Abstract
Flavonoids, due to their pharmacological attributes, have recently become target molecules for metabolic engineering in commonly consumed food crops. Strategies including expression of single genes and gene pyramiding have provided only limited success, due principally to the highly branched and complex biosynthetic pathway of the flavonoids. Transcription factors have been demonstrated as an efficient tool for metabolic engineering of this pathway, but often exhibit variation in heterologous systems relative to that in the homologous system. In the present work, Arabidopsis MYB transcription factor, AtMYB111, has been expressed in tobacco to study whether this can enhance flavonoid biosynthesis in heterologous system. The results suggest that AtMYB111 expression in transgenic tobacco enhances expression of genes of the phenylpropanoid pathway leading to an elevated content of flavonols. However, dark incubation of transgenic and wild type (WT) plants down-regulated both the expression of genes as well as flavonoid content as compared to light grown plants. The study concludes that AtMYB111 can be effectively used in heterologous systems, however, light is required for its action in modulating biosynthetic pathway.
Plants biosynthesize and accumulate a vast array of the small organic molecules which are known as secondary metabolites and primarily meant for the defence against certain biotic and abiotic stresses1,2,3,4. Amongst these, flavonoids, a class of plant phenolics have been extensively studied due to their tremendous chemical diversity, ubiquitous occurrence5 and diverse roles such as signalling molecules, defence chemicals during plant-microbe interaction6,7,8, UV protection2,9, insect resistance4,10, fertility11, and regulation of auxin transport12 etc. Depending upon the oxidation status, hydroxylation, saturation as well as mutual positioning of the phenolic rings in the flavonoid backbone, these compounds can be further classified as flavanones, flavonols, isoflavones, anthocyanins and proanthocyanidins (PA)13,14.
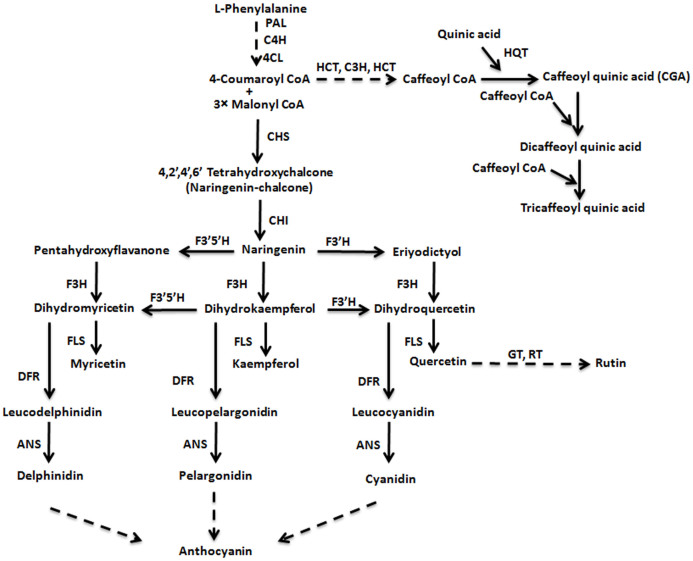
Various studies on the diverse plant species have led to the development of a detailed genetic architecture of the flavonoid biosynthetic pathway providing necessary information about the structural and regulatory genes encoding enzymes and transcription factors, respectively15,16,17. Flavonoids are synthesized from the general phenylpropanoid pathway which is linked to the primary metabolism via phenylalanine ammonia lyase (PAL) enzyme18 (Fig. 1). Chalcone synthase (CHS), the entry step enzyme in flavonoid pathway, catalyzes synthesis of naringenin chalcone using one molecule of coumaroyl Co-A and three molecules of malonyl Co-A as substrates. Further downstream to this step, an assortment of enzymatic activities take place which lead to the biosynthesis of aglycone backbones of diverse flavonoids. The decoration of the aglycone backbones by the modifying enzymes such as acyltransferases (ACTs), glycosyltransferases (GTs) and methyltransferases (MTs) add further chemical diversity to flavonoids and alter their biological properties16,19. Besides acting as a precursor for flavonoid biosynthesis, coumaroyl Co-A synthesized through phenylpropanoid pathway leads to the biosynthesis of chlorogenic acid (CGA) and their derivatives through multiple enzymatic steps (Fig. 1).
Figure 1. Schematic representation of general phenylpropanoid pathway with flavonoid pathway showing biosynthesis of flavonols and caffeoyl quinic acid.
Dashed arrows represent multiple enzymatic steps. ANS, anthocyanidin synthase; CHS, chalcone synthase; CHI, chalcone isomerase; C4H cinnamate 4-hydroxylase; C3H, p-coumaroyl ester 3-hydroxylase; 4CL, 4-coumaroyl CoA ligase; DFR, dihydroflavanol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′5′-hydroxylase; FLS, flavonol synthase; GT, glycosyltransferase; HQT, hydroxycinnamoyl CoA quinate transferase; HCT, p-hydroxycinnamoyl CoA:quinate shikimate p–hydroxycinnamoyltransferase; PAL, phenylalanine ammonia lyase; RT, rhamnosyltransferase.
Transcriptional regulation of the structural genes is the primary mode for the tight control of flavonoid biosynthesis during developmental and other exogenous cues such as light, temperature, biotic and abiotic stresses15,20. The regulatory proteins belonging to the different families such as WD40 repeat, bHLH, MYB, WRKY, Zinc finger and MADS box have been implicated in transcriptional regulation of the flavonoid biosynthesis in the model plant Arabidopsis as well as in other plant species15,20,21,22,23. Biosynthesis of the diverse group of flavonoids remains under the control of a complex consisting of the WD40, bHLH and MYB transcription factor (MBW complex). In monocots e.g. maize, all the genes of the flavonoid pathway are regulated simultaneously as a single unit by this complex. However, the regulation in case of dicots is more complex as entire pathway is regulated in two roughly discrete sets of the co-ordinately regulating units, the early steps (leading to the flavonol and flavones biosynthesis) and the late steps (leading to the anthocyanin and proanthocyanidin production). It has been demonstrated that in contrast to the late steps, the early steps do not require MBW complex15,16,24. Certain MYB transcription factors activate independently distinct set of the structural genes belonging to the early steps (e.g. CHS, F3H, FLS) of flavonoid pathway leading to the flavonol biosynthesis25,26. In addition, various regulatory proteins responsible for the modulation of the expression of genes of the flavonoid biosynthetic pathway and accumulation integrate diverse signalling pathways of the endogenous and exogenous cues.
In past, various efforts have been made to enhance the level of flavonoids in different plants by the genetic engineering approach. The flavonoid-specific MYB transcription factors have emerged as an efficient tool to enhance the level of various class of flavonoids in flavonoid-deficient plant in tissue-specific manner4,27,28. The homologous and heterologous over-expression of MYB transcription factors showed unprecedented effect over the biosynthesis and accumulation of different classes of flavonoids such as anthocyanins, flavonols and PAs24,25,26,29,30. In most of the cases, heterologous expression of MYB transcription factors modulated the flavonoid biosynthetic pathway in a similar manner to homologous system. However, few previous studies suggested certain differences in terms of the target genes and regulation by the MYB transcription factors in heterologous system. On the basis of genome-wide expression studies using AtMYB12 over-expressing tobacco plants, the up-regulation of genes involved in the general phenylpropanoid pathway (C4H and PAL) along with the genes involved in the flavonol biosynthesis (CHS, CHI and FLS) has been reported4. Interestingly, excluding flavonol specific genes, other genes are not conventional targets of AtMYB12 in its homologous system i.e. Arabidopsis. In view of the prospects and potentials of the genetic manipulation of the flavonoids in plants, studies pertaining to the regulation by the heterologous expression of the transcription factors as well as the effects of various exogenous and endogenous cues affecting the flavonoid biosynthesis in transgenic plants are desirable. Such studies will help to ascertain the conditions favourable for enhanced accumulation of the flavonoids. Keeping these factors under consideration, in the present study, modulation of the flavonoid biosynthesis in transgenic tobacco plants expressing Arabidopsis transcription factor AtMYB111 has been undertaken. Our results suggest that the heterologous expression of AtMYB111 in tobacco enhances the biosynthesis of the flavonoids through modulating expression of the structural genes. Our study also suggests that the light regulated factors also play an important role, in addition to AtMYB111, in the regulation of structural genes as well as the flavonoid accumulation.
Results
Development of AtMYB111-expressing transgenic tobacco lines
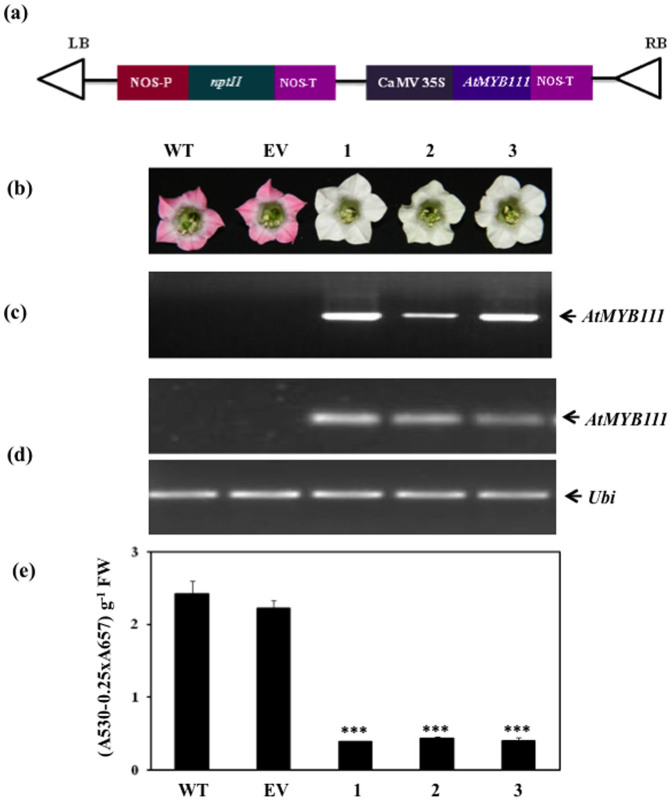
Earlier study demonstrated AtMYB111 as one of the members of MYB transcription factor family which regulate expression of an early set of genes of the flavonoid pathway in Arabidopsis26. To elucidate whether AtMYB111 can modulate flavonoid biosynthesis pathway in a heterologous system, we transformed tobacco plants with the construct carrying AtMYB111 open reading frame under the control of constitutive promoter CaMV35S (Fig. 2a). Several independent transgenic tobacco lines were selected on kanamycin and grown to maturity. Phenotypically, these transgenic lines were indistinguishable from the WT and empty vector (EV) transformed tobacco plants except for the petal pigmentation. Petal pigmentation was reduced at varying levels in most of the AtMYB111-expressing transgenic tobacco lines as compared to the WT and EV transformed tobacco plants (Fig. 2b). The transgenic lines were further confirmed for the presence of the transgene through genomic DNA PCR amplification, using vector-specific primer (CaMV forward) and gene-specific primer (AtMYB111 reverse). All the putative transgenic lines selected for genomic DNA PCR showed amplicon of expected size (Fig. 2c). Selected transgenic lines (line 1, line 2 and line 3) were analyzed for expression of the transgene through semiquantitative RT-PCR using RNA isolated from the young leaves of tobacco. Analysis suggested that selected transgenic lines express AtMYB111. No expression of the AtMYB111 was observed in the WT or EV transgenic lines (Fig. 2d). Reduced petal pigmentation in transgenic lines was further corroborated by analysis of total anthocyanin content. Anthocyanin content in flower petals of AtMYB111-expressing transgenic lines was drastically reduced in comparison to EV and WT plants (Fig. 2e). Similar decrease in the total anthocyanin content in leaf tissue of AtMYB111-expressing transgenic lines was observed in comparison to EV and WT plants (Supplementary Fig. S1). In addition, clearly visible anthocyanin pigmentation over the stamens of WT and EV transformed tobacco plants was absent in stamens of the AtMYB111-expressing transgenic tobacco plants. Together, these results suggest that AtMYB111 can modulate flavonoid biosynthesis pathway even in the heterologous system like tobacco.
Figure 2. Development of AtMYB111-expressing transgenic tobacco lines and their phenotypic characterization.
(a) Schematic representation of T-DNA region of plant expression construct carrying AtMYB111 cDNA in pBI121 vector used for tobacco transformation. (b) Flower color alterations in different transgenic lines. The pigmentation in petals of transgenic lines expressing AtMYB111 (1, 2 and 3) was compared with WT and empty vector (EV) transformed tobacco lines. (c) Confirmation of the presence of the transgene by PCR amplification of transgene using forward and reverse primers of AtMYB111 in different transgenic lines. (d) Expression analysis of transgene through semiquantitative RT-PCR using total RNA from leaves of WT, EV and different transgenic lines. (e) Total anthocyanin content in petals of WT, EV and different transgenic lines. Data are expressed as means ± SE of at least 3 independent experiments, each experiment consisting of 3 technical replicates. In (c) and (d), gels have been cropped for clarity and conciseness of the presentation. *** indicate values that differ significantly from WT at P < 0.001, according to Student's paired t-test.
AtMYB111 expression leads to the elevation in total phenolic and flavonoid content in tobacco
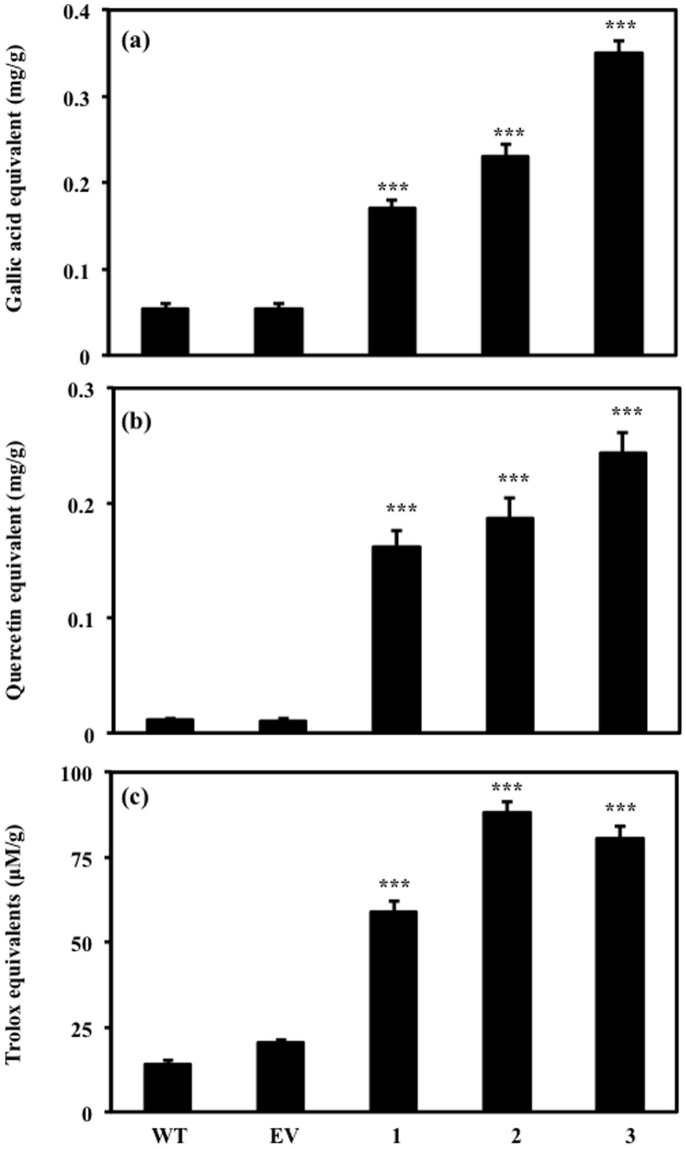
We further studied the modulation by AtMYB111 expression in the phenolic and flavonoid content in the seedlings of tobacco plants. Total phenolic and flavonoid content was significantly enhanced in AtMYB111-expressing transgenic lines in comparison to WT and EV transgenic lines plants (Fig. 3a and b). In addition, total antioxidant activity measured as trolox equivalent of the extracts from seedlings of AtMYB111-expressing tobacco plants was also enhanced as compared to WT and EV transformed tobacco plants (Fig. 3c). These changes can be attributed to the enhanced antioxidant potential of flavonoids.
Figure 3. Phytochemical analysis of methanolic extracts of seedlings in WT, EV and different transgenic lines.
(a) Measurement of total polyphenol content by gallic acid equivalent. (b) Total flavonoid content by quercetin equivalent. (c) Antioxidant activity by trolox equivalent. Data are expressed as means ± SE of at least 3 independent experiments, each experiment consisting of 3 technical replicates. *** indicate values that differ significantly from WT at P < 0.001, according to Student's paired t-test.
Enhanced accumulation of the chlorogenic acid and flavonols in AtMYB111-expressing lines
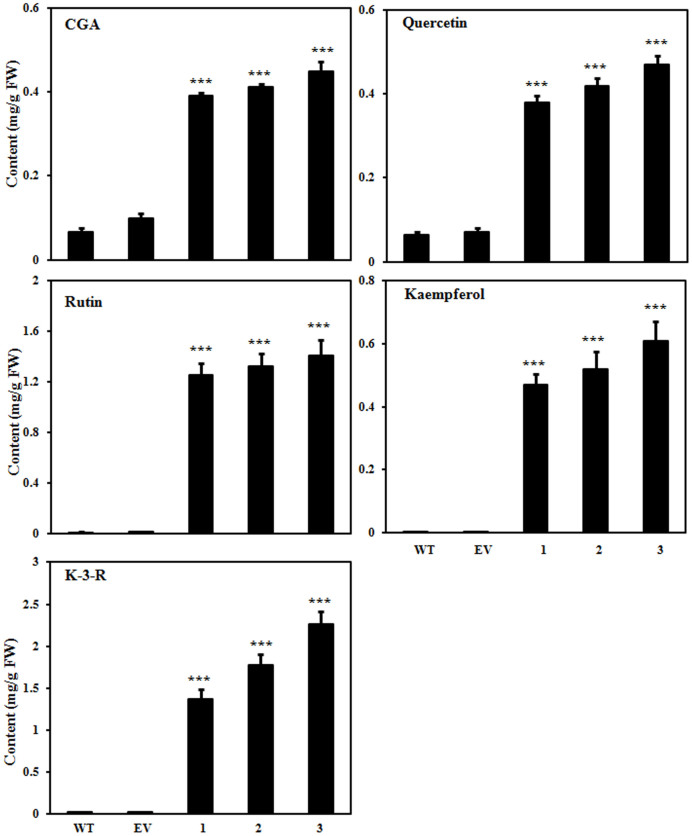
To further quantify major phenolics and flavonoids in tobacco, we carried out LC-MS analysis of methanolic extracts prepared from leaves of transgenic and WT tobacco plants. The analysis suggested significantly higher CGA content in leaves of the AtMYB111-expressing tobacco transgenic lines (up to 0.45 ± 0.022 mg/g FW) in comparison to EV transformed (up to 0.10 ± 0.01 mg/g FW) and WT plants (0.065 ± 0.01 mg/g FW) (Fig. 4 & Supplementary Fig. S2). The presence of CGA as well as other molecules was confirmed through MS analysis (Supplementary Fig. S3). Rutin, a flavonol glycoside, was identified as the major flavonoid in tobacco leaf methanolic extracts and its content was several fold higher in AtMYB111-expressing transgenic tobacco (up to 1.41 ± 0.12 mg/g FW) as compared to WT (up to 0.014 ± 0.001 mg/g FW) and EV (up to 0.016 ± 0.001 mg/g FW) transformed tobacco plants. In addition to CGA and rutin, content of kaempferol 3-O-rutinoside was significantly higher in AtMYB111-expressing transgenic tobacco (2.26 ± 0.15 mg/g FW) as compared to WT (0.022 ± 0.001 mg/g FW) and EV (0.029 ± 0.001 mg/g FW) transformed plants (Fig. 4 and Supplementary Fig. S2).
Figure 4. Qualitative and quantitative estimation of the flavonols and CGA content in plants.
Compounds were quantified by separating methanolic as well as acid hydrolyzed methanolic extracts from the young leaves of WT, EV and transgenic lines using HPLC. A typical HPLC chromatogram is given in Supplementary Fig. S2 & S5. The graph shows values ± SD of three leaves from each of the independent transgenic line. Data are expressed as means ± SE of at least 3 independent experiments, each experiment consisting of 3 technical replicates. *** indicate values that differ significantly from WT at P < 0.001, according to Student's paired t-test.
In order to quantify aglycone forms, the methanolic extracts were acid hydrolyzed and analyzed through LC-MS. The analysis suggested enhanced contents of quercetin and kaempferol (up to 0.47 ± 0.02 mg/g FW and 0.17 ± 0.015 mg/g FW, respectively) in the AtMYB111-expressing transgenic tobacco plants as compared to WT (up to 0.0064 ± 0.0007 mg/g FW and 0.0032 ± 0.0002 mg/g FW, respectively) and EV transformed tobacco plants (up to 0.0071 ± 0.0008 mg/g FW and 0.00328 ± 0.0002 mg/g FW, respectively) (Fig. 4 and Supplementary Fig. S2 & S3). Similar to leaves, petals of the AtMYB111-expressing transgenic tobacco plants accumulated higher contents of the flavonoids in comparison to the WT and EV transformed tobacco plants (Supplementary Fig. S4 & S5). Taken together, these results demonstrate the regulatory role of AtMYB111 transcription factor for enhanced flavonol and CGA biosynthesis in tobacco.
AtMYB111 activates expression of the genes involved in the flavonol biosynthesis
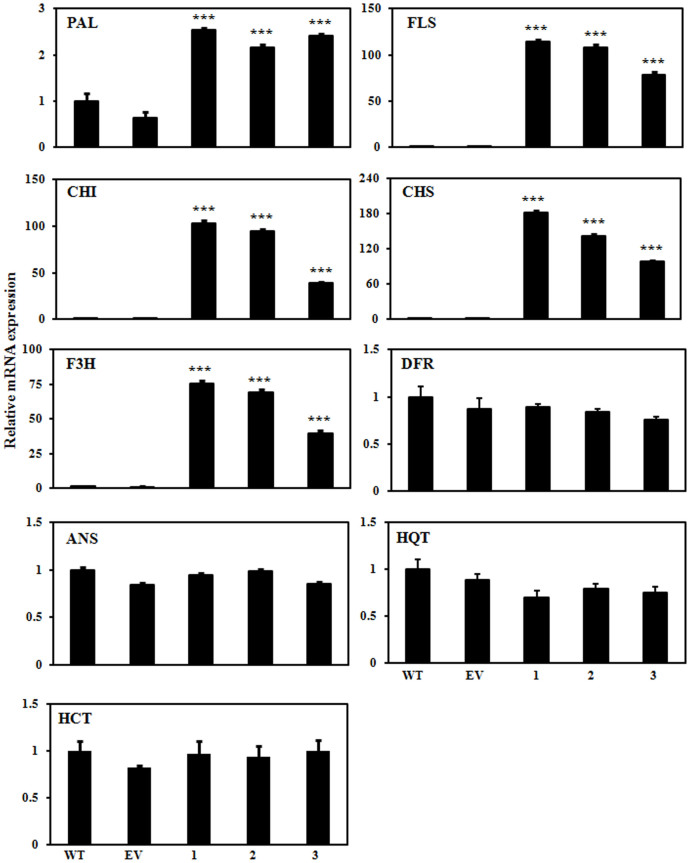
As flavonoid and CGA biosynthesis was modulated in the AtMYB111-expressing transgenic tobacco lines, the expression of genes involved in the flavonol and CGA biosynthesis was assessed in the tobacco plants through real time PCR. Expression of PAL, CHS, CHI, F3H and FLS involved in the flavonol biosynthesis was significantly up-regulated in leaf and petal tissues of AtMYB111-expressing lines in comparison to the WT and EV transformed plants (Fig. 5 & Supplementary Fig. S6). Expression of DFR and ANS genes was not significantly modulated in these transgenic lines. No significant change in transcript levels of genes involved in CGA biosynthesis (e.g. HQT and HCT) was observed between AtMYB111 expressing transgenic lines and the WT or EV transformed tobacco plants. These results suggest that AtMYB111 activates the expression of genes involved specifically in the flavonol branch of phenylpropanoid pathway and thereby acts as flavonol-specific regulator in tobacco.
Figure 5. AtMYB111 expression modulates expression of the genes involved in the CGA and flavonol biosynthesis.
Quantitative real time expression analysis was carried out to study expression of structural genes involved in the phenylpropanoid pathway in leaves of different transgenic lines. The data show values ± SD of three leaves from each of the independent transgenic line. Data are expressed as means ± SE of at least 3 independent experiments, each experiment consisting of 3 technical replicates. *** indicate values that differ significantly from WT at P < 0.001, according to Student's paired t-test.
Dark treatment modulates expression of the genes involved in the flavonoid biosynthesis
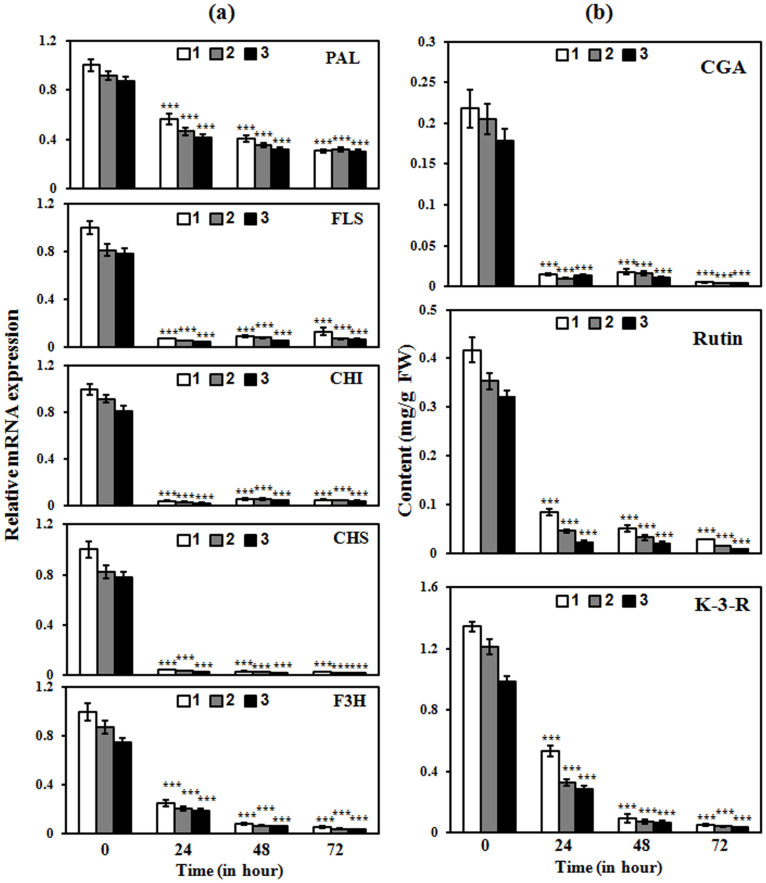
The flavonoid biosynthesis in plants is influenced by various environmental factors including light31. Therefore, we investigated whether enhanced flavonoid biosynthesis in AtMYB111-expressing transgenic plants is further modulated by the light. To study this, the expression of various genes involved in the flavonoid pathway in tobacco plants was assessed after dark incubation of tobacco plants for 0, 24, 48 and 72 h. The real time expression analysis clearly suggested that the dark incubation leads to significantly down regulated expression of flavonol biosynthesis related genes in AtMYB111-expressing transgenic (Fig. 6a and Supplementary Fig. S7a) as well as in WT plants (Supplementary Fig. S8a & S9a). As a reflection of modulation of gene expression, the reduction in the content of CGA, rutin, kaempferol 3-O-rutinoside and anthocyanin was also observed in case of the dark incubated transgenic lines (Fig. 6b & Supplementary Fig. S7b) as well as WT plants (Supplementary Fig. S8b & S9b). The total anthocyanin content of tobacco leaves incubated at the different time points was also reduced significantly (Supplementary Fig. S7 & S9). These results clearly suggest the positive effect of light over the flavonoid biosynthesis and the involvement of light regulated factors in regulation of flavonoid pathway in addition to AtMYB111.
Figure 6. Modulation of flavonoid biosynthesis by light in transgenic tobacco.
(a) Effect of light over the expression of the structural genes involved in flavonoid pathway in the different transgenic lines expressing AtMYB111. Expression of structural genes of phenylpropanoid pathway/flavonoid pathway was analyzed by real time PCR using RNA from the leaves of different transgenic lines expressing AtMYB111. (b) Effect of light over flavonoid biosynthesis in different transgenic lines expressing AtMYB111. Phytochemical analysis of methanolic extracts of leaves in different transgenic lines expressing AtMYB111. Compounds were quantified by separating methanolic extracts from the young leaves of transgenic lines using HPLC. The graph shows values ± SD of three leaves from each of the independent the transgenic line expressing AtMYB111. Data are expressed as means ± SE of at least 3 independent experiments, each experiment consisting of 3 technical replicates. *** indicate values that differ significantly from WT at P < 0.001, according to Student's paired t-test.
Discussion
Flavonoid biosynthesis is primarily regulated at the level of transcription of the structural genes encoding various enzymes of the pathway17. It has been demonstrated that genes involved in the flavonol biosynthesis are transcriptionally activated in Arabidopsis by three functionally redundant transcription factors namely AtMYB12, AtMYB111 and AtMYB11. In Arabidopsis seedlings, MYB12 and MYB111 regulate flavonol biosynthesis primarily in the root and cotyledons respectively. Though transcript of MYB11 was observed in every part of seedlings, expression was significantly lower in comparison to MYB12 and MYB11126. As flavonols are health beneficial compounds, development of strategies for enhanced flavonol biosynthesis in plants through genetic engineering has been a major consideration4,28. In this context, the transcription factors have emerged as an efficient tool owing to their regulatory capability for multiple target genes27,28. Earlier studies pertaining to the AtMYB12 overexpression in tobacco and tomato demonstrated several fold enhanced accumulation of flavonols in different tissues of the transgenic plants4,10,28. The fruit specific expression of AtMYB12 in tomato led to the significant increase in content of flavonols and other phenolics in fruit accompanied by enhanced expression of genes involved in the flavonol biosynthesis28. In the present work, we investigated the impact of AtMYB111, a paralog of AtMYB12, on flavonol biosynthesis in a heterologous system, tobacco, as well as studied effect of light on its regulatory mechanism.
Various transgenic tobacco lines constitutively expressing AtMYB111 gene, developed in this study, accumulated significantly enhanced levels of flavonols such as quercetin, rutin, kaempferol and kaempferol 3-O-rutinoside in both leaf and petal tissues. Previously, Luo et al. (2008) reported that the expression of AtMYB12 in tobacco leads to the several fold higher accumulation of flavonols and CGA in transgenic lines as compared to the WT plants28. Modulation in flavonol content in the transgenic lines developed, in the present study, is comparable to the AtMYB12-expressing transgenic tobacco lines. This suggests functional redundancy between AtMYB111 and AtMYB12 transcription factors for transcriptional activation of flavonol biosynthesis.
We further corroborated high flavonol biosynthesis in the AtMYB111-expressing tobacco plants through gene expression analysis. Expression analysis suggested up-regulation of various genes involved in flavonol biosynthesis in transgenic lines as compared to WT plants. Apart from genes involved in flavonol biosynthesis, the expression of PAL gene was also up-regulated in AtMYB111-expressing transgenic tobacco lines suggesting enhanced flux of substrate towards flavonoid pathway. The expression of DFR gene was not modulated by AtMYB111 which explains the reduced anthocyanin pigmentation in petal tissue. It seems that common precursor for flavonols and anthocyanins may be directed specifically towards flavonol biosynthesis via up-regulated FLS in AtMYB111-expressing transgenic tobacco plants leading to decreased anthocyanin content. The CGA biosynthesis which is also enhanced in AtMYB111-expressing transgenic tobacco plants as compared to WT plants does not seem to be due to the direct regulation of biosynthetic genes by AtMYB111 transcription factor. Instead, enhanced flux of the substrates towards phenylpropanoid pathway due to up-regulated expression of PAL gene in transgenic lines may be responsible for enhanced CGA biosynthesis. These results pertaining to the expression analysis using AtMYB111 transgenic lines are similar to that of AtMYB12 expressing transgenic tobacco lines28.
Among various modulators of the flavonoid biosynthesis, light has been reported to be one of the most important factors10,31,32. The modulatory effect of the light over anthocyanin and condensed tannin accumulation in transgenic plants expressing regulatory genes of flavonoid pathway has already been reported33,34,35. We have earlier reported a decrease in flavonol content in dark incubated AtMYB12-expressing transgenic callus10. In present work, the dark incubation resulted into the down-regulation of genes involved in flavonol biosynthesis in both, WT and transgenic, plants. It suggests that, in addition to MYB111, other light regulated factors may be involved in transcriptional regulation of flavonol biosynthesis in tobacco. In Arabidopsis, the MYB family transcription factors (AtMYB11, AtMYB111 and AtMYB12) and a bZIP family transcription factor, HY5, have been implicated in transcriptional activation of CHS and other early genes of flavonoid pathway. Among these, HY5 is light activated and degraded in dark through proteasome mediated protein degradation pathway36. In addition, the transcription of the MYB12, a paralog of MYB111, is positively regulated by light. During dark, the expression and activity of these orthologs in tobacco might be negatively regulated leading to down-regulated expression of genes involved in flavonol biosynthesis. Our results suggest a possible involvement of similar light regulated transcription factor in regulation of flavonoid biosynthesis in tobacco.
On the basis of gene expression and phytochemical analysis, our results demonstrate that AtMYB111 modulates flavonoid biosynthesis in tobacco in favour of flavonol accumulation in a similar manner to AtMYB12. Earlier, Luo et al. (2008) utilized MYB12 transcription factor for enhancing phenolics and flavonoids in tomato fruits28. Our results demonstrate that AtMYB111 could also be used for manipulation of flavonoid biosynthesis in plants through genetic engineering. In addition, our results also indicate involvement of the light-dependent factor in the activation of structural genes of flavonoid pathway in AtMYB111-expressing transgenic tobacco plants. Therefore, such factors may also be considered while designing strategies for transcription factor mediated manipulation of flavonoid biosynthesis.
Methods
Plant material and growth conditions
Young leaves of Nicotiana tabacum cv. Petit Havana have been used for raising the transgenic plants. Tobacco plants were grown in glass house at 25°C ± 2°C and 16 h/8 h light-dark photoperiods. For dark treatment, plants were shifted in the dark chamber with similar growth conditions as in the glass house. Leaves of AtMYB111-expressing plants were harvested after 0, 24, 48 and 72 h for gene expression as well as phytochemical analysis. Samples were frozen in the liquid nitrogen and kept in −80°C deep freezer until further use. The experiment was performed by using three independent replicates.
Plasmid construction, plant transformation and selection of transgenic lines
The full-length open reading frame of the Arabidopsis MYB111 cDNA, under the control of cauliflower mosaic virus 35S promoter in the binary vector pBI121 (Clontech, USA) was transferred into Agrobacterium tumefaciens strain LBA4404 by electroporation. Tobacco plants were transformed using Agrobacterium-mediated transformation37. Empty vector (EV) was also transformed to generate tobacco plants as EV or vector control. Several transgenic tobacco lines constitutively expressing MYB111 cDNA were selected based on floral color phenotype and RT-PCR. Seeds were harvested, sterilized and plated on solid half strength MS medium38 supplemented with 100 mg/l kanamycin. Antibiotic resistant plants were shifted to the glass house and grown until maturity.
Gene expression analysis
For semi-quantitative expression analysis, total RNA was extracted from the young leaves and flower petals of mature tobacco plants using Spectrum Plant Total RNA kit (Sigma-Aldrich, USA), which was subsequently treated with RNase-free DNase (Fermentas Life Sciences, Ontario, Canada). Total RNA was subjected to the reverse transcription reaction to generate first-strand cDNA using oligo(dT) primers (Fermentas Life Sciences, Ontario, Canada). RT-PCR analysis of a set of selected genes was carried out using 2X PCR Master mix (Fermentas Life Sciences, Ontario, Canada). The lists of selected genes and oligonucleotide primers used in the study are provided in the Supplementary Table S1. The primers for the tobacco ubiquitin gene were used as internal control to ensure that equal amounts of cDNA were used in all the reactions. PCR was carried out using the following cycle conditions: an initial denaturation at 94°C for 2 min, 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final 5 min extension at 72°C.
For real time expression analysis, the PCR mix contained 1 μl of diluted cDNA (10 ng), 10 μl of 2X SYBR Green PCR Master Mix (Applied Biosystems, USA) and 200 nM of each gene-specific primer in a final volume of 20 μl. PCRs with no template were also performed for each primer pair as control. Expression of different genes involved in flavonoid biosynthesis was studied using Fast Real time PCR System (Applied Biosystems, USA). All the PCRs were performed under following conditions: 20 sec at 95°C, 3 sec at 95°C, and 40 cycles of 30 sec at 60°C in 96-well optical reaction plates (Applied Biosystems, USA). The specificity of amplicons was verified by melting curve analysis (60 to 95°C) after 40 cycles. Three technical replicates were analyzed for each gene.
Determination of total anthocyanins
For the estimation of total anthocyanin, 300 mg of fresh tissue was ground into fine powder in liquid N2. Powdered samples were extracted with 1% acidic methanol for 18 h at room temperature with moderate shaking followed by centrifugation to sediment the plant material at 14000 rpm for 1 min. After centrifugation, 400 μl of supernatant was taken and mixed with 600 μl of acidic methanol. Absorbance of the sample was recorded at 530 nm (A530) and 657 nm (A657). Quantification of anthocyanin was carried out as previously described4.
Measurement of total polyphenol content
Total polyphenolic content was determined as described previously39. Briefly, polyphenols were quantified as gallic acid equivalent. Gallic acid was used to make the calibration curve by making different dilutions in ethanol (v/v). 100 μl of the standard solution or plant extracts were mixed with 2.5 ml of diluted Folin-Ciocalteu phenol reagent. The reaction mixture was allowed to react at room temperature for 5 min. After incubation, 2.5 ml of 1 M saturated sodium carbonate was added and again incubated for 1 h at room temperature. The absorbance of the reaction mixture was measured at 725 nm. The experiment was replicated three times with at least three replicates per treatment.
Determination of total flavonoids
Total flavonoid content was determined as described previously39. Briefly, flavonoids were estimated as quercetin equivalent. Quercetin was used to make the calibration curve by making different dilutions in ethanol (80% v/v). The standard solution or plant extracts (0.5 ml) were mixed with 1.5 ml ethanol (95% v/v), 0.1 ml aluminium chloride (10% w/v), 0.1 ml of sodium acetate (1 M) and 2.8 ml water. The volume of aluminium chloride was substituted by the same volume of distilled water in blank. After incubation at room temperature for 30 min, the absorbance of the reaction mixture was measured at 415 nm. The experiment was replicated three times with at least three replicates per treatment.
Determination of total antioxidant activity
The antioxidant activity was determined as described previously40 with slight modifications. The antioxidant activity was determined using DPPH (1, 1-diphenyl-2-picrylhydrazyl free radical) assay. In brief, 0.1 mM solution of DPPH was prepared in methanol. The initial absorbance of DPPH in methanol was measured at 515 nm and did not change throughout the period of assay. An aliquot of the samples (50–100 μl) was used to make up volume to 1.5 ml with DPPH solution. The sample was incubated at room temperature for 30 min. The absorbance of the samples was recorded at 515 nm using Ultraspec 3000 UV/Vis spectrophotometer (Pharmacia Biotech Ltd., Cambridge, CB4, 4FJ, UK). Trolox was used to make the calibration curve using different dilutions. The antioxidant capacity based on the DPPH free radical scavenging ability of the extract was expressed as μM Trolox equivalents per gm of plant material on fresh weight basis.
Preparation of the samples for LC-MS analysis
Extraction of the plant samples were carried out as described previously41. Flavonoids were determined either as aglycones or as flavonol glycosides by preparing acid-hydrolyzed or non-hydrolyzed extracts respectively. Hydrolysis of samples was carried out as described previously42. Briefly, for flavonol detection and quantification, plant material (1 gm) were ground into the fine powder in liquid N2 and were extracted with 80% methanol overnight at room temperature with brief agitation. The extract was filtered and filtrate was evaporated to about 1 ml. Three volume of HCl (1 M) was added into the concentrated extract and the mixture was incubated at 94°C for two h to hydrolyze any conjugate forms of flavonoids. After hydrolysis, samples were extracted with ethyl acetate, evaporated to dry and resuspended in 1 ml 80% methanol. Extracts were filtered through 0.2 μm filter (Millipore, USA) before HPLC. For non-hydrolyzed extracts, samples were extracted as described previously43 with slight modification. Briefly, plant material (1 gm) was ground into fine powder in the liquid N2 and was extracted with methanol: water (80:20) overnight at room temperature with brief agitation. The extract was filtered and filtrate was evaporated. The residue was dissolved in 1 ml methanol. Extracts were filtered through 0.2 μm filter (Millipore, USA) before analysis.
LC-MS analysis for the quantitative estimation of flavonoids
Analysis of samples was carried out as described previously44. Analyses were performed in a liquid chromatograph with Waters (Milford, MA, USA) pumps (Waters 515) equipped with a Waters 2998 photodiode array and 3100 Mass detector with 2767 autosampler. Molecules were separated on a 250 mm × 4.6 (i. d.), 5 μm pore size RP-C18 column (Merck) protected by guard column containing the same packing. The mobile phase was a gradient prepared from 0.05% (v/v) trifluoroacetic acid in HPLC-grade water (component A) and methanol (component B). Before use, the components were filtered through 0.45-μm nylon filters and de-aerated in an ultrasonic bath. The gradient from 25 to 50% B in 0–3 min, 50 to 80% B in 3–18 min, 80 to 25% B in 25 min and 25% B in 30 min was used for conditioning of the column with a flow rate of 1 ml/min. Data were integrated by Mass Lynx software and quantification was carried out at 254 nm by comparison with standards. Quantitative values are mean values from three replicate analyses of the same sample extracted at three different points. All samples and solutions were filtered through 0.45 μm nylon filters (Millipore, USA) before analysis by HPLC. Simple mobile phase was used as control for identification of blank peaks.
Mass spectrometry was carried out using Waters 3100 single quadrupole detector (Milford, MA, USA). A turbo ion-spray source was used in both negative and positive ion mode with using 3 KV capillary voltage, 30 V cone voltage, 3 V extractor voltage, 120°C source temperature, 350°C desolvation temperature using N2 gas.
Statistical analysis
For all the experiments three biological and three technical replicates were used for the analysis. The data were analyzed by Student's paired t test, and the mean values under each treatment were compared at P ≤ 0.05 − 0.001.
Author Contributions
A.P. and P.K.T. designed the experiments. A.P., S.B., P.M. and C.B. performed the experiments. A.P., P.M. and P.K.T. analyse the data and wrote the manuscript. All authors have read and approved the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
Research was supported by Council of Scientific and Industrial Research, New Delhi in the form of Network projects PlaGen (BSC-0107). A.P. acknowledges Council of Scientific and Industrial Research, New Delhi for Senior Research Fellowship. The authors also acknowledge Dr. Pravendra Nath and Dr. Shirish A. Ranade, CSIR-NBRI for editing the manuscript to refine the language.
References
- Thoison O., Sevenet T., Niemeyer H. M. & Russell G. B. Insect antifeedant compounds from Nothofagus dombeyi and N. pumilio. Phytochem. 65, 2173–2176 (2004). [DOI] [PubMed] [Google Scholar]
- Izaguirre M. M., Mazza C. A., Svatos A., Baldwin I. T. & Ballare C. L. Solar ultraviolet-B radiation and insect herbivory trigger partially overlapping phenolic responses in Nicotiana attenuata and Nicotiana longiflora. Ann. Bot. (Lond) 99, 103–109 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Napal G. N., Carpinella M. C. & Palacios S. M. Antifeedant activity of ethanolic extract from Flourensia oolepis and isolation of pinocembrin as its active principle compound. Bioresour. Technol. 100, 3669–3673 (2009). [DOI] [PubMed] [Google Scholar]
- Misra P. et al. Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 152, 2258–2268 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biol. 5, 218–223 (2002). [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W. & Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242 (2005). [DOI] [PubMed] [Google Scholar]
- Hassan S. & Mathesius U. The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp. Bot. 63, 3429–3444 (2012). [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Naciri-Graven Y., Broughton W. J. & Perret X. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol Microbiol. 51, 335–347 (2004). [DOI] [PubMed] [Google Scholar]
- Stapleton A. E. & Walbot V. Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol. 105, 881–889 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Misra P., Chandrashekhar K. & Trivedi P. K. Development of AtMYB12-expressing transgenic tobacco callus culture for production of rutin with biopesticidal potential. Plant Cell Rep. 31, 1867–1876 (2012). [DOI] [PubMed] [Google Scholar]
- Buer C. S., Imin N. & Djordjevic M. A. Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52, 98–111 (2010). [DOI] [PubMed] [Google Scholar]
- Peer W. A. & Murphy A. S. Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12, 556–563 (2007). [DOI] [PubMed] [Google Scholar]
- Marais J. P. J., Deavours B., Dixon R. A. & Ferreira D. The stereochemistry of flavonoids. The Science of Flavonoids (ed. Grotewold, E.) 1–46 (Springer Sci. Business Media, New York, 2006). [Google Scholar]
- Ververidis F. et al. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol. J. 2, 1214–1234 (2007). [DOI] [PubMed] [Google Scholar]
- Lepiniec L. et al. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–30 (2006). [DOI] [PubMed] [Google Scholar]
- Grotewold E. The Genetics and Biochemistry of Floral Pigments. Ann. Rev. Plant Biol. 57, 761–780 (2006). [DOI] [PubMed] [Google Scholar]
- Hichri I. et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in Grapevine. Mol. Plant 3, 509–523 (2010). [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol Plant 3, 2–20 (2010). [DOI] [PubMed] [Google Scholar]
- Winkel B. S. J. The biosynthesis of flavonoids. The Science of Flavonoids (ed. Grotewold, E.) 71–95 (Springer Sci. Business Media, New York, 2006). [Google Scholar]
- Quattrocchio F., Baudry A., Lepiniec L. & Grotewold E. The regulation of flavonoid biosynthesis. The Science of Flavonoids (ed. Grotewold, E.) 97–122 (Springer Sci. Business Media, New York, 2006). [Google Scholar]
- He F., Pan Q.-H., Shi Y. & Duan C.-Q. Biosynthesis and genetic regulation of proanthocyanidins in plants. Molecules 13, 2674–2703 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. et al. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 15, 9057–9091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I. et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483 (2011). [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J. M. & Lloyd A. M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 (2008). [DOI] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P. & Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R. et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E. et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 (2008). [DOI] [PubMed] [Google Scholar]
- Luo J. et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenols. Plant J. 56, 316–326 (2008). [DOI] [PubMed] [Google Scholar]
- Deluc L. et al. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L. et al. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E. et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165, 886–89 (2008). [DOI] [PubMed] [Google Scholar]
- Azuma A., Yakushiji H., Koshita Y. & Kobayashi S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236, 1067–1080 (2012). [DOI] [PubMed] [Google Scholar]
- Albert N. W. et al. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60, 2191–2202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F., Bovone T., Tosti N., Arcioni S. & Damiani F. Light and an exogenous transcription factor qualitatively and quantitatively affect the biosynthetic pathway of condensed tannins in Lotus corniculatus leaves. J. Exp. Bot. 56, 1093–1103 (2005). [DOI] [PubMed] [Google Scholar]
- Ray H. et al. Expression of anthocyanins and proanthocyanidins after transformation of Alfalfa with Maize Lc. Plant Physiol. 132, 1448–1463 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A. et al. Constitutively photomorphogenic1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975–1990 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B. et al. A simple method for transferring genes into plants. Science 227, 1229–123 (1985). [DOI] [PubMed] [Google Scholar]
- Murashige T. & Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- Aslann M. et al. In vivo antidiabetic and antioxidant potential of Helichrysum plicatum ssp. plicatum capitulums in streptozotocin-induced-diabetic rats. J. Ethnopharmacol. 109, 54–59 (2007). [DOI] [PubMed] [Google Scholar]
- Wong S. P., Leong L. P. & Koh J. H. W. Antioxidant activities of aqueous extracts of selected plants. Food Chem. 99, 775–783 (2006). [Google Scholar]
- Misra P., Pandey A., Tewari S. K., Nath P. & Trivedi P. K. Characterization of isoflavone synthase gene from Psoralea corylifolia: a medicinal plant. Plant Cell Rep. 29, 747–75 (2010). [DOI] [PubMed] [Google Scholar]
- Pandey A. et al. Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant biotechnol. 12, 69–80 (2014). [DOI] [PubMed] [Google Scholar]
- Pandey A. et al. Simultaneous separation and quantification of targeted group of compounds in Psoralea corylifolia L. using HPLC-PDA-MS-MS. J. Liq. Chromatogr. & Related Tech. 35, 2567–2583 (2012). [Google Scholar]
- Niranjan A. et al. Development and optimization of HPLC-PDA-MS-MS method for simultaneous quantification of three classes of flavonoids in legume seeds, vegetables, fruits and medicinal plants. J. Liq. Chromatogr. & Related Tech. 34, 1729–1742 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information