Abstract
Umbilical cord blood has been used for a wide variety of immunologic investigations including assessments of developmental perturbations by antenatal exposures. Recent advances in multiparameter flow cytometry have allowed finer characterization of lymphocyte phenotype and function, revealing important differences between the fetal and adult immune systems. The degree of variability between human subjects confounds the ability to draw firm conclusions. Artifacts resulting from processing techniques exacerbate this variability. The unpredictable nature of deliveries, especially of premature infants, makes it difficult to control variables such as timing of umbilical cord mononuclear cell (UCMC) isolation and method of collection. Additionally, in multicenter studies dependent on central processing, delays are inevitable. However, little available literature describes systematic testing of the degree to which processing variations affect UCMC phenotype and function. Using multiparameter flow cytometry, we tested the effect of collection technique and length of time prior to UCMC isolation on T cell phenotype and function, with the goal of creating a standardized operating procedure for a multicenter investigation. The study also provides a benchmark data set including extensive surface and functional phenotyping of umbilical cord T cells. UCMC isolation delay of up to 24 h produced similar T cell phenotype and function as tested by in vitro SEB stimulation. There were few statistically significant differences between time points based on data medians. We conclude that, for the purpose of immunologic investigations, a 24-h time delay from sample collection to mononuclear cell isolation does not introduce a significant degree of variation in T cell phenotype and function when adhering to strict standard operating procedures.
Keywords: umbilical cord blood, T-lymphocytes, phenotype, neonate, immunologic techniques, cell isolation, flow cytometry
Flow cytometry is a powerful tool that can be used to comprehensively profile T cells. Use of higher order multiparameter fluorescence labeling allows selective gating for specific events in the presence of contaminating populations. As a sensitive tool, however, it is prone to high noise-signal ratio, which can be further confounded by subject-to-subject variation (1). It is critical, therefore, to avoid procedures that may introduce cellular changes with nonspecific staining or partial or complete loss of cell subpopulations during T cell phenotyping and functional analysis.
Umbilical cord blood samples are used for many purposes, including research on T cell development and responses in neonates, and cord blood banking for potential transplantation (2–5). This study focuses on optimizing collection and analysis procedures for developmental investigative purposes. One major challenge in analyzing umbilical cord T cells is the unpredictability of deliveries, especially in studies intending to include premature neonates. For most research studies, it is not feasible to maintain availability of technical assistance required for sample processing at all times. The majority of studies investigating umbilical T cell phenotype and function therefore collect only when assistance is available, or alternatively, limit to scheduled cesarean-section deliveries. To overcome subject-to-subject variability seen in human lymphocyte analysis, large numbers of subjects are required to achieve statistical significance, and scheduled processing may preclude efficient subject enrollment. Samples collected in large, multicenter studies are also subject to inevitable delays introduced by shipping for central processing.
There are few studies that address delays in peripheral blood mononuclear cell (PBMC) isolation as an independent variable affecting T cell phenotype or function. In one published study evaluating time delay effects on lymphocyte populations, the authors found more cellular debris after 48 h with both whole blood lysis and density gradient methods, and loss of T lymphocyte subsets with density gradient separation. The investigators attributed T cell subset changes to red blood cell (RBC) and cellular debris contamination (6). Another of the studies standardizing PBMC collection concluded that time between collection and isolation was the variable that contributed most significantly to diminished T cell function. In that study, time zero (T0) and 24-h (T24) isolations were performed in two separate centers, which in itself may have introduced variability (7). A subsequent study demonstrated that whole blood samples left at room temperature for 24 h allowed granulocyte activation, which suppressed T cell function (8). These studies did not include umbilical cord blood samples, which contain a higher fraction of naïve T cells. Naïve T cells are at baseline more quiescent and have higher threshold for activation. A recent publication proposing a protocol for UCB phenotyping by flow cytometry stated that UCMC should be isolated within 12 h, though a supporting reference was not provided. The antibody panel utilized was also not developed to study T cell subtype or function specifically (9). Finally, in a recent systematic review of established techniques aimed at measurement of T cell function, authors questioned the strength of evidence supporting time delay to processing as a critical variable (10).
The purpose of this pilot study was to optimize and standardize umbilical cord blood collection procedures to minimize variation introduced by technical approach. Specifically, we tested time delay to sample processing as a variable affecting T cell phenotype and function in flow cytometric analysis. Additionally, this study generated an extensive surface phenotype and functional analysis of full term umbilical cord T cells, which can be referenced in future studies investigating neonatal T cells.
MATERIALS AND METHODS
Collection Kit Preparation
All procedures were approved by the University of Rochester Research Subjects Review Board (RSRB). Cord blood sampling kits were preassembled to standardize collection procedures, ensuring uniform reagents and labeling was used. The kit included two labeled 10 mL Vacutainer® sodium heparin glass tubes (BD, Cat#366480) aseptically injected with an additional 0.5 mL sterile-filtered lithium heparin (MP Biomedicals, Cat#101929) diluted to 300 USP/mL concentration with 1× PBS (Cellgro, Cat#46-013-CM). The additional heparin was used to reduce coagulation in samples collected in 1× heparin alone (11). Kits also included a Multiple Sample Luer Adapter (BD, Cat#367290) connected to One Use Vacutainer® Holder (BD, Cat#364815), attached to a 23g Vacutainer® Safety Lok Blood Collection Set (BD, Cat#367297).
Umbilical Cord Blood Collection
Umbilical cord blood was collected between fall 2010 and winter 2011, using universal precautions from 15 elective, full term cesarean section deliveries. Clinical information included gestational age, and date and time of collection. Cord blood was first collected for clinical purposes by operating room staff. Cord was then reclamped with a Kelly clamp until a plastic cord clamp could be placed to preserve supply of remaining blood sample within the cord and placenta after delivery. A segment of the cord surface was swabbed with alcohol to reduce maternal blood contamination, and the preassembled collection kit was used to draw a clean sample from the arteries, vein or both by venipuncture. Sample collection by dripping from the severed end of cord was avoided due to high risk of maternal cell and environmental contamination and tendency for the blood collected to clot before processing. Whole blood samples were kept at room temperature until time of processing.
Umbilical Cord Mononuclear Cell Isolation
All processing was completed using BSL 2 procedures. Three aliquots of each sample were designated for processing immediately (T0), at 24 (T24), or at 48 (T48) h following collection. Aliquots at delayed time points were kept at room temperature following sterile transfer to 15 mL conical polypropylene tubes. Transfer of samples was performed to minimize manipulation of glass containers in BSL 2 conditions and to reduce the risk of sample contamination from serial aliquot distribution. Each aliquot was processed using the same standardized operating procedure and by the same technician. Prior to cell isolation, a cord blood aliquot was diluted with two parts 1× Dulbecco’s PBS to 1 part whole blood in a sterile 15 mL conical. Diluted blood was layered over Ficoll-Paque® (Lymphocyte Separation Medium, Cellgro, Cat#25-072-CV) at a ratio of 10-mL blood to 3-mL separation medium. Blood was pipetted slowly down the side of the 15 mL conical to overlay the Ficoll-Paque. Samples were centrifuged at 20°C for 30 min at 1200g with no brake. Following centrifugation, three 1-mL aliquots of plasma (top layer) were recovered and frozen for later use. The buffy coat was siphoned with a 5 mL pipette, placed in a sterile 15 mL conical, and brought to 14 mL with cold HBSS (Cellgro, Cat# 21-022-CM). To remove platelets, isolated cells were washed three times with cold HBSS and centrifugation at 300g for 10 min at 4°C. After the final wash, the cells were resuspended tapping in cold HBSS at a ratio of 1 mL HBSS per 8 mL original blood volume (if volume was <8 mL, 500µL HBSS was used). Ten µL resuspended cells were added to 40µL lysis buffer (ACK Lysing Buffer, Biowhittaker, Cat#10–548E) in a counting tube and incubated at room temperature for 10 min. Trypan blue was added at a 1:10 final dilution. Live and dead cell counts were determined by an automated cell counter (Cellometer® AutoT4, Nexcelom Bioscience) and the remaining UCMC were centrifuged and resuspended in freezing media (10% DMSO, Sigma, Cat#15493-8, 90% sterile-filtered prescreened FBS, Sigma, Cat#F0926) at ≤2.5 × 106 cells mL−1 if the total cell count isolated was ≤5 × 106, 5 × 106 cells mL−1 if total count >5– 150 × 106, or 10 × 106 cells mL−1 if count exceeded 150 × 106. Cells were transferred to cryovials and placed in a Nalgene “Mr. Frosty” 1° freezing container (Cat#5100-0001) in −80°C for 24–72 h, then vials were transferred without warming to vapor-phase liquid nitrogen for long-term storage.
Umbilical Cord Blood Thawing Procedure
All aliquots tested were thawed and prepared for flow cytometry in two batches to avoid variation due to day-to-day changes in cytometer performance or experimental conditions. Tubes were thawed quickly in a 37°C water bath while shaking continuously until the last ice crystal remained. Using a 1-mL pipette, contents were transferred into a 15 mL conical polypropylene tube containing 9 mL complete medium, warmed to 37°C, (RPMI 1640, Cellgro Cat#100-040-CV, 8%FBS, Sigma Cat#F0926) and swirled to mix. Cell samples were centrifuged at 300g for 10 min at 20°C. Supernatant was discarded and pellet was resuspended by finger-tapping. Samples were washed again using above procedure with 10 mL of warm medium. UCMC recovery and viability was determined using trypan blue exclusion assay with RBC lysis buffer as in cell isolation step and counted using an automated cell counter.
UCMC Preparation for T Cell Phenotype Flow Cytometry Panel
Immunocytochemistry was performed with fluorescently tagged antibodies chosen for general cell phenotyping and for analysis of cytokine-producing cells using a micromethod as previously described (12). UCMC were washed twice in PBS, counted and plated at ~1 × 106 UCMC in 100 µL PBS/well of a 96-well v-bottom polystyrene microtiter plate (BD Falcon Cat#353263). Plates were then centrifuged at 400g for 6 min at 4°C, decanted and blotted before turning upright. UCMC were resuspended in 15 µL of a 1:500 dilution of Live/Dead Yellow cocktail (Invitrogen, Cat#L34959) in cold PBS, and incubated at 4°C in the dark for 30 min. After incubation, 200µL cold staining buffer (DPBS, Biowhittaker Cat#17–512F, 1% BSA, Fraction V ICN Cat#160069), and 1 mM EDTA, Sigma, Cat#E6511) was added per well and plates were centrifuged for 6 min at 400g at 4°C. UCMC were then Fc-blocked with 200 µL 5% normal mouse serum (Sigma, Cat#M5905) in staining buffer for 10 min at room temperature. Plates were then centrifuged as described above, resuspended in 10 µL surface staining cocktail (see Supporting Information Table 2) and incubated in the dark at 4°C for 80 min. Surface-stained UCMC were washed with 200 µL staining buffer, decanted and resuspended in 15 µL of a 1:500 streptavidin-fluoro-chrome conjugate diluted in staining buffer and incubated for 20 min in the dark at 4°C. Plates were again washed and pellets resuspended in 220µL fixation buffer (1% formaldehyde, Sigma Cat#F-1635, 1 mM EDTA, in PBS). Stained and fixed UCMCs were kept in the microtiter plate for analysis on BD/LSR II High Throughput Autosampler System. Data was collected and autocompensated using BD FACS Diva Software.
Table 2.
CD4+ cell subset median frequencies and analysis of variance
| MEDIAN (IQR) | P-VALUE | PATIENT LEVEL | TIME LEVEL | REPLICATE LEVEL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD4+ SUBSET | TO T24 T4B |
TO V T24 TO V T4B |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN TIME |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN REPLICATES |
| %CD28 + CD27+ | 73.7 (66.8, 84.5) | 0.003277 | 83.13 | 0.000599 | 15.20 | 0.85 | 0.000066 | 1.67 | 0.98 | |
| 74.9 (66.1, 84.6) | 0.93 | |||||||||
| 67.7 (60.1, 77.9) | 0.24 | |||||||||
| %CCR7 + CD45RA− | 15.4 (11.5, 24.5) | 0.0275 | 90.95 | 0.002327 | 7.70 | 0.92 | 0.000408 | 1.35 | 0.99 | |
| 15.7 (11.0, 20.7) | 0.21 | |||||||||
| 15.0 (11.0, 22.1) | 0.9 | |||||||||
| PreTh1 | 1.50 (0.63, 2.83) | 0.03095 | 83.13 | 0.005031 | 13.51 | 0.86 | 0.001251 | 3.36 | 0.97 | |
| %CXCR3 + CXCR5− | 1.51 (0.53, 2.29) | 0.25 | ||||||||
| 1.71 (0.63,3.14) | 0.76 | |||||||||
| PreTh2 | 5.28 (4.13, 9.27) | 0.1295 | 90.00 | 0.00872 | 6.06 | 0.94 | 0.005673 | 3.94 | 0.96 | |
| %CCR4 + CCR6 − CXCR5− | 5.98 (4.32, 7,56) | 0.64 | ||||||||
| 5.74 (4.01, 9.54) | 0.206 | |||||||||
| %CCR7 − CD45RA+ | 19.2 (20.5, 23.1) | 0.02416 | 92.83 | 0.001663 | 6.39 | 0.94 | 0.000204 | 0.78 | 0.99 | |
| 19.7 (13.7, 21.0) | 0.93 | |||||||||
| 21.6 (13.2, 22.3) | 0.1 | |||||||||
| TH1 | 0.69 (0.46, 1.25) | 0.05748 | 48.85 | 0.03714 | 31.57 | 0.61 | 0.02304 | 19.58 | 0.8 | |
| %CXCR3 + CXCR5− | 0.52 (0.27, 0.95) | 0.04 | ||||||||
| 0.41 (0.30,0.68) | 0.41 | |||||||||
| Th2 | 5.77 (5.56, 7.12) | 0.01115 | 61.36 | 0.006145 | 33.82 | 0.64 | 0.000875 | 4.82 | 0.95 | |
| %CCR4 + CCR6 − CXCR5− | 6.70 (5.04, 7.68) | 0.52 | ||||||||
| 8.34 (5.66, 9.26) | 0.003 | |||||||||
| Th17 | 0.13 (0.11, 0.21) | 0.01334 | 37.84 | 0.01039 | 29.48 | 0.56 | 0.01152 | 32.68 | 0.67 | |
| %CCR4 + CCR6 + CXCR5− | 0.13 (0.09, 0.17) | 0.3 | ||||||||
| 0.12 (0.12, 0.21) | 0.24 | |||||||||
| %CCR7 − CD45RA− | 14.5 (11.2, 19.2) | 0.01905 | 85.45 | 0.002902 | 13.02 | 0.87 | 0.000341 | 1.53 | 0.98 | |
| 15.2 (13.0, 18.7) | 0.07 | |||||||||
| 15.0 (10.0, 17.7) | 0.76 | |||||||||
| TH1 | 0.15 (0.06, 0.26) | 0.08309 | 51.67 | 0.03798 | 23.62 | 0.69 | 0.03974 | 24.71 | 0.75 | |
| %CXCR3 + CXCR5− | 0.11 (0.09, 0.30) | 0.25 | ||||||||
| Th2 | 0.10 (0.07, 0.23) | 0.64 | ||||||||
| %CCR4 + CCR6 − CXCR5− | 6.40 (4.73, 8.48) | 0.02654 | 85.39 | 0.00383 | 12.32 | 0.87 | 0.000712 | 2.29 | 0.98 | |
| 7.75 (5.90, 10.4) | 0.002 | |||||||||
| 6.59 (5.86, 10.3) | 0.003 | |||||||||
| Th17 | 0.17 (0.14, 0.27) | 0.01286 | 37.51 | 0.01036 | 30.22 | 0.55 | 0.01106 | 32.26 | 0.68 | |
| %CCR4 + CCR6 + CXCR5− | 0.18 (0.14, 0.25) | 0.8 | ||||||||
| 0.16 (0.14,0.20) | 0.7 | |||||||||
| %CCR7 + CD45RA+ | 26.2 (15.1,31.7) | 0.03488 | 82.76 | 0.006873 | 16.31 | 0.84 | 0.000391 | 0.93 | 0.99 | |
| 28.8 (20.0, 33.0) | 0.56 | |||||||||
| 17.7 (9.26, 28.7) | 0.01 | |||||||||
| %CD28 − CD27− | 0.64 (0.37, 1.55) | 0.1216 | 68.71 | 0.04737 | 26.77 | 0.72 | 0.007999 | 4.52 | 0.95 | |
| 1.03 (0.44, 1.58) | 0.85 | |||||||||
| 1.41 (0.70, 2.06) | 0.019 | |||||||||
| %CCR7 + CD45RA− | 0.12 (0.02, 0.19) | 0.1984 | 60.13 | 0.1144 | 34.67 | 0.63 | 0.01713 | 5.19 | 0.95 | |
| 0.10 (0.04, 0.34) | 0.055 | |||||||||
| 0.25 (0.14, 0.52) | 0.001 | |||||||||
| PreTh1 | 0.08 (0.01, 0.15) | 0.2688 | 64.75 | 0.1299 | 31.29 | 0.67 | 0.01642 | 3.96 | 0.96 | |
| %CXCR3 + CXCR5− | 0.06 (0.02, 0.22) | 0.12 | ||||||||
| 0.12 (0.10, 0.41) | 0.01 | |||||||||
| PreTh2 | 0.03 (0.01, 0.07) | 0.1334 | 39.71 | 0.1692 | 50.37 | 0.44 | 0.0333 | 9.91 | 0.9 | |
| %CCR4 + CCR6 − CXCR5− | 0.04 (0.01, 0.12) | 0.09 | ||||||||
| 0.10 (0.04, 0.17) | 0.001 | |||||||||
| %CCR7 − CD45RA+ | 0.09 (0.04, 0.13) | 0.07572 | 34.96 | 0.1253 | 57.85 | 0.38 | 0.01556 | 7.18 | 0.93 | |
| 0.12 (0.05, 0.21) | 0.007 | |||||||||
| 0.32 (0.32, 0.41) | 0.001 | |||||||||
| TH1 | 0.004 (0.002, 0.04) | 0.163 | 67.96 | 0.04119 | 17.17 | 0.8 | 0.03567 | 14.87 | 0.85 | |
| %CXCR3 + CXCR5− | 0.005 (0.001, 0.02) | 0.31 | ||||||||
| 0.009 (0.004, 0.03) | 0.005 | |||||||||
| Th2 | 0.02 (0.01, 0.04) | 0.06086 | 23.34 | 0.1575 | 60.40 | 0.28 | 0.04241 | 16.26 | 0.84 | |
| %CCR4 + CCR6 − CXCR5− | 0.03 (0.01, 0.07) | 0.005 | ||||||||
| 0.08 (0.04, 0.17) | 0.001 | |||||||||
| %CCR7 − CD45RA− | 0.38 (0.27, 0.87) | 0.1346 | 73.96 | 0.03883 | 21.34 | 0.78 | 0.008548 | 4.70 | 0.95 | |
| 0.49 (0.35, 0.91) | 0.89 | |||||||||
| 0.50 (0.32, 1.02) | 0.58 | |||||||||
| TH1 | 0.02 (0.004, 0.06) | 0.1727 | 51.57 | 0.09015 | 26.92 | 0.66 | 0.07206 | 21.52 | 0.78 | |
| %CXCR3 + CXCR5− | 0.02 (0.004, 0.05) | 0.39 | ||||||||
| 0.01 (0.004, 0.05) | 0.1 | |||||||||
| Th2 | 0.23 (0.15, 0.45) | 0.169 | 75.52 | 0.04558 | 20.37 | 0.79 | 0.009193 | 4.11 | 0.96 | |
| %CCR4 + CCR6 − CXCR5− | 0.29 (0.13, 0.70) | 0.85 | ||||||||
| 0.28 (0.21, 0.50) | 0.21 | |||||||||
Cord blood CD4+ subpopulations were identified by flow cytometry. Median frequencies with interquartile range are shown for each time point, for each subpopulation. Median population frequency of T0 was compared to T24 and T48 using Wilcoxon signed-rank test; p-values in bold highlight significant differences (p < 0.05). Intraclass correlation coefficient (ICC) was used to determine the relative contribution of variance based on subject-subject, replicate (assay), and time delay to processing variability.
UCMC Preparation for Intracellular Cytokine Staining
Intracellular cytokine staining was performed using a modified immunofluorescent staining method (12,13). Cryovials of UCMC were thawed in two batches to minimize variability in experimental conditions. Samples were thawed as described with the following exception: UCMC were initially thawed into warm complete media with DNAse (RPMI 1640, 10%FBS, 1% Antibiotics/antimycotics, DNAse, 1 mg/100 mL final concentration, Sigma Cat#DN-25). Subsequent washes were performed using complete media without DNAse. DNAse was used prior to intracellular cytokine staining to minimize nonspecific intracellular staining per cited protocol (13). UCMC were incubated overnight in 5 mL warm complete medium in six-well polystyrene tissue culture plates (Costar Cat#3524) at ~2 × 106 cells/1 mL complete medium without DNAse, and 37°C/5% CO2.
After resting, UCMC were transferred from wells and placed in 15 mL conical polypropylene tubes. Plated cells were washed with complete media plus DNAse. UCMC were counted using trypan blue exclusion and RBC lysis buffer, and then centrifuged at 300g for 10 min at 20°C. UCMC were resuspended at 1–2 × 106 cells/100 µL complete media plus DNAse. A 100 µL UCMC suspension was added to each well of a 96-well v-bottom microtiter plate. Staphylococcal Enterotoxin-B (SEB, Sigma Cat#S4881), final concentration 2 µg mL−1 was added to stimulated wells. Final volume was 200 µL. Samples were incubated for 2 h at 37°C, 5% CO2, after which 2 µL Brefeldin A (BD Cat#555029) and 4 µL 1:100 dilution of Monensin (Sigma Cat#M5273) were added for Golgi block. Plates were again incubated for 8 h. Incubation time was optimized for detection of listed cytokines based on previous experiments. Plates were then sealed in parafilm and placed at 4°C overnight.
Immunofluorescence staining with Live/Dead Yellow (Invitrogen, Cat#L34959) and surface markers was performed as described above (see Supporting Information Table 3). After completion of surface staining, plates were washed with staining buffer and centrifuged at 400g for 6 min at 4°C. One hundred µL of Cytofix/Cytoperm reagent (BD Cat#554722) was added to each well and left to incubate at 4°C in the dark for 20 min. Permeabilization buffer (100 µL, BD Cat#554723) with 1 mM EDTA was then added to wells and centrifuged as described above and decanted. UCMC were Fc-blocked using 50 µL of 5% normal mouse serum in permeabilization buffer with EDTA again for 10 min at room temperature. Following blockade of the permeabilized cells, the plated was centrifuged and decanted, prior to adding 20 µL intracellular cytokine staining cocktail to the cells. Plates were incubated at 4°C in the dark for 80 min. Cells were then washed with permeabilization buffer with 1 mM EDTA, and pellets were resuspended in fixation buffer (PBS, 2% paraformaldehyde). Stained samples were kept at 4°C until analyzed.
Table 3.
CD4+ T cell cytokine-positive analysis of variance
| PATIENT LEVEL | TIME LEVEL | REPLICATE LEVEL | ||||||
|---|---|---|---|---|---|---|---|---|
| CD4 CYTOKINE | VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN TIME |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN REPLICATES |
| CD69+ | 0.007492 | 58.80 | 0.003214 | 25.22 | 0.7 | 0.002036 | 15.98 | 0.84 |
| IFN − γ+ | 0.4026 | 89.01 | 0.002746 | 0.61 | 0.99 | 0.04697 | 10.38 | 0.9 |
| TNFα+ | 0.05221 | 63.26 | 0.02324 | 28.16 | 0.69 | 0.007077 | 8.58 | 0.91 |
| IL − 2+ | 0.03741 | 29.35 | 0.06876 | 53.95 | 0.35 | 0.02127 | 16.69 | 0.83 |
| IL − 4/5/13+ | 0.03697 | 55.74 | 0.01234 | 18.61 | 0.75 | 0.01701 | 25.65 | 0.74 |
| MIP − 1β+ | 0.1747 | 48.10 | 0.09303 | 25.61 | 0.65 | 0.09549 | 26.29 | 0.74 |
| IL − 17+ | 0.1347 | 51.95 | 0.0237 | 9.14 | 0.85 | 0.1009 | 38.91 | 0.61 |
Cord blood CD4+ T cell cytokine production following in vitro stimulation was measured using intracellular cytokine staining and flow cytometry. Intraclass correlation coefficient (ICC) measure was used to determine the relative contribution of variance based on subject-subject, replicate (assay), and time delay to processing variability. The majority of cytokines measured had minor variation introduced by time delay to processing. The exception was IL2+ events, for which the majority of variance was based on time.
Instrument and Analysis
Samples were collected on an 18-color BDLSR II (488, 633, 407, and 532 lasers) using BD FACSDiva software (v 6.1.3) for collection and autocompensation. Autocompensation was based on single-antibody-stained Simply Cellular® Compensation Standard beads (anti-mouse IgG, Bangs Laboratories, Cat#550). Instrument configuration details are available in Supporting Information Table 1. Instruments are quality controlled on a daily basis using CS&T beads. “Peak-6” rainbow beads showed no change in instrument sensitivity over the 2 days of event acquisition. List-mode data files were collected using the standard FCS 3.0. All flow cytometry analysis was performed using Flowjo data analysis software (v. 8.8.6, Tree Star, Ashland, OR). Adhering to published approaches to flow cytometry data interpretation and optimal gating, biexponential transformation was performed to create “logicle displays” for event visualization. Fluorescence-minus-one controls were used to accurately distinguish between fluorescence “spillover” and positive events (14,15).
Table 1.
Umbilical cord mononuclear cell type analysis of variance
| PATIENT LEVEL | TIME LEVEL | REPLICATE LEVEL | ||||||
|---|---|---|---|---|---|---|---|---|
| CELL TYPE | VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN TIME |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN REPLICATES |
| Dead | 0.04115 | 66.58 | 0.01862 | 30.13 | 0.69 | 0.002039 | 3.30 | 0.97 |
| CD14+ | 0.04624 | 78.88 | 0.0116 | 19.79 | 0.8 | 0.000783 | 1.34 | 0.99 |
| CD235+ | 0.06874 | 70.35 | 0.02794 | 28.59 | 0.71 | 0.001036 | 1.06 | 0.99 |
| CD56hi | 0.02189 | 28.49 | 0.04384 | 57.05 | 0.33 | 0.01111 | 14.46 | 0.86 |
| CD56lo | 0.02417 | 29.24 | 0.04569 | 55.28 | 0.35 | 0.01279 | 15.47 | 0.85 |
| CD3+ | 0.06002 | 75.74 | 0.01771 | 22.35 | 0.77 | 0.001519 | 1.92 | 0.98 |
| TCRγδ+ | 0.06659 | 88.34 | 0.005556 | 7.37 | 0.92 | 0.003232 | 4.29 | 0.96 |
| CD4+ | 0.001058 | 93.55 | 0.00006 | 5.31 | 0.95 | 0.000013 | 1.15 | 0.99 |
| CD8+ | 0.03147 | 92.97 | 0.002125 | 6.28 | 0.94 | 0.000255 | 0.75 | 0.99 |
Analysis of variance was performed as a complement to standard comparison of medians. Cell types are as listed in the first column. Intraclass correlation coefficient (ICC) measure was used to determine the relative contribution of variance based on subject-subject, replicate (assay), and time delay to processing variability. For all cell types except CD56+, subject variability contributed the greatest to overall result variability.
Statistical Analysis
Flow cytometry data was processed on Microsoft Excel 2008 for Mac v12.2.6, and GraphPad Prism 5 for Mac OS X, version 5.0b. Statistical analysis was performed by the University of Rochester Department of Biostatistics and Computational Biology.
Analyses were performed using version 9.2 of the SAS System for Windows (©2002–2003 SAS Institute, Cary, NC). Descriptive statistics such as mean, standard deviation, median and range were used to summarize the data prior to testing the hypothesis. Variables with distributions violating normality assumption were either logarithm transformed or analyzed with nonparametric procedures. Multilevel data analysis was performed to accommodate the clustered data structure, with patient, time and replicates as three levels of variability. Intercept-only hierarchical linear models (HLM) were fitted (SAS Proc Mixed) to estimate the variability of each level and the intraclass correlation (ICC). Wilcoxon signed-rank test was used to compare medians of T cell phenotype and functions at different time points. A P value of <0.05 was considered statistically significant.
RESULTS
Our study investigated the effect of time delay from umbilical cord blood collection to mononuclear cell isolation on cell count, viability, phenotype and function. Each sample collected was divided into three aliquots processed at either T0, T24, or T48. Analysis was conducted to compare median cell frequencies across time. In addition, we also quantified the variation introduced at the patient level, time delay level and replicate level to determine at which level the variability was the greatest. Umbilical cord blood was collected from 15 healthy, full term, scheduled cesarean-section deliveries. In some samples, there were not enough cells collected to perform surface and intracellular staining at all time points. All 15 samples were included in the analysis for cell counts, recovery and viability. The final number of subjects used for surface phenotyping by flow was 15 for T0 and T24 and 11 for T48. The final number of samples used for intracellular cytokine staining was 14 for T0 and T24 and 10 for T48. (A full description of the flow cytometry panels is provided in Supporting Information Tables 1 and 2).
Viability and Cell Recovery
To evaluate the effect of time delay to processing on cell count, recovery, and viability pre- and postcryopreservation, we first performed UCMC isolation using a standardized protocol by ficoll-hypaque gradient. Following isolation, many samples appeared contaminated with clumped, though not clotted, red blood cells (RBCs). The increased presence of contaminating RBCs in UCB is well established in the literature (11). We therefore included an RBC lysis step prior to counting using the automated counters software program that distinguishes the size of PBMC versus smaller mature RBC to determine isolated cell yields. Prior to cryopreservation, mononuclear cells were counted and viability was calculated using trypan blue dye exclusion after RBC lysis. Recognizing that nucleated red cells, common in newborns, may be resistant to lysis, an RBC surface marker was used in flow cytometry to more accurately assess cell counts.
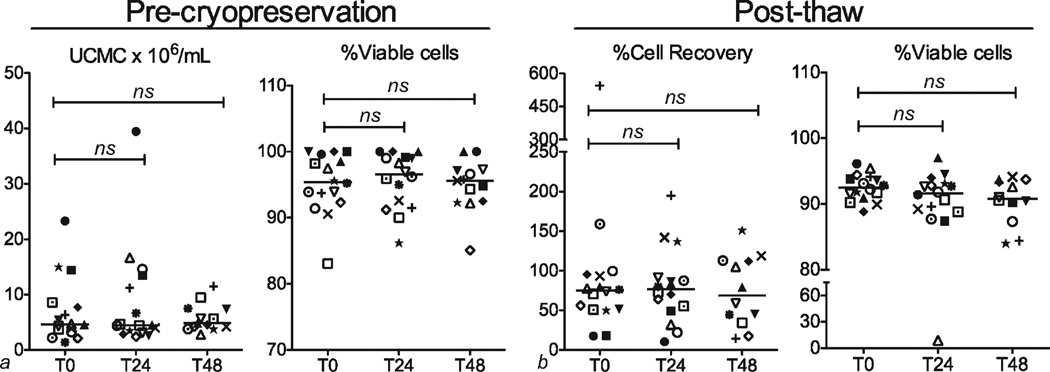
Median and interquartile range (IQR) counts of freshly isolated UCMC × 106/mL UCB at T0, T24, and T48 were 4.65 (3.37, 8.38), 4.45 (3.20, 12.9), and 4.84 (4.0, 7.43), and respective percent viable cells were 95.4 (92.7, 99.3), 96.5 (91.8, 99.1), and 95.6 (92.4, 97.2). Following cryopreservation and upon thawing, median percent recovery ((UCMC after thawing/UCMC prior to cryopreservation) ×100) was 75.1 (50.8, 95.0) for T0, 76.6 (50.6, 90.4) for T24 and 69.1 (36.8, 113) for T48. Median percent viability post-thaw was 92.5 (91.1, 94.0), 91.6 (88.9, 93.0), and 90.1 (88.0, 93.6) for T0, T24, and T48, respectively. Although there was a trend toward lower recovery in samples with delayed processing, analysis by Wilcoxon signed-rank test demonstrated no statistically significant differences for initial counts, precryopreservation viability, postthaw recovery or post-thaw viability (Fig. 1).
Figure 1.
Mononuclear cell count and viability measured prior to cryopreservation (a) and after thawing (b). Cell numbers and percents are shown for processing time points of 0, 24, and 48 h. Each symbol represents a single subject. Comparisons were made between processing delay of 0 h (T0), 24 h (T24), and 48 h (T48) using two-tailed paired t tests. Differences between time points were not significant.
UCMC Subsets
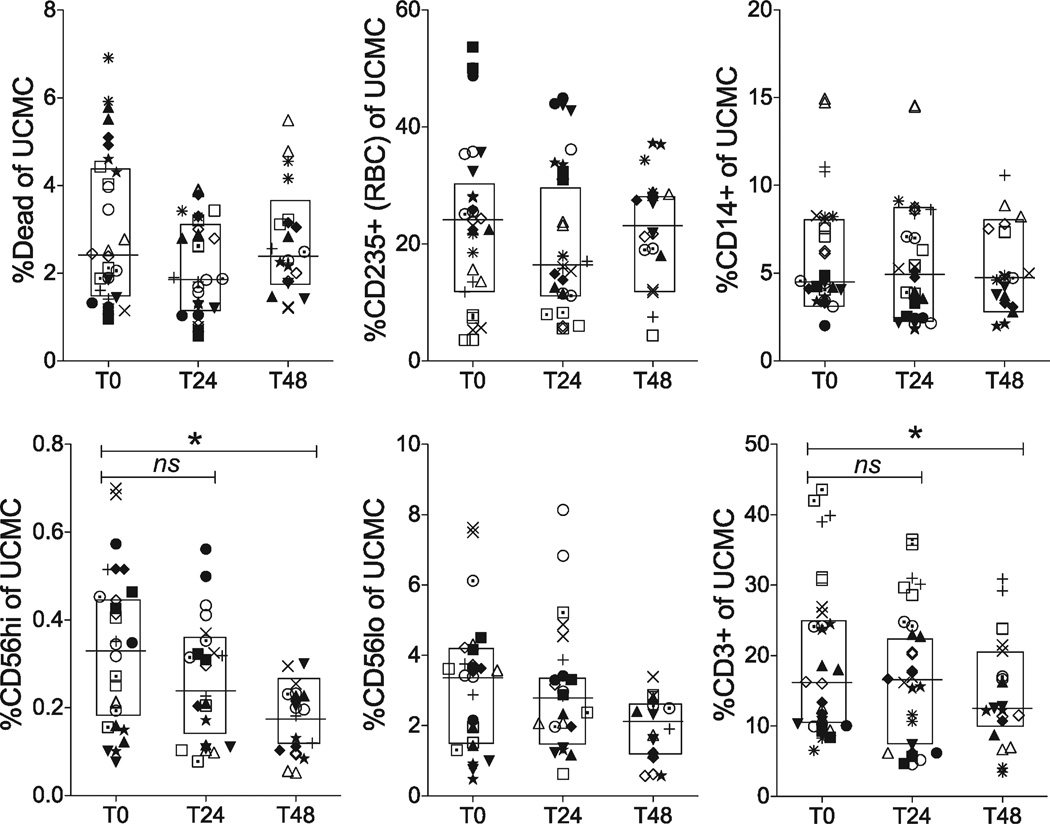
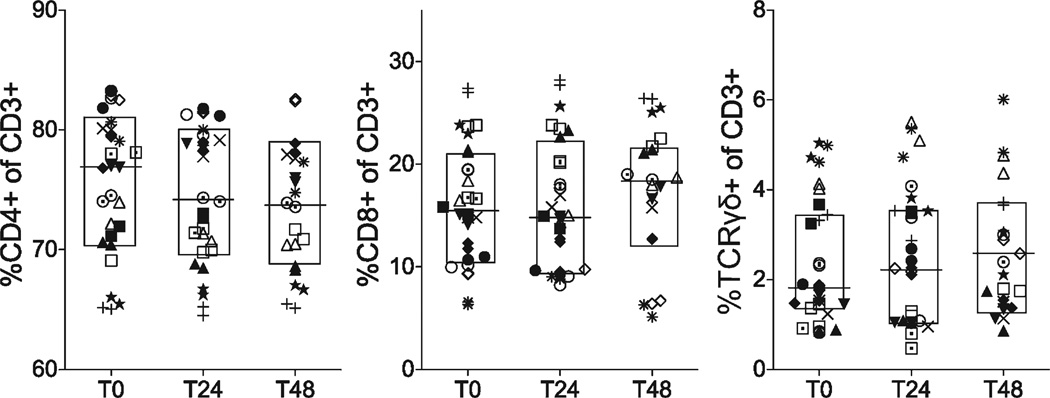
UCMC were thawed and surfaced stained for cell phenotyping. The minimum number of events collected was 250,000, with a maximum of 1,000,000 events. Subsets were identified by FSC/SSC/doublet-/live-dead+ for dead cells, CD14+ (APC subset), CD235+ (RBC contaminants) and CD3+ for T cells. CD3+ events were then subgated using TCRγδ+, and CD4+ or CD8+ on the TCRγδ- events (Supporting Information Fig. 1). Statistical analysis revealed minimal sensitivity of cell populations to time delay before processing (Fig. 2, Table 1). When comparing medians, there were no significant changes in viability (Dead%), RBC contamination (CD235+) or APC (CD14+) or CD56lo markers between time points. However, CD3+ and CD56hi frequencies demonstrated significant decreases by T48. CD56hi variability due to time delay exceeded that due to subject variability (57 and 29%, respectively). Although there was a slight decrease in CD3+ events, proportions of CD3+ cell subsets (CD4+, CD8+, TCRγδ+) did not show significant changes in median frequencies by 24 and 48 h (Fig. 3).
Figure 2.
Mononuclear cell population frequencies measured by flow cytometry. Cord blood mononuclear cells were subtyped and expressed as a percentage of total events using live/dead, anti-CD235 (RBC marker), anti-CD14 (monocytes marker), anti-CD56 (NK marker), and anti-CD3 (T cell marker). Each symbol represents a single subject. Lines show medians within boxed interquartile range. Statistical comparisons were performed using Wilcoxon signed-rank test between samples isolated after delay of 0 h (T0), 24 h (T24), and 48 h (T48). There were no significant differences after a 24-h delay, but a significant decrease in CD56hi and CD3+ frequencies was observed after 48-h delay (P < 0.05).
Figure 3.
T cell subset frequencies measured by flow cytometry. Cord blood T cells were identified using anti-CD3+ antibody. Frequencies of CD3+ T cell subsets were further identified using anti-CD4, anti-CD8, and anti-TCRγδ. Frequencies are expressed as percentages of CD3+ events. Each symbol represents an individual subject. Lines show medians within boxed interquartile range. Statistical comparisons were performed using Wilcoxon signed-rank test between samples isolated after delay of 0 h (T0), 24 h (T24), and 48 h (T48). No significant differences were detected after delay to processing of 24 or 48 h.
CD41 T Cell Subsets
CD4+ T cell subsets were identified according to a scheme of chemokine receptors published in a recent consensus statement summarizing T cell phenotype and function (16). We chose to analyze the combination of surface markers associated with putative polarized CD4+ T cell lymphocytes, including Th1 (CXCR5−, CXCR3+), Th2 (CXCR5−, CCR4+, CCR6−), Th17 (CXCR5−, CCR4+, CCR6+), pre-Th1 (CD45RA−, CCR7+, CXCR5−, CXCR3+), and pre-Th2 (CD45RA−, CCR7+, CXCR5−, CCR4+). Each of these subsets was analyzed within larger, surface marker defined, CD4+ subsets including central memory (CCR7+, CD45RA−), naive (CCR7+, CD45RA+), activated (CCR7−, CD45RA+) and effector memory (CCR7−, CD45RA−). Each of these groups was in turn nested within an analysis of CD27 and CD28 subsets, defined as naïve and early activated (CD27+, CD28+), intermediate (CD27−, CD28+) and late activated (CD27−, CD28−). Data shown includes CD27+CD28+ and CD27−CD28− parent populations only since only rare events were found in the single positive gates.
While the healthy adult donors consistently showed substantial (but minority) frequencies of CD27− events (adult data not shown), the great majority of CD4+ T cells in umbilical blood was seen in the CD27+, CD28+ gate, with fewer events in the CD27−CD28− gate. Overall, most markers showed remarkable stability over time (Table 2). In general, over time, frequencies for CD28+CD27+ cells decreased and CD28−CD27− frequencies increased. However, any changes observed, though significant by comparison of medians, were very small. For most comparisons between median frequencies of subsets, there were no significant differences between T0 and T24, exceptions were in the CD28+CD27+CCR7− CD45RA+CXCR3+CXCR5− (early activated, Th1 effector), CCR7−CD45RA−CCR4+CCR6−CXCR5− (early activated, Th2 effector memory), CD28−CD27−CCR7−CD45RA+ (late activated, effector memory), and CCR7−CD45RA− CCR4+CCR6−CXCR5− (late activated, Th2 effector populations). At T48, however, there were more, small but statistically significant, changes (P <0.05) when comparing median frequencies across time points for two CD28+CD27+ subsets: CCR7−CD45RA−CCR4+CCR6−CXCR5− (early activated Th2 effector memory) and CCR7+CD45RA+ (naïve), and seven populations in the CD27−CD28− subset: CD28− CD27− (late activated), CCR7+CD45RA− (late activated central memory), CCR7+CD45RA−CXCR3+CXCR5− (late activated pre-Th1 central memory), CCR7+CD45RA− CCR4+CXCR5− (late activated pre-Th2 central memory), CCR7−CD45RA+ (late activated, early differentiated) CCR7−CD45RA+CXCR3+CXCR5− (late activated, early differentiated Th1), and CCR7−CD45RA+CCR4+CCR6− CXCR5− (late activated, early differentiated, Th2).
Variation accounted for by time delay exceeded replicate values, though for some populations the percents were close. For all frequencies except CD28−CD27−CCR7−CD45RA+ CCR4+CCR6−CXCR5−, patient variation contributed more heavily to overall variance observed, as expected with human data.
With a few exceptions noted above, a delay of 24 h prior to lymphocyte isolation did not have a major effect on lymphocyte population frequencies or CD4+ T cell subpopulation frequencies. We also did not detect any time dependent change in mean fluorescence intensity for any of the markers included.
CD4+ Intracellular Cytokine Results
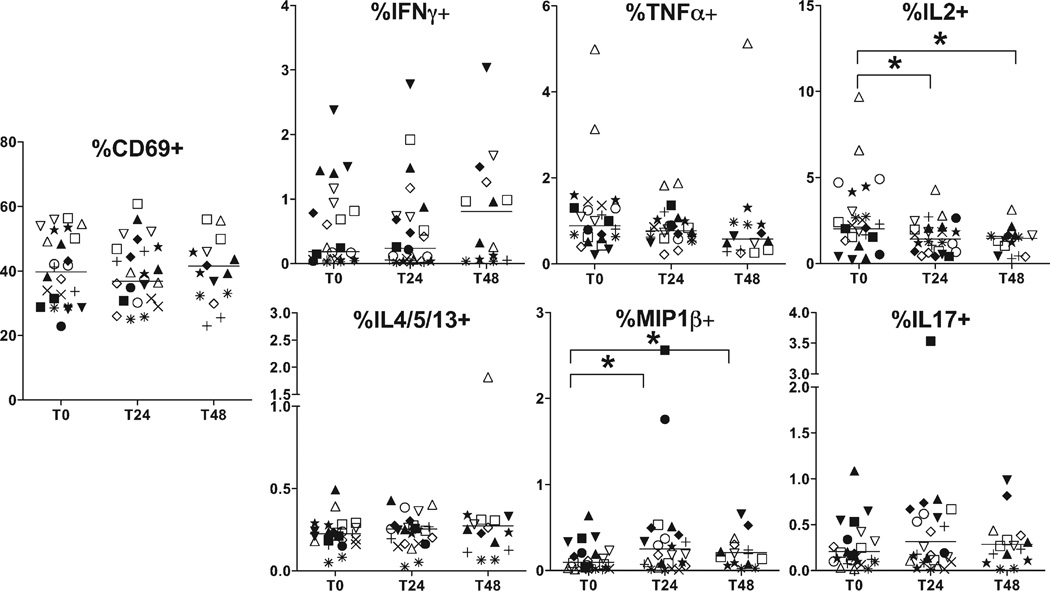
The effect of time delay to processing on CD4+ T cell cytokine production, both spontaneous in unstimulated cells and following SEB-stimulation, was measured using intracellular cytokine staining for IFN-γ, TNF-α, IL-2, IL-4/5/13, IL-17, and MIP-1β. UCMCs were incubated with and without SEB as described above in methods. CD4+ T cells were sequentially identified by FSC/SSC/doublet-/livedead-/CD14−/CD3+/CD8−/CD4+ events. Each cytokine marker was plotted against CD69, an early marker for cell activation. Gating for cytokine-positive cells was verified using healthy adult donor samples with known positive responses to SEB and fluorescence-minus-one controls. Cytokine-positive events were considered within the CD69+ gate only, and CD69+ events were similar to adult levels following SEB stimulation (gating strategy based on representative adult control and umbilical cord samples in Supporting Information Figs. 2 and 3, combined data for adult control not shown). Overall, despite evidence of effective in vitro activation, very low frequencies of cytokine-positive events were observed in UCMC (Fig. 4). Variance at the replicate level was higher than observed in the surface phenotype panel (Table 3) as expected due to the rare event numbers, yet for most cytokines, subject variance contributed the majority of overall variability.
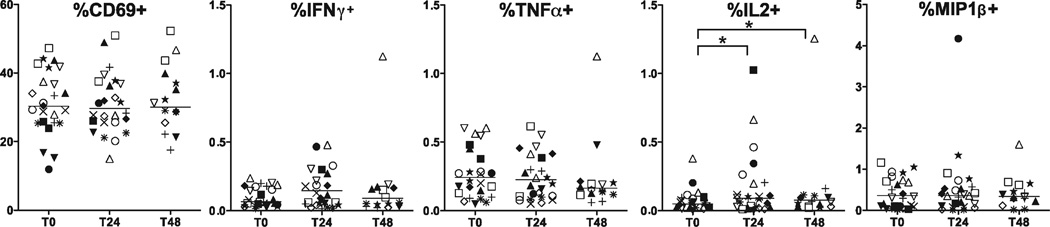
Figure 4.
Cord blood CD41 T cell cytokine production following in vitro stimulation. Cord blood CD4+ T cells were stimulated in vitro with SEB, permeabilized and stained for intracellular cytokines IFNγ, TNFα, IL2, IL4/5/13, MIP1β, and IL17. Only events within the CD69+ gate were considered cytokine-positive. Frequencies are expressed as percentages of total CD4+ events. Each symbol represents a single subject. Statistical comparisons were performed using Wilcoxon signed-rank test between samples isolated after delay of 0 h (T0), 24 h (T24), and 48 h (T48). Significant differences were found at T24 and T48 for IL2 and MIP1β (P < 0.05) only.
Cytokine production in cells from nonstimulated wells was barely detectable, (with the exception of a single outlier in CD8+ IL-2), indicating that there was minimal endogenous activation either by delay to processing or in ICS assay performance (Supporting Information Fig. 3a). Most stimulated CD4+ T cell cytokine frequencies were stable, though a small but significant decrease in IL2-positive events and increase, and then decrease in MIP-1β-positive events was observed at T24 and T48 when comparing medians. This pattern may suggest that the variance seen was random due to experimental staining procedure and not due to time delay to processing. There were no significant time dependent changes in CD4+ activation (CD69+), IFN-γ, TNF-α, IL-4/5/13, or IL-17 for either stimulated or unstimulated conditions. Polyfunctional CD4+ T cell analysis revealed predominance of single-cytokine producing cells (IL_2>TNF-α>IFN-Y), followed by dual-producing IL-2/MIP-1β cells (data not shown). There were only rare triple-positive cytokine events. There was no difference in polyfunctional frequencies over time points measured (data not shown). Overall, there were small but statistically significant differences in CD4+ T cell function after time delay of 24 h, and more detectable changes by 48 h.
CD8+ Intracellular Cytokine Results
CD8+ T cells were sequentially identified by FSC/SSC/ doublet-/live-dead-/CD14−/CD3+/CD4−/CD8+ events. To verify the gating strategy, fluorescence minus one and healthy donor controls were utilized. Cytokine-positive frequencies for IFN-γ, TNF-α, IL-2 and MIP-1β were measured within CD69+ gates only. Though CD69+ events were similar between cord blood samples and adult control, cytokine-positive CD8+ cell frequencies were quite low for all that were measured (Fig. 5, adult data not shown).
Figure 5.
Cord blood CD8+ T cell cytokine production following in vitro stimulation. Cord blood CD8+ T cells were stimulated in vitro with SEB, permeabilized and stained for intracellular cytokines IFNγ, TNFα, IL2, MIP1β. Only events within the CD69+ gate were considered cytokine-positive. Frequencies are expressed as percentages of total CD8+ events. Each symbol represents a single subject. Statistical comparisons were performed using Wilcoxon signed-rank test between samples isolated after delay of 0 h (T0), 24 h (T24), and 48 h (T48). Significant differences were found at T24 and T48 for IL2 (P < 0.05) only.
Spontaneous activation and cytokine production in the unstimulated CD8+ umbilical cord cells was small but detectable for IFN-γ and IL-2 (Supporting Information Fig. 4b). There were no statistically significant differences for IFN-γ, TNF-α, or MIP-1β cytokine-positive frequencies over time (Fig. 5, Table 4). Whereas IL-2 production decreased in CD4+ cells over time, there was a small but significant increase in stimulated IL-2 production for CD8+ T cells at both T24 and T48. Variance for all cytokines based on replicate level was generally higher in CD8+ than for CD4+ cells. For all cytokine-positive frequencies except TNF-α, patient level variance exceeded both time and replicate levels. The majority of cytokine-positive CD8+ T cells were single producers of either IL-2 or TNF-α. Dual cytokine-positive events, when present, produced either MIP-1β/IL-2 or MIP-1β/TNF-α. There were only rare events that were triple-positive, and no detectable quadruple cytokine-producing events. Among the single and dual-producing events, there were no statistically significant differences in population frequencies across time points. In general, CD8+ T cells showed stable function as measured by cytokine production after delay to processing of 24 h.
Table 4.
CD8+ T cell cytokine measures of variance
| PATIENT LEVEL | TIME LEVEL | REPLICATE LEVEL | ||||||
|---|---|---|---|---|---|---|---|---|
| CD8 CYTOKINE | VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN TIME |
VARIANCE ESTIMATE |
VARIANCE EXPLAINED (%) |
ICC WITHIN REPLICATES |
| CD69+ | 0.008174 | 44.04 | 0.005754 | 31.00 | 0.59 | 0.004632 | 24.96 | 0.75 |
| IFN − γ+ | 0.09077 | 50.44 | 0.05438 | 30.22 | 0.63 | 0.0348 | 19.34 | 0.81 |
| TNF − α+ | 0.03605 | 34.67 | 0.03771 | 36.27 | 0.49 | 0.03022 | 29.06 | 0.71 |
| IL − 2+ | 0.1369 | 59.40 | 0.04094 | 17.76 | 0.77 | 0.05264 | 22.84 | 0.77 |
| MIP − 1β+ | 0.2211 | 70.94 | 0.04813 | 15.44 | 0.82 | 0.04243 | 13.61 | 0.86 |
Cord blood CD8+ T cell cytokine production following in vitro stimulation was measured using intracellular cytokine staining and flow cytometry. Intraclass correlation coefficient (ICC) measure was used to determine the relative contribution of variance based on subject-subject, replicate (assay), and time delay to processing variability. For most cytokines, the majority of variance was explained at the patient level. The exception was TNF-α+ events, which showed high variability based on time delay.
DISCUSSION
Logistics for immunologic studies using umbilical cord blood T cells are complicated to manage, due to the uncontrolled timing of deliveries. Often with large neonatal studies, multicenter recruitment of subjects is necessary, requiring shipping of unprocessed cord blood samples. Such unpredictability in processing time is less problematic for studies including only full-term, elective cesarean section deliveries. However, in studies requiring samples from a more heterogeneous population of newborns, including prematurely born infants, quantification of variation introduced by time delay to processing is essential. This is particularly important when analyzing rare T cell populations using sensitive cytometric assays. Intra-assay variation and changes in cell markers based on collection or processing techniques may be attributed erroneously to biologic differences between study subjects.
This study performed multiple statistical comparisons of the effect of time of processing on the identification of T cell populations within a specimen incorporating an assessment of assay variability. Replicates had remarkable overlap within each subject (ICC>0.85 for cell type,>0.75 for CD4 subtype, >0.74 for CD4 cytokines and >0.71 for CD8 cytokines), reflecting the reproducibility of the standardized procedures. For most cell subsets and cytokine-positive frequencies, the order of variables contributing to overall variation was subject > time point > replicate. Markers with higher stain indices and event numbers allow for better separation between positive and negative populations. In newborn samples, chemokine receptor and cytokine staining are more difficult to gate accurately and consistently due to lower receptor expression and lower event numbers, which contribute to increased variance and lower sensitivity of statistical analyses for these populations. Nonetheless, we demonstrated stability of populations at 24 h, with only minor degradation (0.2–5% for chemokine receptor median expression,<0.1% for cytokine medians) or increased variance by 48 h.
Though not tested systematically, our team found several techniques that increased cell recovery and reproducibility of the flow results. Collection technique was designed to maximize cell recovery and minimize maternal cell contamination. Our initial, and common, collection technique of dripping blood from cut end of cord into a standard 150 USP heparin “green top” tube resulted in frequent clotting. Venipuncture of a clamped cord segment, rather than the dripping or stripping techniques, reduced this complication. The cord surface was also rubbed with alcohol prior to venipuncture to eliminate coating maternal cells. We achieved minimal coagulation by using venipuncture and increasing the heparin content per tube to 300 USP based on prior recommendations (11).
For flow analysis, RBC contamination remained a concern. Reduced separation of isolated UCMC from RBC was visibly noted after ficoll centrifugation of samples processed after a 48–72 h delay when compared to a 24-h delay. At all three time points, the RBC were not completely removed by using ammonium chloride lysis buffer. These contaminating RBC, that were not well lysed or removed by ficoll gradient separation, were likely nucleated RBC that also tend to survive cryopreservation. While published protocols suggest repeating the ficoll-paque gradient separation step for umbilical cord samples, we found that this resulted in an unacceptable loss in total cell recovery without adequate removal of the nucleated RBC. Unfortunately, the volume of cord blood available from premature deliveries is commonly <5 mL, which greatly limits the number of mononuclear cells that can be obtained. With an ultimate interest in analyzing rare events (such as polarized CD4+ T cells) in prematurely delivered infants, a repeat ficoll step reduces the recovery to an unacceptable degree. Flow analysis using a CD235 RBC marker showed RBC contamination that was stable over time, and present in overlapping regions with CD4 and CD8 positive events. Backgating analysis by flow cytometry showed that without excluding CD235+ populations, CD3+ (CD4+ and CD8+) events were contaminated with nonspecific staining of RBCs. Because some samples approached 50% contamination of UCMCs by RBCs, inclusion of these events in the final analysis will produce markedly erroneous results in population frequencies if not controlled.
Our study found few differences in median frequencies over time in many cell populations identified by surface markers. Surprisingly, precryopreservation viability and cell counts, and post-thaw viability and cell recovery were not affected significantly by time delay to processing. Among CD14+, CD56+, CD3+, CD4+ and CD8+, CD3+ CD56+(hi) showed significant variation between T0 and T48, but not for T24. There were no differences in the proportions of CD3+ T cell subsets, however, suggesting that changes are distributed equally across cell types. Stability of T cell markers through cryopreservation has been addressed in a previously published study on adult PBMC, which showed poor concordance between fresh and frozen CD4+CD62L+CD45RA+, CD4+CD45RO+, CD8+CD28+CD95−, and CD8+CD28+ CD95+ subpopulations. Other authors have shown loss of CD56 expression after cryopreservation, which may be compounded if there is loss of marker prior to cryopreservation (17,18). One published study examined the effect of cryopreservation on umbilical cord mononuclear cell cytokine secretion (19). Authors found significant blunting of IFN-γ, IL-10, IL-12, and TNF-α to mitogen stimulation, with an unpredictable pattern of cytokine balance (cytokine concentrations neither changed equally nor consistently). Because this study measured only secreted cytokine, we cannot conclude that the changes observed in their study were consistent with our findings that measured negative/positive-cytokine T cell frequency generated from flow cytometry. A separate study comprehensively characterizing the effects of cryopreservation on umbilical cord mononuclear cells is necessary to adequately address this question.
Comprehensive phenotyping on umbilical CD4+ T cells is underrepresented in the literature, although standards have been established for adult peripheral blood samples. While the subsetting of CD4+ T cell using chemokine receptors is a useful tool to aid in understanding broad concepts of T cell differentiation and function, nuances of these populations continue to be redefined. As our knowledge of the various marker combinations expands, the heterogeneity of such populations will be increasingly appreciated. As such, we acknowledge the limitations in assigning eponyms to cell events based on surface markers alone.
Of CD4+ subsets we analyzed, late activated events (CD27−CD28−) appeared to be affected more by time than naïve and early activated (CD27+CD28+). Significant differences in CD4+ subsets were found more frequently at 48 h (8 of 23 subsets identified) with fewer subsets affected at 24 h (4 of 23 subsets). Loss of CD28 expression on CD4+ T cells is associated with cells that have undergone multiple cycles of proliferation and terminally differentiated into IFNγ effectors or into a senescent phenotype (20). Although properties of umbilical cord cells may lend to more rapid cell cycling or down-regulation of CD28 expression, it is unlikely that increases in CD28− frequencies is a result of rapid ex vivo expansion given the short incubation. It is interesting that within the CD28+CD27+ subset, there was a relative loss of chemokine receptor CXCR3 and gain of CCR4+ frequencies at 48 h, with a corresponding gain of both CXCR3 and CCR4 in the CD27−CD28− subset at 48 h. It is possible that there is a fraction of umbilical cord CD4+ T cells that are activated perinatally and differentiate early in vitro.
Time did not have a significant effect on SEB-stimulated intracellular cytokine-positive frequencies for CD4 or CD8+ T cells, except for IL-2, which was affected in opposite directions for the two cell types (decreased from 2.02 to 1.43% CD4 and increased from 0.05 to 0.09% in CD8), and CD4+ MIP-1β (increased from 0.10% at T0 to 0.21% at T48). Umbilical cord cells are known to produce high amounts of IL-2, in both CD4+ and CD8+ T cell subsets (21), and constitutive production in CD4+ T cells may decrease over time as culture substrate is depleted. Brief antigen stimulation also results in increased IL-2 production in CD8+ T cells without concurrent increase in cytotoxic activity (22). It is possible that introduction of antigen through delivery, sample collection or processing could provide enough stimulus to increase background IL-2 production of CD8+ T cells in a the given period of time; this effect may be more evident in umbilical cord samples which produce high amounts of IL-2 at baseline.
While we have demonstrated small changes with delay in processing of UBMC, it is important to recall that our study included blood only from full term scheduled cesarean-section deliveries to control the variable of time delay to processing. It is possible that inflammatory signals present in cord blood collected from premature and/or infected, deliveries after labor or maternal preeclampsia could change the phenotype and function of T cells, changes that may be amplified by delay in processing. In fact, there are studies that support changes in T cells associated with the presence of superantigen (23,24), through toll-like receptors and independent of TCR engagement (25) or physiologic stressors (26,27). Stability of UCMC at 24 h after blood sample collection may be altered in infants whose peripheral T cells were exposed to inflammatory signals in utero and continued to be stimulated while at room temperature prior to processing. The slight decrease in naïve/early activated and increase in late activated CD4+ T cells demonstrated in our study may be more exaggerated following stressful delivery or delivery through infected membranes.
Notably, exclusion of dead cells (live/dead stain positive) did increase the specificity of staining and therefore reproducibility of results with cryopreserved specimens. This study was not designed to test the effects of cryopreservation on the various cell markers, but as all cells were frozen prior to analysis for similar periods of time, any effect is expected to be applied uniformly across time points. Our study did not test T cell functional changes that might affect cord blood cell engraftment, which would require much more comprehensive functional and phenotypic analysis and was not the purpose of the study.
This study does not evaluate the biologic significance of the small time-dependent changes measured. It is not known, for example, whether there is a loss/gain of cell type or if changes across time represent down/upregulation of surface proteins. Surface receptor changes are more likely than differences in cell populations surviving over time, given the absence of differences observed in viability and recovery. When using a combined statistical approach of pair-wise comparisons and measurement of interclass variation, a few cell types are affected at 48 h, and fewer are significantly changed after a 24-h time delay. On the basis of these results, we conclude that our standardized operating procedure for collection and processing of umbilical cord mononuclear cells can generate highly reproducible results, even after a time delay of up to 24 h prior to cell isolation. Of interest, the apparent alterations seen at 24 h, for example in IL-2, suggests that there are caveats associated with delayed processing and that it may indeed by important to optimize processing times to less than a day. For studies in which delay to processing of umbilical cord samples is inevitable however, our data quantifying expected variation and range of results can be applied to appropriately power or correct for changes that may occur secondary to time delay to processing. Our methods can be used by multicenter studies to reliably analyze T cell phenotype and function in umbilical cord blood.
ACKNOWLEDGMENTS
The authors would like to acknowledge Ernest Wang, Sara Misra, Heidie Huyck, Anne Sessler for technical support, and Elizabeth Werner and Deanna Maffet for subject recruitment and sample collection. All flow cytometry was performed in the URMSD Flow Cytometry Core Facility.
Grant sponsor: NIH; Grant numbers: U01 HL101813-01 (Prematurity and Respiratory Outcomes Program [PROP] PI: Pryhuber, Mariani and Ryan), R24 AI054953 (Rochester Human Immunology Center, PI: Quataert), and 1K12HD068373-01 (Translational Molecular Pediatrics, PI: Scheible, Directors: Schor and Gigliotti).
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- 1.Roederer M. How many events is enough? Are you positive? Cytometry A. 2008;73A:384–385. doi: 10.1002/cyto.a.20549. [DOI] [PubMed] [Google Scholar]
- 2.Duijts L, Bakker-Jonges LE, Labout JA, Jaddoe VW, Hofman A, Steegers EA, van Dongen JJ, Hooijkaas H, Moll HA. Fetal growth influences lymphocyte subset counts at birth: The generation R study. Neonatology. 2009;95:149–156. doi: 10.1159/000153099. [DOI] [PubMed] [Google Scholar]
- 3.van Gent R, van Tilburg CM, Nibbelke EE, Otto SA, Gaiser JF, Janssens-Korpela PL, Sanders EA, Borghans JA, Wulffraat NM, Bierings MB, et al. Refined characterization and reference values of the pediatric T- and B-cell compartments. Clin Immunol. 2009;133:95–107. doi: 10.1016/j.clim.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Berrington JE, Barge D, Fenton AC, Cant AJ, Spickett GP. Lymphocyte subsets in term and significantly preterm UK infants in the first year of life analysed by single platform flow cytometry. Clin Exp Immunol. 2005;140:289–292. doi: 10.1111/j.1365-2249.2005.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Alessio F, Mirabelli P, Gorrese M, Scalia G, Gemei M, Mariotti E, Di Noto R, Martinelli P, Fortunato G, Paladini D, et al. Polychromatic flow cytometry analysis of CD34+ hematopoietic stem cells in cryopreserved early preterm human cord blood samples. Cytometry A. 2011;79A:14–24. doi: 10.1002/cyto.a.20989. [DOI] [PubMed] [Google Scholar]
- 6.dos Santos AP, Bertho AL, Martins Rde M, Marcovistz R. The sample processing time interval as an influential factor in flow cytometry analysis of lymphocyte subsets. Mem Inst Oswaldo Cruz. 2007;102:117–120. doi: 10.1590/s0074-02762007000100020. [DOI] [PubMed] [Google Scholar]
- 7.Bull M, Lee D, Stucky J, Chiu YL, Rubin A, Horton H, McElrath MJ. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods. 2007;322:57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna KC, Beatty KM, Vicetti Miguel R, Bilonick RA. Delayed processing of blood increases the frequency of activated CD11b+ CD15+ granulocytes which inhibit T cell function. J Immunol Methods. 2009;341:68–75. doi: 10.1016/j.jim.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Basford C, Forraz N, McGuckin C. Optimized multiparametric immunophenotyping of umbilical cord blood cells by flow cytometry. Nat Protoc. 2010;5:1337–1346. doi: 10.1038/nprot.2010.88. [DOI] [PubMed] [Google Scholar]
- 10.Mallone R, Mannering SI, Brooks-Worrell BM, Durinovic-Bello I, Cilio CM, Wong FS, Schloot NC. Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: Position statement of the T-cell workshop committee of the immunology of diabetes society. Clin Exp Immunol. 2011;163:33–49. doi: 10.1111/j.1365-2249.2010.04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuss IJ, Kanof ME, Smith PD, Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009 Apr;Chapter 7(Unit 7.1) doi: 10.1002/0471142735.im0701s85. [DOI] [PubMed] [Google Scholar]
- 12.Wang JC, Kobie JJ, Zhang L, Cochran M, Mosmann TR, Ritchlin CT, Quataert SA. An 11-color flow cytometric assay for identifying, phenotyping, and assessing endocytic ability of peripheral blood dendritic cell subsets in a single platform. J Immunol Methods. 2009;341:106–116. doi: 10.1016/j.jim.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;1:1507–1516. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 14.Herzenberg LA, Tung J, Moore WA, Parks DR. Interpreting flow cytometry data: A guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 15.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: Unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 16.Appay V, van Lier RA, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: Consensus and issues. Cytometry A. 2008;73A:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 17.Weinberg A, Song LY, Wilkening C, Sevin A, Blais B, Louzao R, Stein D, Defechereux P, Durand D, Riedel E, et al. Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine Immunol. 2009;16:1176–1186. doi: 10.1128/CVI.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marti F, Miralles A, Peiro M, Amill B, de Dalmases C, Pinol G, Rueda F, Garcia J. Differential effect of cryopreservation on natural killer cell and lymphokine-activated killer cell activities. Transfusion. 1993;33:651–655. doi: 10.1046/j.1537-2995.1993.33893342746.x. [DOI] [PubMed] [Google Scholar]
- 19.Shreffler WG, Visness CM, Burger M, Cruikshank WW, Lederman HM, de la Morena M, Grindle K, Calatroni A, Sampson HA, Gern JE. Standardization and performance evaluation of mononuclear cell cytokine secretion assays in a multicenter study. BMC Immunol. 2006;7:29. doi: 10.1186/1471-2172-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilani SR, Vuga LJ, Lindell KO, Gibson KF, Xue J, Kaminski N, Valentine VG, Lindsay EK, George MP, Steele C, et al. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PLoS One. 2010;5:e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasparoni A, Ciardelli L, Avanzini A, Castellazzi AM, Carini R, Rondini G, Chirico G. Age-related changes in intracellular TH1/TH2 cytokine production, immunoproliferative T lymphocyte response and natural killer cell activity in newborns, children and adults. Biol Neonate. 2003;84:297–303. doi: 10.1159/000073638. [DOI] [PubMed] [Google Scholar]
- 22.Usharauli D, Kamala T. Brief antigenic stimulation generates effector CD8 T cells with low cytotoxic activity and high IL-2 production. J Immunol. 2008;180:4507–4513. doi: 10.4049/jimmunol.180.7.4507. [DOI] [PubMed] [Google Scholar]
- 23.Taylor AL, Llewelyn MJ. Superantigen-induced proliferation of human CD4+ CD25i2212; T cells is followed by a switch to a functional regulatory phenotype. J Immunol. 2010;185:6591–6598. doi: 10.4049/jimmunol.1002416. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi N, Kato H, Imanishi K, Miwa K, Yamanami S, Nishida H, Uchiyama T. Immunopathophysiological aspects of an emerging neonatal infectious disease induced by a bacterial superantigen. J Clin Invest. 2000;106:1409–1415. doi: 10.1172/JCI10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLeod H, Wetzler LM. T cell activation by TLRs: A role for TLRs in the adaptive immune response. Sci STKE. 2007;2007:pe48. doi: 10.1126/stke.4022007pe48. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Seidl T, Whittall T, Babaahmady K, Lehner T. Stress-activated dendritic cells interact with CD4+ T cells to elicit homeostatic memory. Eur J Immunol. 2010;40:1628–1638. doi: 10.1002/eji.200940251. [DOI] [PubMed] [Google Scholar]
- 27.Luciano AA, Yu H, Jackson LW, Wolfe LA, Bernstein HB. Preterm labor and chorioamnionitis are associated with neonatal T cell activation. PLoS One. 2011;6:e16698. doi: 10.1371/journal.pone.0016698. [DOI] [PMC free article] [PubMed] [Google Scholar]