agar overlays as described above.
Results & Discussion
For this study, DNA extracted directly from soil collected in the Sonoran Desert (Arizona, USA) was used to construct a cosmid library in a broad host cosmid shuttle vector. This metagenomic library comprised of 700,000 unique clones and was predicted to contain ~22.5 Gb of eDNA. The library was originally constructed in E. coli using lambda phage packaging and transfection and then cosmid DNA from this E. coli-based parent library was electroporated into Ralstonia metallidurans [Fig 1A]. We selected R. metallidurans, a soil dwelling beta proteobacteria, as the library host for this study because it had previously shown improved heterologous expression capabilities in functional screening studies compared to E. coli (Craig et al., 2010).
Previous R. metallidurans-based functional metagenomic studies have focused on the identification of antibacterially active clones producing organic extractable small molecules (Craig et al., 2009; Craig et al., 2010). In light of recent reports highlighting the successful use of bacterial and phage enzymes as potential antibacterial therapies, we sought to explore whether antibacterially hits identified in R. metallidurans-based eDNA library antibiosis screens that did not produce small molecules might be a source of eDNA encoded antibacterially active enzymes. Top agar overlays were carried out on the 700,000 membered R. metallidurans soil eDNA library to identify clones exhibiting antibiosis activities against a B. subtilis assay strain [Fig 1B]. Of the 19 clones we identified as hits in our primary antibiosis assay, cosmid DNA isolated from 6 clones (SZR1, SZR5, WZR9, WZR11, WZR18 and WZR21) showed the ability to confer antibacterial activity to R. metallidurans upon retransformation while DNA from the rest of the clones did not. In small scale fermentation studies none of the clones with reconfirmed antibacterial activity showed the presence of clone specific small molecule in culture broth extracts and these clones were therefore examined both bioinformatically and functionally for the ability to encode for antibacterial active enzymes.
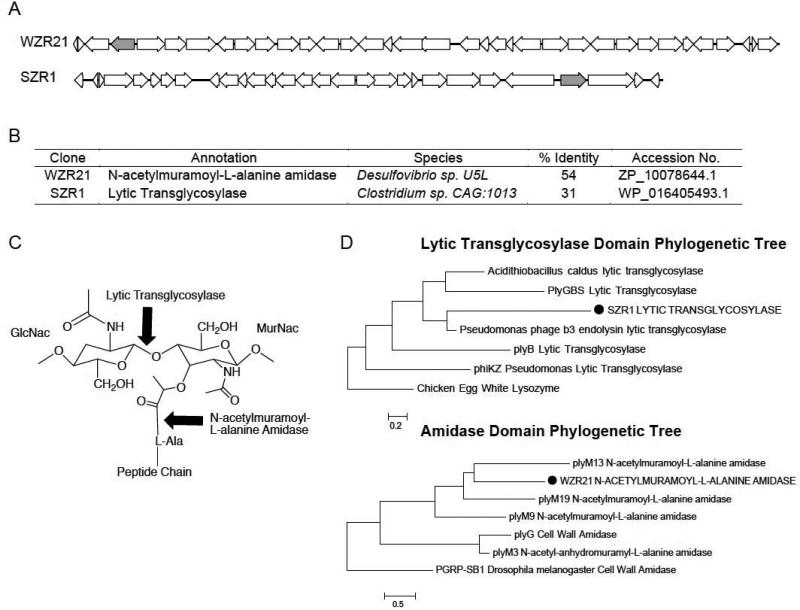
Cosmid DNA from each reproducible antibacterial hit was de novo sequenced and annotated using the online SoftBerry software package to identify open reading frames. Putative gene functions were assigned by alignment to the BLAST and PFAM databases. Based on predicted gene functions, only two clones contained genes encoding obvious antibacterial enzymes. Clones SZR1 and WZR21 encoded enzymes with high similarity to previously well-characterized antibacterially active enzymes targeting peptidogylcan bonds [Fig 2A, B]. The cell wall lytic homolog from the first clone, WZR21, is most closely related (54% identity) to N-acetylmuramoyl-L-alanine-amidases that cleave the amide bond connecting N-acetylmuramic acid to the cross-linked peptides present in the bacterial cell wall. The cell wall lytic homolog from the second clone, SZR1, is most closely related (31% identity) to lytic transglycosylases that cleave the glycosyl bond between N-acetyl glucosamine and N-acetyl muramic acid sugars in the bacterial cell wall [Fig 2C].
Figure 2.
(A) Sequence maps from antibacterially active clones containing putative cell wall degrading enzymes (grey). (B) Annotation of putative cell wall degrading enzymes associated with antibacterial activity. Species and % identity columns describe the top BLAST hit to the predicted cell wall lytic enzyme encoding gene. (C) Repeat structure of the peptidoglycan layer of the cell wall of gram-positive bacteria. Arrows indicate the points of action of the cell wall degrading enzymes found in this study. (D) ClustalW-derived phylogenetic trees comparing functional domains of a representative set of cell wall lytic transglycosylases and amidases with the predicted enzymatic domains found in our phenotypic screen (highlighted in capital letters with black circles). Protein sequences were trimmed to PFAM families PF01464 (SLT Transglycosylase) or PF01520 (Amidase) domains.
PFAM-trimmed functional domains present in the predicted cell wall lytic enzymes from SZR1 and WZR21 were phylogenetically compared to soluble lytic transglycosylase and amidase domains from functionally characterized antibacterial phage endolysins [Fig 2D]. The predicted cell wall lytic domain from clone SZR1 displayed similarity to the lytic transglycosylase domain of the Pseudomonas aeruginosa Bacteriophage B3 endolysin predicted to participate in host cell lysis (Braid et al., 2004). This same analysis indicated high similarity between the predicted N-acetyl-muramic-acid-L-alanine amidase domain from clone WZR21 and the antibacterially active phage amidase domain plyM13. The cell wall lytic amidase plyM13 was found in one of the only other reported metagenomic screens to identify enzymes with cell wall lytic activity, in which metagenomic libraries made from animal feces were screened using a two-step functional assay. In the two-step assay for cell wall lytic activity, phage genes were first identified by their proximity to hemolytic phage holins using a blood agar-based assay and then recombinantly expressed and tested for lytic activity against a heat-killed bacterial assay strain (Schmitz et al., 2010).
For the four antibacterially active clones (WZR9, WZR11, WZR18 and WZR21) where no open reading frames were found to encode enzymes commonly associated with antibacterial activity, the genetic elements required for antibacterial activity were identified through either subcloning (WZR9, WZR11 and WZR18) or transposon mutagenesis (WZR21) experiments, followed by top agar overlay screening to identify either antibacterial active subclones or inactive transposon mutants.
Antibacterial activity assays run on subclone libraries made using sheared cosmid DNA from clones WZR9, WZR11 and WZR18 led to the identification of a single antibacterially active enzyme on each clone. In these studies antibacterially active subclones were recovered and sequenced until the overlapping region on the recovered clones was reduced to the point of a single open reading frame (ORF) [Fig 3A]. The antibacterial associated ORF found on clones WZR11 and WZR18 are predicted to encode for proteases, while the antibacterial ORF from WZR9 was predicted to encode for a lipase [Figure 3A and B]. Antibacterial assays conducted on transposon mutants of SZR5 yielded three unique mutants that lacked the antibacterial phenotype. The transposon insertion in all three mutants was found in an ORF that encodes for a predicted protease.
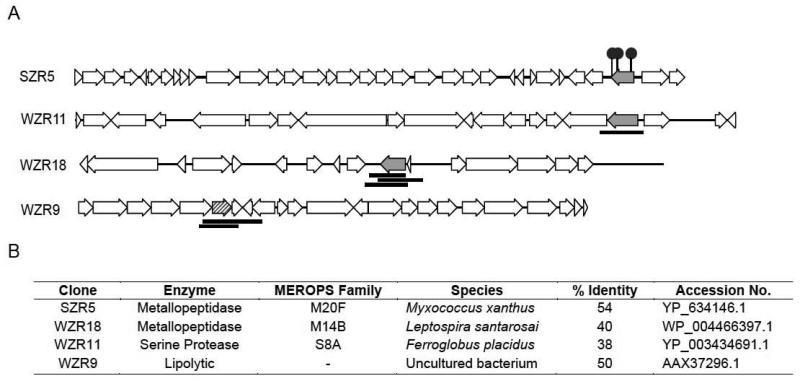
Figure 3.
(A) Sequence maps from clones containing antibacterially active proteases (solid grey ORFs) and lipolytic enzymes (hashed grey ORF). Antibiosis was confirmed either by the insertion of transposons (red circles) leading to loss of activity, or by the expression of the antibiosis phenotype by random shotgun subclones (black lines). (B) Annotation of putative proteases and lipolytic enzymes associated with antibacterial activity. Species and % identity columns describe the closest homolog from the MEROPS database for proteases and from the BLAST database for the lipolytic enzyme.
While proteases and lipases have been found in previous metagenomic screens using E. coli as a heterologous host, they have rarely been identified using direct screens for antibiosis. For example, previous attempts to clone proteases from environmental samples have focused on identifying industrially relevant enzymes for applications such as laundry detergent using milk agar plate assays (Kennedy et al., 2011; Niehaus et al., 2011). Similarly, industrially relevant lipases and esterases have been isolated in previous metagenomic studies using lipase-directed screening methods, including degradation of tributyrin or related techniques (Kennedy et al., 2011; Reyes-Duarte et al., 2012). There is only one case in which lipolytic enzymes conferring antibiosis were identified in an E. coli-based metagenomic study (Yung et al., 2011).
The predicted proteases from clones SZR5, WZR11 and WZR18 were aligned to the MEROPS database – a curated collection of proteases organized into families of related enzymes by sequence similarities (Rawlings et al., 2012). This analysis indicated that the SZR5 and WZR18 proteases belong to the M20F and M14B subfamilies of metallopeptidases, respectively and the WZR11 protease belongs to the S8A subfamily of serine proteases [Fig 3B]. In a BLAST search against GenBank, the predicted lipase from clone WZR9 shows highest similarity (50% identity) to an esterase found in a soil functional metagenomic screen for ester-hydrolysis activity (Kim et al., 2006). The three enzymes found in the only E. coli-based metagenomic screen reporting antibacterial lipases show only low sequence identity (10% identity to CcAb1, 9% to CcAb2, 23 % to UaAb1) to the lipolytic enzyme in clone WZR9 found in our R. metallidurans-based screen.
It is somewhat surprising that enzymes with antibacterial activity have not been reported more frequently in metagenomic screens given that a number of bacteria (e.g Achromobacter lyticus and Myxococcus xanthus) are known to produce bacteriolytic proteases, and genomes of some sequenced bacteria for example predatory bacteria such as Bdellovibrio bacteriovirus are rich in proteolytic enzymes (Sudo & Dworkin, 1972; Li et al., 1998; Rendulic et al., 2004). We hypothesized that this disparity in the finding of antibacterial enzymes in metagenomic screens may be a function of choice of heterologous host. To assess whether or not the putative enzymes identified in our R. metallidurans-based screens could have been discovered using the same techniques but with E. coli as the heterologous host, we performed the same top agar overlay assay on E. coli transformed with each of the six antibacterially active eDNA clones we identified. In these assays, none of the E. coli-based clones displayed the antibacterial phenotype observed in our R. metallidurans-based screens. Potential reasons for the absence of a detectable antibacterial phenotype in E. coli are varied. These include differences in the ability to recognize foreign promoters or ribosome binding sites, differences in codon usage, differences in protein secretion abilities and differences in transcription factors. This experiment underscores the importance of diverse hosts in functional screenings of metagenomic libraries.
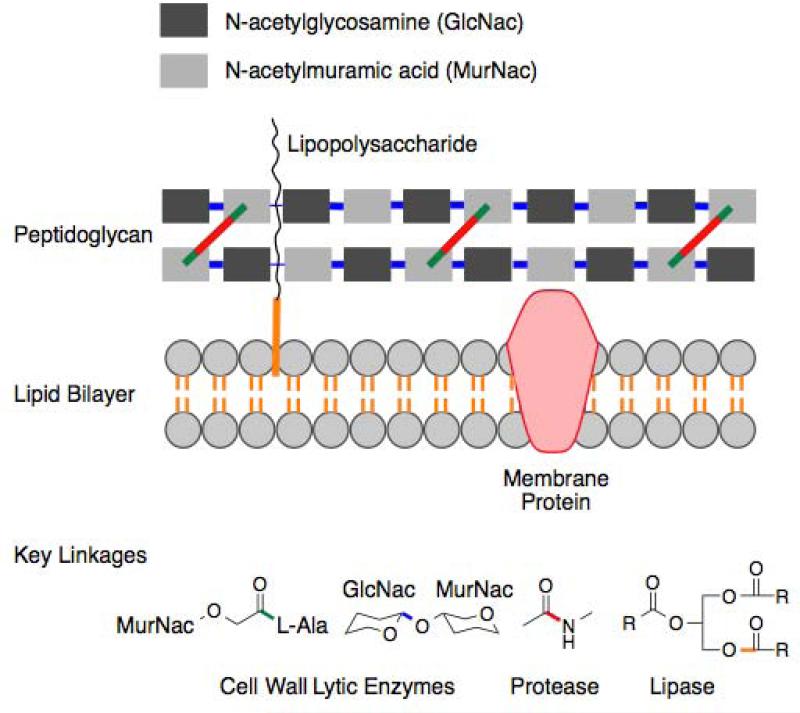
The six clones found in our R. metallidurans-based metagenomic screen for antibacterial activity contain a lipase, proteases and cell wall lytic enzymes that are predicted to hydrolyze the three key linking chemistries (e.g. ester, amide and glycosidic bonds) present in the bacterial cell [Figure 4]. In light of the attention that lytic enzymes have recently garnered as potential therapeutics, obtaining a diverse assortment of novel antibacterial enzymes for development as lead agents has become increasingly relevant. The diversity of antibacterial enzymes found in this study suggests that the phenotypic screening of soil metagenomes using various heterologous hosts, including R. metallidurans, may prove useful for identifying therapeutically relevant antibacterial enzymes in future large-scale screens of environmental samples.
Figure 4.
The enzymes found in our metagenomic screen are predicted to target the key “linking chemistries” (amide, ester and glycolytic bonds) found in bacterial cell walls.
Acknowledgements
This work was supported by NIH GM077516. SFB is a Howard Hughes Medical Institute Early Career Scientist.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc. 2007;2:1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6560–6574. doi: 10.1128/JB.186.19.6560-6574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JW, Chang FY, Brady SF. Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chem Biol. 2009;4:23–28. doi: 10.1021/cb8002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–1641. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL, Sanchez S. Microbial drug discovery: 80 years of progress. The Journal of antibiotics. 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J of Med Microbiol : IJMM. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor EM, Alkema WB, Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol. 2004;6:879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Iqbal HA, Feng Z, Brady SF. Biocatalysts and small molecule products from metagenomic studies. Curr Opin Chemical Biol. 2012;16:109–116. doi: 10.1016/j.cbpa.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J, O'Leary ND, Kiran GS, Morrissey JP, O'Gara F, Selvin J, Dobson AD. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J Appl Microbiol. 2011;111:787–799. doi: 10.1111/j.1365-2672.2011.05106.x. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Choi GS, Kim SB, Yoon GS, Kim YS, Ryu YW. Screening and characterization of a novel esterase from a metagenomic library. Protein Express Purif. 2006;45:315–323. doi: 10.1016/j.pep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li S, Norioka S, Sakiyama F. Bacteriolytic activity and specificity of Achromobacter beta-lytic protease. J Biochem. 1998;124:332–339. doi: 10.1093/oxfordjournals.jbchem.a022116. [DOI] [PubMed] [Google Scholar]
- Niehaus F, Gabor E, Wieland S, Siegert P, Maurer KH, Eck J. Enzymes for the laundry industries: tapping the vast metagenomic pool of alkaline proteases. Microbial biotechnology. 2011;4:767–776. doi: 10.1111/j.1751-7915.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science (New York, NY) 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Reyes-Duarte D, Ferrer M, Garcia-Arellano H. Functional-based screening methods for lipases, esterases, and phospholipases in metagenomic libraries. Methods Mol Biol. 2012;861:101–113. doi: 10.1007/978-1-61779-600-5_6. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Schuch R, Fischetti VA. Identifying active phage lysins through functional viral metagenomics. Appl Environ Microbiol. 2010;76:7181–7187. doi: 10.1128/AEM.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Salamov A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers; 2011. pp. 61–78. [Google Scholar]
- Sudo S, Dworkin M. Bacteriolytic enzymes produced by Myxococcus xanthus. J Bacteriol. 1972;110:236–245. doi: 10.1128/jb.110.1.236-245.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S, van der Lelie D, Mergeay M. Electroporation of Alcaligenes eutrophus with (mega) plasmids and genomic DNA fragments. Appl Environ Microbiol. 1994;60:3585–3591. doi: 10.1128/aem.60.10.3585-3591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thallinger B, Prasetyo EN, Nyanhongo GS, Guebitz GM. Antimicrobial enzymes: an emerging strategy to fight microbes and microbial biofilms. Biotechnology journal. 2013;8:97–109. doi: 10.1002/biot.201200313. [DOI] [PubMed] [Google Scholar]
- Trincone A. Marine biocatalysts: enzymatic features and applications. Marine drugs. 2011;9:478–499. doi: 10.3390/md9040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Crespo P, Ageitos JM, Poza M, Villa TG. Enzybiotics: a look to the future, recalling the past. J of Pharm Sci. 2007;96:1917–1924. doi: 10.1002/jps.20853. [DOI] [PubMed] [Google Scholar]
- Yung PY, Burke C, Lewis M, Kjelleberg S, Thomas T. Novel antibacterial proteins from the microbial communities associated with the sponge Cymbastela concentrica and the green alga Ulva australis. Appl Environ Microbiol. 2011;77:1512–1515. doi: 10.1128/AEM.02038-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]