Abstract
Mammalian genomes are organized into multiple layers of higher-order chromatin structure, and in this organization chromatin looping is a striking and crucial feature that brings together distal genomic loci into close spatial proximity. Such three-dimensional organization of chromatin has been suggested to be functionally important in gene regulation. Many important questions need to be addressed, such as what types of nuclear proteins are responsible for folding chromatin into loops, whether there are any genomic marks that serve as the core sites of chromatin folding events, how distal genomic sites are brought together, and what are the biological consequences for interactions between distal genomic loci. In order to address these fundamental questions, it is essential to devise and employ methods that can capture higher-order structures formed by specific nuclear proteins at high resolution. In this article, in order to describe methods of analyzing protein-mediated chromatin interactions, we will use as an example a global genome-organizer protein, SATB1, which mediates chromatin looping.
Keywords: Chromatin looping, Chromatin immunoprecipitation, SATB1, ChIP-3C, ChIP-4C
1. Introduction
All the cells in our body contain the same genetic information at the level of the primary DNA sequence, yet we are composed of a great diversity of cell types. It is now well known that the activation or repression of specific sets of genes by different combinations of transcription factors plays a crucial role in diverse cellular processes. Epigenetic modifications such as DNA methylation and post-translational histone modifications are another essential component in the control of gene expression. In addition, the three-dimensional organization of chromatin itself (intra- and interchromosomal interactions, or higher-order chromatin organization) is an important component of the epigenome and is now widely accepted as an underlying mechanism for gene regulation [1-13]. This is because chromatin organization influences the accessibility of DNA to gene regulatory components and the way genetic information is utilized.
Chromatin architecture in eukaryotic nuclei is highly dynamic. This finding is in contrast to the classic view that genes are static and that highly mobile, sequence-specific DNA-binding proteins travel to each gene locus to regulate its expression individually. Further, genes from different chromosomes can move within the nucleus to come close together, and, in some cases, assembled with transcription-or other chromatin-factors for transcriptional co-regulation. To understand the function of the mammalian genome from the viewpoint of chromatin architecture, it is important to understand its contemporary concepts as well as the technologies and experimental controls used in chromatin structure analysis. When studying chromatin interaction with specific transcription factors and chromatin looping formation mediated by nuclear proteins in vivo, the quality and reproducibility of the data depend heavily on the way cells are cross-linked, the abundance of factors in the cells used, how they bind chromatin, and the quality of the antibodies used. Due to these many variables, experimentalists need to carefully optimize methods for their biological system of interest. This chapter introduces basic methods of chromatin structure analysis and offers important tips to facilitate studies of chromatin structure.
We will use the genome organizer SATB1 [14, 15] as our example for describing known methods to study chromatin interactions mediated by a nuclear protein, such as chromatin conformation capture (3C) [16] and a combination of chromatin immunoprecipitation (ChIP) and 3C (ChIP-3C) [2, 4]. We will also describe a new method for identifying the interaction of one genomic site of interest with all other genomic sequences genome-wide (ChIP one-to-all, or ChIP-4C). SATB1 functions as a genome organizer by providing a nuclear architectural platform that anchors hundreds of gene loci to regulate their expression, thereby enabling cells to undergo large-scale reprogramming to change their phenotypes[3, 10]. SATB1 has been found to be essential for many biological processes, including T cell and epidermal differentiation, T cell activation, breast cancer progression[2, 15, 17-26] .
The SATB1-target sequences serve as the base of chromatin loops through their association with SATB1. In addition, other genomic regions that contain regulatory sequences can also recruited to the core base[2]. SATB1 binding sequences are characterized, not by primary DNA sequence, but by a specialized DNA context (an ATC sequence context). This sequence context contains a cluster of long stretches of DNA sequences (ATC sequences, typically 25-40bp per stretch) that consists of well mixed A, T, and C -- without G -- on one strand (the other strand consist of A, T, and G without C). The ATC sequence context confers a strong base-unpairing activity when placed under negative-superhelical strain[27, 28]. Therefore, we call these sequences a BUR, or base unpairing region. Mutations that disrupt the ATC sequence context result in loss of unpairing potential. SATB1 was originally cloned by virtue of its high affinity and specificity to BURs when they are forming the double-stranded conformation[14]. SATB1 recognizes the altered phosphate backbone structure of double-stranded BURs[14]. BUR sequences can be detected by the chemical probe chloroacetaldehyde or bromoacetaldehyde, and the method for mapping unpaired bases of BURs at the level of a single-base resolution has been described in detail[29]. BURs are distributed throughout the mouse and human genomes. When one searches for BURs in any given gene locus, several BURs can typically be found when one examines its introns as well as 20kb 3’ and 5’ of the gene locus. When SATB1 regulates gene expression, it is the BURs in these target gene loci that are tethered to the SATB1 nuclear architecture so that these genes are assembled with chromatin remodeling/modifiying enzymes and transcription factors. The SATB1 anchoring BURs folds chromatin into loops, and therefore it is important to map BUR sites and design primers near these sites to examine the chromatin looping events. BURs are not only targeted by SATB1. Several proteins have been identified to display high affinity and specificity to BURs and new BUR-binding proteins are being identified from embryonic stem cells in our laboratory[30-32] (unpublished results). Therefore, BURs may be more general genomic marks interacting with proteins beyond SATB1, depending on the cell type and the status of cells.
We will cover those basic procedures with regard to chromatin cross-linking, purification of cross-linked chromatin, and chromatin immunoprecipitation (ChIP) that are necessary to yield the best results for ChIP-3C and ChIP-4C.
2. Genome-wide mapping of all potential SATB1-binding sites in vitro
Before describing methods to study SATB1-mediated chromatin conformation, it is essential to know beforehand where the SATB1- target sequences are located within the genomic DNA region of interest. This information is critical in designing primers for a ChIP-3C assay or in deciding on a bait sequence for the ChIP-4C assay. Below, we will briefly summarize a method that can be used for genome-wide mapping of BURs in the mouse or human genome, using a recombinant Satb1 (mouse) or SATB1 (human) protein, respectively. (Note: SATB1 and Satb1 proteins are 98% homologous at the amino acid level). We outline the method below for mapping BURs in the mouse genome. The primary advantage to having the genome-wide in vitro SATB1-binding data is that it provides the baseline profile of all potential binding sites. Depending on the cell type, SATB1 binds to a specific subgroup of these sites[2, 18, 24]. This method can be potentially applied to map the binding sites of other DNA-binding proteins whose target sequences are not represented by well-defined primary sequence consensus.
2.1 Methods for mapping of BURs
Genomic DNA is purified from either human fibroblasts or from mouse thymocytes by standard phenol/chloroform extraction and the ethanol precipitation method. Alternatively, urea ultracentrifugation, described below, can be used for purifying genomic DNA (note: in this case, uncrosslinked cells must be used). Genomic DNA can be fragmented for this purpose by either restriction enzyme digestion or sonication. Here, we describe only the sonication method. We use a BioRuptor (UCD-200, Diagenode) for sonication to generate DNA fragments between 300 and 600 bp (30 sec ON, 30 sec OFF, for 30-40 cycles at low power level).
For His-tagged Satb1recombinant protein preparation and quality control:
Prepare His-tagged full-length Satb1 expression vector by inserting mouse Satb1 cDNA into the 6xHis tag vector, pLICTrPC-HA[33] and transformed into Rosetta2 (DE3) pLysS competent cells (Novagen, #71403). (Note: Human SATB1 recombinant protein is prepared in the similar manner using human SATB1 cDNA). The protein is induced by a standard method using 1 mM IPTG at 30°C overnight. The bacterial cells are suspended in Lysis Buffer (25 mM HEPES [pH 7.5], 400 mM NaCl, 10% Glycerol, 20 mM Imidazole, 0.05% Tween-20, 1 mM PMSF, 1 μg/ml Leupeptin, 1 μg/ml Pepstatin) containing 20 mM imidazole, followed by sonication on a Sonifier 150 (Branson) at output power of 10 W for 10 min on ice.
The recombinant His-Satb1 proteins are purified with MagneHis™ Ni-particle (Promega, V8565) and eluted in Lysis Buffer with 400 mM imidazole, after washing three times with buffer containing 100 mM imidazole.
The eluate is dialyzed against Buffer D (10 mM HEPES-NaOH, pH 7.9, 50 mM KCl, 2.5 mM MgCl2, 1 mM DTT) at 4°C overnight, then the purified Satb1 solution is stored in small aliquots at −80°C. Repeated freezing and thawing must be avoided.
For the following assay, one should use a His-tagged Satb1 protein preparation that is higher than 90% purity, which we assess by SDS-PAGE followed by Coomassie staining (Fig. 1). Furthermore, the protein preparation should show high affinity to a positive control BUR, wild-type (25)7 DNA probe [5’-TCTTTAATTTCTAATATATTTAGAA-3’]7, by gel-mobility shift assay (0.1-1 nM Kd)[34]. After meeting the quality control, we use His-Satb1 proteins at 5 nM in the binding assay to capture most of the BURs.
Figure 1.
Purification of recombinant Satb1 protein. Purified full-length his-tagged Satb1 protein is visualized on a SDS-PAGE after staining with Coomassie Blue.
For Satb1 binding to DNA fragments:
Prepare 100 μl of a reaction mixture (designated as Reaction-A mix) containing 5 nM His-Satb1 protein, 10 μg genomic DNA, 1 mg/ml of freshly prepared BSA solution, and 10 μg/ml Poly dI-dC in Buffer D, and incubate for 25 min at 25 °C.
Add 11 μl of 1 M HEPES-NaOH (pH 7.9) and 5.5 μl of 2 M imidazole to Reaction-A mix, resulting in a final concentration of 100 mM HEPES, and 100 mM imidazole. This concentration of imidazole is higher than the typical protocol for capturing His-tagged proteins (10 mM). We found 100 mM imidazole to sufficiently reduce background in His- Satb1preparations.
MagneHis beads (10 μl/sample) are pre-blocked for 1 hr at 25 °C in 1 ml of Binding Buffer (Buffer D containing 100 mM HEPES-NaOH [pH 7.9], 100 mM imidazole [pH 8.0], 1 mg/ml BSA, and 0.4 mg/ml salmon sperm DNA).
His-Satb1-DNA complexes are captured by incubation with the pre-blocked MagneHis beads for 5 min at 25 °C. The beads are then washed three times with Binding Buffer.
His-Satb1-DNA complexes are eluted from the beads by resuspension in 50 μl of Elution Buffer (100 mM HEPES-NaOH [pH 7.9], 500 mM imidazole [pH 8.0], 10 mM EDTA, 1% SDS, and 0.1 mg/ml proteinase K), and incubating for 5 min at 25 °C. The elution is repeated for a total of two elutions (100 μl total). The eluate is then transferred into a new tube and incubated at 45 °C for 1 hour to digest proteins.
The DNA fragments are purified (QIAquick PCR purification Kit (Qiagen, #28106,) or ethanol precipitation is sufficient). Prior to library construction, the purified DNA should be sonicated again to bring all the fragments to lengths shorter than 500 bps. In this example, 30 cycles of sonication on a Bioruptor (30 sec ON, 30 sec OFF) on the low-power setting is sufficient.
Libraries using up to 75 ng of DNA are made using a modified Illumina library construction protocol (original obtained from Illumina). Briefly, the protocol is as follows: end-repair -> A-tailing -> TruSeq adapter ligation -> size-selection on a Pippin Prep (Sage Science; 150-500 bps range) -> PCR amplification (10 cycles).
Using a HiSeq 2000 sequencer (Illumina), normally we obtain 20-50 million reads per sample. The resulting sequence data are aligned to the mouse (mm9) reference genome using Bowtie (0.12.7) [35]. Peaks are called using MACS (1.4.0)[36] with default settings. In the cases where we use enzymatic-digestion of chromatin, we specify the --keep-dup option so as not to remove duplicate reads.
2.2. Results of BUR mapping
As outlined above, naked genomic DNA fragments were reconstituted with in vitro synthesized His-Satb1 protein, and those bound to His-Satb1 protein were isolated and sequenced. These DNA sequences, which we referred to as Satb1 in vitro binding sequences (SATB1-IVB) represent all possible Satb1-binding sites in the entire genome. Distribution of DNA captured by the Satb1-binding assay in a region of mouse chromosome 2 is shown in Figure 2, top (upper panel represents Satb1-IVB-seq, and lower panel represents input DNA-seq). The DNA sequence at one of the peaks (red star on the top panel) represents a Satb1 binding site in vitro and shows a typical ATC sequence context in Figure 2, bottom. Other peaks also show similar ATC sequence context (data not shown). Therefore, large peaks in the Satb1-IVB distribution profile represent BURs of high affinity to Satb1..
Figure 2.
A Snapshot of BURs captured by high-throughput sequencing in a region on chromosome 2. (Top) UCSC Genome Browser tracks showing distribution of Satb1-binding sequences, BURs (top track, Satb1 in vitro binding; bottom track, input control). (Bottom) DNA sequence from the starred region from the top panel. DNA sequence in blue represents ATC sequence stretches, and DNA sequences in red represent ATG sequence stretches. The colored sequences represent the ATC sequence context, typically found in BURs.
3. Chromatin preparation prior to chromatin conformation capture
3.1 Formaldehyde Cross-linking of Chromatin and Purification
Formaldehyde is typically used for cross-linking chromatin at a final concentration of 1%. However, it is important to use formaldehyde that does not contain methanol [e.g., Thermo Scientific (Pierce), #28906, Rockford, IL]. Methanol is added as a stabilizer because it slows down the formation of paraformaldehyde (formaldehyde polymer), though methanol's ability to precipitate proteins is to be avoided in this application. To study in vivo binding of histones with a specific epigenomic mark to genomic DNA, one could find published conditions that work without the need for further optimization [37]. On the other hand, in order to obtain the best results for transcription factors that are expressed at much lower levels than histones, it is important to test a series of cross-linking conditions using the cells that express the factor, by varying temperature and time of incubation of the cells with formaldehyde. In the case of SATB1, we crosslinke with 1% formaldehyde in either a 37°C water bath for 4 min in a 50 ml tube or we place the cells in a 37°C dry incubator for 10 min with rocking/agitation followed by an additional incubation at 4°C for 2 hrs. In case of isolating cells from brain tissue before cross-linking, the brain is first cut into small pieces, homogenized, filtered through a 100 μm cell strainer, suspended in PBS at 25 °C, then filtered a second time through a 70 μm cell strainer to make a single-cell suspension. Note: if formaldehyde cross-linking proves to be insufficient for particular proteins or cells/tissues, one should consider of a combination with another cross-linking reagent, such as 1.5 mM EGS (ethylene glycol-bis[succinic acid] N-hydroxysuccinimide ester) (Thermo Scientific) at 25°C for 20 minutes prior to the 1% formaldehyde treatment as describe above[38].
3.2. Purification of formaldehyde cross-linked chromatin
We have been using a urea-gradient (5-8 M) ultracentrifugation method to purify formaldehyde cross-linked chromatin [39]. However, ultracentrifugation through 8 M urea works just as well as a 5-8 M urea gradient. Therefore, in the remainder of the protocol, we state 8 M urea purification. Formaldehyde cross-linked cells are lysed in 4% SDS (in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1 mM EGTA), and the lysates are loaded on top of 8 M urea in TE (10 mM Tris, pH 8.0, 1 mM EDTA), followed by centrifugation at 30,000 rpm for 16 hrs in a Beckman SW41 rotor (Beckman Coulter, Fullerton, CA). A high concentration of SDS (4%) is necessary to dissolve all formaldehyde cross-linked cells to separate chromatin without mechanical agitation before crosslinked chromatin is purified through 8 M urea. This urea purification method removes all proteins that are not cross-linked to chromatin, leaving on the bottom of the tube a pellet of pure, unsheared, genomic DNA with crosslinked proteins. Therefore, this method cleanly removes non-cross-linked proteins, RNAs, and other cellular debris. ChIP experiments using urea purified cross-linked chromatin as the template have been shown to have a much improved signal-to-noise ratio (2, 3, 4). Without the urea purification step, at least for Satb1 ChIP-3C experiments performed for thymocytes expressing a very large quantity of Satb1, we could not detect any specific chromatin looping events with 3C or ChIP-3C. Also, with this method, it is possible to generate high-quality data with a reduced background for 3C.
The cross-linked chromatin purified by urea ultracentrifugation can be subjected to water-bath sonication (e.g., Bioruptor) or to digestion with restriction enzymes. The pellet formed by urea centrifugation is pure cross-linked chromatin and resembles a transparent soft contact lens. To facilitate solubility of chromatin, this “contact lens” is carefully removed from the bottom of the centrifuge tube and crushed with a Pellet Pestle (Thomas Scientific, #7495211590, Swedesboro, NJ), or disrupted by passing it through an insulin syringe and dissolving (practically, suspending) it in an appropriate buffer for the restriction enzyme to be used. Alternatively, if one chooses sonication to cleave the crosslinked chromatin, the crushed/disrupted “contact lens” is suspended and solubilized in sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 7.5) before sonication. DNA that is either isolated from restriction enzyme-digested cross-linked chromatin or cleaved by sonication must be analyzed by running a reverse-crosslinked aliquot on an agarose gel or DNA Bioanalyzer to confirm the extent of digestion or shearing. Only samples achieving the target fragmentation size range (e.g., 200-500 bps) are considered ready to be processed for further experiments.
Note: If crosslinked cells do not get solubilized completely, the pellet will be opaque (contaminated with cell debris) and the experiment will fail. It is important that the cell suspesion in SDS becomes completely clear before loading onto an 8 M urea centrifugation tube. The purification of cross-linked chromatin through urea-gradient centrifugation, once it is successfully suspended in solution, permits more efficient and quantitative digestion with restriction enzymes (24). This is important because the pattern of ligation products for 3C and ChIP-3C remains unchanged even after an additional 10-20 cycles of PCR amplification.
3.3. Chromatin immunoprecipitation
Before using any antibody for a ChIP experiment, it is essential to confirm that the antibody can immunoprecipitate its antigen efficiently. This property can be tested by immunoprecipitation of the antigen from cell or nuclear extracts using different concentrations of the antibody, followed by Western blot analysis (on immunoprecipitated sample and the soluble fraction). This experiment will determine the optimal level of antibody to use for ChIP. Two important points are that sufficient amounts of initial chromatin samples should be used for immunopr ecipitation, and that control experiments should succeed[39]. For a transcription factor, it is best to use cross-linked chromatin, containing a minimum of 20-40 micrograms of DNA, for immunoprecipitation. For a rabbit antiserum, it is best to use the pre-immune serum as a control from the same rabbit before immunization. If this is not available, one has to obtain nonimmune serum that does not generate nonspecific signals.
Before performing chromatin immunoprecipitation, formaldehyde cross-linked chromatin samples will be precleared twice with either preimmune or nonimmune serum by incubation with protein A-Sepharose 4B beads (Pharmacia, Uppsala, Sweden) or Protein A Dynabeads (Invitrogen, Carlsbad, CA).Precleared samples will be equally divided, to be immunoprecipitated by either antibody or control serum. For an experiment to succeed, using 1/100th of the immunoprecipitated samples obtained from the cross-linked chromatin as template per PCR sample, the control (immunoprecipitated with nonimmune serum) should not give any positive PCR signals after 30-45 cycles of PCR amplification, whereas the ChIP samples (immunoprecipitated with specific antibody) should give strong PCR signals by the 28th PCR cycle (when analyzed on an agarose gel). An example of using this approach for determining in vivo Satb1-binding sites in G10.G4.1 cells (a T-helper-2 [TH2] cell line) within a 200 kb TH2 cytokine cluster region is shown in Figure 3a and 3b [2]. With ChIP followed by quantitative PCR (ChIP-qPCR), it is possible to determine relative fold-difference in quantity of target sequence in immunoprecipitated (IP) DNA using input DNA as a common reference for each primer set. For this purpose, IP DNA and input DNA, after reverse cross-linking, are quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA) or Qubit Quantitation Platform (Invitrogen), followed by real-time PCR on IP DNA. An absolute quantification method determines the amount of the DNA fragments of interest in the target genomic locus in IP DNA using the standard curve generated by real-time PCR for each primer pair from input DNA[40]. We determine the ratio R, which means enrichment of ChIP DNA. R = (moles of target sequence in IP fraction/moles of total IP DNA) / (moles of target sequence in input DNA/moles of total input DNA). The denominator is <1> if the target sequence is unique. The data we obtained using this formula are shown in Figure 3c[2].
Figure 3.
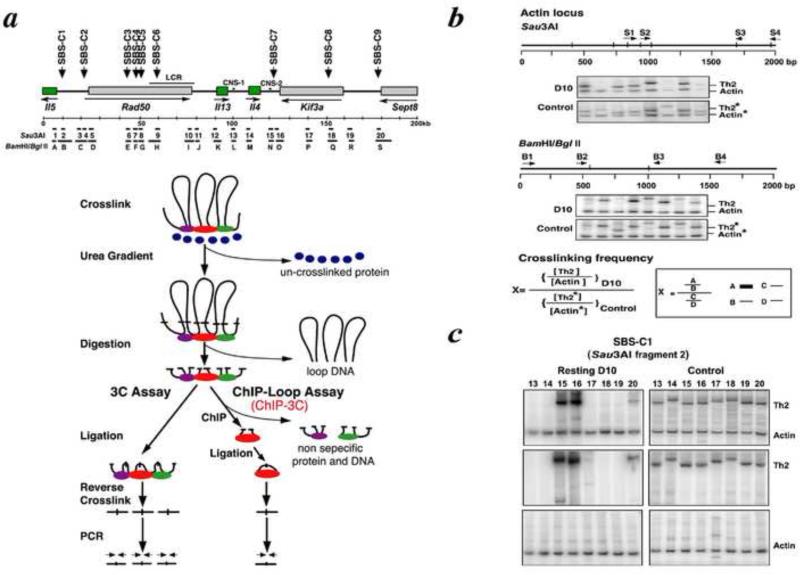
ChIP analysis for Satb1 binding sites in the TH2 cytokine locus. This analysis revealed nine Satb1-binding sequences within the 200 kb TH2 locus (SBS-C1-C9). In addition, two other regulatory sequences (CNS-1 and CNS-2) were bound to Satb1. a, Schematic representation of the TH2 cytokine locus; b, Semiquantitative PCR; and c, Real-time PCR analyses of cross-linked chromatin from resting and activated D10.G4.1 cells purified by urea-gradient centrifugation, digested with Sau3AI, and precipitated with anti-Satb1.
4. Chromatin Conformation Capture (3C) and ChIP-3C assays
4.1. ChIP-3C method
Formaldehyde cross-linking of cells can capture chromatin loops that form in vivo. If remote DNA sequences are brought into close physical proximity by chromatin looping in vivo, they can be found in the same cross-linked chromatin fragment, and the DNA sequences at the stem of chromatin loops can be identified. As shown in Fig. 4a, after digesting cross-linked chromatin with a restriction enzyme that removes the loop portion of chromatin, the two sequences initially located at the stem of a loop can then be ligated intramolecularly, after diluting the chromatin sample to avoid intermolecular ligation events. After reverse cross-linking (to remove proteins), purified genomic DNA that had been intramolecularly ligated (excluding the loop portion of DNA) can be amplified by PCR, using primer pairs designed from the two distal sequences of interest. The ligation products detected by PCR indicate that physical interactions take place between DNA fragments held together by higher-order chromatin structure. The 3C method [16] enables us to estimate the frequency with which two remote genomic sequences interact in space in a given nucleus.
Figgure 4.
3C and ChIP-3C assays to determine higher-order chromatin structure. These assays were applied to determine intrachromosomal interactions at the TH2 cytokine locus upon activation. a, The experimental strategies of ChIP-3C and 3C assays. b, Analysis of 3C and ChIP-3C data. c, Verification that premixing primers from the TH2 cytokine gene locus and the actin locus does not affect the final results of the 3C assay.
The more recently devised ChIP-loop assay (or ChIP-3C)[4] is a modified 3C analysis. ChIP-3C studies chromatin fragments that are immunoprecipitated with an antibody specific for a chromatin-associated protein of interest. The difference between 3C and ChIP-3C is that, whereas whole crosslinked chromatin fragments are used as templates for the 3C assay, the ChIP-3C assay uses immunoprecipitated cross-linked chromatin fragments as templates. Although both assays are used to study long-range DNA interactions that might occur between genomic sites within the same chromosome or between different chromosomes, the ChIP-3C assay allows us to examine a specific group of chromatin loops that are fastened at their base through a specific protein. It also allows for the study of chromatin loops that have histone modifications associated with either transcriptionally active or silent chromatin, depending on antibodies used against specific modifications of histone that are correlated with the transcriptional status of chromatin [4].
As Fig. 4a illustrates, for 3C, experiments include the following steps:
in vivo cross-linking
Urea-gradient ultracentrifugation
Digestion of purified cross-linked chromatin with a restriction enzyme
Ligation of digested cross-linked chromatin overnight at 16 °C using T4 DNA ligase
Reversal of cross-linking
PCR amplification: multiple primers are designed along the regions of interest.
For ChIP-3C, experiments include the following seven steps, as for typical ChIP experiments:
in vivo cross-linking
Urea-gradient ultracentrifugation
Digestion of purified cross-linked chromatin with a restriction enzyme
Ligation of digested cross-linked chromatin overnight at 16 °C using T4 DNA ligase (Note: Alternatively, the ligation step can be done after IP-step 5 without affecting the final results: ChIP-DNA on the beads can be suspended in ligation buffer for ligation)
Immunoprecipitation (note: pre-cleaning of the chromatin is necessary prior to IP).
Reversal of cross-linking
PCR amplification: Multiple primers are designed along regions of interest[2].
We examined the 200 kb murine T helper-2 (TH2) cytokine gene cluster locus (TH2 locus), shown in Fig. 4a. In this case, we used Sau3A restriction enzyme-digested fragments (1–20) as well as BamHI/BglII restriction enzyme-digested fragments (A–S), which were used for the purpose of verification. Two primers (forward and backward) in each of DNA fragments 2–19 were designed, as well as a forward primer in DNA fragment 1 and a backward primer in DNA fragment 2. The primers were designed to test the looping events through Satb1-binding sequences (SBS) and control nonbinding sequences. The genomic DNA prepared for either ChIP-3C or 3C assaying, as described above, was subjected to PCR amplification using various combinations of forward and backward primer pairs derived from the DNA fragments (1–20 or A–S). These PCR products were derived from the 200 kb TH2 cytokine locus using genomic DNA and designated TH2.
Note: The sequences need to be confirmed for all the ligation products by cloning the PCR products, followed by DNA sequencing of the insert. When two fragments of interest are very closely located in a linear configuration, some PCR products are inevitably generated. Therefore, we set up a threshold for looping that two sequences have to be >4 kb apart to assess independent looping events within the genome[2].
4.2. Controls and data analysis for ChIP-3C
The ligation-mediated crosslinking frequencies of any two DNA fragments need to be determined based on the intensity of the PCR signals of a ligation product. Their frequency depends on the relative proximity of the two DNA fragments to each other at a given time-point as well as on various parameters, such as PCR amplification efficiency, enzyme digestion and ligation-mediated crosslinking frequency, and the amount of the template initially used. In order to normalize such factors, relative ligation-mediated crosslinking frequencies of any two DNA fragments must be calculated as described below.
Quantitation of PCR amplification: to calculate and correct the above parameters, a quantitative PCR is performed. The PCR products are labeled by including 0.1 μl [α-32P] dATP (10 mCi/ml) and 0.1 μl [α-32P] dCTP (10 mCi/ml) in each reaction, resolved with 6% PAGE, and quantified using stochastic optical reconstruction microscopy (STORM) phosphoimager and ImageQuant software (GE Healthcare, Uppsala, Sweden).
Correction of genomic DNA amounts: for analysis on the TH2 locus, the β-actin locus is used as an internal control to normalize any difference in the amounts of genomic DNA. First, set up primer pair S2/S3 for Sau3A digestion or B2/B3 for BamH1/BglII digestion in b-actin locus (Fig. 4b). Upon either Sau3A or BamHI/BglII digestion and ligation, a PCR is performed, and the product of 149 bp or 131 bp is generated, respectively, labeled as Actin. To normalize the amount of genomic DNA used for the ChIP-loop assay, we add a known amount of plasmid DNA containing the b-actin locus in the original 3C or ChIP-3C DNA samples as an internal control and perform PCR amplification. Note: Sau3A sites are sometimes methylated and not always 100% digestible. Therefore, BamH1/BglII double digestions may be necessary to verify the DNA digestion efficiency.
Correction of ligation-mediated crosslinking frequency and PCR-amplification efficiency of different primers: in addition to the b-actin locus containing plasmid DNA, set up a control DNA target prepared from two BAC (bacteria artificial chromosome) clones covering the 200 kb TH2 cytokine locus. The BAC clone control DNA is prepared by digesting BAC clones with a restriction enzyme, either Sau3A or BamHI/BglII, and re-ligated so that all possible ligation products are present in the sample. These control templates (from β-actin locus and digested and re-ligated BAC clones) are subjected to PCR amplification with the same series of primer pair combinations from different DNA fragments.
Calculation of ligation frequency: Relative ligation-mediated crosslinking frequency is calculated using the formula shown in Figure 4b. The PCR products derived from 3C or ChIP-3C templates prepared from D10.G4.1 cells (abbreviated as D10) are indicated as TH2 or Actin. The PCR products derived from the BAC clones and Actin plasmid DNA are indicated by TH2* and Actin*, respectively (Fig. 4b). We confirmed that premixing actin-plasmid and BAC clone-TH2 in the sample had no effects on the final PCR products (Fig. 4c). For the ChIP-3C assay, relative ligation-mediated crosslinking frequency is calculated in a similar manner as for the 3C assay, and we use 20 ng of DNA for the 3C assay and ~1 ng of IP DNA for the ChIP-3C assay.
Actual data and the relative ligation-mediated crosslinking frequencies are shown for two Satb1-binding sequences (SBS-C1 and SBS-1C) in Figure 5[2].
Figure 5.
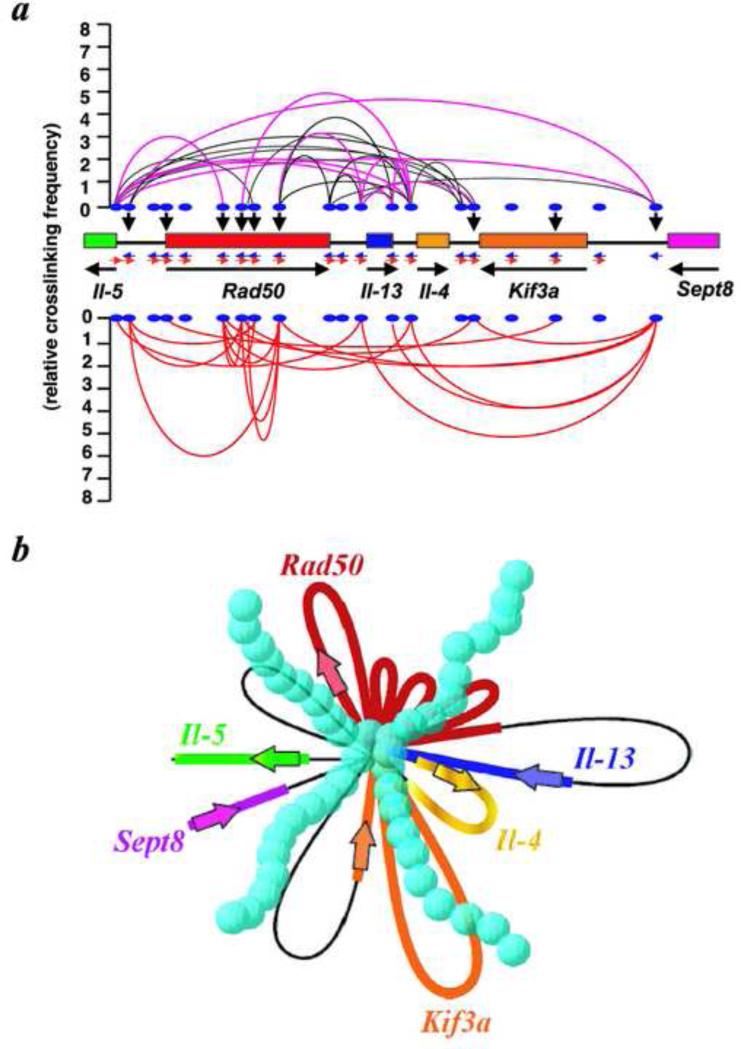
Chromatin looping involving SBS-C1 and SBS-C2 before and after activation of TH2 cells. a, Schematic representation of TH2 cytokine locus and positions of 20 Sau3AI DNA fragments used in the ChIP assay are shown. Black lines indicate chromatin looping detected in resting cells. Pink lines indicate DNA fragments showing enhanced interactions upon cell activation. Red lines indicate positions brought in close proximity after activation. Relative cross-linking frequencies (blue bars in the histograms) are shown between fragment 2 (SBS-C1) as a fixed reference point (red bar in the histogram) or b, fragment 20 (SBS-C9) as a fixed reference point and c, the other fragments of the locus. The experiments are as described in Figure 2a, b, and c.
4.3 Application of ChIP-3C; Satb1- and MeCP2- mediated chromatin looping
By performing the ChIP and ChIP-3C assays, we determined that, upon TH2 cell activation, Satb1 is rapidly induced and forms a unique transcriptionally active chromatin conformation at the 200 kb TH2 cytokine locus on mouse chromosome 11 [2]. The 200kb locus may consist of chromatin folded into many small loops, all anchored to Satb1 at their base, assuming that all looping events occur in a single cell. Even if individual cells did not have all the detected looping events, it still remains true that a large number of new looping forms after activation. Furthermore, transcription factors (GATA3, STAT6, c-Maf), chromatin-remodeling enzyme Brg1, and RNA polymerase I are all bound across the 200 kb region. By knocking down Satb1 using RNA interference, the dense chromatin looping at the TH2 locus is inhibited, and Il-4, Il-5, I1-13, and c-Maf are not induced upon activation. These findings indicate that Satb1 is required for TH2 activation through the establishment of specific transcriptionally competent chromatin architecture at this locus (Fig. 6).
Figure 6.
A three-dimensional, transcriptionally-active complex containing dense looping. a, Summary of 3C and ChIP-3C assays of the cytokine regions in resting and activated D10.G4.1 cells. Black lines connecting two positions indicate juxtaposition of these sites in resting cells. Pink lines represent two positions that show much increased cross-linking frequencies after activation. Red lines represent two positions that are newly brought into close proximity upon activation. Black vertical arrowheads show direct Satb1-binding sites. The cross-linking frequency for each ligation product generated between any two positions is represented by the peak of the parabola connecting the positions. b, A schematic diagram based on the looping events shown in a, assuming that all looping events can occur in a single cell. In this model, all small loops converge onto a common core base bound to Satb1 (blue spheres). As a consequence, the total physical volume of the active transcriptional complex is reduced, enhancing the accessibility of factors to genomic sites.
The advantage of the ChIP-3C assay is enabling us to not only identify chromatin looping fastened at their base with a specific protein of interest, but also to assign chromatin looping at either transcriptionally-silent or active chromatin. This can be achieved by the use of antibodies that are specific for histone modifications well correlated to either active or silent chromatin. We applied the ChIP-3C assay (which we originally called the ‘ChIP-loop’ assay) to study chromatin looping mediated by methyl CpG-binding protein 2 (MeCP2), whose mutations cause RETT syndrome (RTT), an X-linked mental disorder. With the use of antibodies against MeCP2, and two histone H3 modifications (H3K9/14ac and H3K9me2, for transcriptionally-active and silent-chromatin, respectively), MeCP2 was found to form a transcriptionally-silent loop at one of its target loci, Dlx5/6. In RTT model mouse brains which lack MeCP2, the MeCP2-mediated transcriptionally-silent chromatin loop configuration was absent and a different transcriptionally-active chromatin loop was present, linking a defect in chromatin looping to RTT[4].
5. ChIP-One-to-All (ChIP-4C)
In contrast to the ChIP-3C assay, in which one examines the interaction of any two genomic sequences of choice mediated by a protein, ChIP-one-to-all identifies interacting partners genome-wide for a single, specific DNA sequence of choice (as a bait). Several methods for capturing interacting partners from a single bait locus have been published and applied[41-45]. ChIP-one-to-all involves high-throughput sequencing, and the protocol needs to be carefully devised in order to reduce associated background problems. We devised a ChIP-one-to-all assay (we call ChIP-4C), which is designed to avoid false positive signals, so that all captured sequences by high-throughout sequencing represent the interaction partners of the bait sequence of choice. Below, we will describe an experimental approach that can be used to determine the interaction partners of a BUR of choice as a bait (Fig. 7a), in thymocytes (4C), and in Satb1-mediated chromatin interactions involving this locus (ChIP-4C).
Figure 7.
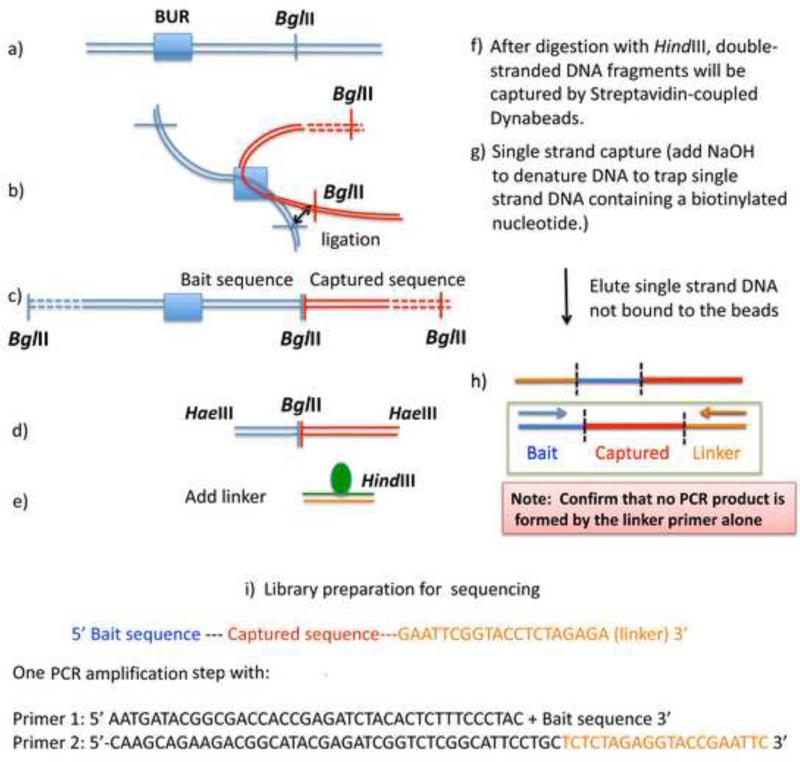
A schematic diagram of the 4C protocol. For ChIP-4C, after digestion of crosslinked chromatin with BglII, one could perform immunoprecipitation either before or after the ligation step in b). See text for details.
5.1 4C and Satb1 ChIP-4C methods
For the 4C assay, one should start with ≥1×107 cells. Formaldehyde-crosslinked chromatin, after purification through urea ultra-centrifugation as described above, is first digested overnight with BglII (NEB) at 1U/μg DNA. Subsequently, the chromatin is ligated with T4 ligase (NEB) at 16 °C overnight, which allows ligation of interacting genomic DNA at the BglII sites (Fig. 7b). (Note: BglII cannot be heat inactivated. However, at 16°C, its activity is greatly reduced, allowing ligation to take place successfully). After heat inactivation of ligase activity at 65 °C for 10 min, the chromatin is reverse-crosslinked (RNase A, Proteinase-K, at 65 °C for 6 hrs), and DNA is purified by phenol-chloroform extraction followed by ethanol precipitation (Fig. 7c). The ligated DNA fragments are further digested with the frequent cutter HaeIII (NEB) (1U/μg DNA at 37°C for 2 hr), heat-inactivated, and DNA is purified by phenol-chloroform extraction followed by ethanol precipitation (Fig. 7d). The purified HaeIII-digested DNA fragments are now ligated to a double-stranded linker containing an internal biotin-conjugated thymidine on one strand, and a HindIII site at its end, as shown in Figure 7e. Blunt-end linker ligation produces four types of ligated products depending on the orientation of ligation at both ends (not shown).
These linker-ligated products need to be digested with HindIII in order to avoid the background of 4C signals (see below for more explanation). After HindIII digestion, the double-stranded (ds)DNA fragments are allowed to bind to streptavidin-coupled Dynabeads-C1 (Invitrogen) in buffer (binding and wash buffer: TE, 1M NaCl and 0.01% Tween 20) for 15 min at 25 oC. The Dynabeads are washed three times with binding and wash buffer. The dsDNA fragments containing biotin are coupled on the Dynabeads-C1 (Fig. 7f). This is followed by single-stranded (ss)DNA isolation. dsDNA coupled with the Dynabeads-C1 is made into single-strands by incubating the Dynabeads-C1 in 40 μl of 0.15 M NaOH for 10 min at room temperature (Fig. 7g). Under this condition, only ssDNA containing biotin coupled with streptavidin will be retained on the Dynabeads-C1, releasing those which are not. The released-ssDNA in the supernatant will be collected and neutralized by adding 4.4 μl of 100 mM Tris (pH 7.6) and 2.6 μl of 1.25 M acetic acid. There are two types of ssDNA found in the supernatant (Fig. 7h), one of which has the captured sequences surrounded by the Bait and the Linker sequences.
This ssDNA will serve as the template for amplifying the captured sequences by PCR only when specific primers designed for the Bait and the Linker are present. At this point, it is of the utmost importance to verify that the ssDNA samples in the supernatant do not get amplified by Linker primer alone. If the ligated fragments had not been digested with HindIII, one would get ssDNA that contains the Linker at both ends in an opposite direction. After successive PCR cycles, due to intermolecular hybridization, this will eventually be amplified by Linker primer alone. Since any products that can be amplified by Linker alone could be derived from any BglII fragments, we need to avoid any amplification in the final sample preparation by Linker alone. The PCR experiment with critical primers (Linker alone versus Linker and Bait primers) is the critical quality-control step to obtain specific interaction partners, avoiding any false negatives.
For the ChIP-4C assay, the 4C method is applied to Satb1 ChIP samples described above for ChIP-3C. The yield of ChIP DNA is relatively small (~1% of input DNA for Satb1 ChIP for thymocytes), and the ChIP DNA has to be processed through multiple steps (Fig. 7). Therefore, it is advisable to start with a greater number of cells (a minimum of 4×107 cells) for ChIP-4C. Also, for BglII digestion of crosslinked chromatin, we performed either ligation first, then ChIP, or ChIP first, then ligation. Either method led to similar results. The subsequent reverse crosslinking and procedures thereafter are as described above for the 4C assay.
5.2. Library construction and sequencing
The sequencing libraries were constructed from one PCR amplification step using the primer 1 and 2 (Fig. 7i), partially overlapping with the Illumina paired-end (PE) PCR primer 1 and 2 sequences. We performed single-end sequencing (75 bp read length) on an Illumina GAII sequencer using a custom primer, which partially overlaps with the bait sequence, allowing the remaining 3’ portion of the bait sequence to be read by the sequencer. This is to verify that the sequence read contains the captured sequence juxtaposed to the bait. Unaligned reads were filtered for the aforementioned portion of the bait, and the processed sequence data (after the removal of the bait sequence) were aligned to the mouse reference genome (mm9) with ELAND, Illumina's short-read aligner. Alternatively, processed, unaligned data can be aligned to a reference genome using Bowtie, SOAP, MAQ, or one of the many other available aligners[46]. In our protocol, there is an important concern about designing the sequencing primer to include the partial bait sequence to be read first. The first four cycles of Illumina's sequencing are used for cluster positioning. Therefore, if clusters are too dense, and the libraries all contain the same initial sequence read by the sequencer, the Illumina image analysis software will have difficulties in resolving the clusters. To compensate for this problem, one has to use lower cluster densities to get good cluster positioning and ultimately good sequence data. This can be achieved by simply adding less sample for sequencing (about one half the DNA amount as would typically be used for ChIPseq). We have sequenced more than forty 4C and ChIP-4C libraries and we obtained 2-4 million captured sequences per sample (run on a single lane of the Illumina GAII sequencer) that could be mapped to the genome. We suggest that this is a sufficient number of reads per 4C experiment. This is because we have observed highly reproducible peaks among biological replicas, and even with duplicate sequencing runs generating two or more times the number of aligned reads, the number of resulting peaks remained largely unchanged. This suggests we have approached saturation with this depth of sequencing.
If multiple samples are to be sequenced in the same lane it is advisable to index the samples by creating an additional unique “barcode” positioned 5’ to the bait sequence, so that the barcode and either the partial or entire bait sequence could be read (note, the longer the barcode and bait, the longer the reads should be: with Illumina's latest sequencing chemistry, 100 and 150 bp reads are possible). This will create heterogeneity between adjacent clusters (due to different barcode sequences) during the first four critical sequencing cycles, and thus will increase the probability of a successful read.
Peaks were called with the ChIP-seq module of GenomeStudio (Illumina) and QuEST (release 2.4)[47] using default settings. There is no need for an input control analysis with the 4C method due to the nature of this method - any read that passes filter and contains the bait sequence is a true interaction. Considering the enzymatic digestion-based nature of 4C, we specified QuEST to keep duplicate reads through the -advanced flag (keeping the duplicates is the default in GenomeStudio). Alternatively, there are many different peak-callers to choose from[48].
Since each mapped read represents a link from one loci to another location in the genome, simply calculating the fraction of the mapped locations to all locations provides a snapshot of the frequency of intra- and inter-chromosomal interactions (also described elsewhere as cis and trans interactions, respectively). In our baseline interaction data we observe that >50% of the mapped reads are interacting with locations within the chromosome containing the bait sequence (cis-interaction). We find the ratio between cis- and trans-interactions is dependent on the state of the cells. When cells respond to external signaling, cells can undergo large-scale rewiring of the interactions, shifting from cis to trans interaction (manuscript in preparation: Cai, S., Richards, H.W., Sankararaman, S., Ayers, S., Jordan, M., Cheng, J-F., Kohwi, Y., Kohwi-Shigematsu, T., SATB1 establishes a network of intra-/inter-chromosomal interactions among c-Myc pathway genes).
5.3 Validation
To validate 4C and ChIP-4C data, one could select several interaction sites identified from 4C experiments and individually perform 3C and ChIP-3C experiments along with non-interaction sites as negative controls. In addition, three-dimensional fluorescence in situ hybridization (3D-FISH) can be done using DNA probes that contain the bait and the interacting sequences and score co-localization frequencies of these signals by confocal or Deltavision microscopes[49]. Most importantly, to validate that the interaction is mediated by a protein of interest, one should perform an additional 4C experiment in parallel using cells that lack the protein using knockout cells or at least knockdown cells in which the protein level is greatly reduced.
5.4. 4C and ChIP-4C results
Examples of peak distributions for 4C and Satb1-ChIP-4C, indicating captured partner sequences from a BUR located 5’ of the c-Myc gene in chromosome 15, are shown for biological replicates of thymocytes isolated from two mice (Fig. 8). The examples shown for 4C and ChIP-4C are trans-interactions. In the 4C example used in Fig. 8a, this single major peak contained ~1.6% of all the mapped reads whereas the minor peak, which we selected, contained ~0.015% of all the mapped reads. In the ChIP-4C example (Fig. 8b) the major peak contains ~1.44% of all mapped reads whereas the minor peak contains ~0.0011% of all mapped reads. In both 4C and ChIP-4C, we find approximately 50 such major peaks. The data shows that even low-ranked interactions (minor peaks) can be faithfully replicated with our method. The 4C and ChIP-4C are not trivial methods. It is recommended to repeat the experiments until one gains the technical expertise until the patterns of peaks are highly reproducible among independent biological replicates.
Figure 8.
Peak calls from 4C and ChIP-4C sequencing. Libraries from 4C (a) and ChIP-4C (b) experiments were sequenced, aligned to the mouse genome, and their peak profiles generated. For 4C and ChIP-4C, peaks profiles viewed by UCSC Genome Browser of two biological replicates (sample #1 and #2) are shown by two tracks with different scales; the upper one being scaled to the major peak, and the lower track scaled to reveal minor peaks. One major and one minor peak (red bar and arrows) chosen from the upper panel are viewed at two levels of higher resolution (lower panels of a and b). For each peak, the position of the single BglII site, which was captured and ligated to the BglII site at the bait, is shown. The RefSeq genes are shown in a dense configuration below the tracks of two biological replicates.
6. Concluding remarks
The 4C and ChIP-4C methods described here can generate remarkably clean and consistent data.
We were able to capture from the whole genome a discrete number of genomic loci near genes that specifically interact with a gene locus of our choice (c-Myc). Such points of genomic interactions can be precisely mapped at high resolution without false positives and in a highly reproducible manner. Furthermore, ChIP-4C can determine which of those interactions identified by 4C are mediated by a nuclear protein of interest. Therefore, these methods allow us to address a myriad of questions to understand chromatin structure and function in a cell type-specific and functional context-specific manner. The molecular mechanisms underlying long-distance gene interactions, the molecular/biochemical events that result as the consequence of those interactions, and their final biological output are not well understood. There are likely to be many unexpected and exciting findings. Methodologies for precisely identifying gene-gene interactions will provide the foundation for investigating these questions.
Acknowledgement
This project was supported by National Cancer Institute R37 CA39681 and Low Dose Radiation Research Program, US Department of Energy (DE-AC02-055CH11231). We thank Mr. Minyong Chung for helpful advise on the sequencing strategy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kosak ST, Groudine M. Genes & development. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 2.Cai S, Lee CC, Kohwi-Shigematsu T. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 3.Cai S, Han HJ, Kohwi-Shigematsu T. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 4.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 5.Fraser P. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Spilianakis CG, Flavell RA. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 7.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 8.de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Curr Top Dev Biol. 2008;82:117–139. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- 9.Wallace JA, Felsenfeld G. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 11.Bantignies F, Cavalli G. Trends Genet. 2011;27:454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misteli T. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. Genes & development. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 17.Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, Kobayashi S, Costa DB, Schulz T, O'Brien KB, Verhaak RG, Delwel R, Haase D, Trumper L, Krauter J, Kohwi-Shigematsu T, Griesinger F, Tenen DG. J Clin Invest. 2007;117:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 19.Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, Grosschedl R. Genes & development. 2009;23:2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A. Dev Cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S. PLoS Biol. 2010;8:e1000296. doi: 10.1371/journal.pbio.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, Cheng C, Zhu S, Yang Y, Zheng L, Wang G, Shu X, Wu K, Liu K, Tong Q. Oncol Rep. 2010;24:981–987. doi: 10.3892/or.2010.981. [DOI] [PubMed] [Google Scholar]
- 23.Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu S, Liu K, Wu K, Tong Q. APMIS. 2010;118:855–863. doi: 10.1111/j.1600-0463.2010.02673.x. [DOI] [PubMed] [Google Scholar]
- 24.Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G, Kohwi Y, Missero C, Kohwi-Shigematsu T, Botchkarev VA. J Cell Biol. 2011;194:825–839. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Takahara M, Oba J, Xie L, Chiba T, Takeuchi S, Tu Y, Nakahara T, Uchi H, Moroi Y, Furue M. J Dermatol Sci. 2011;64:39–44. doi: 10.1016/j.jdermsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Balamotis MA, Tamberg N, Woo YJ, Li J, Davy B, Kohwi-Shigematsu T, Kohwi Y. Mol Cell Biol. 2012;32:333–347. doi: 10.1128/MCB.05917-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohwi-Shigematsu T, Kohwi Y. Biochemistry. 1990;29:9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- 28.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 29.Kohwi-Shigematsu T, Kohwi Y. Methods Enzymol. 1992;212:155–180. doi: 10.1016/0076-6879(92)12011-e. [DOI] [PubMed] [Google Scholar]
- 30.Dickinson LA, Kohwi-Shigematsu T. Molecular and cellular biology. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu WM, Guerra-Vladusic FK, Kurakata S, Lupu R, Kohwi-Shigematsu T. Cancer research. 1999;59:5695–5703. [PubMed] [Google Scholar]
- 32.Galande S, Kohwi-Shigematsu T. The Journal of biological chemistry. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 33.Doyle SA. Methods Mol Biol. 2005;310:115–121. doi: 10.1007/978-1-59259-948-6_8. [DOI] [PubMed] [Google Scholar]
- 34.de Belle I, Cai S, Kohwi-Shigematsu T. The Journal of cell biology. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milne TA, Zhao K, Hess JL. Methods Mol Biol. 2009;538:409–423. doi: 10.1007/978-1-59745-418-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. Biotechniques. 2006;41:694, 696, 698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- 39.Kohwi-Shigematsu T, deBelle I, Dickinson LA, Galande S, Kohwi Y. Methods Cell Biol. 1998;53:323–354. doi: 10.1016/s0091-679x(08)60885-7. [DOI] [PubMed] [Google Scholar]
- 40.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 41.Gondor A, Rougier C, Ohlsson R. Nat Protoc. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 43.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 44.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, Mathe E, Liang G, Cobell J, Nakahashi H, Robbiani DF, Nussenzweig A, Hager GL, Nussenzweig MC, Casellas R. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Homer N. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilbanks EG, Facciotti MT. PloS one. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai S, Kohwi-Shigematsu T. Methods. 1999;19:394–402. doi: 10.1006/meth.1999.0875. [DOI] [PubMed] [Google Scholar]