Abstract
Efficient and functional mitochondrial networks are essential for myocardial contraction and cardiomyocyte survival. Mitochondrial autophagy (mitophagy) refers to selective sequestration of mitochondria by autophagosomes, which subsequently deliver them to lysosomes for destruction. This process is essential for myocardial homeostasis and adaptation to stress. Elimination of damaged mitochondria protects against cell death, as well as stimulates mitochondrial biogenesis. Mitophagy is a tightly controlled and highly selective process. It is modulated by mitochondrial fission and fusion proteins, BCL-2 family proteins, and the PINK1/Parkin pathway. Recent studies have provided evidence that miRNAs can regulate mitophagy by controlling the expression of essential proteins involved in the process. Disruption of autophagy leads to rapid accumulation of dysfunctional mitochondria, and diseases associated with impaired autophagy produce severe cardiomyopathies. Thus, autophagy and mitophagy pathways hold promise as new therapeutic targets for clinical cardiac care.
Keywords: Autophagy, Heart, Mitochondria, Mitophagy, miRNA
Mitochondria are dynamic organelles that use oxidative phosphorylation to supply the energy for myocardial contraction. Efficient clearance of dysfunctional mitochondria prevents activation of cell death pathways, protects against reactive oxygen species (ROS) production, and preserves efficient production of ATP. Mitochondrial clearance is predominantly carried out by mitochondrial autophagy (mitophagy). Autophagy is an evolutionarily conserved cellular recycling process that sequesters cytotoxic protein aggregates, senescent organelles, and other cellular debris in autophagic vesicles and delivers them to lysosomes for destruction.1 Although early work described autophagy as a non-selective, bulk-degradation response during nutrient-deficient conditions, recent studies have demonstrated that autophagy is a tightly regulated process that can selectively target specific organelles, including mitochondria2 and endoplasmic reticulum (ER).3 Damaged mitochondria are labeled and isolated based on reduced membrane potential, enclosed in autophagosomes, and delivered to lysosomes for degradation.4
Multiple Pathways Regulate Myocardial Mitophagy
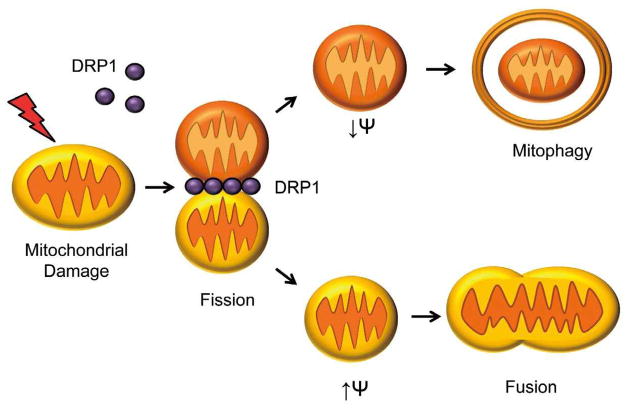
Mitophagy is controlled by proteins that affect mitochondrial morphology, integrity, and ubiquitination. Morphology is regulated by mitochondrial fusion (MFN1, MFN2, OPA1) and fission (DRP1, FIS1) proteins.5,6 Mitochondrial fission has been shown to precede mitophagy, and mitochondrial elongation during starvation prevents mitochondrial destruction by mitophagy (Figure 1).4,7 BCL-2 and BCL-XL are anti-apoptotic proteins that bind BECLIN-1 to prevent its activation, and disruption of this interaction is essential for initiation of autophagy.8 BCL-2 dissociation also allows BECLIN-1 activation by AMBRA1. In order to facilitate phagophore formation, AMBRA1 translocates to the mitochondria and ER after initiation of autophagy. AMBRA1 may also reside on mitochondria in association with BCL-2 until released by autophagic stimuli.9
Figure 1.
Mitochondrial fission precedes mitophagy. Mitochondria undergo fission in response to stress to segregate damaged mitochondrial fragments from healthy mitochondria. These dysfunctional mitochondria are subsequently removed by autophagosomes.
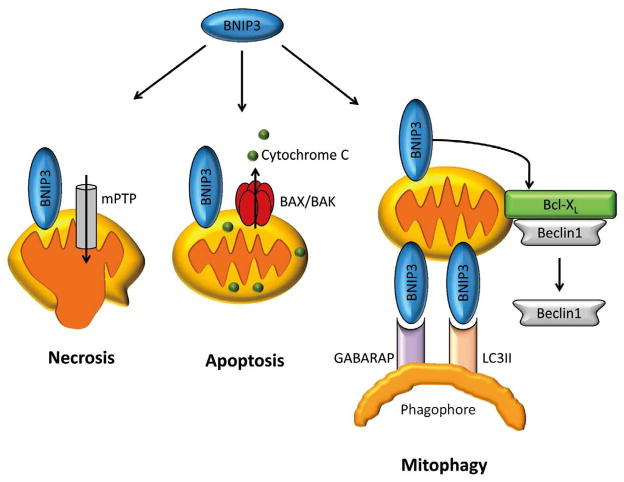
BNIP3 and BNIP3L/NIX are pro-apoptotic BH3-only proteins that cause permeabilization of the mitochondrial membrane via opening of the mitochondrial permeability transition pore or activation of BAX/BAK. These proteins also regulate mitophagy.10,11 BNIP3 resides at mitochondria, and overexpression leads to enhanced mitophagy in myocytes.2,12 BNIP3-mediated mitophagy also occurs in cells lacking BAX/BAK,13 suggesting that the induction of autophagy is separate from its role as a pro-death protein. BNIP3 and NIX interact directly with LC3 and GABARAP on the phagophore to tether mitochondria to forming autophagosomes.3,11 The dual roles of BNIP3 and BNIP3L/NIX emphasize the balance between cell death and mitophagy pathways in the cell (Figure 2). Mice deficient in BNIP3 and NIX accumulate dysfunctional mitochondria in the heart with age, demonstrating the importance of these proteins in normal mitochondrial turnover.14
Figure 2.
Bnip3 can induce both necrotic and apoptotic cell death via opening of the mitochondrial permeability transition pore (mPTP) and activation of BAX/BAK. Bnip3 can also function as receptor for autophagosomes during mitophagy.
In the setting of heart failure, enhanced BNIP3 expression can be detrimental, activating autophagy to the point of cardiac atrophy.15 Mechanical unloading of the heart leads to pronounced upregulation of autophagy and regression of hypertrophy phenotypes that correlate with upregulation of FOXO3 expression. FOXO transcription factors, including FOXO3 and FOXO1, promote autophagy by binding GABARAP and Atg12 promoters.16 Recent studies demonstrate that FOXO3 overexpression can drive autophagy to pathologic levels, upregulating BNIP3 and leading to destruction of cellular components.15 These studies underscore the extensive regulatory network for finely tuned autophagy, and highlight BNIP3 expression as an important autophagic trigger.
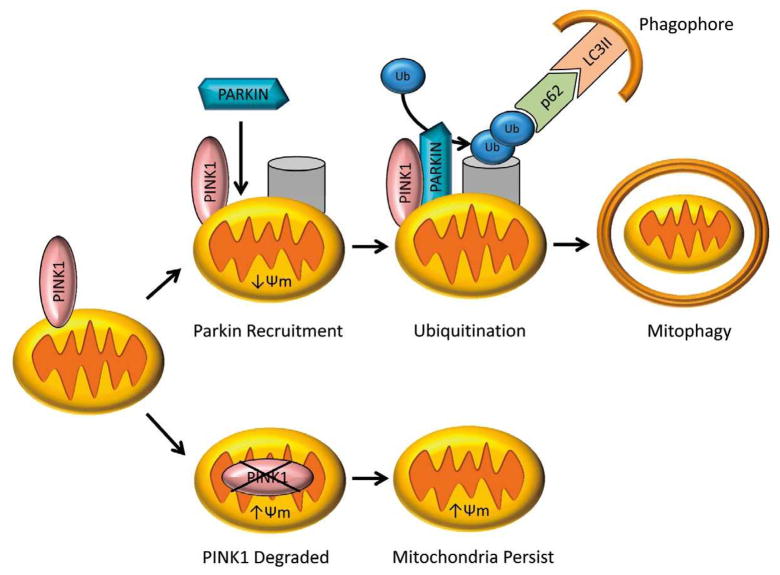
Mitophagy is also regulated by Parkin, an E3 ubiquitin ligase that is mutated in autosomal recessive Parkinson’s disease.17 Recent work demonstrates that Parkin is important for clearance of damaged mitochondria in the heart after myocardial infarction.18 Parkin is localized to the cytosol, but translocates to mitochondria with reduced membrane potential where it ubiquitinates protein targets.19 The adaptor protein p62 then binds ubiquitinated mitochondrial proteins and LC3 on autophagosomes, recruiting autophagic membranes for mitochondrial clearance (Figure 3). PINK1 is a serine/threonine kinase that recruits Parkin to depolarized mitochondria. In mitochondria with intact membrane potential, PINK1 is imported and degraded. In mitochondria with reduced membrane potential, PINK1 breakdown is impaired, causing it to accumulate on the outer mitochondrial membrane and recruit Parkin through direct interaction, Parkin phosphorylation, or phosphorylation of mitochondrial targets.20–23 A recent study by Chen and Dorn reported that PINK1 phosphorylates the mitochondrial fusion protein MFN2, which then acts as a mitochondrial receptor for Parkin.24 PINK1 can also phosphorylate MIRO, an atypical Rho GTPase that tethers mitochondria to the tubulin network. Phosphorylation of MIRO1 by PINK1 leads to ubiquitination by Parkin and proteosomal degradation, isolating damaged mitochondria from tubulin transport and the mitochondrial network.25 Surprisingly, the PINK1/Parkin axis also regulates turnover of specific respiratory chain components, suggesting additional roles for the PINK1/Parkin pathway in regulating mitophagy.26
Figure 3.
Mitophagy is activated by changes in mitochondrial membrane potential (Δ ψ m). Loss of Δ ψ m leads to accumulation of PINK1 and subsequent translocation of Parkin to mitochondria. Parkin ubiquitinates proteins in the outer mitochondrial membrane. The ubiquitin serves as a marker for autophagosomes to degrade these mitochondria.
A new study by Fu et al27 has revealed that other E3 ubiquitin ligases are involved in the regulation of mitophagy. This study demonstrated that glycoprotein 78 (GP78) mediates destruction of mitofusins in the setting of mitochondrial damage/depolarization, leading to mitochondrial fragmentation and autophagy. This E3 ligase pathway operates in Parkin-null cells, showing overlapping patterns of mitophagy regulation by independently functioning ubiquitin ligases.27
Regulation of Mitophagy by miRNAs
Many of the proteins that regulate mitophagy are modulated by microRNAs (miRNAs). These small, non-coding RNA sequences prevent protein translation by binding to complementary messenger RNA (mRNA) in the 3′ untranslated region.28 Post-transcriptional regulation by miRNAs may affect single genes or gene families, ultimately modifying the expression of more than 60% of protein coding genes in animals.28 miRNAs have been implicated in cardiac development, conduction abnormalities, pathologic hypertrophy, and heart failure.29,30 Given their role in cardiac disease, it is not surprising that miRNAs profoundly affect mitophagy in the myocardium.
Reduced Parkin-mediated mitophagy contributes to development of Parkinson’s disease, and Parkin expression can be regulated by miR-34 in affected neurons from Parkinson’s patients.31 In brain regions affected by pathology, miR-34b and miR-34c are downregulated, leading to reduced Parkin expression, mitochondrial dysfunction, increased ROS production, and decreased ATP. Surprisingly, suppression of miR-34 in mice subjected to myocardial infarction (MI) or pressure overload using a nucleic acid antagonist (antagomir) reduced the cardiac damage markers and improved cardiac function.32 This result is unexpected given that genetic ablation of Parkin increased the severity of MI in mice.18 Further studies are needed to determine how miR-34 affects Parkin and its other protein targets in cardiac tissue.
miR-351 plays a critical role in NIX regulation and erythropoiesis. This miRNA antagonizes NIX until repression by a transcription factor called KRAB and its co-repressor KAP1 downregulate miR-351 and allow upregulation of NIX to promote mitophagy. The resulting NIX expression eliminates mitochondria from mature red blood cells. Loss of KAP1 leads to decreased expression of several mitophagy genes, leading to mitochondrial persistence in red blood cells.33 miR-351 is downregulated in the heart by ischemia, but upregulated by hypertrophy.34 These events may correlate with increased mitophagy in ischemic hearts and reduced autophagy in the early phase of cardiac hypertrophy.35
Recent work by Ucar et al demonstrates that miR-212 and miR-132 regulate autophagy and promote cardiac hypertrophy.36 Specifically, the study shows that miR-212/132 knockout mice are protected from pressure overload, whereas animals overexpressing these miRNA proceed rapidly to hypertrophy and failure. These miRNAs activate calcineurin/NFAT hypertrophy signaling and target FOXO3, reducing the heart’s autophagy response.36 Antagonizing these miRNAs with antagomirs rescued the mice from cardiac hypertrophy and may prove therapeutic in the clinical setting. Finally, miRNA are degraded by autophagy, and disruption of autophagy leads to dysregulation in miRNA and their protein targets.37,38 Although these pathways are insufficiently characterized in the heart at present, their effects on mitophagy are likely to significantly modulate cardiac myocyte survival, homeostasis, and stress responses.
Mitophagy in the Myocardium
Basal mitophagy operates constitutively to eliminate damaged/ senescent mitochondria and coordinates with mitochondrial biogenesis to properly balance mitochondrial number with energetic demands.39,40 Under stress such as chronic ischemia, ischemia-reperfusion (IR) injury, and heart failure where there is extensive mitochondrial damage, mitophagy is upregulated to eliminate damaged mitochondria before they cause further damage to the cell. Damaged mitochondria may produce excessive ROS, release pro-apoptotic factors, and trigger necrosis through permeability transition pore opening.41 Depolarized mitochondria may also run in reverse, consuming ATP when cellular stores are already low.42 In addition, autophagy is responsible for clearing mitochondrial DNA (mtDNA) that has been released from ruptured mitochondria. mtDNA that escapes from autophagy can activate the Toll-like receptor 9-mediated inflammatory response.43 Interestingly, disruption of this process can lead to myocarditis and dilated cardiomyopathy.43
Autophagy and mitophagy are important adaptive stress responses that can be rapidly upregulated after cardiac injury. Although electron microscopy has detected occasional mitochondria in autophagosomes in control mice, up to 10% of the autophagosomes in the border zone of the infarct contain mitochondria 8h after injury.44 In addition, Parkin-mediated mitophagy is important for clearance of damaged mitochondria and myocardial recovery after MI. Parkin-deficient hearts rapidly accumulate dysfunctional mitochondria, which lead to cardiac dysfunction and reduced survival.18
Mitophagy is essential in the myocardium, and in mouse models of disrupted mitochondrial turnover, heart failure develops in the absence of other stress. For instance, Nakai et al have demonstrated that loss of ATG5, a critical autophagy protein, in the adult heart leads to rapid cardiac failure and mitochondrial disorganization in adult mice.45 Similarly, loss of MCL-1, an anti-apoptotic BCL-2 protein, in the adult heart leads to impaired autophagy and rapidly culminates in accumulation of dysfunctional mitochondria and heart failure.46 In clinical patients, Danon disease results from a LAMP2 deficiency, which impairs fusion between autophagosomes and lysosomes and causes a lethal cardiomyopathy.47
However, autophagy is not always beneficial. In the setting of increased myocardial demand such as pressure overload, autophagy can lead to pathologic remodeling, contractile dysfunction, and cardiac atrophy.48 Similarly, Matsui et al demonstrated that AMPK-independent autophagy can aggravate cardiac damage during the reperfusion phase of IR injury.49 In summary, context determines whether autophagy promotes survival or cell death. Overlapping regulatory pathways appear to be essential for matching autophagic activity to appropriate cellular conditions.
Mitophagy and Clinical Care
Mitophagy provides a novel therapeutic pathway for cardiac intervention. Cardiovascular disease correlates with energetic and metabolic derangements that severely affect mitochondrial function. These include substrate switching to glycolytic fuel sources, excess ROS production, lipid accumulation, mitochondrial permeabilization, and deficiencies in mitochondrial coupling.50–52 In addition, autophagy is reduced with age and the inadequate removal of dysfunctional mitochondria has been implicated as both cause and consequence of the aging process.53 Novel therapies that maintain mitochondrial integrity and efficiency by facilitating mitophagy have the potential to prevent cardiac damage, heart failure, and age-related cardiomyopathy.
Obesity and type II diabetes lead to cardiac hypertrophy, fibrosis, and dilated cardiomyopathy.54 In the setting of such obesity, cardiac dysfunction is associated with reduced mitochondrial oxidative capacity and increased uncoupling.52 Surprisingly, recent studies reveal that reduced autophagy in the setting of elevated glucose is adaptive, reducing cardiomyocyte injury.55,56 Reduced general autophagy may activate alternative mitophagy pathways, clearing damaged mitochondria to preserve cardiac function.
Recent evidence demonstrates that increased glucose metabolism, mTOR activation, and ER stress are elevated in failing human hearts prior to structural changes such as cardiac hypertrophy. Strikingly, these metabolic changes are reversed by mechanical unloading with a ventricular assist device.57 The role of mitophagy in the clinical setting is still controversial. Restoration of impaired mitophagy may lead to improved mitochondrial quality control and efficiency. In addition, treatments that support mitophagy may alleviate mitochondrial dysfunction caused by current therapies such as nitroglycerin treatment.58 In contexts such as pressure overload, however, unchecked upregulation of autophagy may be detrimental. Future studies should address the pathologic contexts in which autophagy is beneficial or detrimental, as well as differentiating between the effects of general autophagy and targeted mitophagy in disease.
Conclusion
Significant progress has been made in characterizing the pathways that regulate mitophagy in the heart. Researchers have discovered several miRNAs that regulate essential mitophagy proteins. These regulatory miRNAs present new therapeutic targets and clinical biomarkers for heart disease. Although BNIP3/NIX and the PINK1/Parkin pathways are involved in targeting mitochondria to the autophagosomes, it is still unclear how mitochondrial damage leads to upregulation of autophagy. The molecular mechanism underlying activation of mitophagy should be explored by future research. The current methods of assessing mitophagy in cells and tissues are transmission electron microscopy and colocalization between mitochondrial and autophagosomes using immunostaining.2,18,44 Mitophagy can also be assessed indirectly by measuring the levels of Parkin, LC3II and ubiquitin in mitochondrial fractions to determine whether these mitochondria have been marked for degradation.18 These methods are used to measure the level of mitophagy at a single, fixed time point. A major limitation is the lack of a method to measure the rate of mitophagy in tissues. Such a method would allow researchers to unlock new information about mitophagy in the heart. New tools such as GFP/ mCherry LC3 double transgenic mice, which allow researchers to distinguish between autophagosomes and autolysosomes, may also help elucidate individual steps in mitophagy.59 Ultimately, modulating mitophagy in the setting of cardiac stress such as IR and pressure overload may improve clinical outcomes and prevent progression to heart failure.
Acknowledgments
This work was supported by NIH grants R01HL087023 and R01HL101217 (A.B.G.) and a predoctoral fellowship from the American Heart Association’s Western States Affiliate (R.L.T.).
Footnotes
Disclosures
Conflict of Interest: Travel expenses have been provided for Åsa Gustafsson to travel to various scientific meetings to present data.
References
- 1.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 2.Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorn GW., II Mitochondrial dynamism and cardiac fate. Circ J. 2013;77:1370–1379. doi: 10.1253/circj.cj-13-0453. [DOI] [PubMed] [Google Scholar]
- 6.Zungu M, Schisler J, Willis MS. All the little pieces: Regulation of mitochondrial fusion and fission by ubiquitin and small ubiquitin-like modifier and their potential relevance in the heart. Circ J. 2011;75:2513–2521. doi: 10.1253/circj.cj-11-0967. [DOI] [PubMed] [Google Scholar]
- 7.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 13.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorn GW., 2nd Mitochondrial pruning by Nix and BNip3: An essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao DJ, Jiang N, Blagg A, Johnstone JL, Gondalia R, Oh M, et al. Mechanical unloading activates FoxO3 to trigger Bnip3-dependent cardiomyocyte atrophy. J Am Heart Assoc. 2013;2:e000016. doi: 10.1161/JAHA.113.000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 18.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong H, Wang D, Chen L, Choo YS, Ma H, Tang C, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013;24:1153–1162. doi: 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 30.vanRooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 32.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B, Lu Y, Wang Z. Control of cardiac excitability by microR-NAs. Cardiovasc Res. 2008;79:571–580. doi: 10.1093/cvr/cvn181. [DOI] [PubMed] [Google Scholar]
- 35.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 36.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013;14:568–576. doi: 10.1038/embor.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carreira RS, Lee Y, Ghochani M, Gustafsson ÅB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elrod JW, Molkentin JD. Physiologic functions of cyclophilin D and the mitochondrial permeability transition pore. Circ J. 2013;77:1111–1122. doi: 10.1253/circj.cj-13-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grover GJ, Atwal KS, Sleph PG, Wang FL, Monshizadegan H, Monticello T, et al. Excessive ATP hydrolysis in ischemic myocardium by mitochondrial F1F0-ATPase: Effect of selective pharmacological inhibition of mitochondrial ATPase hydrolase activity. Am J Physiol Heart Circ Physiol. 2004;287:H1747–H1755. doi: 10.1152/ajpheart.01019.2003. [DOI] [PubMed] [Google Scholar]
- 43.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, et al. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 46.Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, et al. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 2013;27:1365–1377. doi: 10.1101/gad.215871.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 48.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 50.Luk TH, Dai YL, Siu CW, Yiu KH, Li SW, Fong B, et al. Association of lower habitual physical activity level with mitochondrial and endothelial dysfunction in patients with stable coronary artery disease. Circ J. 2012;76:2572–2578. doi: 10.1253/circj.cj-12-0364. [DOI] [PubMed] [Google Scholar]
- 51.Aubert G, Vega RB, Kelly DP. Perturbations in the gene regulatory pathways controlling mitochondrial energy production in the failing heart. Biochim Biophys Acta. 2013;1833:840–847. doi: 10.1016/j.bbamcr.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudina S, Sena S, O’Neill BT, Tathireddy P, Young ME, Abel ED. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation. 2005;112:2686–2695. doi: 10.1161/CIRCULATIONAHA.105.554360. [DOI] [PubMed] [Google Scholar]
- 53.Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: Mechanisms and therapeutic opportunities. Circ Res. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eguchi K, Boden-Albala B, Jin Z, Rundek T, Sacco RL, Homma S, et al. Association between diabetes mellitus and left ventricular hypertrophyina multiethnic population. Am J Cardiol. 2008;101:1787–1791. doi: 10.1016/j.amjcard.2008.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi S, Xu X, Chen K, Liang Q. Suppression of autophagy is protective in high glucose-induced cardiomyocyte injury. Autophagy. 2012;8:577–592. doi: 10.4161/auto.18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu X, Kobayashi S, Chen K, Timm D, Volden P, Huang Y, et al. Diminished Autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem. 2013;288:18077–18092. doi: 10.1074/jbc.M113.474650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sen S, Kundu BK, Wu HC, Hashmi SS, Guthrie P, Locke LW, et al. Glucose regulation of load-induced mTOR signaling and ER stress in mammalian heart. J Am Heart Assoc. 2013;2:e004796. doi: 10.1161/JAHA.113.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreira JC, Mochly-Rosen D. Nitroglycerin use in myocardial infarction patients. Circ J. 2012;76:15–21. doi: 10.1253/circj.cj-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terada M, Nobori K, Munehisa Y, Kakizaki M, Ohba T, Takahashi Y, et al. Double transgenic mice crossed GFP-LC3 transgenic mice with alphaMyHC-mCherry-LC3 transgenic mice are a new and useful tool to examine the role of autophagy in the heart. Circ J. 2010;74:203–206. doi: 10.1253/circj.cj-09-0589. [DOI] [PubMed] [Google Scholar]