Abstract
Adiponectin has a variety of metabolic effects on obesity, insulin sensitivity, and atherosclerosis. To identify genes influencing variation in plasma adiponectin levels, we performed genome-wide linkage and association scans of adiponectin in two cohorts of subjects recruited in the Genetic Epidemiology of Metabolic Syndrome Study. The genome-wide linkage scan was conducted in families of Turkish and southern European (TSE, n = 789) and Northern and Western European (NWE, N = 2,280) origin. A whole genome association (WGA) analysis (500K Affymetrix platform) was carried out in a set of unrelated NWE subjects consisting of approximately 1,000 subjects with dyslipidemia and 1,000 overweight subjects with normal lipids. Peak evidence for linkage occurred at chromosome 8p23 in NWE subjects (lod = 3.10) and at chromosome 3q28 near ADIPOQ, the adiponectin structural gene, in TSE subjects (lod = 1.70). In the WGA analysis, the single-nucleotide polymorphisms (SNPs) most strongly associated with adiponectin were rs3774261 and rs6773957 (P < 10−7). These two SNPs were in high linkage disequilibrium (r2 = 0.98) and located within ADIPOQ. Interestingly, our fourth strongest region of association (P < 2 × 10−5) was to an SNP within CDH13, whose protein product is a newly identified receptor for high-molecular-weight species of adiponectin. Through WGA analysis, we confirmed previous studies showing SNPs within ADIPOQ to be strongly associated with variation in adiponectin levels and further observed these to have the strongest effects on adiponectin levels throughout the genome. We additionally identified a second gene (CDH13) possibly influencing variation in adiponectin levels. The impact of these SNPs on health and disease has yet to be determined.
INTRODUCTION
Adiponectin is a protein secreted exclusively by adipocytes and is one of the most abundant gene products in adipose tissue, accounting for 0.01% of total plasma protein (1). Plasma adiponectin levels are negatively correlated with BMI, especially visceral adiposity (2), and are lower in subjects with type 2 diabetes mellitus (T2DM) and coronary artery disease than in those without (3,4). Metabolic studies carried out in both animals and humans suggest that adiponectin has a variety of metabolic effects, including antidiabetic, antiatherosclerotic, and antiinflammatory (2), and may partially mediate the relationship between obesity and insulin resistance or T2DM (3,5–9).
The secretion and regulation of plasma adiponectin concentrations is under complex control (10). Its expression and/or secretion is increased by leanness, cold exposure, adrenalectomy, insulin-like growth factor 1, ionomycin, and thiazolidenediones, and decreased by obesity, tumor necrosis factor-α, glucocorticoids, β-adrenergic agonists, and cyclic adenosine monophosphate (11–13). Family studies indicate that genetic factors partly regulate adiponectin concentration (14,15). At least six genome-wide linkage scans for adiponectin levels have been published to date (2,10,16–19), implicating regions on chromosomes 3q, 5p, 8q, 9p, 11q, 14p, and 15q. Although the specific mechanisms through which genes influence variation in adiponectin concentrations have not yet been clarified, strong candidate genes include the adiponectin structural gene (ADIPOQ) as well as genes encoding adiponectin-regulatory proteins. Furthermore, the role of genetic variants regulating adiponectin on health outcomes, including insulin resistance, T2DM, and coronary artery disease, has not been unequivocally determined (12).
The development of low-cost high-throughput genotyping technology has made possible the identification of common genetic variants influencing health outcomes on a genome-wide scale. The Genetic Epidemiology of Metabolic Syndrome (GEMS) Study is a multicenter study designed to identify the genetic determinants of metabolic syndrome and related traits. It is well suited for this purpose because it included two arms: a family-based sample for linkage analysis and a population-based sample for association analysis. In the first arm of the GEMS Study, two sets of families with differing ancestral origins (i.e., families of Turkish and southern European (TSE) origin and of Northern and Western European (NWE) origin) were recruited for the purpose of carrying out linkage analysis of metabolic syndrome and related traits. In the second arm of the GEMS Study, an additional set of unrelated subjects was recruited and a whole genome association (WGA) analysis was carried out using ~500,000 single-nucleotide polymorphisms (SNPs). In this report, we used the rich resources of the GEMS Study to perform a comprehensive assessment of the genetic determinants of adiponectin levels on a genome-wide basis. By conducting both genome-wide linkage and association analyses, we were able to put into context the relative strengths of genetic influences on plasma levels of adiponectin.
METHODS AND PROCEDURES
The GEMS Study was initiated in 1999 for the purpose of identifying genes associated with the metabolic syndrome and related traits. The study included two types of samples: two sets of families recruited for a linkage study and a set of unrelated cases and controls recruited for an association study. Relevant to this report, the linkage study was carried out to determine whether quantitative trait loci (QTLs) having fairly large effects on adiponectin variation could be detected along the genome, while the association study was conducted to identify specific SNPs associated with variation in adiponectin levels and to see if these SNPs colocalized to previously detected linkage regions. Standardized questionnaires were administered to study participants to obtain information about medical history, medication use, and tobacco and alcohol use. Subjects received a physical exam during which height, weight, and waist circumference were measured according to a standardized protocol. Blood samples were collected after a 12-h fast. Serum adiponectin level was measured using an enzyme-linked immunosorbent assay (R&D systems, Minneapolis, MN) (20). For all the following analyses, adiponectin was transformed by its cube root to reduce skewness and normalize the distribution (Supplementary Figure S1). Prior to genetic analyses, mean levels of adiponectin and other traits were compared between groups using “Proc Reg” in SAS version 9 (SAS Institute, Cary, NC).
Linkage study
Families were recruited into the GEMS Study from six study sites, located in Australia, Canada, Finland, Turkey, Switzerland, and the United States. Details of the recruitment procedures, subject characteristics, and inclusion/exclusion criteria have been previously described (20,21). Eligible families consisted of a minimum of two siblings (an affected sib-pair) with atherogenic dyslipidemia (ADL), which was defined by the simultaneous presence of high triglyceride (≥75% percentile for age, sex, and country) and low HDL-cholesterol (HDL-C) (≤25% percentile for age, sex, and country) levels in plasma. Individuals were ineligible for participating in the study if they had a BMI ≥35 kg/m2, were positive for human immunodeficiency virus, were recipients of an organ transplant, were diagnosed with familial hypercholesterolemia, or were heavy users of alcohol (>8 units/day). The mean number of examined individuals within enrolled families ranged from 3.7 (Australia) to 18.9 (Turkey), and the overall family size ranged from 2 to 180. Prior to linkage analysis families were split into two groups based on ancestral origin. All 39 families from Turkey and a few families of Southern European origin from Canada (n = 8) and Australia (n = 12) were considered to be of TSE origin, while the remaining families were considered to be of NWE origin. In total, 3,069 subjects aged 18–70 years from 450 families were phenotyped across all sites (789 subjects from 59 TSE families and 2,280 subjects from 391 NWE families).
DNA was extracted using the Puregene system (Gentra Systems, Minneapolis, MN) at the Center for Human Genetics at Duke University Medical Center. A set of 448 microsatellite markers at an average density of 10 centimorgans (cM) were genotyped on 2,870 individuals at the University of Western Australia in Perth. The average heterozygosity of markers was 0.76. Marker locations were obtained from the Marshfield sex-average genetic map. This analysis is based on 437 markers located across the 22 autosomes. The number of genotyped subjects ranged from 2,057 to 2,827 for each marker. Genotyping call rates ranged from 71.7 to 98.5% for all markers. Two markers had genotyping call rates <80% (71.7 and 79.2% for D6S477 and D18S481, respectively). Ninety-four percent (412/437) of all markers had a genotyping call rate >90%.
Linkage analyses were carried out using a variance decomposition approach as implemented in SOLAR (Sequential Oligogenic Linkage Analysis Routines). This approach partitions the total variance of the quantitatively distributed phenotype (e.g., adiponectin levels) into components attributable to measured environmental effects (e.g., age and sex), additive polygenetic effects, and for linkage, an additive QTL effect. The additive polygenic and QTL effects are parameterized as random effects. The background polygenic effect is measured as a function of the phenotypic covariance among related family members, while the additive QTL effects are measured as the variance attributable to allele-sharing among relative pairs at the specific locus of interest. The hypothesis of linkage is tested by the likelihood-ratio test, in which the likelihood of a full model, which includes the linkage component, is compared to the likelihood of a nested model, in which the linkage effect is constrained to be zero (22). The identity-by- descent probabilities between family members were computed using an MCMC approach as implemented in LOKI. Analyses were conducted using several sets of covariates: age, sex with no additional covariates (model 1); age, sex, and BMI (model 2); and age, sex, BMI, smoking, and alcohol use (model 3).
Because of the sensitivity of the variance component linkage approach to distributional assumptions, we computed P values (and lod scores) empirically by simulating a large number of single unlinked markers to the observed data and evaluating the probability of observing lod scores as high as those detected with the real markers by chance alone. These simulations were conducted using the lodadj module within the SOLAR software program (22). All lod scores presented in this article correspond to the empirical conversions of the nominal lod scores.
Prior to carrying out the linkage analysis, we estimated the power of detecting linkage by simulating QTLs of known effect size within our pedigree structures. Results from our power calculations revealed that our sample provides ~80% power to detect lod scores >3 for QTLs accounting for 35% of the total variance in adiponectin levels in TSE families and 25% of the total variance in adiponectin levels in NWE families. For lod scores >2, our samples would provide ~80% power to detect QTLs accounting for 29% of the total variance in adiponectin levels in TSE and 23% in NWE families.
Association study
In the second arm of the GEMS Study, a set of 1,025 cases with ADL and 1,008 normolipidemic controls were recruited from the five GEMS sites with subjects of NWE origin for a case–control study. Subjects with diabetes were excluded from this arm of the study. Normolipidemic controls were required to have both low triglyceride (lower 50% percentile) and high HDL-C (upper 50% percentile) with adjustment of age and sex. Cases included GEMS probands recruited into the family-based study supplemented by additional individuals with ADL identified from each study site. The controls were frequency matched to cases by sex and study site and had a BMI greater than 25 kg/m2. Controls were 5 years older than cases to reduce the possibility of controls developing dyslipidemia. All controls (and 674 of the cases) were newly recruited into the association arm of the study and did not participate in the family (linkage) study arm.
Genotyping was performed using the Affymetrix 500K chip. After subject QC, 1,847 subjects from the original sample remained. Exclusions (n = 186) were due to a less than 90% sample call rate (n = 161), gender inconsistencies (n = 12) or inconsistent genotypes with a different platform (n = 13). After marker QC, 475,439 markers remained. Excluded markers were either monomorphic (n = 12,441), had a Hardy–Weinberg P < 10−7 (n = 11,352) or had a call rate of <70% (n = 205). Additional SNPs were excluded because of low genotype call rates (<0.90, n = 22,733) and/ or minor allele frequency (<0.01, n = 54,316), leaving a total of 398,625 markers included in our final analysis.
The WGA analysis for adiponectin was performed in an additive linear regression model with adjustment for age, sex, study site, and dyslipidemia status. In addition to reporting marker-wise statistical test results, genome-wide levels of statistical significance were computed by applying a Bonferroni correction. Linkage disequilibrium structure was evaluated at selected chromosomal regions using the Haploview software program (23).
RESULTS
Table 1 shows the baseline characteristics and mean (±s.d.) adiponectin concentrations of GEMS subjects according to ethnicity and sex for the linkage and association samples. In the family-based (linkage) sample, TSE subjects were 3–5 years younger and had lower levels of BMI and lower diabetes prevalence compared to NWE subjects. Plasma adiponectin levels were significantly higher in NWE than TSE subjects and higher in women than men. The ethnicity difference in adiponectin levels persisted even after adjustment of sex and BMI (P < 0.0001). In the association sample, controls were 5 years older than cases and had a significantly higher adiponectin levels and lower BMI.
Table 1.
Baseline characteristics of study subjects according to ethnicity and sex
| Traits | Linkage study
|
Association study
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Turkish and southern European (n = 789)
|
Northern and Western European (n = 2,280)
|
P valuea
|
|||||||
| Men (n = 360) | Women (n = 429) | Men (n = 1,178) | Women (n = 1,102) | Sex | Ethnicity | Case (n = 997) | Control (n = 989) | P (t-test) | |
| Age (years) | 43.4 ± 16.1 | 42.1 ± 16.4 | 47.1 ± 13.7 | 47.3 ± 14.7 | 0.51 | <0.001 | 55.3 ± 9.2 | 59.6 ± 9.2 | <0.0001 |
| BMI (kg/m2) | 26.5 ± 4.6 | 27.3 ± 5.4 | 28 ± 4.1 | 27.2 ± 5.6 | 0.0077 | 0.0004 | 28.7 ± 3.5 | 28.2 ± 3.4 | 0.0012 |
| Adiponectin (μg/ml) (median; 25–75%) | 3,961 (1,987–6,146) | 5,811 (3,437–8,887) | 3,958 (2,417–6,070) | 6,071 (3,792–9,705) | <0.0001 | <0.0004 | 4,545 (3,002–6,791) | 6,599 (4,505–9,931) | <0.01 |
| Adiponectin (cube root) | 15.3 ± 4.5 | 17.3 ± 4.9 | 15.9 ± 4.1 | 18.3 ± 4.8 | <0.0001 | <0.0001 | 16.9 ± 3.9 | 19.2 ± 3.9 | <0.0001* |
| % Diabetes | 4.7 | 4.4 | 7.8 | 6.8 | 0.29 | 0.007 | – | – | – |
| % Current smoker | 49 | 17 | 25 | 18 | <0.001 | <0.001 | 23.7 | 8.5 | <0.0001 |
| % Former smoker | 22 | 7 | 38 | 28 | <0.001 | <0.001 | 37.9 | 37.1 | 0.73 |
| % Alcohol user | 43 | 13 | 79 | 68 | <0.001 | <0.001 | 82 | 88 | <0.0001 |
Values represent (mean ± s.d.) unless otherwise indicated.
Sex effects adjusted for age, ethnicity, and BMI; ethnicity effects adjusted for age, sex, and BMI.
P value <0.0001 after adjustment for age, sex, and BMI.
Linkage analysis results
The heritability of age- and sex-adjusted adiponectin levels was 0.68 ± 0.06 for the TSE and 0.67 ± 0.04 for the NWE populations. These estimates increased slightly with further adjustment for BMI (0.71 ± 0.04 for the TSE and 0.68 ± 0.04 for the NWE populations) and for BMI, smoking, and alcohol consumption (0.73 ± 0.04 for the TSE and 0.69 ± 0.04 for the NWE populations).
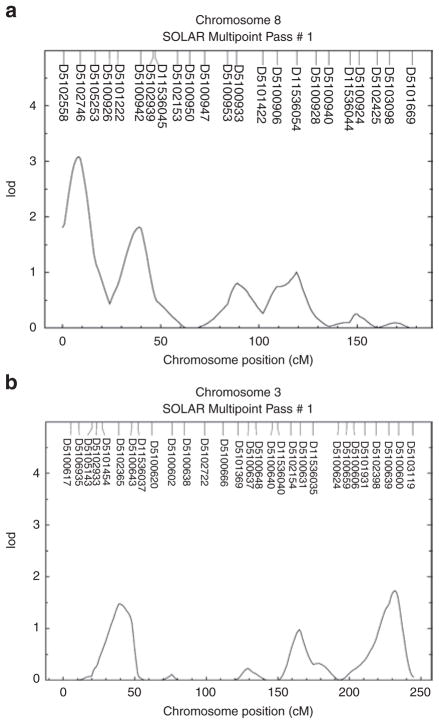
The major results of the genome-wide multipoint linkage analysis of adiponectin levels for each population are shown in Table 2, which summarizes all linkages with lod scores of 1.50 or higher in either the TSE or NWE populations. In the NWE families, the peak lod score (age- and sex-adjusted) was 2.78, occurring at chromosome 8p23 between markers D8S264 (0.7 cM) and D8S277 (8.3 cM). Further adjustment for BMI increased the lod score to 3.10 (Figure 1a) at this locus, although evidence for linkage declined (lod = 2.60) with additional adjustment for smoking and alcohol. The lod scores of 1.80 or higher were also observed slightly centromeric to this locus on chromosome 8p at 39 cM pter (near marker D8S1145, 37cM). In the TSE families, the peak lod score was 1.80, occurring on chromosome 3q28 at D3S2418 (215.8 cM; 187.7 Mb) in the region flanked by markers D3S1262 (201.1 cM) and D3S1311 (224.9 cM) (Figure 1b). Further adjustment for BMI decreased the lod score to 1.70 at this locus, with a further decrease in the evidence for linkage (lod = 1.43) with additional adjustment for smoking and alcohol. This location maps to ~0.23–0.25 Mb from ADIPOQ (188.04–188.06 Mb), the structural gene that encodes the adiponectin protein. An lod score of 1.50 was also observed on chromosome 3p26–3p24 (36.6 cM) in the region flanked by markers D3S4545 (26.3 cM) and D3S3038 (44.8 cM).
Table 2.
Multipoint linkage analysis peaks with lod ≥1.50 (P < 0.005) of adiponectin levels in the TSE and NWE population adjusted for age, sex, and BMI
| Ethnicity | Chr | Position Marshfield map (cM) (1 – lod support interval) | Closest marker(s) | Interval | Lods adjusted for age, sex, and BMI |
|---|---|---|---|---|---|
| TSE | 3 | 36.6 (26.3–47 cM) | D3S1259(SYN2) | 3p25 (3p26–24) | 1.50 |
| 3 | 215.8 (211–220.5 cM) | D3S2418 | 3q28 (3q27–29) | 1.70a | |
| NWE | 8 | 8.3 (1.0–11.8 cM) | D8S277 (D8S264/D8S1825) | 8p23 (8p23–23) | 3.10a |
| 8 | 37 (26.4–42.9 cM) | D8S1145 (D8S1106/D8S828) | 8p12 (8p21–8p11.2) | 1.80 | |
| 4 | 180 | D4S2368/D4S2979 | 4pter–4qter | 1.80 | |
| 2 | 271 | D2S427/D2S2968 | 2q37 | 1.58 | |
| 19 | 51.9 (42.3–68.1 cM) | D19S433 (D19S252–D19S178) | 19q12 | 1.55 |
NWE, Northern and Western European; TSE, Turkish and southern European.
Top lod score in each ethnicity group.
Figure 1.
Multipoint linkage analysis of adiponectin levels with (a) chromosome 8 markers in the NWE sample and (b) chromosome 3 markers in the TSE sample (both adjusted for age, sex, and BMI). NWE, Northern and Western European; SOLAR, Sequential Oligogenic Linkage Analysis Routines; TSE, Turkish and southern European.
WGA results (NWEs)
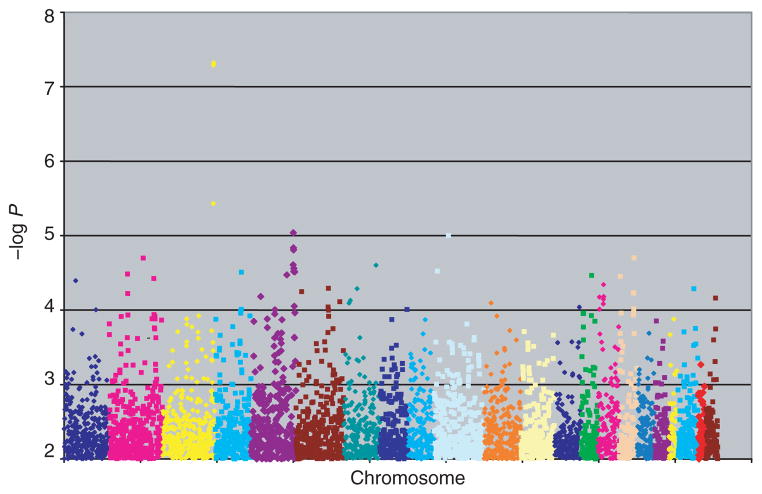
The WGA results for adiponectin in the 1,845 (of 1,847 with genotypes) subjects with measured adiponectin levels are summarized in Figure 2. The 10 most strongly associated SNPs, which fell within five distinct chromosomal regions, are listed in Table 3. Five of these top 10 SNPs are located within genes — three SNPs in ADIPOQ, (P = 3.7 × 10−6 to 4.8 × 10−8), one SNP in LYZL1 (P = 9.9 × 10−6), and one in CDH13, a recently identified receptor for hexameric and high-molecular-weight species of adiponectin (24) (P = 2.0 × 10−5).
Figure 2.
Whole genome association for plasma adiponectin level in the Genetic Epidemiology of Metabolic Syndrome Study association sample.
Table 3.
Top 10 SN Ps from the WGA analysis for adiponectin in NWE population
| SNP | Chromosome | Position | Gene | Point-wise P value |
|---|---|---|---|---|
| rs6773957 | 3 | 188,056,399 | ADIPOQ | 4.78E–08 |
| rs3774261 | 3 | 188,054,253 | ADIPOQ | 5.15E–08 |
| rs17366568 | 3 | 188,053,147 | ADIPOQ | 3.70E–06 |
| rs7722022 | 5 | 172,867,504 | N/A | 9.14E–06 |
| rs1774950 | 10 | 29,622,254 | LYZL1 | 9.96E–06 |
| rs7722165 | 5 | 172,867,576 | N/A | 1.46E–05 |
| rs11738751 | 5 | 172,867,785 | N/A | 1.56E–05 |
| rs7195409 | 16 | 82,085,093 | CDH13 | 1.99E–05 |
| rs1108567 | 2 | 149,000,538 | N/A | 2.01E–05 |
| rs7716959 | 5 | 172,866,448 | N/A | 2.47E–05 |
NWE, Northern and Western European; SNP, single-nucleotide polymorphism; WGA, whole genome association.
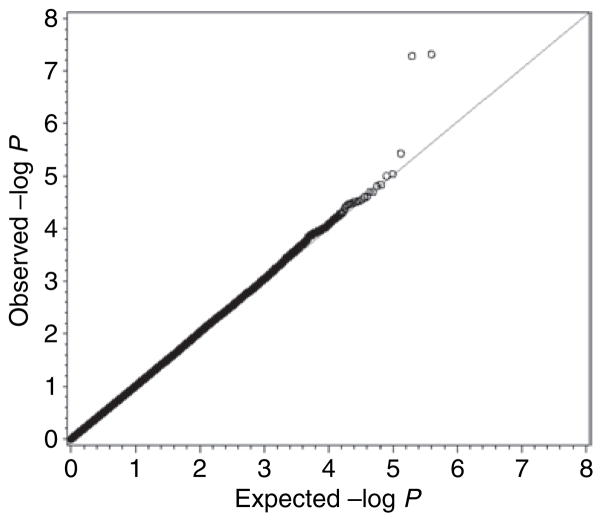
As indicated in the QQ plot (Figure 3), the P values for the two most strongly associated SNPs, rs3774261 and rs6773957, differed markedly from their expectations under the null hypothesis of no association (marker-wise significance levels of 3.44 × 10−8 and 3.02 × 10−8, respectively). Additional adjustment for dyslipidemia status altered these results only slightly (5.15 × 10−8 and 4.78 × 10−8, respectively). Subgroup analyses were further carried out in ADL cases (n = 921) and controls (n = 924) separately. Both rs3774261 and rs6773957 remained strongly associated with adiponectin levels in ADL cases (P = 6.8 × 10−6 and 8.7 × 10−6), and moderately so in controls (P = 0.004 and 0.003) (Table 4). Moreover, as shown in Figure 4, the direction and magnitude of the genotype effect on adiponectin levels were consistent in both cases and controls. Inclusion of BMI in the model did not change the effect of SNP on adiponectin levels (data not shown). An additional analysis was carried out allowing for an SNP × dyslipidemia interaction effect, but this provided no evidence that the effect of SNP was modified in the presence of dyslipidemia (data not shown). The WGA analyses were repeated under both a dominant and a 2 df genotypic model, the latter partitioning the genotype effect into an additive and dominance deviation component. In both cases, SNPs, rs3774261 and rs6773957, remained the most strongly associated SNPs, albeit at slightly reduced levels of statistical significance (P = 2.42 × 10−7 and 2.57 × 10−7 for dominant model, P = 2.16 × 10−7 and 2.24 × 10−7 for genotypic model, for rs3774261 and rs6773957, respectively).
Figure 3.
QQ plot for whole genome association analysis of plasma adiponectin in population-based sample.
Table 4.
WGA analysis results for SNPs in ADIPOQ in NWE population
| SNPs | Start position | MAF |
P value
|
|||
|---|---|---|---|---|---|---|
| Pooled population (n = 1845) | Cases (n = 921) | Controls (n = 924) | ||||
|
| ||||||
| Adjusted for
| ||||||
| Age, sex, site | Age, sex, site, ADL | Age, sex, site | Age, sex, site | |||
| rs864265a | 188,036,986 | T: 0.14 | 0.058 | 0.022 | 0.032 | 0.213 |
| rs182052 | 188,043,476 | A: 0.37 | 0.137 | 0.145 | 0.352 | 0.345 |
| rs16861205 | 188,044,328 | A: 0.09 | 0.607 | 0.573 | 0.721 | 0.331 |
| rs17366568 | 188,053,147 | A: 0.13 | 3.2E–06 | 3.70E–06 | 0.002 | 3.50E–04 |
| rs2241767 | 188,053,890 | G: 0.11 | 0.392 | 0.412 | 0.198 | 0.918 |
| rs3821799 | 188,054,180 | T: 0.40 | 0.015 | 0.026 | 0.037 | 0.500 |
| rs3774261 | 188,054,253 | A: 0.38 | 3.44E–08 | 5.15E–08 | 6.76E–06 | 0.004 |
| rs6444174b | 188,055,883 | C: 0.005 | – | – | – | – |
| rs6773957 | 188,056,399 | A: 0.38 | 3.02E–08 | 4.78E–08 | 8.72E–06 | 0.003 |
| rs11712615a | 188,092,833 | G: 0.30 | 0.958 | 0.932 | 0.484 | 0.46 |
MAF, minor allele frequency; NWE, Northern and Western European; SNP, single-nucleotide polymorphism; WGA, whole genome association.
SNPs on the Affymetrix 500k chip that flank ADIPOQ.
SNP excluded from analysis in Plink as MAF <0.01.
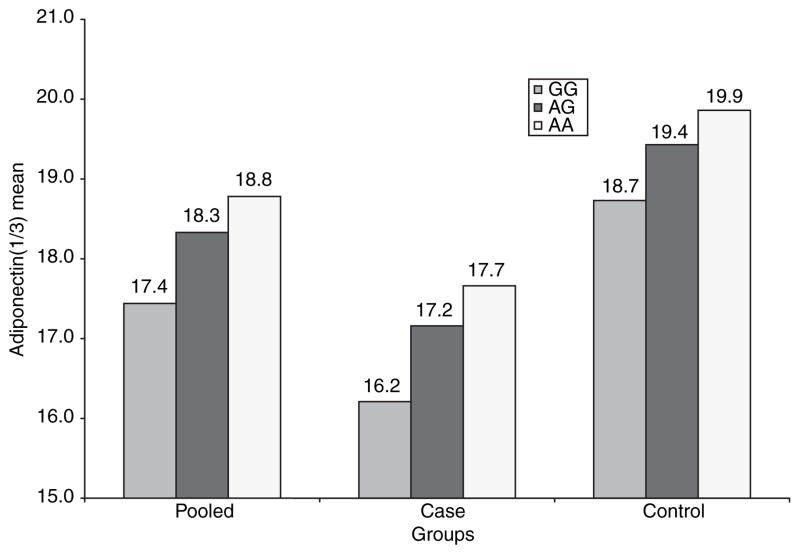
Figure 4.
Mean adiponectin levels (cube root) according to genotype of SNP rs3774261.
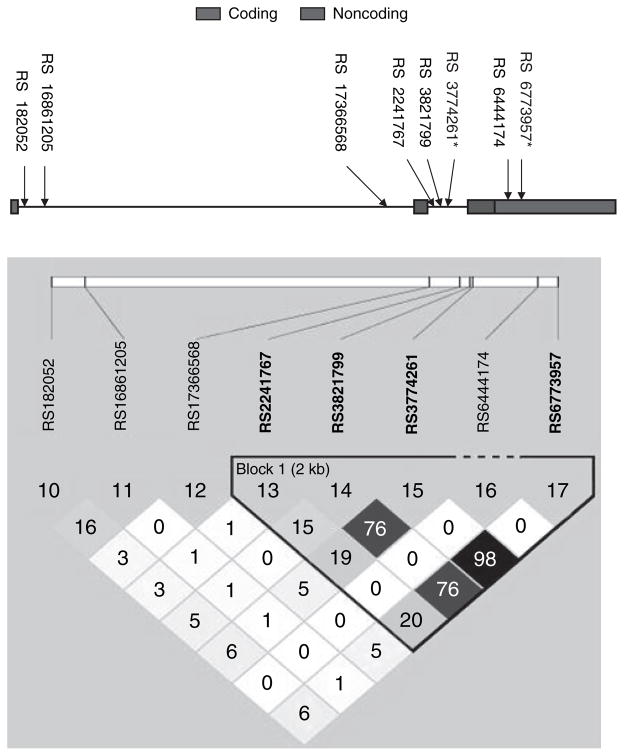
Haploview analyses revealed rs3774261 and rs6773957 to be highly correlated (r2 = 0.98) and located within the adiponectin structural gene ADIPOQ, with one in intron 2 and the other in the 3′-UTR region (Figure 5). SNP rs3774261 accounted for 1.2% of the total variance of plasma adiponectin level in the pooled sample, 2% in cases and 0.9% in controls. We further evaluated the associations of these two ADIPOQ SNPs with other metabolic parameters, including HDL-C, triglyceride, fasting glucose, insulin, and BMI. Neither of the SNPs achieved statistical significance with any of the parameters (all P > 0.01) (data not shown).
Figure 5.
Linkage disequilibrium structure (expressed as r2) of the eight SNPs in the ADIPOQ gene. *SNPs with significant association with plasma adiponectin level. SNP, single-nucleotide polymorphism.
Similar subgroup analyses were carried out for rs7195409, the SNP located within CDH13. As with the ADIPOQ SNPs, there was no strong evidence for a stronger genotype effect on adiponectin in dyslipidemia cases compared to controls (data not shown).
The associations of adiponectin levels with SNPs in the two other adiponectin receptors were also examined. Adiponectin levels were not associated with SNPs within adiponectin receptor 1 (ADIPOR1; 1q32.1) (marker-wise P values = 0.49–0.95) and receptor 2 (ADIPOR2: 12p13.31) (P values 0.35–0.99). SNPs in both of these receptors were reported to be associated with T2DM in an Amish population (25). Adiponectin levels were not associated with any SNPs flanking the linkage peak at 8p21–23.
DISCUSSION
We have carried out a comprehensive analysis of the entire genome to identify the genetic determinants of plasma adiponectin levels using both linkage and association analyses. Although other linkage (10,16–19) and association (26,27) analyses have been carried out on adiponectin, many have focused on particular candidate genes, including ADIPOQ. Our analyses provide evidence of the major loci contributing to adiponectin variation from a genome-wide perspective.
The most striking result from our analyses was the identification of ADIPOQ as the locus having the largest influence throughout the genome on adiponectin variation. Significant associations were observed in the WGA analysis on several SNPs within ADIPOQ, with two of these meeting genomewide levels of statistical significance. In linkage analysis, this region (3q27–29) also showed the strongest evidence for linkage in the TSE families, albeit with only a modest lod score of 1.70. The failure to detect linkage to adiponectin levels in the NWE families in our study may reflect the low power in these families to detect QTLs by linkage analysis.
Genetic studies have previously implicated the ADIPOQ locus for a role in influencing variation in adiponectin levels. Very strong evidence for linkage of serum adiponectin levels to this region has previously been published in Hispanic families from the IRAS Family Study (lod = 6.35) (18) and suggestive/ tentative evidence for linkage has also been reported from at least two other European white populations (2,17). In the Amish population where a peak lod of 2.13 was observed right at the ADIPOQ locus, the linkage peak could be partially explained by two SNPs within the adiponectin structural gene (2), although this was in contrast to the IRAS Family Study, where none of the associated SNPs contributed to the linkage signal (18).
SNPs in ADIPOQ have been associated with T2DM and obesity in several prior studies. In a Japanese population, ADIPOQ SNPs rs2241766 and rs1501299 have been associated with T2DM (28), while SNPs rs266729 and rs1501299 were reported to be associated with obesity in a French population (29), and SNPs rs182052 and CA-11156 were reported to be associated with obesity in Hispanics (30). Kissebah et al. reported linkage of seven metabolic syndrome-related traits to the 3q27 ADIPOQ region, suggesting that this region was also associated with obesity and insulin sensitivity (31). Several prior studies have also reported associations of ADIPOQ SNPs with plasma adiponectin levels, including rs2241766 (exon 2) (2,27), rs1501299 (intron 2) (2,32), and several SNPs in the promoter regions of ADIPOQ (27).
Low adiponectin levels have previously been associated with various components of the metabolic syndrome, including dyslipidemia (33), and indeed HDL-C levels were strongly correlated with plasma adiponectin levels in the GEMS case– control study (r = 0.40, P < 0.0001). However, there was no evidence in our study that the effects of any of the ADIPOQ SNPs on adiponectin levels were modified in the presence of dyslipidemia because the effect sizes of the SNPs did not differ significantly between subjects with and without dyslipidemia.
An additional intriguing result from the WGA analysis was the association observed between rs7195409, located in intron 7 of CDH13, and adiponectin levels. CDH13 is a member of the cadherin superfamily and was recently found to be expressed in vascular endothelial cells and smooth muscle cells, where it interacts with hexameric and high-molecular weight species of adiponectin but not trimeric or globular species (24,34). The magnitude of association (P = 2.0 × 10−5) did not achieve conventional genome-wide thresholds for statistical significance, but the identification of this gene is intriguing in light of recent evidence that T-cadherin, the product of CDH13, specifically binds with hexamer and high-molecular weight isoforms of adiponectin at vascular endothelial and smooth muscle cells (24). The significance of these isoforms of adiponectin are not entirely clear, but they may activate NF-κB pathways, therefore, regulating antiapoptotic activity (34).
Despite the moderately high heritability of adiponectin (68–70% in our study), our genetic analyses did not uncover any additional genes contributing to variation in this trait. It is sobering that the most strongly associated SNPs in our analysis accounted for <2% of the variance in adiponectin levels in this population. Possibly, the associated SNPs are in linkage disequilibrium with a true functional SNP in the region and this functional variant has a larger effect. More broadly, however, it seems likely that simple sequence variants at a single locus capture only a small part of the total genetic contribution to trait variation—other genetic effects may stem from interaction among single loci and between individual loci and environmental factors, or from epigenetic factors that influence gene expression. Additionally, it is possible that much of the genetic effect on adiponectin is attributable to multiple rare variants, each associated with a relatively large effect. GWAS studies are not well suited for detecting rare variants and sequencing of candidate regions (e.g., ADIPOQ) may be required. We did, however, observe strong evidence for linkage in the NWE families to chromosome 8p23 (BMI-adjusted lod = 3.10; D8S264/ D8S1825). To our knowledge, no previous studies have detected significant linkage (lod >3) to adiponectin levels on chromosome 8p23; however, linkages to BMI have been reported within or around this region, including in Nigerians (35), in European whites from the Framingham Heart Study (36), and in a meta-analysis of five independent genome-wide scans (37). An attractive candidate gene in this region is lipoprotein lipase (LPL, 8p22, 39.3 cM, physical map position: 19.9 Mb), which encoded an enzyme that plays a critical role in transporting fats and breaking down fat-carrying particles and preheparin LPL mass. Although LPL mass has been found to be significantly correlated with insulin sensitivity (38–40) and adiponectin levels (41,42), we did not observe any association between SNPs in LPL and adiponectin in this population.
Our observation of higher plasma adiponectin levels observed in subjects from NWE families compared to subjects from TSE families is intriguing and cannot be explained simply by differences in BMI between the two populations (20,43). Differences in adiponectin levels across ethnic groups have previously been reported—e.g., Japanese > Chinese (10) and Hispanic Americans > African Americans (18). It is possible that these differences stem from differences in other metabolic measures between populations or that they result from differences in other lifestyle factors (20). Higher adiponectin levels observed in women compared to men are also widely observed, and this sex dimorphism might be partially explained by the selective inhibition effect from testosterone in both human and mice (44,45). In the population-based study, although BMI is higher in cases than in controls, the difference of adiponectin levels between cases and controls could not be explained by age, sex, and BMI (P < 0.001). This is consistent with a prior study showing that adiponectin concentrations are correlated with serum lipids independently of BMI and that following weight loss, changes in adiponectin are significantly correlated with changes in HDL-C and triglyceride, independently of changes in BMI and insulin sensitivity (46).
In conclusion, our results support an effect of DNA variation at the ADIPOQ locus influencing plasma adiponectin levels. However, the degree to which DNA sequence variants at this locus influence health and disease remains to be seen. Furthermore, our analyses indicated that SNPs at the ADIPOQ locus were the most strongly associated with adiponectin variation throughout the entire genome. The association of adiponectin with CDH13 is a novel observation and of particular interest in light of recent studies identifying this gene as a third adiponectin receptor.
Supplementary Material
Acknowledgments
This research was supported by GlaxoSmithKline. The genotyping was carried out at the Australian Neuromuscular Research Institute in Perth.
Footnotes
Disclosure
The authors declared no conflict of interest.
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
References
- 1.Arita Y, Kihara S, Ouchi N, et al. Adipocyte-derived plasma protein adiponectin acts as a platelet-derived growth factor-BB-binding protein and regulates growth factor-induced common postreceptor signal in vascular smooth muscle cell. Circulation. 2002;105:2893–2898. doi: 10.1161/01.cir.0000018622.84402.ff. [DOI] [PubMed] [Google Scholar]
- 2.Pollin TI, Tanner K, O’Connell JR, et al. Linkage of plasma adiponectin levels to 3q27 explained by association with variation in the APM1 gene. Diabetes. 2005;54:268–274. doi: 10.2337/diabetes.54.1.268. [DOI] [PubMed] [Google Scholar]
- 3.Hotta K, Funahashi T, Arita Y, et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol. 2000;20:1595–1599. doi: 10.1161/01.atv.20.6.1595. [DOI] [PubMed] [Google Scholar]
- 4.Weyer C, Funahashi T, Tanaka S, et al. Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab. 2001;86:1930–1935. doi: 10.1210/jcem.86.5.7463. [DOI] [PubMed] [Google Scholar]
- 5.Ouchi N, Kihara S, Arita Y, et al. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999;100:2473–2476. doi: 10.1161/01.cir.100.25.2473. [DOI] [PubMed] [Google Scholar]
- 6.Arita Y, Kihara S, Ouchi N, et al. Paradoxical decrease of an adiposespecific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 7.Fruebis J, Tsao TS, Javorschi S, et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA. 2001;98:2005–2010. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi T, Kamon J, Waki H, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 9.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 10.Chuang LM, Chiu YF, Sheu WH, et al. Biethnic comparisons of autosomal genomic scan for loci linked to plasma adiponectin in populations of Chinese and Japanese origin. J Clin Endocrinol Metab. 2004;89:5772–5778. doi: 10.1210/jc.2004-0640. [DOI] [PubMed] [Google Scholar]
- 11.Guerre-Millo M. Adipose tissue hormones. J Endocrinol Invest. 2002;25:855–861. doi: 10.1007/BF03344048. [DOI] [PubMed] [Google Scholar]
- 12.Stefan N, Stumvoll M. Adiponectin – its role in metabolism and beyond. Horm Metab Res. 2002;34:469–474. doi: 10.1055/s-2002-34785. [DOI] [PubMed] [Google Scholar]
- 13.Haluzik M, Parizkova J, Haluzik MM. Adiponectin and its role in the obesity-induced insulin resistance and related complications. Physiol Res. 2004;53:123–129. [PubMed] [Google Scholar]
- 14.Maeda N, Takahashi M, Funahashi T, et al. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50:2094–2099. doi: 10.2337/diabetes.50.9.2094. [DOI] [PubMed] [Google Scholar]
- 15.Yang WS, Tsou PL, Lee WJ, et al. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med. 2003;81:428–434. doi: 10.1007/s00109-002-0409-4. [DOI] [PubMed] [Google Scholar]
- 16.Lindsay RS, Funahashi T, Krakoff J, et al. Genome-wide linkage analysis of serum adiponectin in the Pima Indian population. Diabetes. 2003;52:2419–2425. doi: 10.2337/diabetes.52.9.2419. [DOI] [PubMed] [Google Scholar]
- 17.Comuzzie AG, Funahashi T, Sonnenberg G, et al. The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab. 2001;86:4321–4325. doi: 10.1210/jcem.86.9.7878. [DOI] [PubMed] [Google Scholar]
- 18.Guo X, Saad MF, Langefeld CD, et al. Genome-wide linkage of plasma adiponectin reveals a major locus on chromosome 3q distinct from the adiponectin structural gene: the IRAS Family Study. Diabetes. 2006;55:1723–1730. doi: 10.2337/db05-0428. [DOI] [PubMed] [Google Scholar]
- 19.Tejero ME, Cai G, Goring HH, et al. Linkage analysis of circulating levels of adiponectin in Hispanic children. Int J Obes (Lond) 2007;31:535–542. doi: 10.1038/sj.ijo.0803436. [DOI] [PubMed] [Google Scholar]
- 20.Stirnadel H, Lin X, Ling H, et al. Genetic and phenotypic architecture of metabolic syndrome-associated components in dyslipidemic and normolipidemic subjects: The GEMS Study. Atherosclerosis. 2008;197:868–876. doi: 10.1016/j.atherosclerosis.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 21.Wyszynski DF, Waterworth DM, Barter PJ, et al. Relation between atherogenic dyslipidemia and the Adult Treatment Program-III definition of metabolic syndrome (Genetic Epidemiology of Metabolic Syndrome Project) Am J Cardiol. 2005;95:194–198. doi: 10.1016/j.amjcard.2004.08.091. [DOI] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Hug C, Wang J, Ahmad NS, et al. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci USA. 2004;101:10308–10313. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damcott CM, Ott SH, Pollin TI, et al. Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the Old Order Amish. Diabetes. 2005;54:2245–2250. doi: 10.2337/diabetes.54.7.2245. [DOI] [PubMed] [Google Scholar]
- 26.Fumeron F, Aubert R, Siddiq A, et al. Adiponectin gene polymorphisms and adiponectin levels are independently associated with the development of hyperglycemia during a 3-year period: the epidemiologic data on the insulin resistance syndrome prospective study. Diabetes. 2004;53:1150–1157. doi: 10.2337/diabetes.53.4.1150. [DOI] [PubMed] [Google Scholar]
- 27.Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- 28.Hara K, Boutin P, Mori Y, et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 29.Bouatia-Naji N, Meyre D, Lobbens S, et al. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes. 2006;55:545–550. doi: 10.2337/diabetes.55.02.06.db05-0971. [DOI] [PubMed] [Google Scholar]
- 30.Sutton BS, Weinert S, Langefeld CD, et al. Genetic analysis of adiponectin and obesity in Hispanic families: the IRAS Family Study. Hum Genet. 2005;117:107–118. doi: 10.1007/s00439-005-1260-9. [DOI] [PubMed] [Google Scholar]
- 31.Kissebah AH, Sonnenberg GE, Myklebust J, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara M, Maruoka S, Katayose S. Decreased plasma adiponectin concentrations in women with dyslipidemia. J Clin Endocrinol Metab. 2002;87:2764–2769. doi: 10.1210/jcem.87.6.8550. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi T, Adachi Y, Ohtsuki Y, Furihata M. Adiponectin receptors, with special focus on the role of the third receptor, T-cadherin, in vascular disease. Med Mol Morphol. 2007;40:115–120. doi: 10.1007/s00795-007-0364-9. [DOI] [PubMed] [Google Scholar]
- 35.Adeyemo A, Luke A, Cooper R, et al. A genome-wide scan for body mass index among Nigerian families. Obes Res. 2003;11:266–273. doi: 10.1038/oby.2003.40. [DOI] [PubMed] [Google Scholar]
- 36.Atwood LD, Heard-Costa NL, Cupples LA, et al. Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am J Hum Genet. 2002;71:1044–1050. doi: 10.1086/343822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson L, Luke A, Adeyemo A, et al. Meta-analysis of five genome-wide linkage studies for body mass index reveals significant evidence for linkage to chromosome 8p. Int J Obes (Lond) 2005;29:413–419. doi: 10.1038/sj.ijo.0802817. [DOI] [PubMed] [Google Scholar]
- 38.Hanyu O, Miida T, Obayashi K, et al. Lipoprotein lipase (LPL) mass in preheparin serum reflects insulin sensitivity. Atherosclerosis. 2004;174:385–390. doi: 10.1016/j.atherosclerosis.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Miyashita Y, Shirai K, Itoh Y, et al. Low lipoprotein lipase mass in preheparin serum of type 2 diabetes mellitus patients and its recovery with insulin therapy. Diabetes Res Clin Pract. 2002;56:181–187. doi: 10.1016/s0168-8227(01)00369-2. [DOI] [PubMed] [Google Scholar]
- 40.Miyashita Y, Shirai K. Clinical determination of the severity of metabolic syndrome: preheparin lipoprotein lipase mass as a new marker of metabolic syndrome. Curr Med Chem Cardiovasc Hematol Agents. 2005;3:377–381. doi: 10.2174/156801605774322292. [DOI] [PubMed] [Google Scholar]
- 41.Saiki A, Oyama T, Endo K, et al. Preheparin serum lipoprotein lipase mass might be a biomarker of metabolic syndrome. Diabetes Res Clin Pract. 2007;76:93–101. doi: 10.1016/j.diabres.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi J, Kusunoki M, Murase Y, et al. Relationship of lipoprotein lipase and hepatic triacylglycerol lipase activity to serum adiponectin levels in Japanese hyperlipidemic men. Horm Metab Res. 2005;37:505–509. doi: 10.1055/s-2005-870318. [DOI] [PubMed] [Google Scholar]
- 43.Barter P, Gotto AM, LaRosa JC, et al. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357:1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- 44.Berra M, Armillotta F, D’Emidio L, et al. Testosterone decreases adiponectin levels in female to male transsexuals. Asian J Androl. 2006;8:725–729. doi: 10.1111/j.1745-7262.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- 45.Xu A, Chan KW, Hoo RL, et al. Testosterone selectively reduces the high molecular weight form of adiponectin by inhibiting its secretion from adipocytes. J Biol Chem. 2005;280:18073–18080. doi: 10.1074/jbc.M414231200. [DOI] [PubMed] [Google Scholar]
- 46.Baratta R, Amato S, Degano C, et al. Adiponectin relationship with lipid metabolism is independent of body fat mass: evidence from both cross-sectional and intervention studies. J Clin Endocrinol Metab. 2004;89:2665–2671. doi: 10.1210/jc.2003-031777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.