Abstract
Siderophores are high-affinity iron chelators produced by microorganisms and frequently contribute to the virulence of human pathogens. Targeted inhibition of the biosynthesis of siderophores staphyloferrin B of Staphylococcus aureus and petrobactin of Bacillus anthracis hold considerable potential as a single or combined treatment for methicillin-resistant S. aureus (MRSA) and anthrax infection, respectively. The biosynthetic pathways for both siderophores involve a nonribosomal peptide synthetase independent siderophore (NIS) synthetase, including SbnE in staphyloferrin B and AsbA in petrobactin. In this study, we developed a biochemical assay specific for NIS synthetases to screen for inhibitors of SbnE and AsbA against a library of marine microbial-derived natural product extracts (NPEs). Analysis of the NPE derived from Streptomyces tempisquensis led to the isolation of the novel antibiotics baulamycins A (BmcA, 6) and B (BmcB, 7). BmcA and BmcB displayed in vitro activity with IC50 values of 4.8 µM and 19 µM against SbnE and 180 µM and 200 µM against AsbA, respectively. Kinetic analysis showed that the compounds function as reversible competitive enzyme inhibitors. Liquid culture studies with S. aureus, B. anthracis, E. coli and several other bacterial pathogens demonstrated the capacity of these natural products to penetrate bacterial barriers and inhibit growth of both Gram-positive and Gram-negative species. These studies provide proof-of-concept that natural product inhibitors targeting siderophore virulence factors can provide access to novel broad-spectrum antibiotics, which may serve as important leads for the development of potent anti-infective agents
Keywords: high throughput screening, Staphylococcus aureus, Bacillus anthracis, natural product, antibiotic
INTRODUCTION
The rapid ability of pathogens to develop resistance to antibiotics is endangering the management of a multitude of serious infections.1–3 In alarming contrast to the increase of bacterial adaptation to currently marketed drugs, the discovery of novel classes of antimicrobials is on the decline.4 Although the majority of pharmaceutical efforts during the past six decades have focused on the synthetic enhancement of a limited set of unique core scaffolds, a more sustainable route to combat antibiotic resistance is the discovery of novel chemical structures possessing unique microbial targets.2,5 Iron acquisition mechanisms in particular may represent effective antimicrobial targets that present substantial hurdles to bacterial antibiotic resistance.6 Iron is required for growth and survival of bacteria but remains tightly regulated in the mammalian host. Many pathogenic Gram-positive and Gram-negative bacteria utilize virulence-associated siderophores to scavenge iron from this restricted environment and return it to the microbial cell.7 Although previous studies have corroborated siderophore biosynthetic enzymes as effective drug targets through the tailoring of established chemical scaffolds,8–11 no novel chemical structures have been identified.
In our efforts to identify new structural antibiotic classes with inhibitory activity against siderophore biosynthetic enzymes, we selected the Gram-positive bacteria methicillin-resistant Staphylococcus aureus (MRSA) and Bacillus anthracis as model systems. The “superbug” MRSA is a major public health concern, attributed to more than 18,000 deaths a year in the United States.2,12 In contrast, the spore-forming microorganism B. anthracis is the causative agent of anthrax. The ability of the bacterium to quickly achieve high concentrations within infected hosts makes it a serious bioterrorism threat, with mortality rates for inhalational infection historically reaching as high as 94%.13
Both pathogens are strongly associated with antimicrobial resistance,14 and their siderophore biosynthetic pathways have been extensively characterized.15,16 The siderophores staphyloferrin B (2) of S. aureus17–22 and petrobactin (5) of B. anthracis23–30 in particular have been shown to be critical for survival in iron-limited environments.
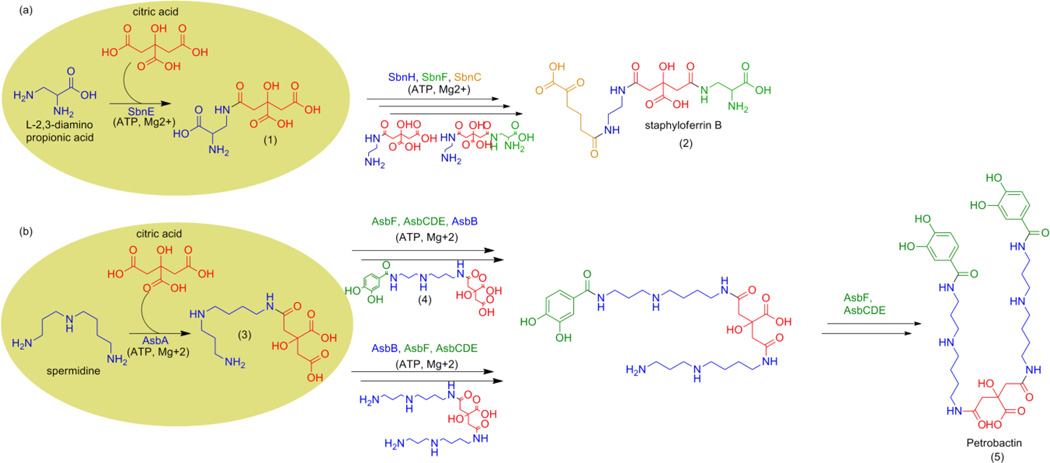
The biosynthetic pathways for both siderophores involve a type A nonribosomal peptide synthetase independent siderophore (NIS) synthetase, SbnE (Figure 1A) in staphyloferrin B and AsbA (Figure 1B) in petrobactin. Type A NIS synthetases are a unique class of enzymes found within siderophore biosynthetic pathways of a number of pathogenic bacteria, including Shigella flexneri, Escherichia coli, and Salmonella typhimurium.31,32 These enzymes catalyze the condensation of citric acid with either a polyamine or amino alcohol substrate in an ATP-dependent reaction.31,32 Since type A NIS synthetases share similar catalytic mechanisms and substrate preferences, we sought to identify novel antibiotics against S. aureus or B. anthracis that could also serve as broad-spectrum antibiotics against other NIS synthetase-containing pathogens.
Figure 1.
Biosynthesis of the virulence-associated siderophore (A) staphyloferrin B in S. aureus (B) petrobactin in B. anthracis. Target biosynthetic enzymes SbnE and AsbA are highlighted by the yellow circles.
RESULTS AND DISCUSSION
High Throughput Screening for Inhibitors of Siderophore Biosynthesis Derived from Natural Product Extracts
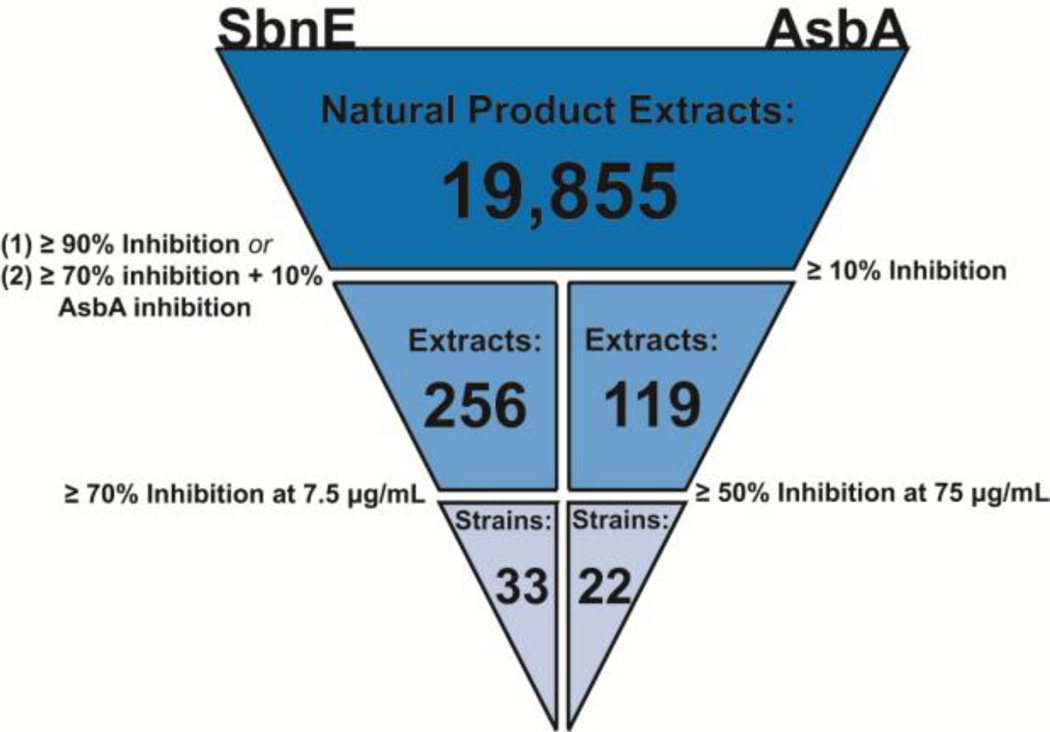
Fueled by the need to identify a new structural class of antibiotics, we selected a marine microbial-derived natural product extract (NPE) library to identify inhibitors of NIS synthetases in S. aureus and B. anthracis. The majority of currently marketed drugs are derived from natural products,33,34 and the marine environment in particular is thought to be an underexplored source of novel chemical structures.35 We developed an enzymatic high throughput assay specific to SbnE and AsbA and adapted it to a malachite green reporter system.36–38 The assay was then queried against a library of 19,855 marine microbial-derived NPEs to identify inhibitors of enzyme activity (Figure 2). The NPEs in this library are primarily from marine-derived microorganisms collected in Costa Rica, Panama, and Papua New Guinea. Screening SbnE and AsbA in parallel limited the number of false positives and provided a convenient method for prioritization of active extracts. Two rounds of screening yielded 33 strains with greater than 70% inhibition against SbnE at 7.5 µg/mL and 22 strains with extracts showing greater than 50% inhibition against AsbA at 75 µg/mL (Figure 2). The higher frequency and activity of extracts against SbnE is likely due to the lower enzyme concentration 5 nM) compared to the AsbA (100 nM) utilized in screening. These were the lowest enzyme concentrations for each respective assay that produced a response fit for screening (Figure S2). Low enzyme concentrations were desired to maximize the likelihood of detecting extracts possessing low-abundance bioactive components.
Figure 2.
Triage of hits derived during high throughput screening of SbnE and AsbA against the full microbial NPE library.
Among active extracts, Streptomyces tempisquensis (Figure S4) was of particular interest due to its high activity against both SbnE (95.9%) and AsbA (90.2%) (Figure S3). The strain was originally isolated from sediments collected in Playa Grande, Costa Rica (−85°49’39.8”, 10°18’39.8”) near Las Baulas National Marine Park.
Isolation and Structural Elucidation of the Baulamycins (6–7)
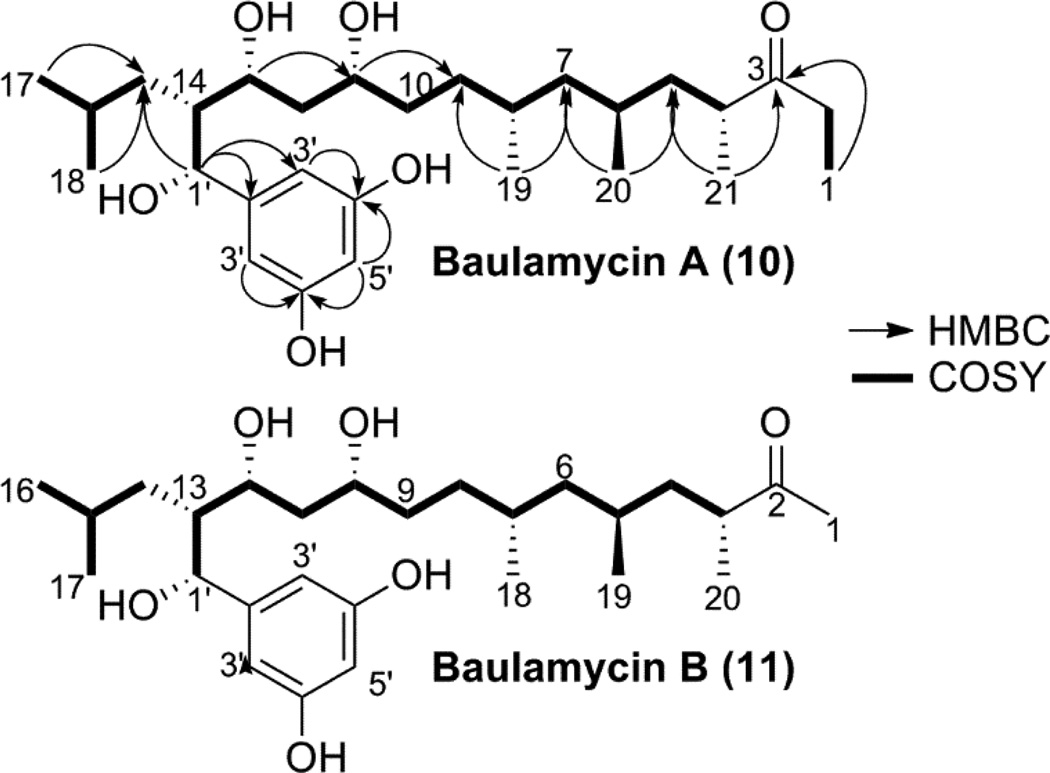
An iterative bioassay guided C18 fractionation (Figure S5) and subsequent RP-18 HPLC purification (Figure S6) yielded two novel bioactive molecules (Figure 3), baulamycins A (BmcA, 6) and B (BmcB, 7), whose structures are consistent with biogenesis from a type I polyketide synthase pathway. BmcA (6) was purified as a light yellow amorphous solid and possesses a molecular formula of C28H48O6 as suggested by HRAPCIMS based on [M+H]+ ion peak at m/z 481.3530 (Figure S7). The 1H (Figure S8) and 13C NMR (Figure S9) data, recorded in CD3OD indicated the polyketide nature of 6 and indicated the presence of at least three hydroxyl group bearing methines with chemical shifts at δ 4.47 (76.5), 4.01 (73.3) and 3.69 (72.5). Further analysis of the 1H NMR spectrum of 6 identified 12 aliphatic protons in the region of δH 0.95–2.77 and six methyl groups at δH 0.77 (d), 0.83 (d), 0.86 (d), 0.88 (d), 1.02 (t) and 1.06 (d). The 13C NMR and HSQCAD spectra (Figure S10) revealed the presence of four quaternary carbons at δC 148.6 (an aromatic carbon), 159.1 (two chemically equivalent aromatic carbons) and at δC 218.7 (a carbonyl carbon). The gCOSY (Figure S11) correlations between two equivalent aromatic protons with a signal at δH 6.33, 6.15 and HMBCAD (Figure S12) correlation between δH 6.33, 6.15 to the equivalent carbons at δC 159.1, 105.9 and carbon at δC 148.6 clearly suggested the presence of resorcinol moiety (Table 1). Similarly, the connectivity from C-1 to C-17 was confirmed by an array of COSY and HMBC couplings in both CD3OD and DMSO-d6 to construct a 17-carbon aliphatic straight chain with a characteristic carbonyl carbon at δC 218.7. In addition, COSY correlation was observed between H-1 and protons at C-2 along with their HMBC connection to C-3 suggesting an ethyl-ketone terminus for molecule 6. Moreover, the COSY and HMBC correlations indicated the branching of aliphatic chain through a methyl group at C-4 (δH 2.77, δC 44.6), C-6 (δH 1.42, δC 29.1), C-8 (δH 1.53, δC 30.9), and C-16 (δH 1.38, δC 26.6) positions. The position of the hydroxyl group at C-11 and C-13 were consistent with the distinctive 1H and 13C chemical shifts at δH 3.69, δC 72.5 and δH 4.01, δC 73.3, respectively. Furthermore, HMBC showed a correlation from H-1’ (δH 4.47) to C-2’ (δC 148.6) along with its contiguous COSY correlation to H-14 (δH 1.88) indicating a branch-point for the aliphatic chain at C-14 through a pendant attachment of 1’-(hydroxymethyl)-resorcinol moiety to complete the structure of BmcA (6) (Table 1).
Figure 3.
Planar structure of baulamycins A (6) and B (7) showing COSY correlation with bold bonds and HMBC correlations with arrow.
Table 1.
NMR spectroscopic data for baulamycins A (6) and B (7) in CD3OD at 700 MHz.
| Baulamycin A (6) | Baulamycin B (7) | |||||
|---|---|---|---|---|---|---|
| δC | δH, multi (J in Hz) | COSY | HMBC | δC | δH | |
| 1 | 35.1 | 2.57, dp | 2" | 1, 2", 2 | 27.4 | 2.15 |
| 2.49, dp | 2" | 1, 2", 2 | ||||
| 2 | 7.8 | 1.02, t (6.9, 7.3) | 1" | 1, 1", 2 | ||
| 3 | 218.7 | 214.6 | ||||
| 4 | 44.6 | 2.77, ddq (3.2, 6.7, 8.0) | 3 | 1, 3, 19 | 43.9 | 2.70 |
| 5 | 41.7 | 1.73, ddd (3.2, 5.0, 11.0) | 2 | 1, 2, 4 | 41.8 | 1.72, |
| 0.98, ddd (6.7, 8.0, 11.0) | 4 | 0.97 | ||||
| 6 | 29.1 | 1.42, ddddq (3.0, 3.2, 5.0, 13.0) | 3, 5 | 6, 18 | 29.2 | 1.44 |
| 7 | 46.3 | 1.22, ddd (3.0, 11.0, 13.0) | 6 | 17, 18 | 46.0 | 1.22 |
| 0.95, ddd (3.2, 5.0, 11.0) | 3 | 17, 18 | 0.95 | |||
| 8 | 30.9 | 1.53, ddddq (3.0, 5.0, 8.5, 13.0) | 5, 7 | 5, 7, 17 | 31.0 | 1.55 |
| 9 | 33.0 | 1.33, dddd (3.0, 3.0, 10.6) | 6, 8 | 9, 17 | 33.4 | 1.34 |
| 1.19, dddd (8.5, 9.0, 10.6) | 6, 8 | 7, 5 | 1.21 | |||
| 10 | 35.4 | 1.41, dddd (9.0, 11.0, 11.6) | 7, 9 | 11, 17 | 35.1 | 1.40 |
| 1.39, dddd (3.0, 4.0, 11.6) | 7, 9 | 1.38 | ||||
| 11 | 72.5 | 3.69, dddd (8.0, 11.0, 12.0) | 8, 10 | 7, 8. 11 | 72.2 | 3.72 |
| 12 | 40.6 | 1.79, ddd (8.0, 7.2, 11.0) | 9, 11 | 40.2 | 1.80, | |
| 1.55, ddd (3.1, 3.2, 11.0) | 1.50 | |||||
| 13 | 73.3 | 4.01, dt (9.9, 3.4) | 10, 12 | 1', 9, 13 | 73.0 | 3.99 |
| 14 | 48.5 | 1.88, ddq (7.7, 7.0) | 11, 13 | 1', 11, 14 | 48.7 | 1.80 |
| 15 | 37.3 | 1.21, m | 12, 14 | 37.0 | 1.20 | |
| 16 | 26.6 | 1.38, m | 13, 15 | 26.6 | 1.38 | |
| 17 | 22.5 | 0.83, d (6.5) | 14 | 13, 14 | 22.4 | 0.82 |
| 18 | 23.3 | 0.77, d (6.5) | 14 | 13, 14 | 23.3 | 0.77 |
| 19 | 20.3 | 0.86, d (6.6) | 6 | 5, 6, 7 | 20.1 | 0.85 |
| 20 | 20.5 | 0.88, d (6.5) | 4 | 3, 4, 5 | 20.2 | 0.88 |
| 21 | 17.9 | 1.06, d (6.9) | 2 | 1, 2, 3 | 18.0 | 1.01 |
| 1' | 76.5 | 4.47, d (7.0) | 12 | 2', 3', 8, 13, 11 | 76.4 | 4.50 |
| 2' | 148.6 | 148.2 | ||||
| 3' | 105.9 | 6.33, d (2.2) | 5' | 1', 2', 3', 4', | 106.1 | 6.35 |
| 4' | 159.1 | 158.5 | ||||
| 5' | 101.9 | 6.15, t (2.2) | 3' | 3', 4' | 101.8 | 6.16 |
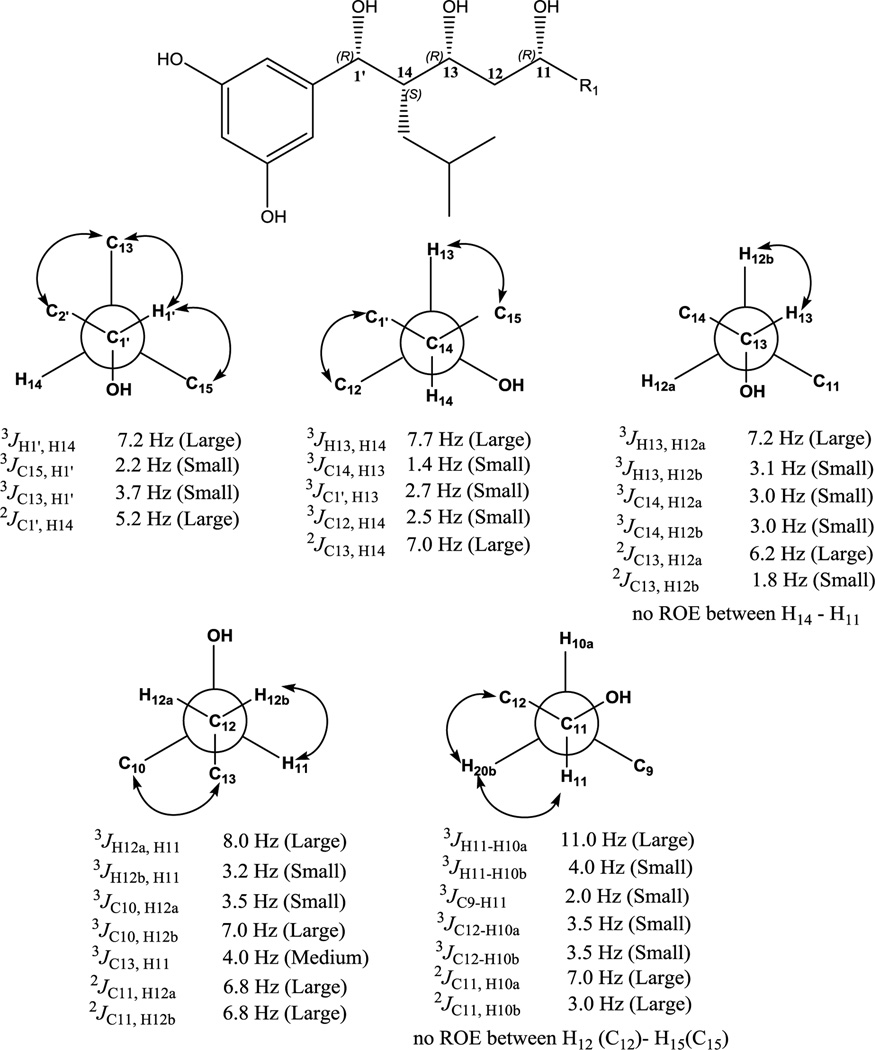
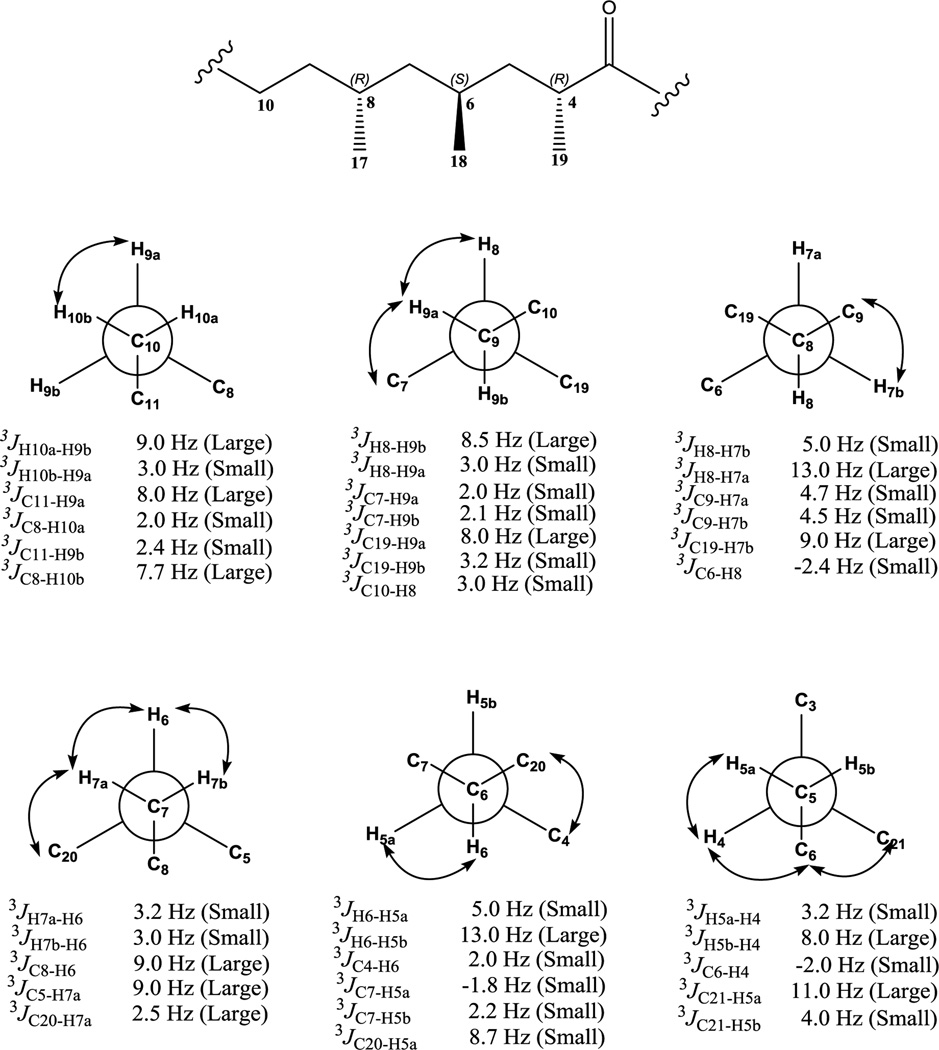
BmcA (6) consists of seven stereocenters with three hydroxyl bearing carbons, three methyl bearing carbons and a 2-methylpropane containing stereocenter. Initial attempts were made to obtain absolute stereochemical information through chemical manipulations for at-least hydroxyl bearing chiral centers. However, the relatively low yield of baulamycins rendered this approach impractical. Therefore, a nondestructive J based configuration analysis (JBCA)39 was employed to propose the relative configuration of BmcA (6).40–42 We calculated 3JH-H, 2JC-H, and 3JC-H values using a combination of phase sensitive double quantum filtered (PS-DQF)-COSY, homonuclear 2DJ spectroscopy (HOMO2DJ), gradient- and sensitivity-enhanced hetero (ω1) half filtered TOCSY (HETLOC-gse) and heteronuclear 2DJ (HET2DJ) spectral analyses (Figures S13–15).
The relative configurations of stereocenters (C11-C13-C14-C1’) in 6 established by JBCA are as follows. At the C1’-C14 axis, a large coupling constant 3JH1’-H14 ~ 7.2 Hz and 2JC1’-H14 ~ 5.2 Hz, inferred their anti and gauche orientations, respectively39 (Figure 4). For the C14-C13 bond, the large 3JH14-H13 ~ 7.7 Hz suggested anti orientation, which was also supported by the observed ROESY correlations (Figure S15). An additional large 2JC13-H14 ~ 7.0 Hz was indicative of the gauche orientation between O-13 and H-1439 (Figure 4). Further, we sliced the HOMO2DJ spectrum at δH 4.01 (H-13) suggesting a large coupling constant between H13-H12a and a small value between H13-H12b indicative of anti and gauche orientations, respectively (Figure 4). For the C12-C11 moiety, we sliced the HOMO2DJ spectrum at δH 3.69 (H-11) to yield 3JH12a-H11 ~ 8.0 Hz and 3JH12a-H11 ~ 3.2 Hz suggesting anti and gauche orientation, respectively (Figure 4). Furthermore, large coupling constants were recorded for 2JC11-H12a ~ 6.8 Hz and 2JC11-H12b ~ 7.2 Hz clearly indicating gauche orientations between O-11 and H-12a/H-12b39(Figure 4). Moving down the axis over C11-C10 moiety, large coupling constant between H-11/H-10a, H-11/H10b and C11/H10a suggested gauche orientation between O-11 and H-10a. Hence, the relative configurations at C1’, C14, C13 and C11 were proposed as 1’R*, 14S*, 13R* and 11R*, respectively (Figure 3).
Figure 4.
H-H, C-H coupling values and relative configuration determined for C1’-C11. Arrows showing ROESY correlations.
Establishment of the relative orientation of protons associated with non-chiral C-10 and C-9 was necessary to measure stereochemistry of subsequent chiral carbons (C8-C4). We sliced both HOMO2DJ and PS-DQF-COSY spectrum at δH 1.41, 1.39 (H-10a, H-10b) and δH 1.19, 1.33 (H-10a, H-10b), respectively. The slicing provided large coupling constants between H-10a/ H-9b and small between H-10a/ H-9a suggesting anti and gauche orientations, respectively (Figure 5). Correspondingly, for the C9-C8 axis, calculations yielded 3JH9b-H8 ~ 8.5 Hz (large) and 3JH9a-H8 ~ 3.0 Hz (small) indicating anti and gauche conformations, respectively (Figure 5). Furthermore, the ROESY correlations and small coupling values obtained from 3JC7-H9a ~ 2.0 Hz and 3JC7-H9b ~ 2.1 Hz confirmed the gauche orientations between C7 and H-9a/H-9b facilitating assignment of the relative conformation of H-9a and H-9b at C9 (Figure 5).
Figure 5.
H-H, C-H coupling values and relative configuration determined for C10-C4. Arrows showing ROESY correlations.
For C9-C8 axis, the observation of a large 3JH8-H9b (8.5 Hz) and 3JC19-H9a (8.0 Hz) values indicated that H-8 and H9b as well as C-19 and H-9a are in the anti orientation. The relative configuration at C8 was established to be R* according to the ROE between H-8 and H-9a (Figure 5). For the C8/C7 bond, the relationship of C19/H-7b was revealed as anti based on a large 3JC19-H7b ~ 9.0 Hz value. Another large coupling value (3JH7a-H8 ~ 13.0 Hz) and corresponding small coupling value between C6 and H-8 (3JC6-H8 ~ -2.4 Hz) led us to assign the relative conformation of H-7a and H-7b at C7 (Figure 5). For the C7/C6 bond, the observation of small coupling values (3JH7a-H6 ~ 3.0 Hz and 3J7b-H6 ~ 3.2 Hz) along with apparent ROE between H-6 and H-7a/H-7b suggested gauche orientation between H-6 and H-7a/H7b, establishing relative configuration at C6 as S* (Figure 5).
For C6 and C5, a large 3JH6-H5b value (~ 13.0 Hz) and small 3JH6-H5a value (~5.0 Hz) revealed that H-6/H-5b and H-6/H-5a possess anti and gauche relationships, respectively. Additionally, small 3JC4-H6 ~ 2.0 Hz and 3JC7-H5a ~ −1.8 Hz values as well as observed ROE between H-5a and H-6 revealed the relative conformation of H-5a and H-5b at C5 (Figure 5). For the C5/C4 axis, a large 3JH5b-H4 ~ 8.0 Hz and small 3JH5a-H4 ~ 3.2 Hz coupling values was obtained from HOMO2DJ and PS-DQF-COSY indicating anti and gauche conformations, respectively. In addition, an anti orientation of the C21 and H-5a atoms was deduced from the large 3JC21-H15a ~ 11.0 Hz value obtained from HETLOC-gse spectrum. Further, in ROESY spectrum, ROEs between H-5a/H-4, H-4/H-6 and H-6/H-21 was observed. Taking these data into consideration, the relative configuration at C4 was proved to be R*. Therefore, the relative configuration for C10-C4 moiety was anticipated as 8R*, 6S* and 4R*, respectively, in BmcA (6).
BmcB (7) was isolated by RP-18-HPLC from the same C18 fraction containing compound 6. The HRESIMS of the molecule provided a molecular formula of C27H46O6 showing a [M+H]+ ion peak at 467.3171 (Figure S20). Moreover, baulamycin B (7) had high structural similarity to 6, as evidenced by nearly identical 1H and 13C NMR chemical shifts when measured in CD3OD (Table 1). However, it displayed a subtle difference towards the carbonyl terminus, revealing the absence of terminal ethyl protons and substituting with a terminal methyl singlet at δH 2.18 in the 1H NMR spectrum of compound 7 (Figure S21). The relative stereochemistry from C1’-C10 and C5-C3 were proposed to be identical to those of 6 on the basis of highly comparable NMR spectroscopy data (Figures S21–25).
Assessment of the in vitro Biological Activity of Compounds against Purified NIS Enzymes
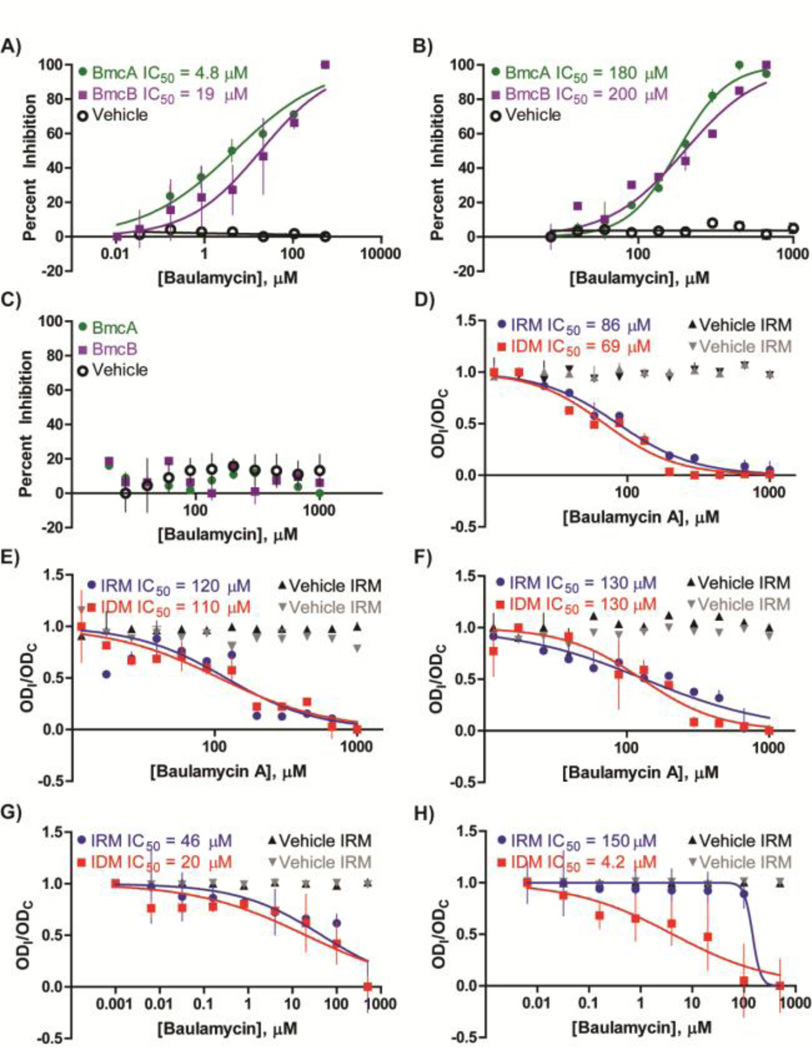
The malachite green-based bioassay was next employed to assess the dose response of novel compounds BmcA (6) and BmcB (7) against purified SbnE and AsbA (Table 2). Both compounds exhibited in vitro bioactivity against SbnE, with IC50 values of 4.8 µM and 19 µM for BmcA and BmcB, respectively (Figure 6A). Inhibition was also observed against AsbA, with IC50 values of 180 µM and 200 µM for BmcA and BmcB, respectively (Figure 6B). The significant difference in apparent inhibition is likely due to the different enzyme concentrations used in the dose response assays, 25 nM for SbnE and 100 nM for AsbA (Figure S2). The assays were originally optimized at these enzyme concentrations to maximize the detection of extracts containing low levels of bioactive components during screening.
Table 2.
In vitro bioactivity analysis of the baulamycins against purified enzymes. The IC50 values were determined through the NIS-tailored malachite green assay while the Ki values were found through steady state kinetic studies. The Ki value for AsbB was not determined (ND).
| Enzymatic Target |
NIS Synthetase Classification |
Associated Microbial Strain | Associated Siderophore | BmcA IC50 (µM) |
BmcB IC50 (µM) |
|---|---|---|---|---|---|
| SbnE | Type A | S. aureus | Staphyloferrin B | 4.8 | 19 |
| AsbA | Type A | B. anthracis | Petrobactin | 180 | 200 |
| AsbB | Type C | B. anthracis | Petrobactin | > 1000 | > 1000 |
Figure 6.
(A–C) In vitro bioactivities against purified enzyme recorded for BmcA and BmcB against the NIS synthetase (A) SbnE, (B) AsbA, or (C) AsbB. (D–H) BmcA inhibition against live cultures of microbial strains (D) S. aureus (Newman), (E) B. anthracis (Sterne 34F2), F) MRSA (USA 300), G) S. flexneri (BS103), or (H) E. coli (MC 1061) in iron-depleted (IDM) or iron-rich (IRM) media. The y-axis represents a comparison of the optical densities of inhibitor- (ODI) and DMSO control-treated (ODC) cultures. Assays were conducted in duplicate due to the current limited availability of the baulamycins.
As a next step to address whether the baulamycins selectively inhibited type A over other NIS synthetase subfamilies, we explored the activity of both compounds against the petrobactin biosynthetic type C NIS synthetase AsbB. While type A NIS synthetases prefer citric acid as a substrate, type C enzymes generally utilize citric or succinic acid derivatives often found as complex intermediates within their respective siderophore biosynthetic pathways.31 In the petrobactin pathway, AsbB catalyzes condensation of a second molecule of spermidine with either N8-citryl-spermidine (3) or N1-(3,4-DHB)-N8-citryl spermidine (4).30 Interestingly, recent work in our laboratory also demonstrates AsbB to be more flexible in substrate selection and capable of partial compensation for AsbA activity in vitro.30 Despite these findings, dose response studies revealed inhibition against AsbB to be negligible for both BmcA and BmcB (Figure 6C). These data indicate that both natural products are more potent in vitro against the type A NIS synthetases SbnE and AsbA than the type C subfamily member AsbB.
Initial Enzyme Mechanism of Inhibition Studies
To investigate how the more potent of the two compounds, BmcA (6), inhibits SbnE and AsbA enzymatic activity, we monitored NIS synthetase ATP turnover using a previously described 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG) kinetic pyrophosphate detection assay.43,44 These experiments were conducted by varying a single substrate and holding remaining reaction components at a constant saturating level in the presence and absence of inhibitor. The resulting double reciprocal (Lineweaver-Burk) plots confirmed that kinetic parameters closely matched previously reported values for both enzymes against varied citrate concentrations (Table S1).20,30 The plots suggested that BmcA inhibits both enzymes in a reversible, competitive manner with respect to citric acid, the corresponding polyamine, and ATP (Figure S26, Table S1). Although kinetic assays were only conducted with BmcA due to its higher potency and availability (obtained in substantially higher yield from Streptomyces tempisquensis), it is reasonable to expect that BmcB also possesses these inhibition patterns.
A replot of slopes derived from the Lineweaver-Burk plot versus inhibitor concentration also enabled an initial investigation into the inhibition constants (Ki values) for BmcA against both enzymes (Table S1).45 Interestingly, the inhibitor constants were lowest for citric acid (SbnE: 50 µM; AsbA: 110 µM) followed by the polyamine substrate (SbnE: 210 µM; AsbA: 170 µM) and ATP (SbnE: 680 µM; AsbA: 230 µM). This is surprising given the polar nature of citric acid in comparison to the baulamycins. The explanation for this likely depends on the structure and substrate binding sites in the proteins. Although no structural studies have yet been conducted with SbnE or AsbA, the crystal structures of AsbB and the achromobactin type A NIS synthetase in Pectobacterium chrysanthemi have been determined.30,46 Both reveal a single binding pocket that houses substrates and intermediates during catalysis. Elucidation of the differences in the structures of SbnE, AsbA, and these other NIS synthetases could shed light on the exact mechanism of inhibition exerted by the baulamycins and their complete lack of activity against AsbB. Efforts to acquire X-ray crystal structures of both SbnE and AsbA in the presence and absence of baulamycin compounds are expected in due course. Additionally, mutagenesis and selection of S. tempisquensis should improve the yield of the baulamycins, enabling a sufficient amount of these molecules for comprehensive kinetic analysis.
Assessment of Biological Activity against Microbial Cultures
Since siderophore biosynthesis is required for bacterial survival in iron-limited environments, a selective siderophore synthetase inhibitor would be expected to limit growth only under these conditions. Therefore, we monitored bacterial growth in both iron-depleted (IDM) and iron-rich (IRM) media conditions in the presence of 6 and 7. We also conducted live culture studies to demonstrate the ability of both natural products to penetrate bacteria, an established advantage of natural products over many synthetic chemicals.47 However, due to the extremely low yield of the baulamycins from S. tempisquensis, experiments could only be completed in duplicate and the following studies represent an initial assessment of microbial culture bioactivity.
BmcA was found to inhibit growth of S. aureus (Newman) in both iron-depleted (IC50 = 69 µM) and iron-rich (IC50 = 86 µM) conditions, (Figure 6D, Table 3). Similar inhibition was observed with B. anthracis (Sterne 34F2) under iron-depleted (IC50 = 110 µM) and iron-rich (IC50 = 120 µM) conditions (Figure 6E, Table 3), suggesting possible secondary targets in the cell. In agreement with in vitro findings against purified enzyme, BmcB was significantly less potent against S. aureus (Newman) and B. anthracis (Sterne 34F2) (Figure S27).
Table 3.
Bioactivity analysis of the baulamycins against Gram-positive and Gram-negative microbial cell cultures. Dose response assays were conducted in iron-repleated (IRM) and iron-depleted (IDM) media.
| Microbial Strain | Classification | Targeted Siderophore Path- way |
Associated NIS Synthetase | BmcA IRM IC50 (µM) |
BmcA IDM IC50 (µM) |
|---|---|---|---|---|---|
| S. aureus (Newman) | Gram-positive | Staphyloferrin B | SbnE (A), SbnC (B), SbnF (C) | 86 | 69 |
| MRSA (USA 300) | Gram-positive | Staphyloferrin B | SbnE (A), SbnC (B), SbnF (C) | 130 | 130 |
| B. anthracis (Sterne 34F2) | Gram-positive | Petrobactin | AsbA (A), AsbB (C) | 120 | 110 |
| S. typhimurium | Gram-negative | Aerobactin | IucA (A), IucC (C) | > 1000 | > 1000 |
| E. coli (MC 1061) | Gram-negative | Aerobactin | IucA (A), IucC (C) | 150 | 4.2 |
| S. flexneri (BS103) | Gram-negative | Aerobactin | IucA (A), IucC (C) | 46 | 20 |
We next tested the potency of BmcA against a clinically isolated MRSA strain (USA 300). Similar to S. aureus (Newman) and B. anthracis (Sterne 34F2), BmcA also inhibited MRSA (USA 300) in iron-depleted (IC50 = 130 µM) and iron-rich (IC50 = 130 µM) conditions (Figure 6F, Table 3), again suggesting secondary cellular targets.
Encouraged by the potency of these compounds on S. aureus and B. anthracis strains, we also tested inhibition of the more active BmcA on additional microorganisms possessing NIS synthetase siderophore biosynthetic pathways. Aerobactin was the first discovered siderophore to be assembled by an NIS synthetase system.31,48 It contributes to the virulence of a multitude of bacteria, including E. coli, and multiple species of Salmonella, Yersinia, and Shigella.31,49 The pathway involves the type A and C NIS synthetases IucA and IucC, respectively,50,51 which share sequence similarity with NIS synthetases found in most other siderophore biosynthetic pathways.31,32 Indeed, SbnE and AsbA appear to be relatives of IucA.31,32 Furthermore, staphyloferrin B biosynthetic enzyme SbnF and petrobactin biosynthetic enzyme AsbB share sequence similarity with IucC,31,32 collectively suggesting that the baulamycins could also inhibit aerobactin-producing bacteria. Thus, we monitored the ability of BmcA to inhibit bacterial growth of the aerobactin-producing Gram-negative strains of Shigella flexneri, E. coli, and Salmonella typhimurium in iron-depleted and iron-rich conditions (Figure 6G,H). BmcA inhibited growth of S. flexneri and E. coli cultures in iron-depleted conditions (S. flexneri: IC50 = 20 µM; E. coli: IC50 = 4.2 µM) with lower potency in iron-rich conditions (S. flexneri: IC50 = 46 µM; E. coli: IC50 = 150 µM). Although increased potency of BmcA against E. coli in IDM suggests that this microbe could possess fewer nonspecific cellular drug targets, additional studies are required to draw any firm conclusions. While BmcA also appeared to inhibit growth of S. typhimurium cultures, the large standard deviations acquired during the assay led us to classify the IC50 as above 1 mM in both iron-depleted and rich conditions (Figure S27). As improved access to baulamycins becomes possible, follow up studies will enable more extensive analysis of its broad-spectrum antibiotic activity.
The current analysis provides an initial assessment of the ability of BmcA to inhibit growth and demonstrates that the molecule is cell permeable. However, inhibition of all target strains under both growth conditions suggests that BmcA is acting on some additional cellular targets. Indeed, SbnE and AsbA seem to share varying levels of sequence identity with a number of enzymes in S. aureus and B. anthracis, including aminoacyl-tRNA synthetases, CoA synthetases, and fatty-acid CoA ligases in addition to critical enzymes involved in cell envelope biogenesis, inhibition of apoptosis, and spore formation (Figure S28).
Finally, the ability of BmcA to inhibit bacterial growth of both Gram-positive and Gram-negative strains (Table 3) classifies it as a broad-spectrum antibiotic. Although BmcB was not included in these experiments due to low production by Streptomyces tempisquensis, the compound is of a comparable size and possesses similar functional groups, suggesting it also could be capable of inhibiting microbial growth and infiltrating the bacteria. On the other hand, the decreased chain length and potency of BmcB suggests that longer terminal carbon chains may be important in future structure-activity relationship (SAR) studies.
CONCLUSION
We have identified a new structural class of broad-spectrum antibiotics isolated from a marine microbial derived NPE library. BmcA and BmcB inhibit siderophore biosynthesis, a crucial virulence factor associated with iron sequestration in pathogenic bacteria. Although previous studies have validated siderophore biosynthesis as an effective synthetic drug target,8–11 the aforementioned study is the first to demonstrate its potential as a target for discovering novel chemical scaffolds. Structurally unique drugs like the baulamycins present hurdles for the bacterial development of antibiotic resistance. High throughput screening for natural product inhibitors of virulence factors including those for siderophore biosynthesis, could provide a fresh arsenal of chemical scaffolds to combat drug-resistant pathogens.
The baulamycin natural products represent promising lead structures that can be further manipulated to improve both potency and target selectivity if required. We have illustrated that the compounds are efficacious in vitro and are capable of penetrating bacterial barriers to inhibit growth of cultures, including B. anthracis, MRSA, S. flexneri, and E. coli. The ability of the drug to inhibit growth of both Gram-positive and Gram-negative bacteria suggests its potential use as a broad-spectrum antibiotic. These growth inhibition studies further suggest that the compounds possess multiple targets in the microbial cell. Given the simultaneous decline in antibiotic drug discovery and increase of multi-drug resistant bacteria, the baulamycins may represent an auspicious starting-point for expanding discovery efforts against significant human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martha J. Larsen and Tom McQuade in the Center for Chemical Genomics, Life Sciences Institute, University of Michigan for high throughput screening automation and NPE sample management. We also thank Eugenio Alvarado in the NMR core facility, Department of Chemistry, University of Michigan for assistance with acquiring spectra for stereochemical studies. We also thank Eli Eisman for initial assistance with mass spectrometry, Andrew Lowell for IR spectroscopy and Dr. Masato Koreeda for access to a polarimeter. The S. aureus Newman was a generous gift from Dominique Missiakas (University of Chicago). The B. anthracis Sterne 34F2 strain was provided by Phillip Hanna (University of Michigan). The S. typhimurium strain was a donation of Brenda Franklin (University of Michigan). This research was supported by the International Cooperative Biodiversity Groups initiative (U01 TW007404) at the Fogarty International Center, the Great Lakes Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research Grant (U54 AI57153), and the Hans W. Vahlteich Professorship (D.H.S.). Additional funding for M.M.S was provided by the National Science Foundation Graduate Research Fellowship Program (1256260). We thank the Technical Office, CONAGEBIO, Ministry of the Environment and Telecommunications, Costa Rica for providing sample collection permits.
ABBREVIATIONS
- MRSA
methicillin-resistant S. aureus
- NIS
nonribosomal peptide synthetase independent siderophore
- NPE
natural product extract
- MESG
2-amino-6-mercapto-7-methylpurine ribonucleoside
- IDM
Iron-depleted media
- IRM
iron-rich media
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental details, 1D and 2D NMR spectral data, MS fragmentation patterns, high throughput screening campaigns, phylogenetic analysis, kinetic analysis, and bioactivity data of 6 and 7. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Andersson DI, Hughes D. FEMS Microbiol. Rev. 2011;35:901. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 2.Fischbach Ma, Walsh CT. Science. 2009;325:1089. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nathan C. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livermore DM. J. Antimicrob. Chemother. 2011;66:1941. doi: 10.1093/jac/dkr262. [DOI] [PubMed] [Google Scholar]
- 5.Moir DT, Opperman TJ, Butler MM, Bowlin TL. Curr. Opin. Pharmacol. 2012;12:535. doi: 10.1016/j.coph.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Quadri LEN. Infect. Disord. Drug Targets. 2007;7:230. doi: 10.2174/187152607782110040. [DOI] [PubMed] [Google Scholar]
- 7.Miethke M, Marahiel MA. Microbiol. Mol. Biol. Rev. 2007;71:413. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupte A, Boshoff HI, Wilson DJ, Neres J, Labello NP, Somu RV, Xing C, Barry CE, Aldrich CC. J. Med. Chem. 2008;51:7495. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreras Ja, Ryu J-S, Di Lello F, Tan DS, Quadri LEN. Nat. Chem. Biol. 2005;1:29. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 10.Neres J, Labello NP, Somu RV, Boshoff HI, Wilson DJ, Vannada J, Chen L, Barry CE, Bennett EM, Aldrich CC. J. Med. Chem. 2008;51:5349. doi: 10.1021/jm800567v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, Aldrich CC. J. Med. Chem. 2006;49:31. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 12.Klevens RM, Morrison Ma, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. JAMA. 2007;298:1763. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney DA, Hicks CW, Cui X, Li Y, Eichacker PQ. Am. J. Respir. Crit. Care Med. 2011;184:1333. doi: 10.1164/rccm.201102-0209CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.As defined by the US Center for Disease Control, see: http://www.cdc.gov/drugresistance/diseasesconnectedar.html
- 15.Haley KP, Skaar EP. Microbes Infect. 2012;14:217. doi: 10.1016/j.micinf.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotta K, Kim C-Y, Fox DT, Koppisch AT. Microbiology (Reading, U.K.) 2010;156:1918. doi: 10.1099/mic.0.039404-0. [DOI] [PubMed] [Google Scholar]
- 17.Haag H, Fiedler HP, Meiwes J, Drechsel H, Jung G, Zähner H. FEMS Microbiol. Lett. 1994;115:125. doi: 10.1111/j.1574-6968.1994.tb06626.x. [DOI] [PubMed] [Google Scholar]
- 18.Drechsel H, Freund S, Nicholson G, Haag H, Jung O, Zahner H, Jung G. Biometals. 1993;6:185. doi: 10.1007/BF00205858. [DOI] [PubMed] [Google Scholar]
- 19.Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE. Infect. Immun. 2004;72:29. doi: 10.1128/IAI.72.1.29-37.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE. Mol. Microbiol. 2009;74:594. doi: 10.1111/j.1365-2958.2009.06880.x. [DOI] [PubMed] [Google Scholar]
- 21.Cheung J, Murphy MEP, Heinrichs DE. Chem. Biol. 2012;19:1568. doi: 10.1016/j.chembiol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Beasley FC, Cheung J, Heinrichs DE. BMC Microbiol. 2011;11:199. doi: 10.1186/1471-2180-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cendrowski S, MacArthur W, Hanna P. Mol. Microbiol. 2004;51:407. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 24.Koppisch AT, Browder CC, Moe AL, Shelley JT, Kinkel BA, Hersman LE, Iyer S, Ruggiero CE. Biometals. 2005;18:577. doi: 10.1007/s10534-005-1782-6. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Janes BK, Passalacqua KD, Pfleger BF, Bergman NH, Liu H, Håkansson K, Somu RV, Aldrich CC, Cendrowski S, Hanna PC, Sherman DH. J. Bacteriol. 2007;189:1698. doi: 10.1128/JB.01526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfleger BF, Lee JY, Somu RV, Aldrich CC, Hanna PC, Sherman DH. Biochemistry. 2007;46:4147. doi: 10.1021/bi6023995. [DOI] [PubMed] [Google Scholar]
- 27.Pfleger BF, Kim Y, Nusca TD, Maltseva N, Lee JY, Rath CM, Scaglione JB, Janes BK, Anderson EC, Bergman NH, Hanna PC, Joachimiak A, Sherman DH. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17133. doi: 10.1073/pnas.0808118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oves-Costales D, Kadi N, Fogg MJ, Song L, Wilson KS, Challis GL. J. Am. Chem. Soc. 2007;129:8416. doi: 10.1021/ja072391o. [DOI] [PubMed] [Google Scholar]
- 29.Fox DT, Hotta K, Kim C-Y, Koppisch AT. Biochemistry. 2008;47:12251. doi: 10.1021/bi801876q. [DOI] [PubMed] [Google Scholar]
- 30.Nusca TD, Kim Y, Maltseva N, Lee JY, Eschenfeldt W, Stols L, Schofield MM, Scaglione JB, Dixon SD, Oves-Costales D, Challis GL, Hanna PC, Pfleger BF, Joachimiak A, Sherman DH. J. Biol. Chem. 2012;287:16058. doi: 10.1074/jbc.M112.359349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oves-Costales D, Kadi N, Challis GL. Chem. Commun. (Cambridge, U.K.) 2009;43:6530. doi: 10.1039/b913092f. [DOI] [PubMed] [Google Scholar]
- 32.Challis GL. Chembiochem. 2005;6:601. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 33.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 34.Newman DJ, Cragg GM. J. Nat. Prod. 2012;75:311. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montaser R, Luesch H. Future Med Chem. 2012;3:1475. doi: 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itaya K, Ui M. Clin. Chim. Acta. 1966;14:361. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- 37.Pegan SD, Tian Y, Sershon V, Mesecar AD. Comb. Chem. High Throughput Screening. 2010;13:27. doi: 10.2174/138620710790218203. [DOI] [PubMed] [Google Scholar]
- 38.McQuade TJ, Shallop AD, Sheoran A, Delproposto JE, Tsodikov OV, Garneau-Tsodikova S. Anal. Biochem. 2009;386:244. doi: 10.1016/j.ab.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Matsumori N, Kaneno D, Murata M, Nakamura H, Tachibana K. J. Org. Chem. 1999;64:866. doi: 10.1021/jo981810k. [DOI] [PubMed] [Google Scholar]
- 40.Morinaka BI, Molinski TF. Org. Lett. 2011;13:6338. doi: 10.1021/ol2024778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S-X, Wang X-J, Yan Y, Wang J-D, Zhang J, Liu C-X, Xiang W-S, Shen B. Org. Lett. 2012;14:1254. doi: 10.1021/ol300074d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng J, Place AR, Yoshida W, Anklin C, Hamann MT. J. Am. Chem. Soc. 2010;132:3277. doi: 10.1021/ja9091853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb MR. Proc. Natl. Acad. Sci. U. S. A. 1992;89:4884. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson DJ, Aldrich CC. Anal. Biochem. 2010;404:56. doi: 10.1016/j.ab.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Brandish PE, Kimura K-i, Inukai M, Southgate R, Lonsdale JT, Bugg TDH. Antimicrob. Agents Chemother. 1996 doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmelz S, Botting CH, Song L, Kadi NF, Challis GL, Naismith JH. J. Mol. Biol. 2011;412:495. doi: 10.1016/j.jmb.2011.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roemer T, Xu D, Singh SB, Parish Ca, Harris G, Wang H, Davies JE, Bills GF. Chem. Biol. 2011;18:148. doi: 10.1016/j.chembiol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Gibson F, Magrath DI. Biochim. Biophys. Acta. 1969;192:175. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- 49.Warner PJ, Williams PH, Bindereif A, Neilands JB. Infect. Immun. 1981;33:540. doi: 10.1128/iai.33.2.540-545.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Lorenzo V, Neilands JB. J. Bacteriol. 1986;167:350. doi: 10.1128/jb.167.1.350-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986;165:570. doi: 10.1128/jb.165.2.570-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.