Abstract
Specification of both neural crest cells and Rohon–Beard (RB) sensory neurons involves a complex series of interactions between the neural and non-neural ectoderm. The molecular mechanisms directing this process are not well understood. The zebrafish narrowminded (nrd) mutation is unique, since it is one of two mutations in which defects are observed in both cell populations: it leads to a complete absence of RB neurons and a reduction in neural crest cells and their derivatives. Here, we show that nrd is a mutation in prdm1, a SET/zincfinger domain transcription factor. A Morpholino-mediated depletion of prdm1 phenocopies the nrd mutation, and conversely overexpression of prdm1 mRNA rescues the nrd RB sensory neuron and neural crest phenotype. prdm1 is expressed at the border of the neural plate within the domain where neural crest cells and RB sensory neurons form. Analysis of prdm1 function by overexpression indicates that prdm1 functions to promote the cell fate specification of both neural crest cells and RB sensory neurons, most likely as a downstream effector of the BMP signaling pathway.
Keywords: Narrowminded, Neural crest, Rohon–Beard sensory neurons, Blimp-1
Introduction
Neural crest cells, RB sensory neurons and cranial placodes form at the border between the neural and nonneural ectoderm in response to signaling mechanisms along the mediolateral axis of the embryo, including factors such as BMPs, Wnts, and FGFs. Although these proteins are required to specify a narrow region that is permissive for formation of the cell types at the neural plate border (Garcia-Castro, 2002; Garcia-Castro and Bronner-Fraser, 1999; Lewis et al., 2004; Liem et al., 1995; Mayor et al., 1997; Nguyen et al., 1998, 2000); for review, see Knecht and Bronner-Fraser, 2002), it is not clear how these signals are integrated to specify these cell types or to define their stereotypic position in the embryo. Neural crest cells are specified early in embryogenesis and migrate extensively to form the peripheral nervous system (PNS), craniofacial structures as well as other derivatives (Le Douarin, 1982). RB sensory neurons transduce mechanosensory stimuli in response to touch and remain within the central nervous system (CNS; Lamborghini, 1980). How these two populations are specified within a specific domain at the lateral neural plate remains an interesting question. Evidence from lineage experiments suggest that both neural crest cells and RB sensory neurons can arise from a common progenitor population but typically do not share a common precursor (Cornell and Eisen, 2000; Jacobson and Moody, 1984). Because these two cell types can arise from one precursor, it remains an unanswered question if the characteristic mechanism for neural crest and RB sensory neuron formation is to be generated from one precursor or two separate precursors within the same domain. But if they can share a common precursor, it is interesting to consider how a precursor cell is capable of giving rise to very different populations of neurons. One possibility is that a similar set of combinatorial transcription factors provides the information for the differentiation of neural crest and RB sensory neurons. We have determined that the nrd mutation is an excellent model for the study of questions related to cell fate specification between progenitors for RB sensory neurons and neural crest cells, since these populations are absent or reduced in mutant embryos. nrd was isolated as part of a small scale ENU-based mutagenesis screen in which mutations that affect neurogenesis and neural crest formation were isolated (Artinger et al., 1999; Kim et al., 2000). We have determined that nrd acts cell-autonomously to specify RB sensory neurons (Artinger et al., 1999), suggesting that this molecule integrates several signals that specify cell fate at the border of the neural plate. While RB sensory neurons fail to form at all, neural crest cells do arise in nrd mutant embryos but at reduced numbers as compared to wildtype in all neural crest derivatives studied including PNS neurons, melanocytes and craniofacial cartilage. Thus, knowledge of nrd will shed light on the specification of both of these very different cell types that may share a common precursor.
The cell fate apportionment within the region involves the combinatory expression of several transcription factors, such as neurogenins, components of the notch pathway, dlx and msx, that refine the initial pattern established by the inductive signals and restrict the region competent to form the cell types at the neural plate border (Cornell and Eisen, 2000, 2002; Tribulo et al., 2003; Woda et al., 2003). prdm1 transcription factors have been shown to be involved in cell fate specification in both the mammalian and zebrafish embryonic development. In the mouse embryo, prdm1 appears to function primarily in the immune system by promoting B-cell differentiation (Shaffer et al., 2002; Shapiro-Shelef et al., 2003; Turner et al., 1994). Expression is observed within the embryonic epidermis, not the CNS (Chang et al., 2002). In Xenopus, the prdm1 orthologue is expressed within the anterior endomesodermal cells and lateral stripes in the neural plate (de Souza et al., 1999) and functions in the formation of the head structures. In zebrafish, prdm1 functions by regulating cell fate between fast and slow twitch muscle fibers (Baxendale et al., 2004). However, the function of prdm1 in the nervous system has not been specifically described. Here, we have shown that zebrafish nrd is a mutation in prdm1. prdm1 is required for the specification of neural crest cells and RB sensory neurons, acting cell autonomously in RB sensory neuron and most likely in neural crest specification, downstream of initial BMP inductive signal.
Materials and methods
Zebrafish strain and maintenance
The zebrafish were maintained according to Westerfield (1993). nrdm805 was isolated from a small scale in situ hybridization based screen described in our previous publication (Artinger et al., 1999), and is in the EKK background. To produce a polymorphic strain for genetic mapping, fish with the nrd mutation were crossed into WIK.
Cloning of nrd(prdm1)
Genomic DNA was isolated from a large number of homozygous nrdm805 embryos. Bulk segregant analysis with SSLP markers was performed to identify the linkage group. Analysis of SSLP markers was used to fine map the mutation. We identified linkage with nrdm805 in a 3-cM interval between z21155 and z6365 on linkage group 16. Technical difficulties required us to regenerate the line, using both a new cross of EKK nrdm805 into WIK and EKK nrdm805:HuC:GFP (generated by A. Chitnis). This allowed us to position nrdm805 closer to z19602. From the Sanger Institute Zebrafish Sequencing Project (http://www.sanger.ac.uk/Projects/D_rerio/), we were able to identify zebrafish prdm1 as a candidate in the region between z6365 and z19602. cDNA was isolated from nrdm805 and wildtype siblings at 48 hpf and prdm1 cDNA was cloned by PCR using Platinum Taq High Fidelity polymerase (Invitrogen) and primers: forward 5′-GGATCCATGTGTGGCTGGGACCAG-3′ and reverse 5′-CTCGAGCTAGGTATCCATGGCCTCCT-3′. Both a wildtype and nrdm805 2.4 kb cDNA fragment was inserted into pCS2+ between BamHI and XhoI and sequenced by ABI-prism (Barbara Davis Center Sequencing Core, UCHSC) for direct comparison. A stop codon denoting a truncation was observed in nrdm805 at amino acid 154 was confirmed in multiple PCR reactions within the two nrdm805 strains.
Whole-mount in situ hybridization and immunohistochemistry
Whole-mount in situ hybridization was performed as described by Thisse (1998). The following DIG-labeled antisense RNA probes (Roche Diagnostics) and Alkaline phosphatase using NBT/BCIP color reaction were used for in situ hybridization: prdm1, HuC (Kim et al., 1996), foxd3 (Odenthal and Nusslein-Volhard, 1998), crestin (Rubinstein et al., 2000). Additionally, fluorescent-labeled probes utilizing Alkaline phosphatase and fast red were used in double in situ hybridization. The fast red reaction was performed first and inactivated with 0.1 M glycine–HCL, followed by the second color reaction. The HNK-1 antibody (Sigma) was used at the dilution 1:1000. Immunohistochemistry was performed as described by Solnica-Krezel and Driever (1994).
Antisense morpholino oligonucleotide and RNA injections
An antisense Morpholino Oligonucleotide (MO) was designed against exon2–intron2 splice site (E2I2) of prdm1 and has the sequence of 5′-TGGTGTCATACCTCTTTGGAGTCTG-3′. The control MO has a sequence of 5′-CCTCTTACCTCAGTTACAATTTATA-3′. The oligonucleotides were dissolved in distilled water. Three to 24 ng prdm1-MO were injected into the yolk of 1- to 8-cell-stage embryos together with 10 kDa lysinated Fluorecein dextran (LFD; Molecular Probes) for a total volume of 6–14 nl. Sense prdm1 RNA was synthesized with Ambion Message Machine kit from PCS2+-prdm1 for rescue experiments. One hundred to 700 pg was injected along with a fluorescent tracer, as for the MO injections.
Nomenclature note
According to the Zfin zebrafish nomenclature guide, narrowminded will now be referred to as prdm1 or U-boot (prdm1).
Results
narrowminded is a mutation in prdm1
To clone the nrd mutation, we first established the position on the genetic map using bulk segregant analysis with single-strand length polymorphic (SSLP) markers (Fig. 1). This analysis initially placed nrd on linkage group 16 between SSLP markers z21155 and z6365 (Fig. 1A). Subsequent analysis revealed that nrd was more closely linked to z19602. Further examination of candidate genes within this interval drew our attention to prdm1, which had recently been positioned by the Sanger Institute Zebrafish Sequencing Project (Zv3 ensembl). prdm1 is a SET/zinc finger transcription factor that is known to be critical for cell fate determination in the mammalian immune system where the protein is called Blimp-1. In the case of mammals, prdm1 acts as a repressor of genes such as beta-interferon (Gyory et al., 2004; Tamura et al., 2003), c-myc (Lin et al., 1997) and pax-5 (Klein et al., 2003), to drive differentiation of a B-cell to a plasma cell. Analysis of prdm1 cDNA from homozygote nrd zebrafish revealed a point mutation that results in a premature stop codon, consistent with ENU-based mutagenesis, and causes a truncation of the protein at amino acid 154 within exon 4 (Figs. 1B, C). The predicted nrd (prdm1) protein contains only part of the SET domain and none of the five zinc finger DNA binding domains. Another alteration in the nrd allele (698 V to M) encodes a missense mutation at the 3′ end of the protein. Since this change is downstream of the mutation that created the stop codon, it would not be represented in the nrd protein. In addition to these mutations, we identified two substitutions in the wildtype EK/WIK strain prdm1 sequence that differ from the published sequence (95 amino acid H to R; and 260 D to G). These differences are seen within the EK/WIK wildtype or heterozygote EK/WIK strain, suggesting that these are polymorphisms between this strain and the published strain (Tubingen). Although we believe that a truncation at position 154 would eliminate DNA binding, and thus create a complete null allele, we cannot rule out the possibility that nrd retains residual activity or harbors neo- or antimorphic activity, perhaps because of cryptic splicing or transcriptional reinitiation from a downstream start codon.
Fig. 1.
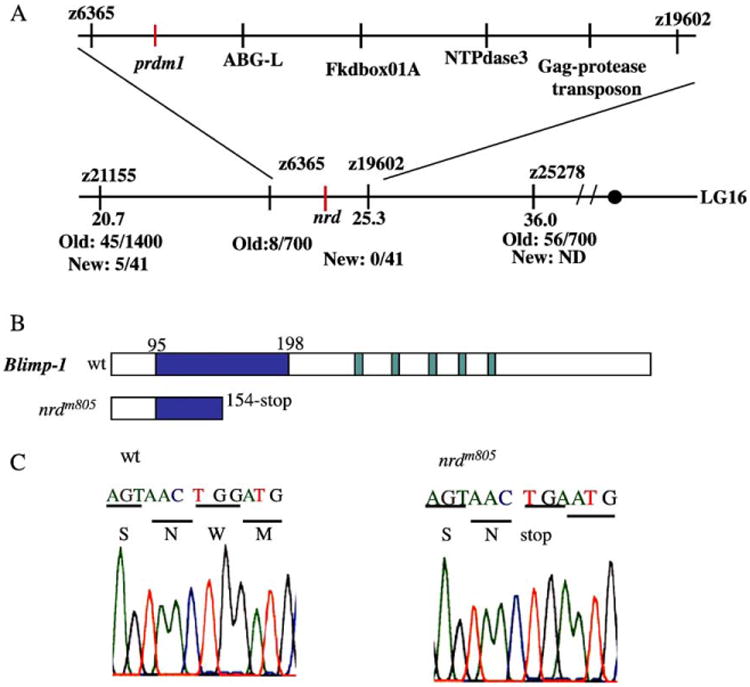
Linkage between nrd and prdm1. (A) Physical and genetic maps of zebrafish linkage group (LG) 16. The genomic interval that contains nrd was determined using bulk segregant analysis with single strand length polymorphic (SSLP) markers (“z” positioned before the number). We positioned nrd between z6365 and z19602. Recombination frequency is listed as old and new. Old is from the original nrd strain nrdm805EK/WIK and New is nrdm805HuC:GFP. Several genes are located in this interval, including prdm1, which we identified as a candidate (Sanger Zv3 ensembl; other genes in the region include ABG-L, Fkdbox01A, NTPdase3, Gag-protease transposon). (B, C) Sequence comparison of wildtype (wt) and nrdm805 prdm1. (B) Diagrams illustrate conceptual wildtype and nrdm805 Blimp-1 (prdm1) protein products. The wildtype sequence contains a SET domain (blue) between amino acid 95–198 and 5 zinc finger DNA binding domains (green). The nrdm805 sequence has a stop codon (TGA) at amino acid 154 (exon 4) within the SET domain and before the zinc finger domains. (C) Sequence comparison between wildtype (wt; left) and nrdm805 (right). Sequence was confirmed in multiple PCR reactions within the two nrdm805 strains, and both contained the stop at position 154.
prdm1 is required for neural crest and Rohon–Beard sensory neuron development
We confirmed that loss of prdm1 is responsible for nrd phenotype by phenocopying the defect by antisense Morpholino (MO) depletion. The Morpholino is predicted to disrupt normal splicing of the prdm1 RNA at the exon2–intron2 boundary (E2I2-MO; Gene Tools, LLC; Draper et al., 2001). The nrd phenotype is characterized by the complete absence of RB sensory neurons, and a reduction in the number of neural crest cells and their derivatives. As visualized with the same panel of markers used to analyze the nrd mutants, injection of prdm1 MO replicated this phenotype: RB sensory neurons were absent and neural crest cell numbers were reduced (prdm1 MO in Figs. 2C, I as compared to wildtype in Figs. 2 A, G and nrd in Figs. 2B, H; Table 1). Embryos injected with standard control MO at the same concentration displayed the wildtype RB sensory neuron and neural crest cell phenotype (data not shown). Similarly, HNK-1 and crestin expression are identical in nrd and prdm1-depleted embryos (Figs. 2F, L as compared to wildtype in Figs. 2D, J and nrd in Figs. 2E, K). The large HNK-1 expressing RB sensory neurons are absent from both nrd and prdm1 MO injected embryos. Often, the nrd mutant embryos maintain projections along dorsal lateral fasciculas (DLF), as well as interneurons cell bodies, which are not observed to the same extent in the prdm1 MO injected embryos (smaller stained neurons in Fig. 2E as compared to 2F). Trunk neural crest cell migration is reduced and disorganized in homozygous nrd and prdm1 MO-injected embryos. Cranial neural crest numbers in nrd embryos are comparable to wild-type levels by 14 hpf of development (Artinger et al., 1999). However, the trunk region remains depleted of neural crest at 24 hpf, suggesting that the mechanism by which neural crest cells are induced in the absence of prdm1 is different along the anterior–posterior axis or that prdm1 is involved in the maintenance of one population over the other (Figs. 2K, L as compared to wildtype in 2J; for cranial regions see (Artinger et al., 1999). In addition to the migrating neural crest cells, we observed a reduction in neural crest derivates, including PNS neurons, melanocytes, and craniofacial cartilage, in the prdm1 MO-injected embryos (similar to the nrd phenotype; data not shown). Interestingly, a higher dose of MO (16–18 ng) had to be injected to generate the RB sensory neuron phenotype than was required for the neural crest phenotype (12 ng), suggesting differential sensitivity to prdm1 dosage (Table 1). Low concentrations of prdm1 may suffice to specify the RB sensory neurons while a higher concentration, more readily reduced by the MO, might be needed to specify the neural crest cells.
Fig. 2.
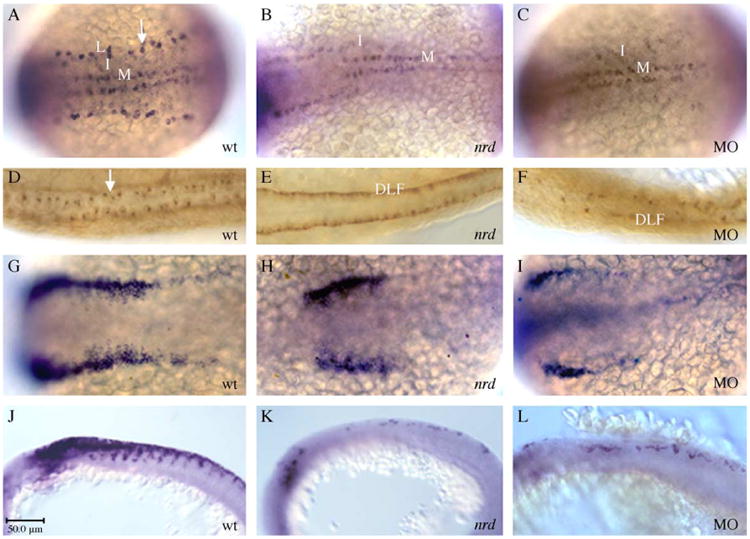
prdm1 MO injection reduces neural crest and eliminates Rohon–Beard sensory neurons, reproducing the nrd phenotype. (A–C, G–I) 11 hpf dorsal views, anterior is to the left. (D–F) Twenty-four hpf dorsal views, anterior to the left. (J–L) Eighteen hpf lateral views, anterior is to the left. (A–C) In situ hybridization with HuC showing the three lateral stripes of primary neurons: lateral (L), intermediate (I) and medial (M). The neurons in the lateral stripe form RB sensory neurons (arrow in A) and are completely absent in nrd (B). Following prdm1 MO injection (C), there is a loss of RB sensory neurons, reproducing the nrd phenotype (B). (D–F) HNK-1 immunohistochemistry at 24 hpf, showing the characteristically large RB sensory neurons in a wildtype embryo (arrow in D). Both nrd (E) and prdm1 MO-injected embryos (F) lack RB sensory neurons. Consistent with a loss of RB sensory neurons, both nrd embryos (H) and prdm1 MO embryos (I) display a reduction in foxd3 expression at 11 hpf as compared to wildtype (G). This early reduction of neural crest remains in older embryos shown with crestin (J–L). Abbreviations: wt, wildtype; nrd, narrowminded mutant embryos; MO, prdm1 MO; DLF, dorsal lateral fasciculas; Scale bar is 50 μm.
Table 1. prdm1 MO and rescue effects in zebrafisha.
| Injection | Amounts (ng/embryo) | N | N nrd −/− | Phenotypeb (% Reduced or absent) | Phenotypeb (% Rescued) | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| NC | RBSNc | NC | RBSNc | ||||
| Control MO | 20 | 22 | 0 | 0 | |||
| prdm1-MO | 6 | 15 | 13 | ||||
| 12 | 47 | 56 | 40 | ||||
| 16–18 | 37 | 57 | 100 | ||||
| 24 | 8 | 100 | |||||
| nrd + prdm1-RNA rescue | 0.1–0.3 | 145 | 32 | 35 | |||
| 0.3–0.6 | 103 | 25 | 20 | 49 | |||
All sets of injection experiments were done at least three times.
Analyzed at the 24 hpf.
RBSN = Rohon–Beard sensory neuron, NC = neural crest.
To provide further support for the hypothesis that nrd is a mutation in prdm1, we injected nrd embryos with prdm1 mRNA. This rescued both the RB sensory neuron and the neural crest phenotype observed in nrd embryos. HNK-1-positive RB sensory neurons appeared in nrd mutant embryos injected with wildtype prdm1 mRNA (Fig. 3E; compared to wildtype in A and nrd in C), and neural crest cells expressing crestin were similar to wildtype levels (Fig. 3F; compared to wildtype in B and nrd in D). Additionally, in the nrd background following injection of prdm1 RNA, we also observed neurons were positioned more ventrally that have the morphology of a RB sensory neuron, which peripheral processes, except they were smaller in size. Thus, prdm1 mRNA is sufficient to drive RB sensory neuron and neural crest cell differentiation in an nrd (prdm1)−/− background. In both cell types, however, we were unable to drive a complete rescue of the nrd mutant phenotype with injection of prdm1 mRNA. This may be due to inadequate levels of prdm1 at the appropriate time in development. We did not observe any difference in dosage sensitivity in regard to the prdm1 RNA injections as with the MO experiments.
Fig. 3.
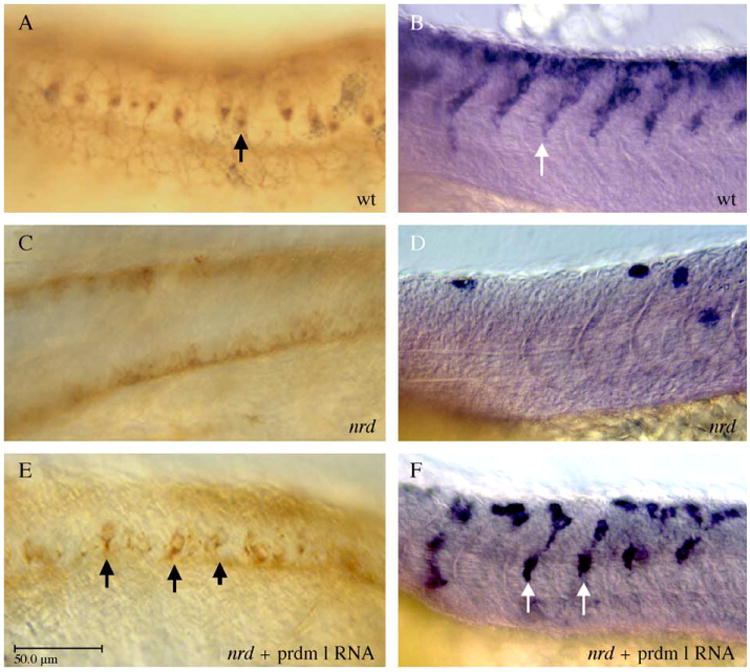
prdm1 mRNA injection can rescue Rohon–Beard sensory neurons and neural crest cells in nrd mutant embryos. HNK-1 immunohistochemistry, dorsolateral views; anterior is to the left. (A) Expression pattern of HNK-1 in RB sensory neurons in a wildtype embryo. (C) nrdm805 embryos lack RB sensory neurons and do not express HNK-1 in the dorsal neural tube. (E) RB sensory neurons are induced to form in nrdm805 mutant embryos after injection of prdm1 mRNA (arrows point to several RB sensory neurons). Crestin expression in lateral views, anterior to the left. (B) Wildtype expression of crestin at 24 hpf. (D) nrd embryos have very little trunk neural crest migration (F) After injection of prdm1 mRNA, numerous crestin-positive neural crest cells can be seen in the migration pathway (arrows). Abbreviations: wt, wildtype; nrd, narrowminded mutant embryos. Scale bar is 50 μm.
prdm1 is expressed at the neural plate border
Given the phenotype of the nrd mutation, we sought to determine if expression of prdm1 is consistent with a role in cell fate specification in the nervous system. At 90% epiboly and continuing to tail bud stage, we find that prdm1 RNA is expressed within a region that spans the border of the neural plate, with more pronounced expression anteriorly (Figs. 4A–C; arrows). By 11 hpf (2–3 somite stage), prdm1 is expressed in a broad domain spanning the neural plate border and also at the midline in the adaxial cells (Fig. 4D; higher magnification in E). Adaxial cells (labeled “A”) form adjacent to the axial hypochord and contain muscle pioneer cells (Thisse et al., 1993). At 13 hpf, expression is decreased within the neural plate border region (Fig. 4F; arrow), but later at 24 hpf, expression is strong in the median fin fold, somites, and posterior fin bud (not shown). An RNA probe to the 3′end of prdm1 revealed reduced prdm1 expression in homozygous nrd (prdm1) mutant embryos (Figs. 4G, H; arrowheads show neural plate border). The observed decrease in prdm1 mRNA could occur for several reasons including absence of prdm1-expressing neural crest cells and RB sensory neurons, non-sense-mediated decay of the RNA, or feedback auto-regulation of prdm1 gene in the wildtype.
Fig. 4.
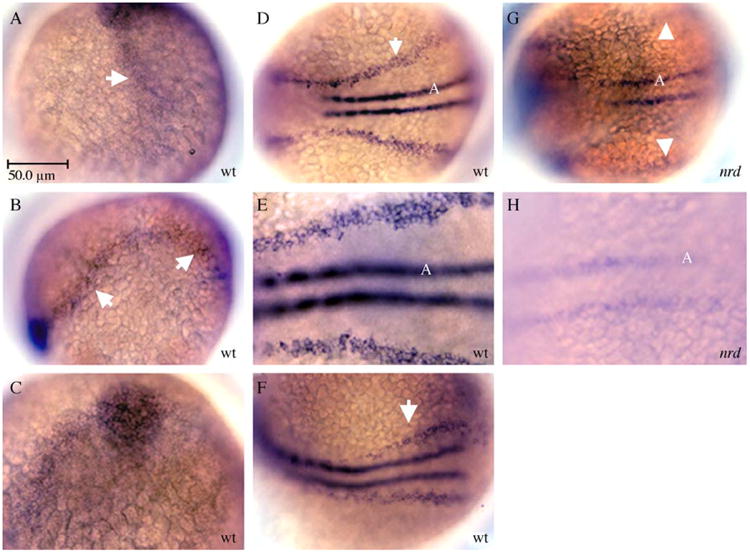
Expression of prdm1 in the nervous system of wildtype and mutant nrd zebrafish embryos. Lateral views (A, B) and dorsal view(C–H) where anterior is to the left, except in A and C where anterior is to the top. (A) At 90% epiboly, expression is observed along the border of the neural plate where neural crest cells and RB sensory neurons are being specified (arrow), more strongly at the anterior-most border. At tailbud stages (B), expression remains high at the neural plate border (arrows). The expression is particularly strong at the anterior border at tail bud stages (C). (D) At 11 hpf, expression is high at the lateral edge of the neural plate (arrow; higher magnification in E), and also is expressed ventrally in the adaxial cells (A) adjacent to the midline. (F) By 13 hpf, the expression at the border of the neural plate remains only in the most posterior region (arrow). (G, H) Analysis of prdm1 expression in nrd mutant embryos at 11 hpf. Low (G) and high (H) magnification images of nrdm805 embryos showing reduced expression of prdm1, especially at the neural plate border (arrowheads in G). A slight reduction in expression is also observed in the ventral adaxial cells (A). Abbreviations: wt, wildtype; nrd, narrowminded mutant embryos; A, adaxial cells. Scale bar is 50 μm.
To confirm that the expression of prdm1 is within the domain where neural crest and RB sensory neurons arise, we performed double in situ hybridization analysis with Foxd3 and HuC. prdm1 is expressed in a broad domain at the lateral edge of the neural plate, which includes neural crest, RB sensory neurons, and cranial placodes. At 12 hpf, HuC is expressed in all primary neurons, including the RB sensory neurons (Figs. 5A, B; blue), and overlaps with the prdm1 expression (Fig. 5B; shown overexposed at higher magnification in red). prdm1 is also expressed within the same domain as Foxd3-positive neural crest cells (blue with prdm1 in red; Figs. 5C, D). Based on this co-expression analysis, prdm1 is expressed within the same domain as forming neural crest cells and RB sensory neurons.
Fig. 5.
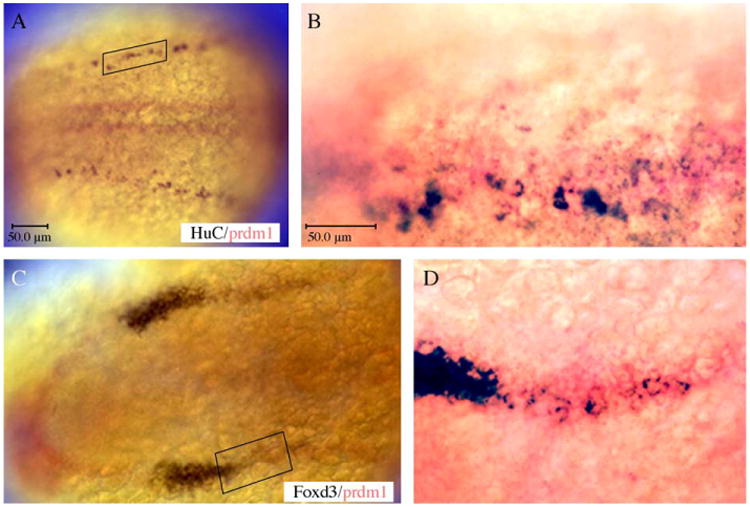
prdm1 is expressed in the domain where neural crest and Rohon–Beard sensory neurons form. Double in situ hybridization of prdm1 with Foxd3 and HuC at 12 hpf; dorsal views, anterior to the left. (A, B) Low and high magnification views of prdm1 (red) and HuC (blue) expression. HuC-positive cells are co-expressed in the prdm1 expression domain. Boxes indicate region viewed under higher magnification in (B, D). (C, D) prdm1 (red) and Foxd3 (blue) expression indicate that neural crest cells are formed within the prdm1-expressing domain. Scale bar is 50 μm.
prdm1 functions downstream of the initial BMP inducing signal
Previous studies have implicated the BMP signaling pathway in the initial induction of neural crest cells. To determine how prdm1 fits into the hierarchy of the BMP pathway during specification of cells at the neural plate border, we examined its expression in several BMP mutations: swirl (swr) which encodes zebrafish BMP2b (Kishimoto et al., 1997), somitabun (sbn) which encodes the BMP mediator Smad5 (Hild et al., 1999), and snailhouse (snh), which encodes BMP7 (Dick et al., 2000). Each of these BMP signaling components have previously been shown to be required for specification of neural crest cells and dorsal and intermediate neurons in the spinal cord in zebrafish (Nguyen et al., 1998, 2000), and their homologues also function in the induction of neural crest cells in chick and Xenopus (Liem et al., 1995; Marchant et al., 1998). prdm1 expression is eliminated at the neural plate border in swr (Bmp2b), expanded in sbn (Smad5) and reduced in snh (BMP7) mutant embryos (Figs. 6B–D, ventral views; higher magnification in Figs. 6F–H as compared to wildtype in Figs. 6A, E). This is consistent with the reduction of the domain of neural crest and RB sensory neurons in swr (Bmp2b) and snh (BMP7), and an expansion in sbn (Smad5), shown schematically below the images in Fig. 6 (as observed by Nguyen et al., 1998, 2000). Dorsal views are shown in Figs. 6J–L, as compared to wildtype in Fig. 6I, indicating the maintenance of adaxial prdm1 expression (A) in all BMP pathway mutants. Thus, the neural plate border corresponds to the prdm1 expression domain: In swr, the neural plate border region does not form, and in sbn and snh, the border is shifted to the ventral side (arrows; Figs. 6F–H). These data suggest that prdm1 acts downstream of the initial BMP inducing signal or is dependent on BMP signaling for its expression.
Fig. 6.
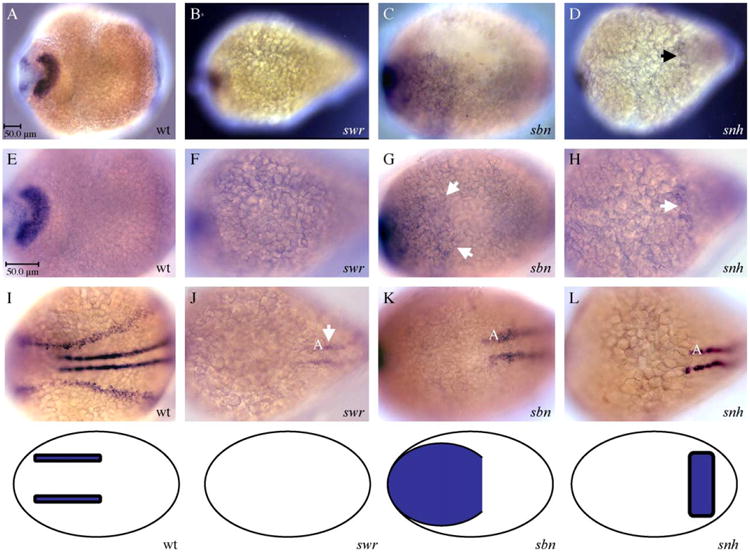
Expression of prdm1 in the nervous system of BMP mutant zebrafish embryos. BMP mutants are visualized from the ventral side, where the reduction or expansion of the neural crest/RB sensory neuron domain is observed. Ventral (A–H) views of 12 hpf wildtype and BMP mutant embryos. swr (Bmp2b) mutant embryos (B, F) show an absence of prdm1 mRNA at the lateral border of the neural plate (compare to wildtype in A, E). The domain of expression of prdm1 is expanded in sbn (Smad5) embryos at 12 hpf (Low (C) and high (G) magnification; ventral views, compare to ventral view in wildtype in E), consistent with an increase of the neural crest/RB sensory neuron domain. Low (D) and high (H) magnification views of 12 hpf snh (Bmp7) showing a reduction in expression in the neural crest/RB sensory neuron domain of these embryos (arrows in D, H; compare to wildtype in A, E). Dorsal view of wildtype embryo in (I) as compared to dorsal views of swr (J), sbn (K), and snh (L) show that the adaxial expression (arrow in J) remains. Below, images are schematic representations of the neural crest domain in each of the mutant embryos as compared to wildtype. Since the BMP mutants are ventralized and the neural plate domain is expanded, the neural plate border form on the ventral side in sbn and snh and is completely absent in swr. Abbreviations: wt, wildtype; swr, swirl; sbn, somitabun; snh, snailhouse; A, adaxial cells. For (A–D), scale bar is shown in A and is 50 μm; for (E–L), scale bar is in E and is 50 μm.
prdm1 functions to promote both neural crest and Rohon–Beard sensory neurons
To determine how prdm1 functions in the specification and differentiation of neural crest cells and RB sensory neurons, we performed ectopic expression of prdm1. If neural crest cells and RB sensory neurons derive from a common precursor, prdm1 might mediate a switch between these two cell types within the precursor population. To examine this possibility, we overexpressed prdm1 RNA in wildtype embryos. Overexpression of prdm1 increased the number of RB sensory neurons as shown by the increased number of HNK-1-positive cell bodies at 24 hpf (Fig. 7B as compared to wildtype in 7A.) Wildtype embryos averaged 54 HNK+ cell bodies/20 somite length compared to 58–60 in prdm1 overexpressing embryos (Table 2). A noticeable difference observed between wildtype and embryos overexpressing prdm1 is the spacing between the cell bodies. In wildtype embryos, there is one cell body in each segment per side, with a space between the neurons (Fig. 7A arrow). The embryos overexpressing prdm1 have more cell bodies per segment and the spacing between the cell bodies is reduced or absent (Table 2; Fig. 7B arrows). Although the wildtype cell bodies are close together, they typically do not touch each other as seen in the embryos overexpressing prdm1. In addition to the increased number of cell bodies, the processes that intervate the skin appear to have increased, with a thicker diameter of the axon in prdm1-overexpressing embryos (Fig. 7B as compared to wildtype in 7A; arrowheads). The increase in the number of RB sensory neurons was often observed to be one sided, which can be explained by a sidedness of the injection. Analysis of crestin expression at 24 hpf shows that neural crest cell numbers also increased after ectopic expression of prdm1 (Fig. 7C as compared to D). In wildtype embryos, neural crest cells begin their migration from the dorsal aspect of the neural keel and migrate along defined pathways through the somite, with a characteristic single line of cells in each pathway at one time (Fig. 7C arrows). After ectopic expression of prdm1, the numbers of migrating cells appear to have increased such that many more cells are exiting the neural keel (Fig. 7D, arrowheads), with the appearance of more than one cell diameter in the somite at one time (Fig. 7D, arrows). Interestingly, in the injected embryos, the normal migration seems to be accelerated to a more ventral location as compared to wildtype. These data suggest that prdm1 can promote the formation of both Rohon–Beard sensory neurons and neural crest cells.
Fig. 7.
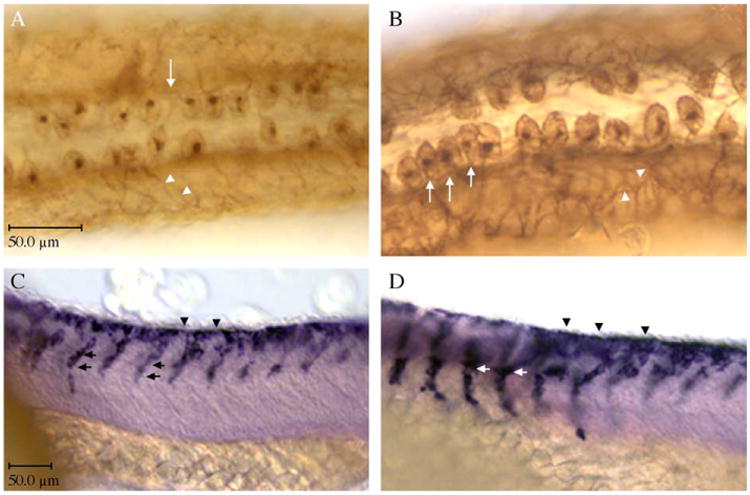
Ectopic expression of prdm1 increases neural crest and Rohon–Beard sensory neurons. (A, B) HNK-1 immunohistochemistry at 24 hpf; dorsal views, anterior to the left. (A) Wildtype expression of HNK-1, (B) Subsequent to prdm1 mRNA injections, RB sensory neurons are increased (arrows). (C, D) Crestin expression at 24 hpf, lateral views, anterior to the left. (C) Wildtype expression of crestin, (D) Following injection of prdm1 mRNA, the neural crest emigration and migration streams appear thick with cells (arrows) as compared with the same region in wildtype embryos. The migration of the cells also appears premature in the prdm1 expressing embryos. At the same time point in development, cells in prdm1-expressing embryos have migrated down to the yolk, whereas wildtype cells are only half way down their migration path. Scale bar for A, B is in A and is 50 μm; for C, D the scale bar is in C and is 50 μm.
Table 2. Overexpression effect of prdm1 on the numbers of RB sensory neuronsa.
| Injection | Amounts (ng/embryo) | N | Average number of RB sensory neuronsb |
|---|---|---|---|
| Uninjected | 0 | 12 | 54.8 ± 1.3 |
| prdm1 RNA | 0.25 | 10 | 59.2 ± 2.4 |
| 0.7 | 22 | 58.7 ± 2.0 | |
| 0.91 | 6 | 60 ± 4.8 |
All sets of injection experiments were done at least three times.
The number of Rohon–Beard sensory neurons (RBSN) were counted on each side of a 24-hpf embryo, visualized with HNK-1 immunohistochemistry.
Discussion
Taken together, our analysis indicates that zebrafish prdm1 is required for RB sensory neuron and neural crest cell specification. Several studies have shown that prdm1 functions in cell fate specification during vertebrate embryogenesis. Mammalian prdm1 is necessary and sufficient to drive differentiation of plasma cells from B-cells (Shaffer et al., 2002; Shapiro-Shelef et al., 2003; Turner et al., 1994). The targeted disruption of prdm1 in mouse results in an early embryonic lethal suggesting that prdm1 functions in multiple cells types during development (Shapiro-Shelef et al., 2003). A nervous system defect has not specifically been noted, but the possibility remains that a phenotype exists but is partially masked by redundant or compensatory mechanisms. Thus, it would be interesting to determine in detail the nervous system phenotype in the prdm1 null mice. With respect to the zebrafish, we have not examined nrd (prdm1) mutant embryos for immune deficits. Since nrd mutant zebrafish die during early embryogenesis, they might not survive long enough to visualize defects in B-cell development. It is known that, in addition to the neural phenotype described here, zebrafish and Xenopus prdm1 functions during gastrulation in the formation of the head structures and to control cell movements (de Souza et al., 1999; Bischof and Driever, personal communication; Solnica–Krezel, personal communication).
Another zebrafish mutation, U-boot, has recently been shown to alter the prdm1 coding region. U-boot (prdm1) mutant embryos display a U-shaped somite phenotype and lack slow twitch muscle fibers (Baxendale et al., 2004). The sequence differences between nrd and U-boot may explain the differing phenotypes. As shown here, nrd truncates prdm1 at amino acid 154 within the SET domain, whereas U-boot is a missense mutation within two sites, one within the second zinc finger domain and the other between the SET domain and the first zinc finger (Baxendale et al., 2004). Whereas nrd appears to be a null allele, U-boot is predicted to be a hypomorphic allele that would elicit a milder phenotype. Consistent with this possibility, neural crest cell, RB sensory neuron or cranial placode deficits have not been described in U-boot embryos, although pigment and fin defects exist (Kelsh et al., 1996). Therefore, it would be interesting to determine if similar, but subtler, nervous system defects exist in U-boot (prdm1) mutant embryos. We have primarily focused on the nervous system defects in nrd (prdm1). However, we have observed a U-shaped somite phenotype in nrd (data not shown), consistent with those observed in the U-boot mutation, suggesting that similar muscle defects exist.
We have shown that prdm1 RNA expression is altered in mutants that affect BMP signaling, indicating that prdm1 likely functions downstream of BMPs to establish mediolateral pattern at the border of the neural plate. If neural crest cells and RB sensory neurons derive from a common precursor, prdm1 might promote the differentiation of these two cell types within the precursor population. Consistent with this possibility, ectopic expression of prdm1 increased the number of RB sensory neurons and neural crest cells, suggesting that prdm1 function may be more complex than a simple switch between these cells types. The increase in RB sensory neurons after ectopic expression of prdm1 is subtle and shows only a trend towards an increase in the number of neurons. Thus, prdm1 may promote the formation of RB sensory neurons but is not sufficient to override Notch-Delta signaling, which regulates the final cell number via lateral inhibition (Chitnis, 1995; Cornell and Eisen, 2000). Conversely, since both neural crest and RB sensory neurons are deficient in nrd (prdm1) mutants, prdm1 might be required for RB sensory neuron fate and one of several signals that can specify neural crest fates. Previous mosaic analysis suggests that prdm1 functions cell autonomously in the specification of RB sensory neurons (Artinger et al., 1999). Results from coexpression experiments suggest that prdm1 also functions cell autonomously for neural crest cells, since the expression is observed within the neural crest forming domain. Although we cannot rule out the possibility that prdm1 also has a non-cell autonomous role, this suggests that within the precursor cell population, cell intrinsic prdm1-dependent signals instruct precursors to become both neural crest cells and RB sensory neurons, rather then promoting one fate versus another. Our hypothesis is that the precursor population persists in nrd since some neural crest cells still form in mutant embryos. This suggests that these precursors are capable of responding to prdm1-independent signals that also promote the neural crest lineage. Detailed lineage labeling of the precursor populations at the nascent lateral border of the neural plate in wildtype and mutant embryos is needed to resolve this issue. The use of nrd as a probe of prdm1 function will provide a powerful method for investigating the diversification of neurogenic cell fates at the lateral border of the neural plate.
Note added in proof
While this paper was in revision, the identification of a neural crest and Rohon-Beard sensory neuron phenotype in the U-boot mutation, a narrowminded allele, was published by Roy and Ng in Current Biology, Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo, Current Biology 2004 (14), 1772–1777. Ubo does seem slightly less severe then nrd in regards to the neural crest, but very similar in RB sensory neuron phenotype. Interestingly, using a heat shock construct to overexpress Ubo mRNA beginning at 80% epiboly, an increase in RB sensory neurons is demonstrated, suggesting that a high level of expression at the appropriate time may be critical for prmd1/Blimp-1 function.
Acknowledgments
K.B.A would like to thank members of the Mercola and Zon labs, especially N. Trede and B. Paw, for technical and emotional support in regards to mapping; D. Riedel for excellent technical assistance; A. Chitnis for nrdm805:HuC:GFP and discussions; W. Driever and L. Sonica-Kretzel for sharing data prior to publication; and T. Williams and M. Klymkowsky for critically reading the manuscript. We gratefully acknowledge the support of NIH 5 R37 DK055381-05 to L.Z., NIH RO1 HL59502 and RO1 HL67079 and American Cancer Society RPG-97-121-DDC to M.M, and NIH K22 DE14200 to K.B.A. K.B.A. is a recipient of the Basil O'Connor Starter Scholar Research Award from the March of Dimes.
References
- 1.Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126(18):3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36(1):88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 3.Chang DH, Cattoretti G, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech Dev. 2002;117(12):305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 4.Chitnis AB. The role of Notch in lateral inhibition and cell fate specification. Mol Cell Neurosci. 1995;6(6):311–321. [PubMed] [Google Scholar]
- 5.Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127(13):2873–2882. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- 6.Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Integr Annu Index. 2002;129(11):2639–2648. doi: 10.1242/dev.129.11.2639. erratum appears in Development 2002 Jul; 129 13:3279. [DOI] [PubMed] [Google Scholar]
- 7.de Souza FS, Gawantka V, Gomez AP, Delius H, Ang SL, Niehrs C. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann's organizer. EMBO J. 1999;18(21):6062–6072. doi: 10.1093/emboj/18.21.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick A, Hild M, Bauer H, Imai Y, Maifeld H, Schier AF, Talbot WS, Bouwmeester T, Hammerschmidt M. Essential role of Bmp7 (snailhouse) and its prodomain in dorsoventral patterning of the zebrafish embryo. Development. 2000;127(2):343–354. doi: 10.1242/dev.127.2.343. [DOI] [PubMed] [Google Scholar]
- 9.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30(3):154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Castro MI. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297(5582):848–851. doi: 10.1126/science.1070824. see comments. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Castro M, Bronner-Fraser M. Induction and differentiation of the neural crest. Curr Opin Cell Biol. 1999;11(6):695–698. doi: 10.1016/s0955-0674(99)00038-1. [DOI] [PubMed] [Google Scholar]
- 12.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5(3):299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 13.Hild M, Dick A, Rauch GJ, Meier A, Bouwmeester T, Haffter P, Hammerschmidt M. The smad5 mutation somitabun blocks Bmp2b signaling during early dorsoventral patterning of the zebrafish embryo. Development. 1999;126(10):2149–2159. doi: 10.1242/dev.126.10.2149. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson M, Moody SA. Quantitative lineage analysis of the frog's nervous system: I. Lineages of Rohon–Beard neurons and primary motoneurons. J Neurosci. 1984;4(5):1361–1369. doi: 10.1523/JNEUROSCI.04-05-01361.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, Haffter P, Odenthal J, Mullins MC, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Kane DA, Warga RM, Beuchle D, Vogelsang L, Nusslein-Volhard C. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 16.Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Mikki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216(2):109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- 17.Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407(6806):913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124(22):4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- 19.Klein B, Tarte K, Jourdan M, Mathouk K, Moreaux J, Jourdan E, Legouffe E, De Vos J, Rossi JF. Survival and proliferation factors of normal and malignant plasma cells. Int J Hematol. 2003;78(2):106–113. doi: 10.1007/BF02983377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3(6):453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- 21.Lamborghini JE. Rohon–Beard cells and other large neurons in Xenopus embryos originate during gastrulation. J Comp Neurol. 1980;189(2):323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- 22.Le Douarin NM. The neural crest. Cambridge Univ. Press; New York: 1982. [Google Scholar]
- 23.Lewis JL, Bonner J, Modrell M, Ragland JW, Moon RT, Dorsky RI, Raible DW. Reiterated Wnt signaling during zebrafish neural crest development. Development. 2004;131(6):1299–1308. doi: 10.1242/dev.01007. [DOI] [PubMed] [Google Scholar]
- 24.Liem KF, Jr, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82(6):969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276(5312):596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 26.Marchant L, Linker C, Ruiz P, Guerrero N, Mayor R. The inductive properties of mesoderm suggest that the neural crest cells are specified by a BMP gradient. Dev Biol. 1998;198(2):319–329. [PubMed] [Google Scholar]
- 27.Mayor R, Guerrero N, Martinez C. Role of FGF and noggin in neural crest induction. Dev Biol. 1997;189(1):1–12. doi: 10.1006/dbio.1997.8634. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen VH, Schmid B, Trout J, Connors SA, Ekker M, Mullins MC. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev Biol. 1998;199(1):93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen VH, Trout J, Connors SA, Andermann P, Weinberg E, Mullins MC. Dorsal and intermediate neuronal cell types of the spinal cord are established by a BMP signaling pathway. Development. 2000;127(6):1209–1220. doi: 10.1242/dev.127.6.1209. [DOI] [PubMed] [Google Scholar]
- 30.Odenthal J, Nusslein-Volhard C. Fork head domain genes in zebrafish. Dev Genes Evol. 1998;208(5):245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- 31.Rubinstein AL, Lee D, Luo R, Henion PD, Halpern ME. Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis. 2000;26(1):86–97. doi: 10.1002/(sici)1526-968x(200001)26:1<86::aid-gene11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 32.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 34.Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120(9):2443–2455. doi: 10.1242/dev.120.9.2443. [DOI] [PubMed] [Google Scholar]
- 35.Tamura T, Kong HJ, Tunyaplin C, Tsujimura H, Calame K, Ozato K. ICSBP/IRF-8 inhibits mitogenic activity of p210 Bcr/Abl in differentiating myeloid progenitor cells. Blood. 2003;102(13):4547–4554. doi: 10.1182/blood-2003-01-0291. [DOI] [PubMed] [Google Scholar]
- 36.Thisse C, T B. High resolution whole-mount in situ hybridization. Zebrafish Science Monitor. 1998;15:8–9. [Google Scholar]
- 37.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119(4):1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 38.Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130(26):6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- 39.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77(2):297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 40.Westerfield M. The Zebrafish Book. The University of Oregon Press; Eugene, OR: 1993. [Google Scholar]
- 41.Woda JM, Pastagia J, Mercola M, Artinger KB. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130(2):331–342. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]


