Abstract
In this article, we first review some aspects of functional magnetic resonance imaging (fMRI) paradigm designing for major cognitive functions by using stimulus delivery systems like Cogent, E-Prime, Presentation, etc., along with their technical aspects. We also review the stimulus presentation possibilities (block, event-related) for visual or auditory paradigms and their advantage in both clinical and research setting. The second part mainly focus on various fMRI data post-processing tools such as Statistical Parametric Mapping (SPM) and Brain Voyager, and discuss the particulars of various preprocessing steps involved (realignment, co-registration, normalization, smoothing) in these software and also the statistical analysis principles of General Linear Modeling for final interpretation of a functional activation result.
Keywords: Functional magnetic resonance imaging, general linear modeling, statistical parametric mapping
Introduction
Functional magnetic resonance imaging (fMRI) is an effective tool for analyzing brain functions. It is a breakthrough imaging modality which is non-invasive and is quite precise in tracking functional responses of brain through Blood Oxygen Level Dependent (BOLD) contrast.[1] The BOLD effect was first explained by Seiji Ogawa.[2] This technique relies on difference in the magnetic properties of oxyhemoglobin and deoxyhemoglobin (dHb).[3] When we perform a task, the neuronal activity in the brain area specific to that particular task increases. Enhanced neuronal activity leads to a local increase in energy and oxygen consumption in functional brain areas, which is followed by an increase of regional cerebral blood volume (rCBV) and regional cerebral blood flow (rCBF). Since increased neuronal activity results in an increase in blood flow beyond the exact demand, more oxygenated hemoglobin appears in the venous capillaries. This shifts the relation between oxygenated and deoxygenated hemoglobin.[4] BOLD contrast relies on variations in dHb, which acts as an endogenous contrast agent. Therefore, changes in the local dHb concentration in the brain lead to changes in the signal intensity of magnetic resonance images (MRIs).[5] This effect is reflected in the fMRI BOLD contrast images. To comprehend, BOLD effect can be described as a positive contrast caused by relative decrease in the magnetic field inhomogeneities due to relative increase in the oxyHb level contributed by the blood flowing in the region of brain with increased activity. Compared to other imaging techniques,[6,7] fMRI is more powerful with relatively good spatial and temporal resolution.[8,9] Since fMRI is quite efficient in elucidating neural correlates associated with specific brain functions, the technique has got wide acceptance in functional brain mapping both clinically and in research setting.
fMRI Paradigm Designing Tools
A paradigm is a temporal allocation of stimuli to acquire BOLD responses from the subject. During a fMRI experiment, specific paradigms with stimuli or events are used to evoke hemodynamic response or brain activation in the subject. An entire fMRI experiment relies on precise and effective paradigm design. At present, there are many software packages available for designing fMRI paradigms. Each one of them has its own pros and cons. Here, we provide a comparative description of three prominent stimulus delivery packages – Cogent (Laboratory of Neurobiology, Wellcome Trust, London, UK;), E-Prime (Psychology Software Tools, Inc.; Pennsylvania, USA), and Presentation (Neurobehavioral Systems, Inc.; California, USA). At the end of this section, mention is also made of fMRI experimental design and the tool developed in-house at our institute for stimulus presentation.
Cogent
Cogent is an open-source Matlab toolbox for delivering experimental stimuli as well as to collect response from brain imaging equipments. Cogent program is available in two forms – Cogent Graphics and Cogent 2000.[10] Cogent Graphics is a graphical toolbox for Matlab which facilitates the generation of real-time graphical animations. Cogent 2000 is a software program which can effectively deliver different types of stimuli, synchronized with the scanner. Cogent is completely programmable and is based on Matlab commands.
Cogent program uses Matlab and Cogent 2000 commands sequentially, and therefore, it is essential that the user must be familiar with Matlab scripting as well as scripting in Cogent. The main drawback of cogent program is that when network connections are available, it gets a hold-up which results in a delayed stimulus delivery, especially in auditory stimulus, and creates timing inaccuracies. There are also issues in establishing serial and parallel port communication. Cogent runs in Windows platform together with Microsoft Application Programming Interface called DirectX and Matlab version 6.0 or above. The software is made by Laboratory of Neurobiology, University College London and can be downloaded freely from www.vislab.ucl.ac.uk.
E-prime
The name E-Prime stands for experimenter's prime. It is quiet easy to learn and use E-Prime suit. There are five programs (subdivisions) that constitute E-Prime; they are E-Studio, E-Run, E-DataAid, E-Merge, and E-recovery.[11,12] Among these, E-Studio is the major program and it aids in the creation of experiments. The structure of the paradigm is described in E-studio. A basic experiment is represented as frames, trials, and blocks. It is very user friendly and has drag and drop graphical interfaces for experimental design. E-Run is used to run an experiment. One can also use this module for experiment testing, i.e. the use of licence key is not essential for the working of E-Run since it can be used to test the same paradigm in different systems, different labs, and different users. E-DataAid program gives the recorded data in a tabular form. The user can modify this data and it can also be exported to other formats compatible with other data analysis packages. E-Merge utility facilitates group analysis of data and gives a single output file with results from multiple subjects. E-Recovery program is meant for data recovery. It can recover an aborted experiment text file and convert the same into a new data file.
The major advantage of using E-Prime is that it provides an easy-to-use environment for experimental design, data collection, and analysis of cognitive experiments. Most of the interfaces in E-Prime Graphical User Interface (GUI) have “drag and drop” option, which makes it very user friendly. Compared to other stimulus delivery programs, it is quite fast and easy to use. A user can generate programs without a deep understanding about the tool. One needs to know only what is required to be done in their own paradigm/task. E-Prime is a commercial software package. One requires a license key to activate and use it. In order to run E-Prime, one requires a Windows OS (XP/vista/7) and DirectX video card. It is developed by Psychology Software Tools, Inc., Pennsylvania, USA. The download option and more information can be obtained from www.pstnet.com.
Presentation
Presentation (Neurobehavioral Systems, Inc., California, USA) is a software application for handling psychological and neurobehavioral experiments. Presentation tool is comparatively more precise and accurate; it tracks stimulus response, reaction time, and different performance measures with sub-millisecond temporal accuracy.[13] Presentation facilitates effective control of parallel and serial port, which enables the communication to and from fMRI system and other devices. To enable synchronization with the scanner, presentation has an fMRI mode which facilitates reception of pulses from the scanner and also enables start of stimulus sequence on specific pulses.
Presentation experiments are subdivided into units called Scenarios. Scenarios are sequence of actions that presentation performs without interruption. They are specified using text description, and the main text file is called Scenario file. Programmability enables effective creation, manipulation, delivery, and control of stimuli in different neurobehavioral experiments. This tool has its own built-in programming language and is subdivided into (i) Scenario Description Language (SDL) and (ii) Program Control Language (PCL). To simplify the concept of programming language, we can assume the Presentation paradigm to be our favourite cuisine. To prepare it, we need to know the ingredients and the method of preparation. Here, SDL refers to the ingredients (the stimuli which need to be included in the paradigm). PCL can be related to the method of preparation (describes the order/sequence in which each stimulus has to appear).
Presentation program is more accurate and powerful among other programs.[14] It has precise timing accuracy (less than a millisecond) in stimulus delivery and response recording. But learning presentation is quite difficult if we do not have enough background in programming. Presentation is Windows-based (XP/Vista/7) and uses standard personal computer (PC) hardware. It is commercial software and can only be used with a licence key. Presentation runs in a PC installed with Windows OS and DirectX interface. The software is developed by Neurobehavioral Systems, Inc. More information about Presentation can be obtained at www.neurobs.com.
The software for designing paradigms is not limited to the three which are mentioned above. In fact, there are other softwares which serve the same purpose. SuperLab, Matlab Psychophysics Toolbox, Paradigm, PsychoPy, PyEPL, Inquisit, PEBL, Experiment Builder, Neuroscan, etc., are the other examples. For stimulus delivery and response recording in fMRI experiments, there are no hard rules that we have to stick to a specific stimulus delivery package. We can generate our own program for presenting stimuli and get it synchronized with the scanner trigger.
fMRI - Experimental Design and Setup
The fMRI experiments mainly fall under two design categories: (i) block design and (ii) event-related design.[15] The block designs use interleaved and equitemporal blocks of activity and rest to elucidate the actual response.[16,17] In an event-related design, the trials are presented randomly and a specific cognitive event is in focus. It is often used to establish neurobehavioral tasks, since it provides a subjective response to the stimuli being presented.[18]
The fMRI procedure is being utilized for clinical examinations as well as for research investigations. In clinical practice, fMRI mainly aids in surgical planning, for analyzing the localization of language, sensory and motor function, and to follow-up functional rehabilitation after neurological disorders.[19,20] Unlike a neurobehavioral experiment laboratory, in a hospital setup, we need to have routinely accessible fMRI setting and the delivery of results needs to be quick. Modern scanners have inbuilt real-time processing option. The real points to be looked into are paradigm presentation and scanner synchronization. Laborious setup procedures and time-consuming processing techniques may be difficult in a clinical setting. Therefore, clinical procedures require custom-designed fMRI setting. Our institute has developed an in-house fMRI setting for carrying out clinical investigation. The visual and auditory paradigms are programmed with the help of Visual Basic and presented in coordination with the scanner trigger. This is achieved by engineering a synchronization box that tracks the scanner pulses. The whole setup includes a stimulus presenting computer, a synchronization box, a projector (kept in the console room), an earphone for auditory stimulus delivery, and a screen (in the scanner room) to project visual stimuli [Figure 1]. The cost of making such a system is quite low compared to the cost quoted by the vendors. The processing of data is done in real time with the inline BOLD. The results obtained from the inline BOLD had significant correlation with the results from Statistical Parametric Mapping (SPM) image processing software. Both the results maintained a degree of similarity in indicating the region as well as the amount of BOLD activation.[21] Here, variation was made only in specifying the threshold values, i.e. the inline bold required comparatively higher threshold than that of SPM. The promising point is that the inline bold system is less time consuming, which is a demanding aspect in clinical procedures.
Figure 1.
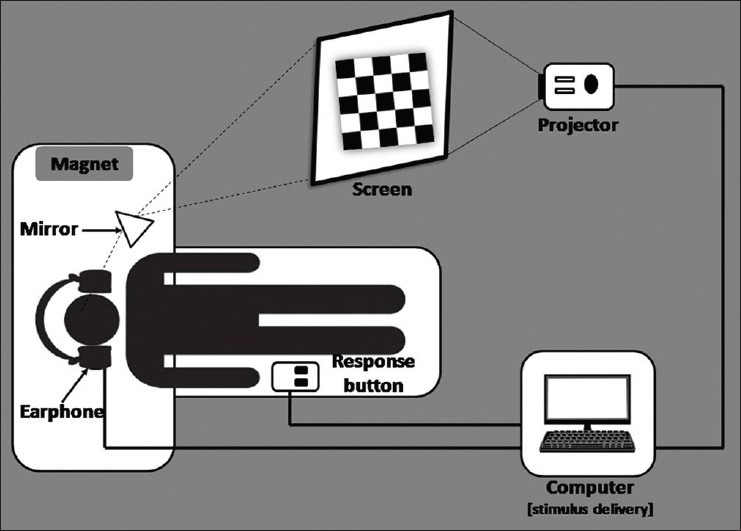
Layout of the fMRI setting
The research setting requires more sophisticated paradigms and processing techniques and probably combinations of different brain mapping techniques.[22] Here, the processing and data analysis may be time consuming. The experiments need to have time-bound and response-dependent stimulus presentation. Accurate logging of timings corresponding to stimulus as well as response may be essential for data processing. Research investigations also require advanced image processing tools.
fMRI Data Analysis Tools
Basic concepts of fMRI data analysis
The principal aim of fMRI data analysis is to determine cortical brain regions, where the signal changes occur during the presentation of a passive, sensory, motor, or cognitive task (paradigm) designed to specific brain functions. Three main stages [Figure 2] are involved in the analysis of the data from any fMRI experiment.[23,24] They are as follows: (a) the preprocessing steps which can be applied to the data to improve the detection of activation events; these include image registration to correct for subject movement during the experiment, normalization of subject head to a standard stereotactic template [e.g. Talairach template, International Consortium for Brain Mapping (ICBM) template, Montreal Neurological Institute (MNI) template], and smoothing to improve the signal to noise ratio; (b) performing statistical analysis which determines the voxels in the image that show a response to the stimulus; and (c) displaying the activation images and probability values which give the statistical confidence. In a typical fMRI experiment, whole-brain functional images are acquired every 2-3 s resulting in several images (time series data), which include roughly about 100,000 voxels. This data needs to be further in a statistically meaningful way. A general issue in fMRI data analysis is to understand the relationship between the neurobiological hypothesis and the statistical models adopted to test that hypothesis.[25]
Figure 2.
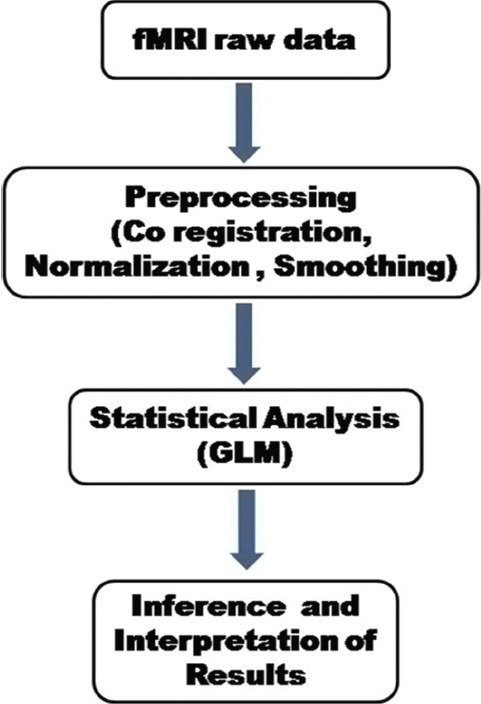
Steps involved in the fMRI data processing
fMRI data analysis can be done by two ways: (a) offline processing using a popular Matlab-based software package called SPM or similar software packages (Brain Voyager, AFNI, FSL, etc.) working on the same principle (General Linear Modeling) and (b) real-time fMRI (Rt-fMRI) data analysis which can be done by using the module provided by MRI vendor. Using Rt-fMRI, the imaging data can be analyzed, and displayed more interactively, as it is acquired from the scanner with a latency of only a few seconds. The major advantages of this method are immediate confirmation of the experimental results from simple block design paradigms, real-time monitoring of “resting-state fMRI,” and tracking of the subject head movement and giving instant feedback to the subject, enabling clinicians to investigate the dynamic nature of the human brain. Most of the MRI vendors have an Rt-fMRI analysis tool which can be used in busy clinical settings for eloquent cortex mapping and language lateralization. Earlier studies have shown that the Rt-fMRI post-processing gives similar results as offline processing for simple block design paradigms in the clinical setting.[26]
In this review, we discuss the fMRI data processing using SPM and BrainVoyager, the two techniques that we use in our practice. The modules used for basic data analysis of brain images in BrainVoyager are similar to those of SPM: preprocessing, co-registration, spatial normalization, and statistical analysis.
Statistical Parametric Mapping
The SPM tool was originally developed for the statistical analysis of functional neuroimaging data from Positron Emission Tomography (PET) by Karl Friston,[27,28] along with conceptual and technical help from John Ashburner[29] (Functional Imaging Laboratory, Wellcome Trust Institute, London, UK). This software was made available to the emerging functional imaging community in 1991.[30] SPM’94 was the first major version of the SPM software. The revised versions are SPM’95, SPM′96, SPM′99, SPM2, SPM5 and SPM8, which are exactly based on SPM’94 and represent the ongoing technical improvements and theoretical advances.
SPM is a voxel-based technique which makes inference about regionally specific responses of cortical brain areas to experimental tasks (paradigms). In order to observe the response of a particular brain region, the data must conform to a standard anatomical space.[31] Usually a univariate approach is followed in which the parametric map is computed by examining every voxel location across all images, meaning that a statistical value (e.g. t-value) is calculated for every voxel using a statistical approach called “General Linear Model (GLM).”
Steps involved in SPM
Preprocessing of fMRI data - preparing data for analysis
The essential preprocessing steps are: (a) realignment, (b) co-registration, (c) normalization, and (d) smoothing.
Realignment
During the scanning session, subjects may move inside the scanner. Even small head movements can cause movement artifacts, which may add up to the residual variance and reduce sensitivity. Data may be lost if sudden movements occur during a single volume and it may be correlated with the task performed. So, movement-related variance components in fMRI present one of the most serious confounds of analysis.[32] A rigid body registration with six parameters, three translations (X, Y, Z), and three rotations (pitch, roll, and yaw) are used for realignment in SPM. It minimizes the squared sum of differences between each successive scan and reference scan (usually the first or the average of all scans in the time series) and resamples the data.
Co-registration - within-subject registration
By means of co-registration between two modalities [a structural image (e.g. T2-weighted) to a functional image series, (echo planar imaging)], one can overlay functional activations onto an individual's own anatomy and it is also possible to overlay group-level functional activations onto an average structural scan. Co-registration gives a better spatial image for further use in normalization step, as warps derived from the higher-resolution structural image can be applied to the functional image. This is again a rigid body transformation, but the registration cannot be simply performed by minimizing the residual sum of squares due to different imaging modalities. The 12 parameters affine transformation [3 translations, 3 rotations (rigid-body), 3 shears, and 3 zooms] step registers the structural image and the first image of the functional image series to template images. These transformations are constrained in such a way that only the parameters that describe the rigid body transformation are allowed to differ. Next the images are segmented using tissue probability maps of gray matter, white matter, and cerebrospinal fluid. At last, the image partitions can be simultaneously co-registered to produce the final solution.
Normalization - between-subject registration
In order to average the signal across different subjects, it is important to warp brain images into roughly the same stereotactic space. The advantage of spatially normalized images is that areas of functional brain activation can be reported within this standard space,[33] in SPM′94–SPM′99, according to their spatial coordinates. The SPM2 uses MNI template, an average of 152 brain images, and hence is more representative of the population, as compared to the Talairach and Tournoux atlas.[34] Later versions of SPM use ICBM template. The normalization step not only considers the six rigid-body transformations, but also considers three shears and three zooms to match the individual subject's images to the template. A nonlinear transformation is also required for accurate normalization which would correct gross differences in head shapes that cannot be accounted for by the affine transformation. The normalization step need not be done while doing a single-patient fMRI analysis in clinical setting. The technique is used more often while analyzing group data in a research setting.[35]
Smoothing
Functional images needs to be smoothed prior to the statistical analysis, especially in group-level analysis, so that corresponding sites of activation from the different brains are superimposed. Smoothing is generally done by convoluting the data with a Gaussian kernel and it potentially increases the signal-to-noise ratio according to the matched filter theorem.[36] Since hemodynamic response functions (HRFs) are modeled to have the shape of a Gaussian filter, we need to use a Gaussian kernel of size at least twice the voxel size [Full Width Half Maximum (FWHM) of about 6 or 8 mm] for smoothing the functional images.[37] During smoothing, the intensity value within each voxel is replaced with a weighted average that incorporates the intensity values of the adjacent voxels, which corrects the inter-subject variability between individuals after normalization. Smoothing allows the application of Gaussian random field theory to make inference at statistical analysis stage.
Statistical analysis - using GLM
The GLM is used to specify the different conditions/blocks in the form of a design matrix, which defines the experimental task and the nature of hypothesis to be tested. It provides a framework that allows us to make refined statistical inferences after taking into account (1) the (preprocessed) 3D MRI images, (2) BOLD time series, (3) user-defined experimental conditions, (4) HRF, and (4) technical/noise corrections.
The basic idea behind GLM is that the observed data (y) is equal to a weighted combination of several model factors (x) plus an additive error term (ε). It is a model (i.e. an equation: Y = X · β + ε), where Y is the BOLD signal at various time points at a single voxel (observed data), X represents several components which describe the observed data, i.e. the BOLD time series for a particular voxel (design matrix), β denotes the contribution of each component of the design matrix to the value of Y (parameter estimates), and Є is the difference between the observed data, Y, and that predicted by the model, Xβ (error). The design matrix has one row for each scan and one column for each effect one has built into the experiment. These are referred to as explanatory variables, covariates, or regressors. Each column of the design matrix corresponds to experimental conditions of interest (the hypothesis under test) and a set of columns corresponds to model with effects of no interest. In this stage, the groups designated for the images are specified. This stage represents modeling the data to observe neurophysiologic responses, confounds, and error [Figure 3].
Figure 3.
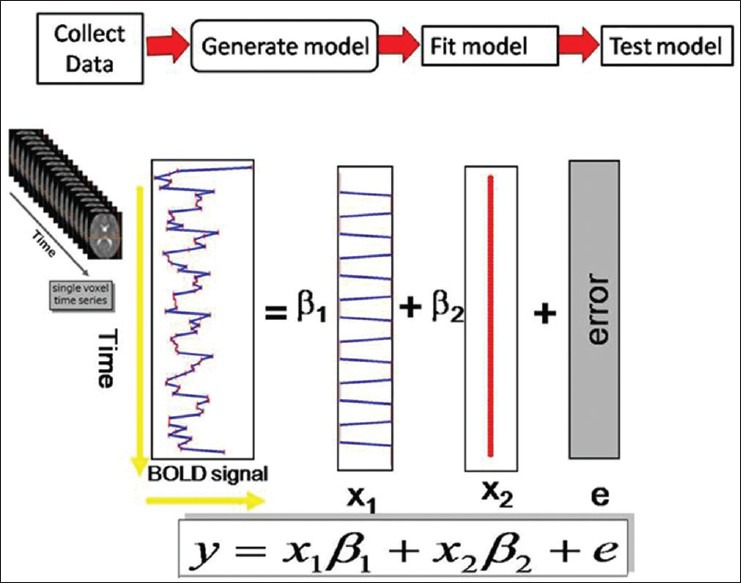
General linear modeling
Inference and interpretation of results
The neuronal activity in response to an experimental task is obtained by specifying linear contrasts (T or F contrast). Cognitive subtraction logic[38] is applied to predict brain activity scales in a linear fashion. The conditions of interest (active condition) are given a positive value, such as 1, and conditions (baseline condition) that are to be subtracted from these conditions of interest take on a negative value, such as −1. The end result is a statistical parametric map.[39] The functional activations (blobs) obtained can be further overlaid or rendered onto the high-resolution structural image of the subject in order to accurately locate the neural activity. Since SPM follows a univariate approach,[40] each voxel should be separately analyzed. So, for a statistical threshold of P < 0.05, 5% of the voxels would show activation by chance alone (false activation – type I error), which means a multiple comparison correction is required.[41] Bonferroni correction is the traditional way of doing this. However, due to involvement of a huge number of voxels, direct implementation would severely reduce the estimated number of degrees of freedom. Hence, to the extent that the image data approximate a random Gaussian field[42] correction for multiple comparison need to be only made for number of voxels that can be resolved independently (resells or resolution elements). The multiple comparisons correction is controlled for family-wise error (FWE) rate.[43,44] This assumption of random Gaussian field is assured by applying a Gaussian smoothing filter in the preprocessing stages. A serious limitation of correcting for multiple comparisons is that the number of false negatives (type II error) is greatly increased. Further approach is to define the false discovery rate (FDR), that controls for about 5% (P < 0.05) of false positive activation. The FWE method controls for a 5% chance of a single false positive. To correct for multiple comparisons, some alternative approaches have been also used: (i) using a strict uncorrected threshold (e.g. P < 0.001), (ii) using an inference over the cluster size, (iii) small volume corrections in regions where a prior hypothesis exists, and (iv) a region of interest (ROI) analysis in which the average signal for all voxels in an anatomical or functional ROI is used, thereby decreasing the number of multiple comparison voxel space to the number of ROIs [Figure 4].
Figure 4.
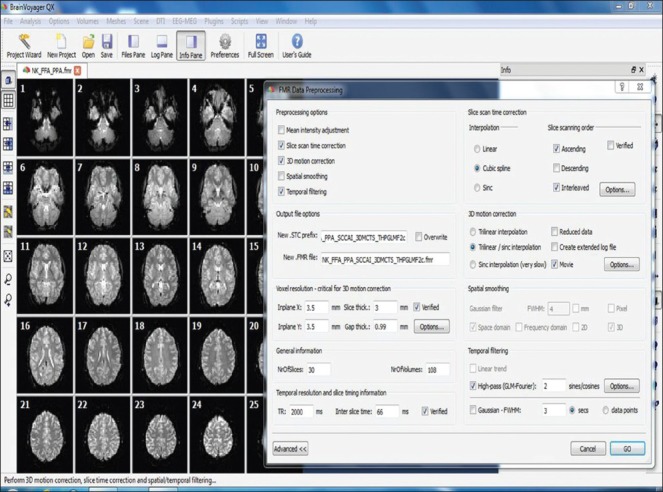
BrainVoyager software GUI shows functional data as FMR project format and FMR data preprocessing dialog window
Brain voyager
BrainVoyager QX (Brain Innovation, Maastricht, The Netherlands) is a software package for the analysis and visualization of MRI data sets. It supports on all major computer platforms including Windows XP/Vista/7, Linux, and Mac OS X. It is a multimodal analysis tool used for fMRI, diffusion tensor imaging (DTI), electroencephalogram (EEG), magnetoencephalography (MEG), and transcranial magnetic stimulation (TMS) data. BrainVoyager QX has been completely programmed in C++ and provides easy-to-use interactive GUI for all platforms.[45] Automatic brain segmentation, surface reconstruction, cortex inflation and flattening can also be done by using this software. BrainVoyager also supports advanced data analysis of fMRI brain images, including Multi-Voxel Pattern Analysis (MVPA), Independent Component Analysis (ICA), and Probabilistic Maps. Hypothesis-driven statistical data analysis is an important tool to identify activations in brain regions exhibiting increased or decreased responses in specific experimental conditions as compared to others. OpenGL (Open Graphics Library), the most widely used cross-language for 2D and 3D graphics rendering, is implemented in BrainVoyager software for rendering 3D computer graphics (surface module). BrainVoyager is a commercial software and requires a Hardware against Software Piracy (HASP; a USB dongle that protects software vendors from unauthorized use or distribution of their software) key for a single computer or network dongle for many computers that works as a floating license. The steps for processing functional data include the following.
Creation of functional MR project
Functional MR (FMR) project is created by loading the raw functional data and converting it into FMR data format.[46] FMR is the 2D visualization of functional data. BrainVoyager supports various types of data formats including DICOM, ANALYZE, and PHILIPS_REC. MR scanner names the DICOM files in a complex way. Before creating FMR project, BrainVoyager renames the DICOM files using header information. This will avoid the problems during importing and further analysis of the data.
Preprocessing of FMR project
Preprocessing is used to improve the image quality by suppressing undesired distortions or enhancing some image features made to be more suitable for further processing of the images.
Preprocessing of functional data includes several steps like mean intensity adjustment, slice scan time correction, 3D motion correction, spatial smoothing, and temporal filtering. This software provides a single window for all these preprocessing options and can run preprocessing steps at once.
Creation of stimulation protocol
Stimulation protocol allows defining the conditions (block or event-related design) used for the presentation of stimuli. BrainVoyager saves this data as protocol (PRT) file. The same protocol file can be used for different FMR projects, if the same conditions are used for different subjects.
Statistical analysis of functional data
The statistical analysis test is performed to determine which voxels in the brain are significantly activated by a certain type of stimulus. BrainVoyager software provides options for single subject and multi-subject statistical analysis. This software uses GLM for single subject analysis specifying statistical models. It is obtained by adding several explanatory variables known as predictors, which give precise activations. GLM analysis is a univariate method performed independently on each voxel time course and beta values are estimated for each voxel. Multi-subject data is analyzed by using fixed-effects group analysis and random-effects group analysis.[47] BrainVoyager also supports multivariate approaches like ICA[48,49] and MVPA.[50] The MVPA tools include multivariate searchlight mapping and support vector machine. The end result of the statistical analysis is a statistical map that shows which voxels are significantly activated given a specified statistical threshold. The thresholded statistical map can be overlayed directly to the functional (FMR) data, co-registered anatomical [volumetric MR (VMR)] data, or surface module for visualization.
Anatomical Data Processing
The steps may include the following:
Creation of VMR projects
Anatomical data is loaded into BrainVoyager and converted into BrainVoyager-supported VMR data format. VMR is the 3D graphical representation of anatomical data. This data may exhibit inhomogeneous intensity and can be removed by using intensity inhomogeneous correction tool in BrainVoyager software.
Iso-voxel
Before FMR–VMR alignment, resolution of VMR data set is needed to match with the resolution of the functional data. In order to do this, VMR data set is iso-voxel to a resolution of 1 × 1 × 1 mm.
Talairach transformation
For multi-subject analysis, data set is transformed into a standard space. BrainVoyager provides two options for Talairach transformation: manual and automatic. In manual Talairach transformation, we need to find anterior commissure (AC), posterior commissure (PC), anterior point of the cerebrum (AP), posterior point (PP), superior point (SP), inferior point (IP), most right point (RP), and most left point (LP) manually by dragging the mouse pointer to the above specified region or entering the coordinate values. In automatic Talairach transformation, the above Talairach reference points are set automatically by the software. The Talairach transformation is needed for both anatomical and functional data for only multi-subject analysis.[51]
Co-registration of Functional and Anatomical Data
Co-registration allows alignment of functional and anatomical data for the purpose of overlaying the brain activity precisely. Co-registration has been divided into two steps:
Initial alignment (IA)
Fine-turning alignment (FA)
The IA brings the functional and anatomical data into same orientation. Head movements are rectified by using FA. FMR data is set as the “source” and VMR data is set as the “target.” FMR data is fixed and the VMR data is scaled, translated, and rotated with respect to FMR data.
Turbo-brain voyager
Turbo-BrainVoyager (TBV; Brain Innovation, Maastricht, The Netherlands) is a software package for the real-time analysis and visualization of fMRI data sets.[52] TBV performs data preprocessing in real time, including 3D motion correction and spatial smoothing. It visualizes the data in multi-slice or single-slice functional data view and anatomical volume view. ROIs can easily be defined in TBV by dragging the mouse pointer around the activated cluster in functional data and also shows ROI time course window for the corresponding ROI. TBV is not a replacement for BrainVoyager Q ×. TBV does not have some functionality like segmentation, Talairach transformation, between-subject statistical data analysis, etc., TBV runs on Microsoft Windows 98/NT/2000/XP, Linux, and Mac OS X. TBV is a commercial software and requires a HASP USB dongle.
Conclusion
There are many choices available to a research group for analysis of fMRI data. Discussion in the current article was exclusive to the softwares and techniques familiar to the authors. We have provided sufficient information about different fMRI post processing tools that are of particular interest in cognitive and clinical neuroscience research.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Ogawa S, Sung Y. Functional magnetic resonance imaging. Scholarpedia. 2007;2:3105. [Google Scholar]
- 2.Ogawa S, Lee TM. Magnetic resonance imaging of blood vessels at high fields: In vivo and in vitro measurements and image simulation. Magn Reson Med. 1990;16:9–18. doi: 10.1002/mrm.1910160103. [DOI] [PubMed] [Google Scholar]
- 3.Pauling L, Coryell CD. The magnetic properties and structure of hemoglobin, oxyhemoglobin and carbonmonoxy hemoglobin. Proc Natl Acad Sci U S A. 1936;22:210–6. doi: 10.1073/pnas.22.4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekstrom A. How and when the fMRI BOLD signal relates to underlying neural activity: The danger in dissociation. Brain. 2010;62:233–44. doi: 10.1016/j.brainresrev.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews PM, Jezzard P. Functional magnetic resonance imaging. J Neurol Neurosurg Psychiatry. 2004;75:6–12. [PMC free article] [PubMed] [Google Scholar]
- 6.Feng CM, Narayana S, Lancaster JL, Jerabek PA, Arnow TL, Zhu F, et al. CBF changes during brain activation: fMRI vs. PET. Neuroimage. 2004;22:443–4. doi: 10.1016/j.neuroimage.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Carroll TJ, Teneggi V, Jobin M, Squassante L, Treyer V, Hany TF, et al. Absolute quantification of cerebral blood flow with magnetic resonance, reproducibility of the method, and comparison with H215O positron emission tomography. J Cereb Blood Flow Metab. 2002;22:1149–56. doi: 10.1097/00004647-200209000-00013. [DOI] [PubMed] [Google Scholar]
- 8.Mandeville JB, Marota JJ, Ayata C, Moskowitz MA, Weisskoff RM, Rosen BR. MRI measurement of the temporal evolution of relative CMRO2 during rat forepaw stimulation. Magn Reson Med. 1999;42:944–7. doi: 10.1002/(sici)1522-2594(199911)42:5<944::aid-mrm15>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 9.Volkow ND, Rosen, Farde L. Imaging the living human brain: Magnetic resonance imaging and positron emission tomography. Proc Natl Acad Sci U S A. 1997;94:2787–8. doi: 10.1073/pnas.94.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. [Last accessed on 2013 Sep 25]. Available from: http://www.vislab.ucl.ac.uk/cogent.php .
- 11. [Last accessed on 2013 Sep 25]. Available from: http://www.pstnet.com/eprime.cfm .
- 12.Laurence R, Dominic C. An introduction to E-Prime. Tutor Quant Methods Psychol. 2009;5:68–78. [Google Scholar]
- 13. [Last accessed on 2013 Sep 25]. Available from: http://www.neurobs.com .
- 14.Plong M, Shen K, Van Vliet M, Robben A, Van Hulle M, Geurts L. Asian Conference on Information Systems; 2012. Accurate visual stimulus presentation software for EEG experiments; p. 1. [Google Scholar]
- 15.Aguirre GK, D’Esposito M. Berlin: Springer-Verlag; 2000. Experimental design for brain fMRI. Functional MRI; pp. 369–71. [Google Scholar]
- 16.Amaro E, Jr, Barker GJ. Study design in fMRI: Basic principles. Brain Cogn. 2006;60:220–32. doi: 10.1016/j.bandc.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Buckner RL. Event-Related fMRI and the hemodynamic response. Hum Brain Mapp. 1998;6:373–7. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<373::AID-HBM8>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen SE, Dubis JW. The mixed block/event-related design. Neuroimage. 2012;62:1177–84. doi: 10.1016/j.neuroimage.2011.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutten GJ, Ramsey NF. The role of functional magnetic resonance imaging in brain surgery. Neurosurg Focus. 2010;28:E4–13. doi: 10.3171/2009.12.FOCUS09251. [DOI] [PubMed] [Google Scholar]
- 20.Engström M, Ragnehed M, Lundberg P, Söderfeldt B. Paradigm design of sensory -motor and language tests in clinical fMRI. Neurophysiol Clin. 2004;34:267–77. doi: 10.1016/j.neucli.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Kesavadas C, Thomas B, Sujesh S, Ashalata R, Abraham M, Gupta AK, et al. Real-time functional MR imaging (fMRI) for presurgical evaluation of paediatric epilepsy. Pediatr Radiol. 2007;37:964–74. doi: 10.1007/s00247-007-0556-4. [DOI] [PubMed] [Google Scholar]
- 22.Smits M, Visch-Brink E, Schraa-Tam CK, Koudstaal PJ, Van der Lugt A. Functional MR imaging of language processing: an overview of easy-to-implement paradigms for patient care and clinical research. Radiographics. 2006;26:S145–58. doi: 10.1148/rg.26si065507. [DOI] [PubMed] [Google Scholar]
- 23.Amaro E, Jr, Barker GJ. Study design in fMRI: Basic principles. Brain and Cognition. 2006;6:220–2. doi: 10.1016/j.bandc.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Casey BJ, Davidson M, Rosen B. Functional magnetic resonance imaging: Basic principles of and application to developmental science. Dev Sci. 2002;5:301–8. [Google Scholar]
- 25.Buckner RL. Event-Related fMRI and the Hemodynamic Response. Hum Brain Mapp. 1998;6:373–4. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<373::AID-HBM8>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engström M, Ragnehed M, Lundberg P, Söderfeldt B. Paradigm design of sensory–motor and language tests in clinical fMRI. Neurophysiol Clin. 2004;34:267–10. doi: 10.1016/j.neucli.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Friston KJ, Frith CD, Liddle PF, Dolan RJ, Lammertsma AA, Frackowiak RS. The relationship between global and local changes in PET scans. J Cereb Blood Flow Metab. 1990;100:458–66. doi: 10.1038/jcbfm.1990.88. [DOI] [PubMed] [Google Scholar]
- 28.Friston KJ, Frith CD, Liddle PF, Frackowiak RS. Comparing functional (PET) images: The assessment of significant change. J Cereb Blood Flow Metab. 1991;110:690–9. doi: 10.1038/jcbfm.1991.122. [DOI] [PubMed] [Google Scholar]
- 29.Ashburner J, Neelin P, Collins DL, Evans A, Friston K. Incorporating prior knowledge into image registration. Neuroimage. 1997;60:344–8. doi: 10.1006/nimg.1997.0299. [DOI] [PubMed] [Google Scholar]
- 30.Ashburner J. SPM: A history. Neuroimage. 2012;62:791–9. doi: 10.1016/j.neuroimage.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD. 1st ed. London: Academic Press; 2007. Statistical Parametric Mapping: The Analysis of Functional Brain Images; pp. 49–13. [Google Scholar]
- 32.Friston KJ, Jezzard P, Turner R. Analysis of functional MRI time-series. Hum Brain Mapp. 1994;1:153–8. [Google Scholar]
- 33.Crinion J, Ashburner J, Leff A, Brett M, Price C, Friston K. Spatial normalization of lesioned brains: Performance evaluation and impact on fmri analyses. Neuroimage. 2007;370:866–9. doi: 10.1016/j.neuroimage.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talairach J, Tournoux P. New York: Thieme Medical Publishers; 1988. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging. [Google Scholar]
- 35.Kasess CH, Stephan KE, Weissenbacher A, Pezawas L, Moser E, Windischberger C. Multi-subject analyses with dynamic causal modeling. Neuroimage. 2010;49:3065–9. doi: 10.1016/j.neuroimage.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worsley KJ, Friston KJ. Analysis of fMRI time-revisited- again. Neuroimage. 1995;2:173–8. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- 37.Polzehl J, Voss HU, Tabelow K. Structural adaptive segmentation for statistical parametric mapping. Neuroimage. 2010;52:515–8. doi: 10.1016/j.neuroimage.2010.04.241. [DOI] [PubMed] [Google Scholar]
- 38.Price CJ, Moore CJ, Friston KJ. Subtractions, conjunctions, and interactions in experimental design of activation studies. Hum Brain Mapp. 1997;5:264–72. doi: 10.1002/(SICI)1097-0193(1997)5:4<264::AID-HBM11>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Martin A, Lindquist The statistical analysis of fMRI data. Stat Sci. 2008;23:439–24. [Google Scholar]
- 40.Abramovich F, Bailey T, Sapatinas T. Wavelet analysis and its statistical applications. Statistician. 2000;49:1–29. [Google Scholar]
- 41.Bennett CM, Wolford GL, Miller MB. The principled control of false positives in neuroimaging. Soc Cogn Affect Neurosci. 2009;4:417–22. doi: 10.1093/scan/nsp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenfeld A, Kak AC. Digital Picture Processing. Orlando: Academic Press. 1982;1:435. [Google Scholar]
- 43.Van De Ville D, Unser M. False discovery rate for wavelet-based statistical parametric mapping. IEEE J Sel Top Signal Process. 2008;2:897–10. [Google Scholar]
- 44.Worsley K, Marrett S, Neelin P, Vandal AC, Friston K, Evans AC. A unified statistical approach for determining significant voxels in images of cerebral activation. Hum Brain Mapp. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 45. [Last accessed on 2013 Sep 23]. Available from: http://www.brainvoyager.com .
- 46.Goebel R, Jansma H, Eck J. BrainVoyager QX Getting Started Guide Version 2.8 for BVQX 2.1. 2009 [Google Scholar]
- 47.Heller R, Golland Y, Malach R, Benjamini Y. Conjunction group analysis: An alternative to mixed/random effect analysis. Neuroimage. 2007;37:1178–85. doi: 10.1016/j.neuroimage.2007.05.051. [DOI] [PubMed] [Google Scholar]
- 48.Formisano E, Esposito F, Di Salle F, Goebel R. Cortex-based independent component analysis of fMRI time-series. Magn Reson Imaging. 2004;22:1493–504. doi: 10.1016/j.mri.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 49.Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–52. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 50.De Martino F, Valente G, Staeren N, Ashburner J, Goebel R, Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. Neuroimage. 2008;43:44–4. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- 51.Lindquist MA. The Statistical Analysis of fMRI Data. Stat Sci. 2008;23:439–26. [Google Scholar]
- 52.Caria A, Sitaram R, Birbaumer N. Real-Time fMRI: A tool for local brain regulation. Neuroscientist. 2012;18:487–501. doi: 10.1177/1073858411407205. [DOI] [PubMed] [Google Scholar]


