Abstract
Background
The ciliate Paramecium bursaria harbors several hundred cells of the green-alga Chlorella sp. in their cytoplasm. Irrespective of the mutual relation between P. bursaria and the symbiotic algae, both cells retain the ability to grow without the partner. They can easily reestablish endosymbiosis when put in contact with each other. Consequently, P. bursaria is an excellent model for studying cell–cell interaction and the evolution of eukaryotic cells through secondary endosymbiosis between different protists. Despite the importance of this organism, no genomic resources have been identified for P. bursaria to date. This investigation compared gene expressions through RNA-Seq analysis and de novo transcriptome assembly of symbiont-free and symbiont-bearing host cells.
Results
To expedite the process of gene discovery related to the endosymbiosis, we have undertaken Illumina deep sequencing of mRNAs prepared from symbiont-bearing and symbiont-free P. bursaria cells. We assembled the reads de novo to build the transcriptome. Sequencing using Illumina HiSeq2000 platform yielded 232.3 million paired-end sequence reads. Clean reads filtered from the raw reads were assembled into 68,175 contig sequences. Of these, 10,557 representative sequences were retained after removing Chlorella sequences and lowly expressed sequences. Nearly 90% of these transcript sequences were annotated by similarity search against protein databases. We identified differentially expressed genes in the symbiont-bearing P. bursaria cells relative to the symbiont-free cells, including heat shock 70 kDa protein and glutathione S-transferase.
Conclusions
This is the first reported comprehensive sequence resource of Paramecium – Chlorella endosymbiosis. Results provide some keys for the elucidation of secondary endosymbiosis in P. bursaria. We identified P. bursaria genes that are differentially expressed in symbiont-bearing and symbiont-free conditions.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-183) contains supplementary material, which is available to authorized users.
Keywords: Paramecium bursaria, Chlorella variabilis, Secondary symbiosis, Transcriptome analysis
Background
As demonstrated by the evolution of mitochondria and chloroplasts, endosymbiosis is a major driving force behind eukaryotic cell evolution leading to acquisition of new intracellular components and cell diversity [1, 2]. Although endosymbiosis is an important and widespread phenomenon, the mechanisms controlling the establishment of endosymbiosis between different eukaryotic cells are not well understood. In fact, P. bursaria cells harbor about 700 symbiotic algae in their cytoplasm [3]. Each alga is enclosed in a perialgal vacuole (PV) membrane derived from the host digestive vacuole (DV) membrane, which protects the alga from the host’s lysosomal fusion [4–6]. Irrespective of the mutual relations between P. bursaria and symbiotic algae [7–11], the symbiont-free cells and the symbiotic algae retain the ability to grow without a partner. Symbiont-free cells can be prepared by various means: cultivation under constant dark conditions [12–14], treatment with cycloheximide [3, 15, 16], and treatment with the photosynthesis inhibitor dichlorophenyl dimethylurea (DCMU) [17]. However, symbiotic algae can be isolated by homogenization or by sonication or by the treatment of symbiotic cells with detergent. They can grow outside host cells [18]. Symbiont-free cells are easily reinfected with symbiotic algae by mixing the two together. Therefore, P. bursaria has been considered an excellent model for studying cell–cell interaction and the evolution of eukaryotic cells through secondary endosymbiosis between different protists [19]. However, neither genomic nor transcriptomic information has been available to elucidate the establishment of endosymbiosis in P. bursaria to date. To expedite the process of gene discovery related to the endosymbiosis, we have undertaken Illumina deep sequencing of mRNAs prepared from symbiont-bearing and symbiont-free P. bursaria cells in this study. Our data provide a comprehensive sequence resource for the advancement of P. bursaria study.
Results and discussion
Deep-sequencing and assembly
We constructed three RNA-seq libraries from mRNA of P. bursaria harboring symbiotic alga, Chlorella variabilis, and three libraries from symbiont-free P. bursaria. Sequencing using Illumina HiSeq2000 platform yielded 232.3 million 101-by-101 bp paired-end sequence reads. After trimming the low-quality parts and removing reads of less than 50 bp, 436.9 million reads (42.9 Gb) remained. To obtain a comprehensive sequence set of the P. bursaria transcriptome, all the clean reads of symbiont-bearing and symbiont-free libraries were assembled together using the Trinity program [20]. The de novo assembly produced 68,175 contigs, clustering into 40,805 subcomponents (i.e. unigenes). We selected the longest transcript as the representative for each cluster. The unigene sizes were 200 bp up to 22,858 bp, with mean length of 904 bp, N50 of 1,832 bp totaling 36,894,860 bp for all unigenes; 9,620 (23.6%) of unigenes were longer than 1,000 bp.
We excluded unigenes derived from the symbiotic Chlorella and other contaminants. Of the 68,175 contig sequences, 11,256 were matched to the C. variabilis sequences, and were therefore removed. Unigenes lowly expressed with log-counts-per-million (logCPM) < 0 were also discarded because they are likely to be contaminant sequences or poor assembly models. Based on the database search, the small amount of the contaminant sequences appears to be derived from some bacteria such as Methylobacterium and Burkholderiales, which are likely to be included in the culture media in which we grew P. bursaria. These procedures produced P. bursaria transcript reference sequences composed of 10,557 unigenes.
Annotation of the assembled contigs
We performed similarity searches of the 10,557 P. bursaria unigenes against the Swiss-Prot and UniRef90 protein sequence databases [21] using BLASTX [22] with the E-value cutoff of 1e-5 and assigned the functional annotations of the most similar protein sequences. Of the 10,557 unigenes, 7,051 (67%) had matches with 4,102 unique records in the Swiss-Prot database; 9,536 (90%) had matches with 8,189 unique records in the UniRef90 database. The species distribution of the BLASTX best hits in the UniRef90 database showed that 8,710 (91.7%) of the 9,502 hits had top matches with sequences from P. tetraurelia, followed by Tetrahymena thermophila with 153 (1.6%) best BLASTX hits.
We predicted open reading frames (ORFs) from the 10,557 P. bursaria unigene sequences using OrfPredictor [23]. Of the 10,557 ORFs, 10,535 were longer than 50 amino acids, 10,134 were longer than 100 amino acids, and 3,425 were longer than 500 amino acids. Although whole genome sequences have been clarified in P. tetraurelia[24] and T. thermophila[25], endosymbiotic algae including Chlorella species have not yet been detected in these ciliates. Therefore, we tried to compare their ORFs length, GC%, and shared gene clusters among these two ciliates and P. bursaria to elucidate the genomic features of P. bursaria as a potential host cell for the symbiotic algae.
We compared ORFs (>100 amino acids) of P. bursaria with those of its close relatives P. tetraurelia and T. thermophila. The maximum values for lengths of ORFs for P. bursaria, P. tetraurelia, and T. thermophila were, respectively, 19,640, 21,570, and 34,740. The median values for lengths of ORFs for P. bursaria, P. tetraurelia, and T. thermophila were, respectively, 1,161, 1,155, and 1,503 (Additional file 1). The corresponding values for G + C contents (%) of ORFs were 30.1, 29.7, and 27.8 (Additional file 2).
We identified 29,835 orthologous gene clusters containing 70,114 ORFs from the three organisms: i.e. 10,134, 37,382, and 22,598 ORFs longer than 100 amino acids long, respectively, from P. bursaria, P. tetraurelia, and T. thermophila. Figure 1 shows the number of orthologous gene clusters shared among the three organisms. Of the 29,835 orthologous gene clusters, 3,421 were common to all three organisms, 2,854 were unique to Paramecium species, and 2,330 were unique to P. bursaria.
Figure 1.
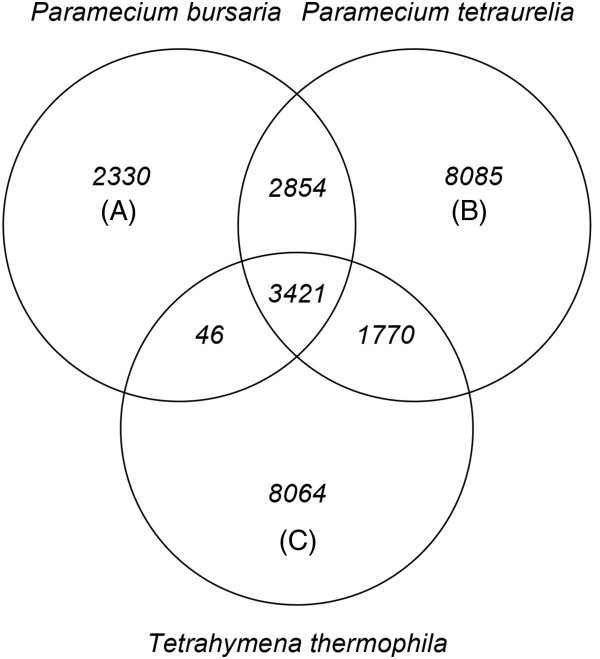
Venn diagram showing the number of orthologous gene clusters shared among three organisms: P. bursaria (A), P. tetraurelia (B), and T. thermophila (C). Protein coding sequences from the three organisms were grouped into orthologous gene clusters using OrthoMCL [26].
Differential gene expression between symbiont-bearing and symbiont-free conditions
We compared gene expressions of symbiont-bearing and symbiont-free P. bursaria to elucidate the genetic control for establishment of secondary symbiosis. Of the 10,557 transcripts, 6,698 were significantly differentially expressed between symbiont-bearing and symbiont-free cells with false discovery rates (FDR) < 0.05 (Additional file 3).
The positive and negative values of log2 Fold Change (logFC) show that the sequences were up-regulated and down-regulated in symbiont-bearing cells compared to symbiont-free cells. The parametric analysis of gene set enrichment (PAGE) [27] detected enrichment in the 17 gene ontology (GO) terms, i.e. 8 biological processes, 3 cellular components, and 6 molecular functions based on the logFC between symbiont-bearing and symbiont-free cells with false discovery rates (FDR) < 0.05 (Table 1). Figure 2 portrays a parent–child relation between GO molecular function terms including the six highest ranking GO terms based on PAGE P-values: oxidoreductase activity (Z = -6.879), structural constituent of ribosome (Z = -4.153), pyridoxal phosphate binding (Z = -4.015), phosphorelay response regulator activity (Z = 3.310), chromatin binding (Z = 3.107), and phosphorelay sensor kinase activity (Z = 2.901). A closer examination of individual proteins assigned to these GO terms revealed that trans-2-enoyl-CoA (oxidoreductase activity), aminotransferases (pyridoxal phosphate binding) and ribosomal proteins tended to be down-regulated, whereas transcriptional activator Myb-related proteins (chromatin binding), and signal transduction histidine kinase (phosphorelay response regulator activity and phosphorelay sensor kinase activity) tended to be up-regulated in symbiont-bearing cells relative to symbiont-free cells.
Table 1.
Enrichment of gene ontology terms in differentially expressed sequences in P. bursaria detected by PAGE
| GO_name | GO_id | Number of sequences | Z score | P -value | False discovery rate |
|---|---|---|---|---|---|
| BP oxidation–reduction process | GO:0055114 | 135 | -6.992 | 2.71E-12 | 3.28E-10 |
| MF oxidoreductase activity | GO:0016491 | 92 | -6.879 | 6.04E-12 | 3.65E-10 |
| BP metabolic process | GO:0008152 | 104 | -5.659 | 1.52E-08 | 6.15E-07 |
| BP carbohydrate metabolic process | GO:0005975 | 33 | -4.656 | 3.23E-06 | 9.76E-05 |
| CC integral to membrane | GO:0016021 | 196 | -4.399 | 1.09E-05 | 0.0003 |
| BP translation | GO:0006412 | 56 | -4.187 | 2.83E-05 | 0.0006 |
| MF structural constituent of ribosome | GO:0003735 | 57 | -4.153 | 3.29E-05 | 0.0006 |
| CC ribosome | GO:0005840 | 54 | -4.044 | 5.25E-05 | 0.0008 |
| MF pyridoxal phosphate binding | GO:0030170 | 13 | -4.015 | 5.96E-05 | 0.0008 |
| MF phosphorelay response regulator activity | GO:0000156 | 40 | 3.310 | 0.0009 | 0.0102 |
| BP phosphorelay signal transduction system | GO:0000160 | 40 | 3.310 | 0.0009 | 0.0102 |
| BP ATP hydrolysis coupled proton transport | GO:0015991 | 16 | -3.239 | 0.0012 | 0.0121 |
| MF chromatin binding | GO:0003682 | 21 | 3.107 | 0.0019 | 0.0176 |
| BP DNA replication | GO:0006260 | 18 | 3.008 | 0.0026 | 0.0227 |
| CC intracellular | GO:0005622 | 95 | -2.953 | 0.0031 | 0.0254 |
| MF phosphorelay sensor kinase activity | GO:0000155 | 26 | 2.901 | 0.0037 | 0.0281 |
| BP biosynthetic process | GO:0009058 | 16 | -2.798 | 0.0051 | 0.0366 |
Notes: GO, gene ontology; PAGE, parametric analysis of gene set enrichment; BP, biological process; MF, molecular function; and CC, cellular component. We used log2 Fold Change values between symbiont-bearing and symbiont-free cells to calculate Z scores and corresponding P-values for each GO term.
Figure 2.
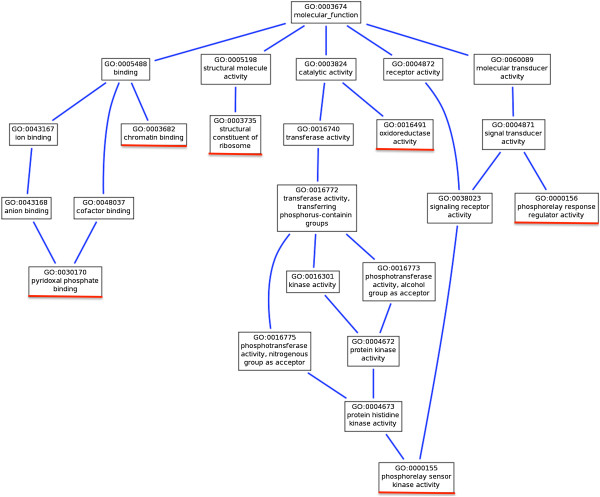
Parent–child relation between gene ontology (GO) molecular function terms viewed by AmiGO ( http://amigo.geneontology.org/ ). Red lines show the six highest ranking GO molecular function terms based on P-values in Table 1.
Based on the knowledge of P. bursaria accumulated to date, functions can be inferred for some of the six highest ranking GO terms. Down-regulation of ribosomal proteins in symbiont-bearing P. bursaria cells suggests that algal proteins with functions equivalent to those of the host Paramecium cells are transferred to the host through the PV membrane. Consequently transcriptional activity of the host was reduced. However, no report to date has described a demonstration showing that the proteins synthesized by the algae and transferred to the host cell through photosynthetic products, mainly maltose, are transferred from the algae [7, 9, 11].
Up-regulation of signal transduction histidine kinase in symbiont-bearing P. bursaria cells suggests that the histidine kinases play an important role in signal perception in this secondary symbiosis, as shown in the primary symbiosis by bacteria [28] for the adaptation and survival of various organisms to harsh environmental conditions [29]. Sensor histidine kinases are known to play important roles in several eukaryotic species including yeasts, fungi, slime molds, and higher plants [30–32]. Symbiont-bearing P. bursaria cells acquire various stress resistance through endosymbiosis with Chlorella spp [33–36].
In addition to the genes of enriched GO terms discussed above, heat shock 70 kDa protein (Hsp70) and glutathione S-transferase (GST) genes were up-regulated and down-regulated as shown by the positive and negative values of logFC, respectively, in symbiont-bearing cells compared to symbiont-free cells (Table 2). Of the 10,557 unigenes, 8 were annotated as Hsp70 with logFC of -1.3 to 5.6, with a median of 0.92. Symbiont-bearing P. bursaria cells are known to show a higher survival ratio against nickel chloride, high temperature, and hydrogen peroxide than the symbiont-free cells show [35, 36]. Furthermore, P. caudatum cells reportedly acquire heat-shock resistance by infection of endonucler symbiotic bacteria Holospora[37–39], and osmotic-shock resistance [40]. Hori and Fujishima [39] reported that H. obtusa-bearing paramecia expressed high levels of Hsp70 mRNA even at 25°C. The up-regulation of the transcripts encoding Hsp70 might be related to the host’s tolerance to environmental fluctuations. Of the 10,557 transcripts, 7 were annotated as GST and tended to be down-regulated with logFC of -5.7 to -0.12, with a median of -0.85 (Table 2). This enzyme is related to protection of cells from oxidative stress, as shown by McCord and Fridovich [41] and by Veal et al. [42]. Although it was conceivable that photo-oxidative stress in symbiont-bearing P. bursaria cells is greater than that in symbiont-free ones, our data showed opposite results from the prediction. A similar result was obtained by Hörtnagl and Sommaruga [33]. They suggested that the presence of algal symbionts minimizes photo-oxidative stress [33]. Consequently, different expression levels in these genes between symbiont-free and symbiont-bearing P. bursaria agree well with differences in cytological phenomena observed in these paramecia, suggesting that these proteins appear to be involved in the symbiosis. Immunological detections of the gene products and comparisons of the amount of the antigens or qualitative PCR between the symbiont-free and the symbiont-bearing paramecia are necessary for future experiments. To ascertain the molecular mechanisms controlling the secondary symbiosis between Paramecium and Chlorella cells, transcriptome analyses and proteome analyses of the symbiotic Chlorella alone cultivated in algal culture medium and the same Chlorella inside the host Paramecium are necessary for future studies.
Table 2.
Transcripts encoding glutathione S-transferase and heat shock 70 kDa protein in P. bursaria
| Trinity transcript name | Annotation from the SwissProt database | logFC |
|---|---|---|
| Heat shock 70 kDa protein | ||
| comp43044_c0 | sp|P09446|HSP7A_CAEEL|Heat shock 70 kDa protein A OS = Caenorhabditis elegans | 5.601 |
| comp36402_c4 | sp|P09446|HSP7A_CAEEL|Heat shock 70 kDa protein A OS = Caenorhabditis elegans | 4.183 |
| comp36402_c6 | sp|P14834|HSP70_LEIMA|Heat shock 70 kDa protein (Fragment) OS = Leishmania major | 1.975 |
| comp36402_c1 | sp|Q9S9N1|HSP7E_ARATH|Heat shock 70 kDa protein 5 OS = Arabidopsis thaliana | 1.555 |
| comp37280_c1 | sp|P37899|HSP70_PYRSA|Heat shock 70 kDa protein OS = Pyrenomonas salina | 0.287 |
| comp43771_c0 | sp|P09446|HSP7A_CAEEL|Heat shock 70 kDa protein A OS = Caenorhabditis elegans | -0.594 |
| comp41901_c0 | sp|Q9S7C0|HSP7O_ARATH|Heat shock 70 kDa protein 14 OS = Arabidopsis thaliana | -1.076 |
| comp41912_c0 | sp|F4JMJ1|HSP7R_ARATH|Heat shock 70 kDa protein 17 OS = Arabidopsis thaliana | -1.337 |
| Glutathione S-transferase | ||
| comp37410_c0 | sp|P78417|GSTO1_HUMAN|Glutathione S-transferase omega-1 OS = Homo sapiens | -0.119 |
| comp32377_c0 | sp|Q9ZRT5|GSTT1_ARATH|Glutathione S-transferase T1 OS = Arabidopsis thaliana | -0.288 |
| comp36943_c0 | sp|Q9ZVQ3|GSTZ1_ARATH|Glutathione S-transferase Z1 OS = Arabidopsis thaliana | -0.748 |
| comp37841_c0 | sp|Q9ZRT5|GSTT1_ARATH|Glutathione S-transferase T1 OS = Arabidopsis thaliana | -0.851 |
| comp36483_c0 | sp|Q9ZRT5|GSTT1_ARATH|Glutathione S-transferase T1 OS = Arabidopsis thaliana | -1.557 |
| comp35816_c1 | sp|P78417|GSTO1_HUMAN|Glutathione S-transferase omega-1 OS = Homo sapiens | -1.564 |
| comp36242_c0 | sp|P16413|GSTMU_CAVPO|Glutathione S-transferase B OS = Cavia porcellus | -5.749 |
Conclusion
This study is the first whole transcriptome analysis conducted between symbiont-free and symbiont-bearing P. bursaria. Results showed P. bursaria genes that differentially expressed in symbiont-bearing and symbiont-free conditions. We showed that genes for glutathione S-transferase, trans-2-enoyl-CoA, aminotransferases, and ribosomal proteins are down-regulated, and that genes for Hsp70, transcriptional activator Myb-related proteins, and signal transduction histidine kinase are up-regulated in the symbiont-bearing P. bursaria. Our results enable us to understand the molecular mechanism for establishment of the secondary symbiosis and for the host evolutionary adaptation to global climate change.
Methods
Strains and cultures
Symbiont-free P. bursaria strain Yad1w was produced from Chlorella sp.-bearing P. bursaria strain Yad1g cells (syngen 3, mating type I) through repeated cloning and cultivation under dark conditions. The Yad1g cell strain was collected in Yamaguchi, Japan in 2004. Symbiont-bearing strain of Yad1g1N cells was used for symbiont-bearing cells. The Yad1g1N strain was produced by infection of cloned symbiotic Chlorella variabilis (formerly C. vulgaris) strain 1 N cells to the Yad1w cell [15]. Paramecium strains used for this study were provided by Symbiosis Laboratory, Yamaguchi University with support in part by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan. The culture medium used was 1.25% (w/v) fresh lettuce juice in modified Dryl’s solution (MDS) [43] (KH2PO4 was used instead of NaH2PO4 · 2H2O), inoculated with a non-pathogenic strain of Klebsiella pneumoniae one day before use [44]. In ordinary cultures, several hundred cells were inoculated into 2 ml culture medium. Then 2 ml of fresh culture medium was added on each of the next 12 days. One day after the final feeding, the cultures were in the early stationary phase of growth. All cells used in the present experiments were at this phase. Cultivation and all experiments were performed at 25 ± 1°C. In the case of the symbiont-bearing strain of Yad1g1N, the cells were cultivated under constant light conditions: 20–30 μmol photon/m2/s.
Transcriptome sequencing
Total RNA was isolated from 400,000 cells of symbiont-bearing and symbiont-free P. bursaria cells using Trizol reagent (Invitrogen Corp.) according to the manufacturer’s protocol. To construct three RNA-seq libraries from mRNAs of P. bursaria, the total RNA was isolated from three independent cultures of symbiont-free and symbiont-bearing P. bursaria. After suspension in Trizol reagent, the symbiont-bearing and symbiont-free cells were stirred in the presence of 600 μl of 0.5 mm zirconia/silica beads (BioSpec Products Inc.) to break cell walls of the symbiotic algae. The RNA was treated with RNase-free DNase and was cleaned up using RNeasy Mini (Qiagen Inc.) according to the manufacturer’s protocol. RNA integrity was confirmed (2100 Bioanalyzer; Agilent Technologies Inc.) with a minimum RNA integrated number value of 7.6. The samples for transcriptome analyses were prepared using Illumina’s kit following the manufacturer’s recommendations. First, mRNA was purified from 0.5 μg of total RNA of symbiont-bearing or symbiont-free cells using oligo (dT) magnetic beads. Following purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were used for first strand cDNA synthesis using SuperScript II Reverse Transcriptase (Invitrogen Corp.) and random primers. Second strand cDNA synthesis was conducted next. These cDNA fragments then went through an end repair process and ligation of adapters. These products were purified and enriched with PCR to create the final cDNA library. Multiplex sequencing of paired-end reads was performed on an Illumina Hiseq2000 instrument at NIBB Core Research Facilities, followed by raw data processing, base-calling, and quality-control by manufacturer’s standard pipeline using RTA, OLB, and CASAVA. The output sequence quality was inspected using the FastQC program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/).
De novoassembly
The reads were cleaned up with cutadapt program by trimming low-quality ends (<QV30) and adapter sequences, and by discarding reads shorter than 50 bp. De novo assembly of the clean reads was conducted using Trinity [20] in the paired-end mode with an option ‘–min_kmer_cov = 2’.
Differential expression analysis
Data of two biological replicates were used in this analysis for each condition. Using scripts included the Trinity package suite [20], cleaned reads were aligned to the Trinity-assembled transcripts using Bowtie [45]. Then transcript abundance was estimated using RSEM [46]. We used the edgeR package [47] of Bioconductor to identify genes that are differentially expressed between the conditions. To adjust for library sizes and skewed expression of transcripts, the estimated abundance values were normalized using the Trimmed Mean of M-values (TMM) normalization method [48] included in the edgeR package. Based on a negative binomial model implemented in edgeR, genes that were differentially expressed between symbiont-bearing and symbiont-free P. bursaria samples were identified.
Functional annotation of assembled contigs
Nucleotide sequences of Chlorella variabilis NC64A were obtained from the DOE Joint Genome Institute (JGI) site (http://genome.jgi.doe.gov/ChlNC64A_1/ChlNC64A_1.home.html). The assembled contigs that matched the Chlorella sequences indicated by MEGABLAST search (E-value cutoff of 1e-40) were excluded from analyses. We performed a BLASTX search (with the ciliate nuclear genetic code and E-value cutoff of 1e-5) of the P. bursaria transcripts against the UniProtKB Swiss-Prot and Uniref90 protein sequence databases [21] and assigned the functional annotations of the most similar protein sequences.
The protein-coding region of RNA sequences was predicted using OrfPredictor with the ciliate nuclear genetic code [23]. For sequences having BLASTX hits, the frames used by BLASTX were used for identifying the coding regions of the sequences. For sequences without BLASTX hits, the coding regions were predicted based on the intrinsic signals of the sequences. For comparative analysis, protein-coding sequences of P. tetraurelia were obtained from ParameciumDB (http://paramecium.cgm.cnrs-gif.fr). Those of T. thermophila were retrieved from Tetrahymena Genome Database (http://www.ciliate.org/). Proteins from the three organisms (P. bursaria, P. tetraurelia, and T. thermophila) were grouped into ortholog clusters using OrthoMCL [26] with BLASTP E-value cutoff of 1e-5 and inflation parameter of 1.5.
Gene ontology (GO) enrichment analysis
We conducted a domain search of the P. bursaria transcripts against the Pfam database release 26.0 [49]. Gene ontology (GO) terms were assigned to each transcript using the pfam2go conversion table (http://www.geneontology.org/external2go/pfam2go) [50]. We performed parametric analysis of gene set enrichment or PAGE [27] to test over-representation and under-representation of GO terms based on the logFC between symbiont-bearing and symbiont-free cells.
Availability of supporting data
The data sets supporting the results of this article are available in the DDBJ Sequence Read Archive (DRA) (accession number DRA000907, http://trace.ddbj.nig.ac.jp/DRASearch/submission?acc=DRA000907).
Electronic supplementary material
Additional file 1: Histogram showing the distribution of lengths for protein-coding sequences in P. bursaria (A), P. tetraurelia (B), and T. thermophila (C). The x-axis shows the length (bp) of the sequences. The y-axis shows their frequencies. (PDF 372 KB)
Additional file 2: Histogram showing the distribution of G + C contents for protein-coding sequences in P. bursaria (A), P. tetraurelia (B), and T. thermophila (C). The x-axis shows the G + C content (%) of the sequences. The y-axis shows their frequencies. G + C content is defined as 100 × (G + C)/(A + T + G + C). (PDF 264 KB)
Additional file 3: Data for Paramecium bursaria transcripts. The first five columns show the Trinity sequence name, length (bp), and functional annotations from different databases: Swiss-Prot, UniRef90, and gene ontology (GO). The remaining four columns show data of differential expression analysis using edgeR: log2 Fold Changes (logFC), log-counts-per-million (logCPM), P-values, and False Discovery Rate (FDR) adjusted P-values. (CSV 3 MB)
Acknowledgements
We thank Dr. Motohide Aoki, School of Life Sciences, Tokyo University of Pharmacy and Life Sciences, for his kind technical assistance in isolation of Chlorella RNAs. This work was supported by a Grant-in-Aid for Research Activity Start-up (No. 22870023) and by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS), by a grant from the Inoue Foundation for Science (Inoue Research Award for Young Scientists), by a Narishige Zoological Science Award, and by a Grant for Basic Science Research Projects from The Sumitomo Foundation to Y. Kodama. This work was also supported by a Grant-in-Aid for Scientific Research (B) (No. 22370082) and a Grant-in-Aid for Challenging Exploratory Research (No. 23657157) from JSPS to M. Fujishima.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YK and MF cultured paramecia and isolated RNA from paramecia. TK, YK, and KY prepared RNA for RNA-seq. HS and SS conducted the bioinformatics analyses. HD and MS participated in the data analysis and interpretation. YK and HS drafted the manuscript. All authors contributed to editing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yuuki Kodama, Email: kodama@life.shimane-u.ac.jp.
Haruo Suzuki, Email: haruo@yamaguchi-u.ac.jp.
Hideo Dohra, Email: gihdour@ipc.shizuoka.ac.jp.
Manabu Sugii, Email: manabu@yamaguchi-u.ac.jp.
Tatsuya Kitazume, Email: tat.anthrax@gmail.com.
Katsushi Yamaguchi, Email: kyamaguc@nibb.ac.jp.
Shuji Shigenobu, Email: shige@nibb.ac.jp.
Masahiro Fujishima, Email: fujishim@yamaguchi-u.ac.jp.
References
- 1.Fujishima M. Infection and maintenance of Holospora species in Paramecium caudatum. In: Fujishima M, editor. Endosymbionts in Paramecium vol. 12. Heidelberg: Springer Berlin; 2009. pp. 201–225. [Google Scholar]
- 2.Nowack ECM, Melkonian M. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodama Y, Fujishima M. Cycloheximide induces synchronous swelling of perialgal vacuoles enclosing symbiotic Chlorella vulgaris and digestion of the algae in the ciliate Paramecium bursaria. Protist. 2008;159(3):483–494. doi: 10.1016/j.protis.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Gu F, Chen L, Ni B, Zhang X. A comparative study on the electron microscopic enzymo-cytochemistry of Paramecium bursaria from light and dark cultures. Eur J Protistol. 2002;38(3):267–278. doi: 10.1078/0932-4739-00875. [DOI] [Google Scholar]
- 5.Karakashian SJ, Rudzinska MA. Inhibition of lysosomal fusion with symbiont-containing vacuoles in Paramecium bursaria. Exp Cell Res. 1981;131(2):387–393. doi: 10.1016/0014-4827(81)90242-1. [DOI] [PubMed] [Google Scholar]
- 6.Kodama Y, Fujishima M. Localization of perialgal vacuoles beneath the host cell surface is not a prerequisite phenomenon for protection from the host’s lysosomal fusion in the ciliate Paramecium bursaria. Protist. 2009;160(2):319–329. doi: 10.1016/j.protis.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Brown JA, Nielsen PJ. Transfer of photosynthetically produced carbohydrate from endosymbiotic Chlorellae to Paramecium bursaria. J Protozool. 1974;21(4):569–570. doi: 10.1111/j.1550-7408.1974.tb03702.x. [DOI] [PubMed] [Google Scholar]
- 8.Reisser W. The metabolic interactions between Paramecium bursaria Ehrbg. and Chlorella spec. in the Paramecium bursaria-symbiosis. I. Nitrogen and the carbon metabolism (author’s transl) Arch Microbiol. 1976;107(3):357–360. doi: 10.1007/BF00425352. [DOI] [PubMed] [Google Scholar]
- 9.Reisser W. Endosymbiotic Associations of Freshwater Protozoa and Algae. Bristol: Biopress Ltd; 1986. [Google Scholar]
- 10.Albers D, Wiessner W. Nitrogen nutrition of endosymbiotic Chlorella spec. Endocytobio Cell Res. 1985;2:55–64. [Google Scholar]
- 11.Reisser W. The metabolic interactions between Paramecium bursaria Ehrbg. and Chlorella spec. in the Paramecium bursaria-symbiosis. II. Symbiosis-specific properties of the physiology and the cytology of the symbiotic unit and their regulation (author’s transl) Arch Microbiol. 1976;111(1–2):161–170. doi: 10.1007/BF00446564. [DOI] [PubMed] [Google Scholar]
- 12.Karakashian SJ. Growth of Paramecium bursaria as influenced by the presence of algal symbionts. Physiol Zool. 1963;36:52–68. [Google Scholar]
- 13.Pado R. Mutual relation of protozoans and symbiotic algae in Paramaecium bursaria. I. The influence of light on the growth of symbionts. Folia Biol. 1965;13(2):173–182. [PubMed] [Google Scholar]
- 14.Weis DS. Regulation of host and symbiont population size in Paramecium bursaria. Experientia. 1969;25(6):664–666. doi: 10.1007/BF01896584. [DOI] [PubMed] [Google Scholar]
- 15.Kodama Y, Nakahara M, Fujishima M. Symbiotic alga Chlorella vulgaris of the ciliate Paramecium bursaria shows temporary resistance to host lysosomal enzymes during the early infection process. Protoplasma. 2007;230(1–2):61–67. doi: 10.1007/s00709-006-0193-z. [DOI] [PubMed] [Google Scholar]
- 16.Weis DS. The effect of accumulation time of separate cultivation on the frequency of infection of aposymbiotic ciliates by symbiotic algae in Paramecium bursaria. J Protozool. 1984;31:14A. [Google Scholar]
- 17.Reisser W. Die stoffwechselphysiologischen Beziehungen zwischen Paramecium bursaria Ehrbg. und Chlorella spec. in der Paramecium bursaria-Symbiose. I. Der Stickstoff- und der Kohlenstoff-Stoffwechsel. Arch Microbiol. 1976;107(3):357–360. doi: 10.1007/BF00425352. [DOI] [PubMed] [Google Scholar]
- 18.Siegel R, Karakashian SJ. Dissociation and restoration of endocellular symbiosis in Paramecium bursaria. Anat Rec. 1959;134:639. [Google Scholar]
- 19.Kodama Y, Fujishima M. Secondary symbiosis between Paramecium and Chlorella cells. In: Jeon KW, editor. International Review of Cell and Molecular Biology vol. 279. 2010. pp. 33–77. [DOI] [PubMed] [Google Scholar]
- 20.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UniProt_Consortium Update on activities at the universal protein resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33:677–680. doi: 10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 25.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4(9):e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 29.Ueguchi C, Koizumi H, Suzuki T, Mizuno T. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001;42(2):231–235. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- 30.Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262(5133):539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 31.Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117(3):723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurgler-Murphy SM, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22(5):172–176. doi: 10.1016/S0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 33.Hörtnagl PH, Sommaruga R. Photo-oxidative stress in symbiotic and aposymbiotic strains of the ciliate Paramecium bursaria. Photochem Photobiol Sci. 2007;6(8):842–847. doi: 10.1039/b703119j. [DOI] [PubMed] [Google Scholar]
- 34.Summerer M, Sonntag B, Hortnagl P, Sommaruga R. Symbiotic ciliates receive protection against UV damage from their algae: a test with Paramecium bursaria and Chlorella. Protist. 2009;160(2):233–243. doi: 10.1016/j.protis.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita H, Oomori S, Nozaki M, Miwa I. Timing of establishing symbiosis during the re-infection of Chlorella sp. in Paramecium bursaria. Jpn J Protozool. 2009;42:88–89. [Google Scholar]
- 36.Miwa I. Regulation of circadian rhythms of Paramecium bursaria by symbiotic Chlorella species. In: Fujishima M, editor. Endosymbionts in Paramecium vol. 12. Berlin Heidelberg: Springer; 2009. pp. 83–110. [Google Scholar]
- 37.Fujishima M, Kawai M, Yamamoto R. Paramecium caudatum acquires heat-shock resistance in ciliary movement by infection with the endonuclear symbiotic bacterium Holospora obtusa. FEMS Microbiol Lett. 2005;243(1):101–105. doi: 10.1016/j.femsle.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 38.Hori M, Fujii K, Fujishima M. Micronucleus-specific bacterium Holospora elegans irreversibly enhances stress gene expression of the host Paramecium caudatum. J Eukaryot Microbiol. 2008;55(6):515–521. doi: 10.1111/j.1550-7408.2008.00352.x. [DOI] [PubMed] [Google Scholar]
- 39.Hori M, Fujishima M. The endosymbiotic bacterium Holospora obtusa enhances heat-shock gene expression of the host Paramecium caudatum. J Eukaryot Microbiol. 2003;50(4):293–298. doi: 10.1111/j.1550-7408.2003.tb00137.x. [DOI] [PubMed] [Google Scholar]
- 40.Smurov AO, Fokin SI. Resistance of Paramecium caudatum infected with endonuclear bacteria Holospora against salinity impact. Proc Zool Inst RAN. 1998;276:175–178. [Google Scholar]
- 41.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244(22):6049–6055. [PubMed] [Google Scholar]
- 42.Veal EA, Toone WM, Jones N, Morgan BA. Distinct roles for glutathione S-transferases in the oxidative stress response in Schizosaccharomyces pombe. J Biol Chem. 2002;277(38):35523–35531. doi: 10.1074/jbc.M111548200. [DOI] [PubMed] [Google Scholar]
- 43.Dryl S. Antigenic transformation in Paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J Protozool. 1959;6(Suppl):25. [Google Scholar]
- 44.Fujishima M, Nagahara K, Kojima Y. Changes in morphology, buoyant density and protein composition in differentiation from the reproductive short form to the infectious long form of Holospora obtusa, a macronucleus-specific symbiont of the ciliate Paramecium caudatum. Zoolog Sci. 1990;7:849–860. [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Histogram showing the distribution of lengths for protein-coding sequences in P. bursaria (A), P. tetraurelia (B), and T. thermophila (C). The x-axis shows the length (bp) of the sequences. The y-axis shows their frequencies. (PDF 372 KB)
Additional file 2: Histogram showing the distribution of G + C contents for protein-coding sequences in P. bursaria (A), P. tetraurelia (B), and T. thermophila (C). The x-axis shows the G + C content (%) of the sequences. The y-axis shows their frequencies. G + C content is defined as 100 × (G + C)/(A + T + G + C). (PDF 264 KB)
Additional file 3: Data for Paramecium bursaria transcripts. The first five columns show the Trinity sequence name, length (bp), and functional annotations from different databases: Swiss-Prot, UniRef90, and gene ontology (GO). The remaining four columns show data of differential expression analysis using edgeR: log2 Fold Changes (logFC), log-counts-per-million (logCPM), P-values, and False Discovery Rate (FDR) adjusted P-values. (CSV 3 MB)


