Abstract
Biofilms are highly structured, surface-associated communities. A hallmark of biofilms is their extraordinary resistance to antimicrobial agents that is activated during early biofilm development of Pseudomonas aeruginosa and requires the regulatory hybrid SagS and BrlR, a member of the MerR family of multidrug efflux pump activators. However, little is known about the mechanism by which SagS contributes to BrlR activation or drug resistance. Here, we demonstrate that ΔsagS biofilm cells harbor the secondary messenger c-di-GMP at reduced levels similar to those observed in wild-type cells grown planktonically rather than as biofilms. Restoring c-di-GMP levels to wild-type biofilm-like levels restored brlR expression, DNA binding by BrlR, and recalcitrance to killing by antimicrobial agents of ΔsagS biofilm cells. We likewise found that increasing c-di-GMP levels present in planktonic cells to biofilm-like levels (≥55 pmol/mg) resulted in planktonic cells being significantly more resistant to antimicrobial agents, with increased resistance correlating with increased brlR, mexA, and mexE expression and BrlR production. In contrast, reducing cellular c-di-GMP levels of biofilm cells to ≤40 pmol/mg correlated with increased susceptibility and reduced brlR expression. Our findings suggest that a signaling pathway involving a specific c-di-GMP pool regulated by SagS contributes to the resistance of P. aeruginosa biofilms.
Keywords: c-di-GMP, biofilm tolerance, small colony variants, SCV, Psl polysaccharide, Pel
INTRODUCTION
Pseudomonas aeruginosa ranks second among the most common human pathogens isolated from surgical sites, chronic and burn wounds, and is the most frequent gram-negative etiologic agent associated with infections of indwelling catheters and foreign body implants. This occurs due to the ability of P. aeruginosa to form biofilms, or highly structured, sessile microbial communities exhibiting surface-associated growth. Biofilm cells differ from their planktonic (free floating) counterparts in the genes that they express and the proteins that they produce, as well as in the synthesis and levels of signaling molecules such as c-di-GMP. High concentrations of this molecule correlate with a sessile lifestyle (e.g. biofilm formation), while its absence favors motility (e.g. swarming, swimming) and the free-swimming lifestyle (D’Argenio & Miller, 2004). These molecular differences result in distinct biofilm-specific phenotypes that include altered resistance to antibiotics and the human immune system (Costerton et al., 1999). It is thus not surprising that biofilm bacteria are a source of many recalcitrant and chronic infections (Costerton et al., 1999).
Resistance, indicated by an increased minimum inhibitory concentration (MIC), refers to the ability of a microorganism to grow in the presence of an elevated level of an antimicrobial agent, to which they were once sensitive, due to mechanisms such as the presence of plasmid-borne resistance markers or resistance conferred by mutation (Donlan & Costerton, 2002, Gilbert et al., 2002, Lewis, 2001, Mah & O’Toole, 2001, Stewart & Costerton, 2001). Many reports have shown that by this convention biofilm cells do not display growth in the presence of an antimicrobial at concentrations inhibitory to planktonic cell growth (Lewis, 2001, Spoering & Lewis, 2001). Instead, the observed reduced susceptibility of biofilm cells to antimicrobial agents, also known as biofilm tolerance, is distinct from commonly known mechanisms conferring resistance, indicating that the mechanisms involved in resilience of biofilms against antimicrobials may differ from the mechanisms responsible for antimicrobial resistance in planktonic bacteria. Instead, the reported “resistance” or biofilm tolerance describes an increased resistance of cells to killing. Indeed, this is what biofilm cells are good at: while not growing in the presence of antibiotics, they are recalcitrant to eradication by bactericidal antimicrobials. This is further supported by the findings that biofilm cells can be 10 – 1000 fold less susceptible to various antimicrobial agents than their planktonic counterparts, and are extremely problematic to eradicate by conventional antimicrobial treatment strategies. The nature of this biofilm tolerance has been deemed multifactorial with contributing factors including slow growth, reduced metabolic rates, increased stress tolerance, reduced diffusion rates, and the presence of extracellular polymeric matrices (Keren et al., 2004, Nguyen et al., 2011, Lewis, 2001, Gilbert et al., 2002, Mah & O’Toole, 2001, Mah et al., 2003, Stewart & Costerton, 2001, Colvin et al., 2011, Khan et al., 2010). Additionally, differences in the intracellular level of the secondary messenger molecule c-di-GMP present in biofilm and free-floating, dispersed cells have been shown to correlate with differences in the production of proteins involved in antimicrobial peptide resistance and resistance towards colistin (Chua et al., 2013). Recent findings further suggest that in P. aeruginosa biofilms, drug tolerance is a function of the progression of biofilm development. Biofilm development occurs as a sequential process, with at least four two-component regulatory systems, namely SagS, BfiRS, BfmRS, and MifRS, being required to coordinate the progression of P. aeruginosa biofilm formation in a stage-specific manner. Through phosphorelay events and regulation of gene expression, these systems form a coordinated signaling network that regulates committed biofilm developmental steps following attachment in response to environmental cues. The four committed steps of the P. aeruginosa biofilm life cycle are initial attachment, enabling the transition from reversible to irreversible attachment and initiation of biofilm formation (SagS, BfiRS), biofilm maturation (BfmRS), and microcolony formation (MifRS) (Fig. 1). SagS coordinates the transition from the reversible to the irreversible attachment stage, via direct interaction with and modulation of the phosphorylation state of BfiS (Petrova & Sauer, 2011) (Fig. 1). BfiS likewise contributes to surface associated bacteria transitioning to the irreversible attachment stage (Petrova & Sauer, 2010). In addition, the SagS-dependent transition to the irreversible attachment stage also marks the switch to the high-level resistance phenotype, as inactivation of sagS rendered biofilms but not planktonic cells more susceptible to tobramycin, norfloxacin and hydrogen peroxide (Gupta et al., 2013) (Fig. 1). It is of interest to note that inactivation of bfiS, bfiR, or bfmR did not affect the susceptibility of the respective mutant biofilm cells to antimicrobial agents (Gupta et al., 2013). Inactivation of sagS also eliminated the recalcitrance of biofilm cells to killing by the bactericidal antimicrobial agents norfloxacin and tobramycin. Intriguingly, the susceptibility of biofilms formed by the ΔsagS mutant to antibiotics was comparable to that observed upon inactivation of brlR, encoding a MerR-like transcriptional regulator. Inactivation of brlR was shown by Liao et al. (Liao et al., 2013, Liao & Sauer, 2012) to render biofilm but not planktonic cells grown to exponential or stationary phase significantly more susceptible to hydrogen peroxide and five different classes of antibiotics, by contributing to the expression of at least two MDR pumps, thus affecting the minimum inhibitory concentration (MIC) and recalcitrance to killing by bactericidal antimicrobial agents. Moreover, BrlR and SagS were found to be linked, with sagS inactivation correlating with reduced expression of brlR and BrlR being impaired in binding to its target MDR promoters PmexA and PmexE (Gupta et al., 2013, Liao et al., 2013) (Fig. 1).
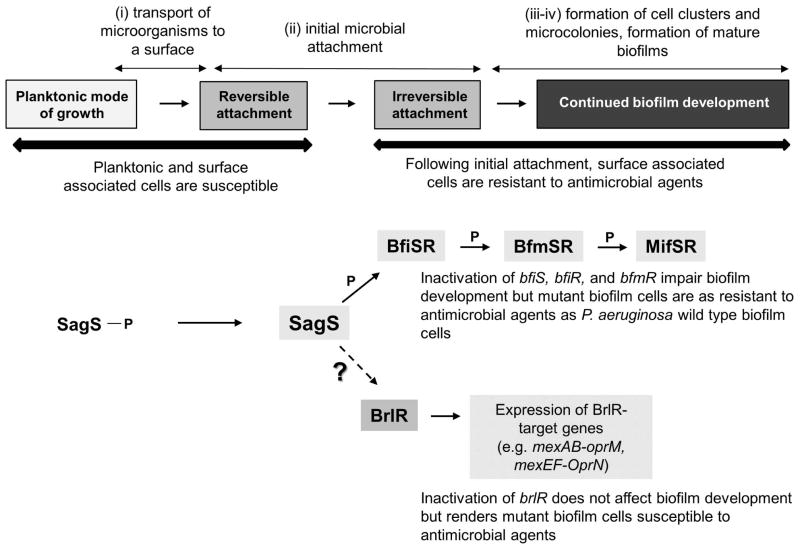
Figure 1. Overview of the contribution of the two-component hybrid SagS to the motile-sessile and susceptible-resistance switches by P. aeruginosa cells.
Under planktonic conditions, SagS is phosphorylated. Upon transition to surface associated growth, SagS directly interacts with and phosphorylates the TCS BfiSR, thus enabling surface associated cells to transition to the irreversible attachment stage (Petrova & Sauer, 2011). The TCS BfiSR, BfmSR and MifSR are sequentially required to enable the formation of P. aeruginosa biofilms in a stage-specific manner (Petrova et al., 2012, Petrova & Sauer, 2009, Petrova et al., 2011, Petrova & Sauer, 2010). Moreover, transition to the irreversible attachment stage, regulated by SagS, marks the timing when surface associated cells gain their heightened resistance to antimicrobial agents, with inactivation of sagS having been previously demonstrated to correlate with biofilm cells but not planktonic cells being more susceptible to antimicrobial agents (Gupta et al., 2013). Resistance of biofilm cells to antimicrobial agents is conferred, in part, by BrlR, a member of the MerR family of multidrug efflux pump activators, by affecting MIC, increased expression of multidrug efflux pump genes (mexAB-OprM, mexEF-OprN), and recalcitrance to killing by bactericidal antimicrobial agents (Liao et al., 2013, Liao & Sauer, 2012). Dashed arrow indicates that SagS indirectly contributes to the expression of brlR and BrlR function (Gupta et al., 2013). Question mark (“?”) indicates unknown factor(s) required for the susceptible-resistance switch by P. aeruginosa cells. P, protein phosphorylation or phosphotransfer reaction.
The findings suggested that SagS acts as a molecular switch in the transition of P. aeruginosa (i) from the free floating to the sessile lifestyle by activating BfiSR to enable biofilm development, and (ii) from a susceptible to a highly resistant biofilm phenotype, by contributing to brlR expression and BrlR function (Fig. 1). However, with the exception of SagS contributing to brlR expression and enhancing the DNA binding capability of BrlR to its target promoters (Gupta et al., 2013), little is known about the mechanism by which SagS confers resistance to surface associated cells. Likewise, the conditions or factors contributing to BrlR activation, and thus to tolerance of biofilm cells to antimicrobial agents, are not known. To elucidate the mechanism by which SagS contributes to biofilm tolerance via BrlR activation, we first asked how biofilms formed by ΔsagS and ΔbfiS mutants differed, considering that SagS activates BfiS via direct interaction and modulation of the phosphorylation state of BfiS, and that inactivation of sagS but not bfiS renders mutant biofilms more susceptible to antimicrobial agents.
RESULTS
Inactivation of sagS correlates with significantly reduced biofilm c-di-GMP levels
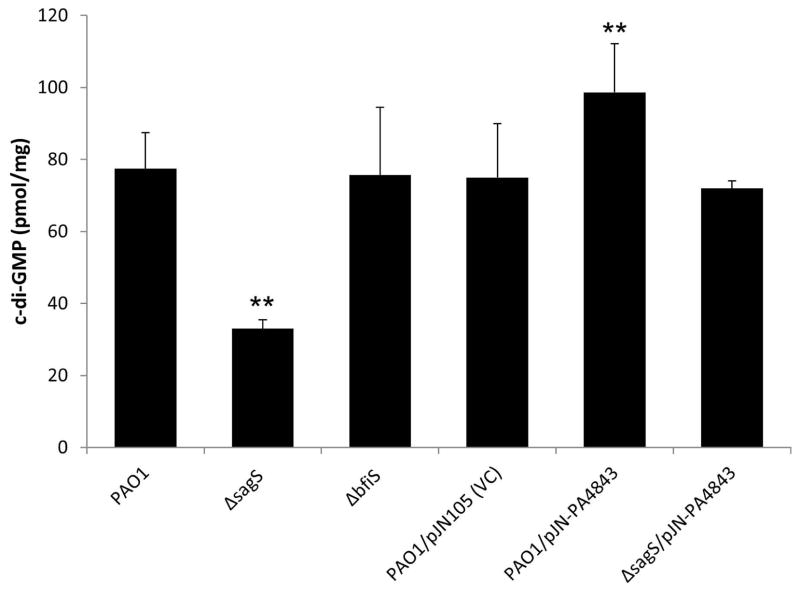
P. aeruginosa biofilm cells inactivated in bfiS (here referred to as ΔbfiS biofilms) and sagS (ΔsagS biofilms) have been described to be arrested in biofilm development at the transition from the planktonic lifestyle to the irreversible attachment stage, with SagS acting upstream of BfiS (Fig. 1). Despite ΔsagS and ΔbfiS biofilms being arrested at a similar biofilm stage, biofilm cells inactivated in bfiS are resistant to antimicrobial agents, while ΔsagS biofilm cells are susceptible (Gupta et al., 2013). To elucidate the mechanism by which SagS contributes to the resistance of biofilm cells towards antimicrobial agents, we asked whether ΔsagS biofilms differ from wild-type and ΔbfiS biofilms in the level of the secondary messenger molecule c-di-GMP that has been associated with controlling the transition between planktonic and biofilm lifestyles (D’Argenio & Miller, 2004). Previous reports indicated mature P. aeruginosa biofilms to contain on average 84–110 pmol c-di-GMP per mg total protein, while planktonic cells have been described to contain less than 30 pmol/mg (Basu Roy et al., 2012, Barraud et al., 2009). Here, wild-type P. aeruginosa and ΔbfiS biofilm cells were found to contain on average 75–78 pmol/mg c-di-GMP (Fig. 2). In contrast, c-di-GMP levels in ΔsagS biofilms were found to be significantly reduced compared to wild-type biofilms, with ΔsagS biofilms only containing 33±2 pmol/mg c-di-GMP (Fig. 2). These findings suggest that the steady state level of c-di-GMP is altered in ΔsagS biofilms relative to wild-type and ΔbfiS biofilm cells, resulting in a reduction in the overall cellular levels of this signaling molecule.
Figure 2. c-di-GMP levels present in wild-type and mutant biofilm cells, and biofilm cells overexpressing PA4843 encoding a predicted diguanylate cyclase.
The strains were grown in tube reactors under flowing conditions for 3 days prior to c-di-GMP extraction and quantitation by HPLC analysis. pmol/mg refers to c-di-GMP levels (pmol) per total cell protein (in mg). P. aeruginosa PAO1 harboring the empty vector pJN105 was used as vector control (VC). Experiments were repeated at least 3 times. Error bars denote standard deviation. **, significantly different from PAO1 biofilm cells and vector control (PAO1/pJN105); p < 0.01 as determined by ANOVA and SigmaStat.
The susceptibility of ΔsagS biofilm cells to antimicrobial agents can be rescued by elevating c-di-GMP levels
Considering the presence of reduced c-di-GMP levels in ΔsagS biofilms compared to wild-type and ΔbfiS biofilms, we next asked whether the low antibiotic resistance phenotype of ΔsagS biofilms is linked to the reduced c-di-GMP levels, and whether restoration of c-di-GMP levels to those observed in wild-type biofilms would result in reduced susceptibility of ΔsagS biofilm cells. In an attempt to restore the c-di-GMP levels to wild-type levels, we cloned PA4843 and PA4929 predicted to encode diguanylate cyclases (Kulasekara et al., 2006, Petrova & Sauer, 2012), under the control of a PBAD promoter and transferred the plasmid-borne constructs into the ΔsagS strain, thus generating strains ΔsagS/pJN-PA4843 and ΔsagS/pJN-PA4929. Biofilms formed by these strains differed in the concentration of c-di-GMP per biofilm biomass (Fig. 2). C-di-GMP levels present in biofilms formed by strain ΔsagS/pJN-PA4843 were significantly increased compared to ΔsagS biofilms but comparable to those detected in wild-type biofilms (Fig. 2). Moreover, biofilms formed by PAO1/pJN-PA4843 demonstrated significantly icnreased c-di-GMP levels compared to biofilms formed by wild type or vector control. In contrast, expression of PA4929 had very little effect on the c-di-GMP levels present in biofilms formed by ΔsagS/pJN-PA4929 compared to ΔsagS biofilms (not shown). The difference in cellular c-di-GMP upon overexpression of PA4843 or PA4929 may be due to the two predicted diguanylate cyclases exhibiting different levels of specific activity.
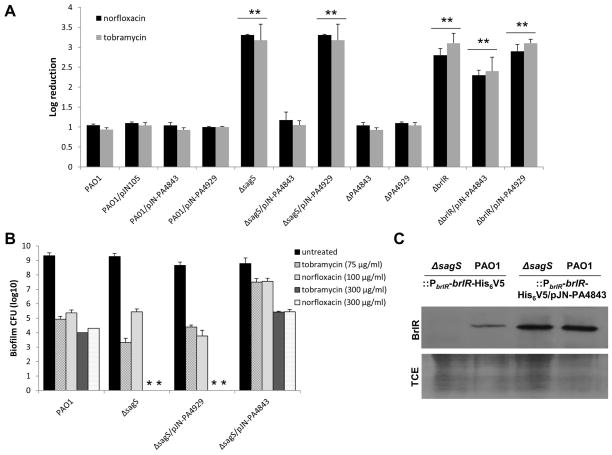
Given the restoration of c-di-GMP levels in ΔsagS/pJN-PA4843 biofilm cells, the susceptibility of these cells was compared to that of ΔsagS biofilm cells. ΔsagS biofilms do not develop high-level biofilm-specific drug resistance compared to the wild type as evidenced by treatment with tobramycin (150 μg/ml) or norfloxacin (450 μg/ml) for 1 hr resulting in a 3.3-fold and 3.2-fold log reduction, respectively. In contrast, ΔsagS/pJN-PA4843 biofilms were as resistant to tobramycin and norfloxacin as wild-type biofilms (Fig. 3A). Biofilm cells of the ΔsagS/pJN-PA4929 strain, however, were as susceptible to tobramycin and norfloxacin as ΔsagS biofilms (Fig. 3A). No difference between PAO1 and vector control was noted (Fig. 3A). It is of interest to note that inactivation of PA4843 and PA4929 had no effect on the susceptibility of biofilm cells to tobramycin and norfloxacin (Fig. 3A). This suggested that PA4843 or PA4929 are not specifically required for biofilm resistance and that the effects observed following PA4843 overexpression are likely the result of elevated c-di-GMP levels. The finding furthermore indicated that PA4843 and PA4929 are not the diguanylate cyclases contributing to the elevated intracellular c-di-GMP levels present in P. aeruginosa wild-type biofilm cells compared to ΔsagS biofilm cells. However, while our findings suggested resistance of P. aeruginosa to be linked to elevated c-di-GMP levels, the effect of c-di-GMP on resistance was BrlR-dependent as overexpression of PA4929 or PA4843 in a ΔbrlR mutant did not result in significantly decreased susceptibility (Fig. 3A).
Figure 3. Multicopy expression of PA4843 encoding a predicted diguanylate cyclase restores BrlR production and recalcitrance of ΔsagS biofilm cells to killing by bactericidal antimicrobial agents.
(A) Indicated P. aeruginosa strains were grown in tube reactors for 3 days and subsequently treated with tobramycin (150 μg/ml) or norfloxacin (450 μg/ml) for 1 hr under flowing conditions. P. aeruginosa harboring the empty vector pJN105 was used as a vector control. (B) Multicopy expression of PA4843 restores recalcitrance of ΔsagS biofilm cells to killing by antimicrobial agents as determined using biofilm-MBC assays. P. aeruginosa PAO1, ΔsagS and ΔsagS cells overexpressing PA4843 and PA4929 were grown for 3 days as biofilms and subsequently treated for 24 hr under continuous flowing conditions before recovering and enumerating surviving cells. Biofilm susceptibility to tobramycin (75 and 300 μg/ml) and norfloxacin (100 and 300 μg/ml) was determined by viable counts (CFU). The concentrations were chosen based on findings by Gupta et al. (Gupta et al., 2013) indicating complete killing of ΔsagS mutant biofilm cells following treatment with tobramycin concentrations exceeding 100 μg/ml and norfloxacin concentrations exceeding 200 μg/ml. (C) Total cell extracts obtained from P. aeruginosa wild-type and ΔsagS biofilms expressing a chromosomally located His6/V5-tagged brlR construct under the control of its own promoter (PbrlR-brlR-V5/His6) in the absence of presence of plasmid-borne PA4843 and PA4929 were probed for the presence of BrlR by immunoblot analysis using anti-V5 antibodies (anti-V5, see “BrlR” and top panel). Lower panel shows Coomassie-stained gel after immunoblotting demonstrating similar loading of all four total cell extract (TCE) used for immunoblotting and probing for BrlR using anti-V5 antibodies. A total of 15μg total cell extract was loaded. All experiments were done in triplicate and a representative image is shown. Error bars denote standard deviation. **, Significantly different from PAO1, p < 0.01 as determined by ANOVA and SigmaStat.
Elevated levels of c-di-GMP restore the recalcitrance of ΔsagS biofilm cells to killing by bactericidal antibiotics
While the hallmark characteristic of biofilm populations is their increased antimicrobial tolerance, recent reports suggest that biofilm drug tolerance describes an increased recalcitrance of biofilm cells to killing (Lewis, 2008, Spoering & Lewis, 2001). Previous findings indicated SagS to contribute to the recalcitrance of biofilm cells to killing by bactericidal antibiotics (Gupta et al., 2013). In contrast to wild -type biofilm cells, inactivation of sagS correlated with complete killing of biofilms cells following 24 hr treatment by tobramycin and norfloxacin at concentrations exceeding 100 μg/ml (Gupta et al., 2013). To determine whether restoration of the c-di-GMP abundance in ΔsagS biofilms to wild-type levels also restored the resistance to killing of biofilms by bactericidal antibiotics, we made use of Biofilm-MBC assays using tube reactor grown biofilms and the two antibiotics tobramycin and norfloxacin. P. aeruginosa wild type, ΔsagS mutant, ΔsagS/pJN-PA4929, and ΔsagS/pJN-PA4843 were grown for 3 days as biofilms, after which time the medium was switched to the same medium containing 75 μg/ml or 300 μg/ml tobramycin and 100 or 300 μg/ml norfloxacin. The concentrations were chosen as the higher concentrations previously demonstrated to eradicate both ΔsagS and ΔbrlR biofilm cells but not wild-type biofilm cells (Gupta et al., 2013), while the lower concentrations were found to only eradicate ΔbrlR biofilm cells (Gupta et al., 2013). Following 24 hr of exposure to the antibiotic under continuous flow, biofilms were harvested and the surviving bacteria enumerated. Multi-copy expression of PA4929 had no effect on ΔsagS biofilm cells, as no viable cells were recovered following 24 hr treatment with norfloxacin and tobramycin, respectively (Fig. 3B). In contrast, expression of PA4843 in ΔsagS biofilms resulted in significantly reduced susceptibility to tobramycin compared to ΔsagS biofilms, with PA4843 expression rendering ΔsagS biofilms comparable in resistance to wild-type biofilms (Fig. 3B). Similar results were obtained when biofilms were treated with norfloxacin (Fig. 3B). The findings indicated that restoration of c-di-GMP levels due to multicopy expression of PA4843 rendered ΔsagS/pJN-PA4843 biofilm cells more resistant to killing by tobramycin and norfloxacin than ΔsagS mutant biofilms, for which no viable cells were recovered at concentrations exceeding the biofilm-MBCs (Fig. 3B).
Multicopy expression of PA4843 correlates with restoration of brlR, mexA, and mexE expression in ΔsagS biofilm cells
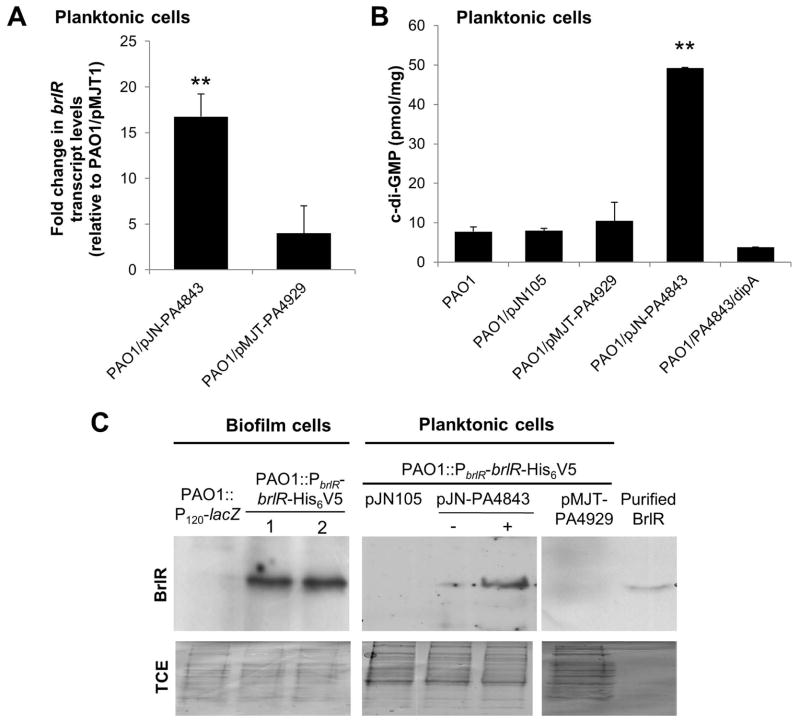
Previous findings linked the low-antibiotic resistance phenotype of ΔsagS mutant biofilm cells not only to the reduced expression of brlR, encoding the MerR-like regulator BrlR, but also to the reduced transcript abundance of the BrlR-target genes encoding the multidrug efflux pumps MexAB-OprM and MexEF-OprN (Gupta et al., 2013). To determine whether the heightened resistance of P. aeruginosa ΔsagS biofilm cells upon increasing c-di-GMP levels is due to increased transcript levels of brlR and BrlR-target genes, qRT-PCR was performed. Overexpression of PA4843 in ΔsagS biofilm cells resulted in mexA and mexE transcript levels being comparable to those of wild-type biofilms (Table 2). Likewise, brlR transcript levels increased significantly in ΔsagS biofilm cells upon overexpression of PA4843 compared to ΔsagS biofilms alone, resulting in brlR transcript levels being comparable to those of wild-type biofilms (Table 2). We furthermore determined whether increased brlR transcript abundance correlated with increased BrlR production. To do so, we generated a recombinant BrlR protein by grafting a His6V5 to the C-terminus of BrlR (referred to as His6/V5-tagged BrlR). The construct was placed under the control of the brlR promoter and integrated into the chromosome (PAO1::PbrlR-brlR-His6V5, ΔsagS::PbrlR-brlR-His6V5). This construct ensured expression of brlR at native levels. Immunoblot analysis indicated that while BrlR protein abundance was below detectable levels in ΔsagS biofilms, PA4843 overexpression in these biofilm cells resulted in restoration of BrlR protein production to detectable levels comparable to those observed for wild-type biofilm cells (Fig. 3C).
Table 2. qRT-PCR analysis.
Transcript levels of brlR, mexA and mexE present in ΔsagS and ΔsagS/pJM-PA4843 biofilm cells were determined relative to wild-type biofilms (PAO1 or PAO1/pJN105). Biofilms were grown for 3 days under flowing conditions. Transcript levels of brlR, mexA and mexE present in P. aeruginosa PAO1/pJN-PA4843 cells grown planktonically to exponential phase were determined relative to wild-type cells (PAO1/pJN105). Experiments were carried out 5 times. Transcript levels of mreB were used as control.
| Transcript/strain | Fold-change in transcript levels relative to wild-type biofilm cells
|
||
|---|---|---|---|
| brlR | mexA | mexE | |
| ΔsagS | −25.3±0.4* | −2.7±0.2* | −3.7±0.3* |
| ΔsagS/pJN-PA4843 | 0.88±0.17 | 0.96±0.2 | 0.87±0.19 |
|
Fold-change in transcript levels relative to wild-type cells grown planktonically
|
|||
| PAO1/pJN-PA4843 | 17.0±4.5** | 8.8±0.2** | 7.4±1.3** |
transcript levels were significantly different compared to wild-type biofilms; p < 0.01.
transcript levels were significantly different compared to exponential phase planktonic wild-type cells; p < 0.01.
Restoration of DNA binding by BrlR
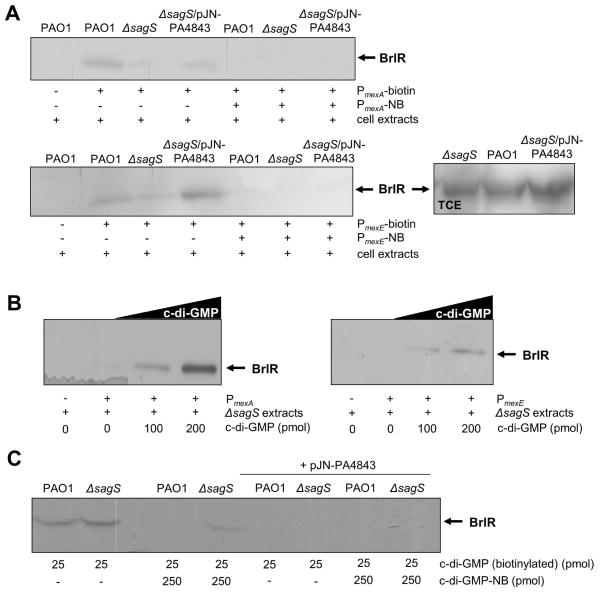
BrlR purified from total extracts obtained from ΔsagS biofilm cells (referred to as ΔsagS-produced BrlR) was previously shown to be unable to bind to its target promoters PmexA and PmexE, resulting in significantly reduced mexA and mexE transcript levels (Gupta et al., 2013). Considering that overexpression of PA4843 restored not only the c-di-GMP levels (Fig. 2) and the resistance phenotype (Fig. 3) of this mutant strain but also mexA, mexE and brlR transcript abundance to wild-type levels (Table 2), we next determined whether expression of PA4843 could restore BrlR function. To analyze the DNA binding capabilities of BrlR, we made use of a recombinant BrlR protein by grafting a His6V5 sequence to the C-terminus of BrlR (referred to as His6/V5-tagged BrlR). The gene encoding the recombinant protein was cloned into pMJT1 under the control of the arabinose inducible PBAD promoter and transferred into P. aeruginosa wild type and ΔsagS in the presence or absence of the PA4843 overexpression vector pJN-PA4843. The resulting strains (PAO1/pMJT-brlR, PAO1/pJN-PA4843/pMJT-brlR, ΔsagS/pMJT-brlR, ΔsagS/pJN-PA4843/pMJT-brlR) were grown as biofilms for 3 days after which time the biofilm cells were harvested and lysed to obtain total cell extracts. The resulting cell extracts harboring tagged BrlR protein were used in DNA binding assays with the BrlR-target promoter DNA PmexA and PmexE. While wild-type produced BrlR did bind to the PmexA and PmexE target promoter DNA, very little or no binding was noted for BrlR obtained from total cell extracts of ΔsagS biofilm cells (Fig. 4A). In contrast, His6/V5-tagged BrlR obtained from ΔsagS/pJN-PA4843 biofilm extracts demonstrated DNA binding to PmexA and PmexE, with this DNA binding by BrlR being reduced in the presence of non-biotinylated competitor DNA (Fig. 4A). Enhanced DNA binding by BrlR to the biotinylated DNA probes PmexA and PmexE was likewise observed in the presence of exogenously added c-di-GMP, with increasing concentrations of c-di-GMP correlating with increased DNA binding by BrlR (Fig. 4B).
Figure 4. Exogenous addition of c-di-GMP in vitro and in vivo elevated levels of c-di-GMP upon multicopy expression of PA4843 restore DNA-binding capability of BrlR obtained from total cells extracts of ΔsagS biofilm cells.
(A) Streptavidin magnetic bead DNA-binding assays using cell extracts obtained from ΔsagS and PAO1 biofilms harboring pMJT-brlR-V5/His6 in the absence of presence of PA4843 expressed in trans. Binding assays were carried out using a total of 5pmol the BrlR-V5/His6 protein and 1 pmol biotinylated PmexA or 1 pmol of biotinylated PmexE in wild-type and ΔsagS strains. Non-biotinylated PmexA and PmexE (PmexA-NB, PmexE-NB) were used as specific competitor DNA in 20-fold excess. BrlR binding to PmexA and PmexE was detected by immunoblot analysis using anti-V5 antibodies. +, indicates presence of specified components. Images on the right demonstrate BrlR to be present in similar amounts in aliquots removed prior to addition of streptavidin magnetic beads in ΔsagS and PAO1 biofilm samples (loading control). (B) Streptavidin magnetic bead DNA-binding assays using cell extracts obtained from ΔsagS and PAO1 harboring pMJT-brlR-V5/His6 in the presence of increasing concentrations of c-di-GMP (0–200 pmol). Binding assays were carried out using a total of 5pmol the BrlR-V5/His6 protein and 1 pmol biotinylated PmexA or 1 pmol of biotinylated PmexE in wild-type and ΔsagS strains. (C) BrlR-c-di-GMP binding assays. Pulldowns using biotinylated c-di-GMP immobilized to streptavidin magnetic beads demonstrate that BrlR produced by wild-type and ΔsagS biofilms is capable of c-di-GMP binding. Binding to biotinylated c-di-GMP was reduced by the addition of non-biotinylated c-di-GMP (c-di-GMP-NB). No binding was detected when BrlR produced by wild-type and ΔsagS biofilms overexpressing PA4843 was used. BrlR-c-di-GMP binding was detected by immunoblot analysis using anti-V5 antibodies. Experiments were repeated at least 4 times and representative images are shown.
Reduced DNA binding by BrlR correlates with reduced binding of c-di-GMP to BrlR
Our findings indicated a role of elevated c-di-GMP in biofilm resistance by contributing to brlR expression and restoring the DNA binding capabilities of BrlR in ΔsagS biofilms. To determine whether the difference in DNA binding was due to the absence of c-di-GMP in ΔsagS biofilms rather than BrlR being produced in a non-functional manner, we determined the c-di-GMP binding capabilities of BrlR obtained from total cell extracts of wild-type and ΔsagS biofilm cells. To do so, we made use of a pulldown assays using biotinylated c-di-GMP, with BrlR binding to c-di-GMP being detected by immunoblot analysis using anti-V5 antibodies. BrlR produced by ΔsagS biofilms was equally capable to binding biotinylated c-di-GMP. Binding of biotinylated c-di-GMP by wild-type-produced BrlR was outcompeted by non-biotinylated c-di-GMP (Fig. 4C) indicating a specificity of BrlR for c-di-GMP. The same concentration of non-biotinylated c-di-GMP reduced but was unable to fully outcompete the binding to biotinylated c-di-GMP by ΔsagS-produced BrlR. The finding indicated ΔsagS-produced BrlR to potentially have a higher capacity for c-di-GMP binding in vitro than wild-type BrlR, likely due to lower levels of pre-bound c-di-GMP in the ΔsagS strain. In other words, ΔsagS biofilms may have more c-di-GMP-free BrlR due to reduced c-di-GMP levels. To test for the contribution of pre-bound c-di-GMP affecting the ability of BrlR to bind to biotinylated c-di-GMP in our pulldown assays, we made use of BrlR obtained from biofilm cells overexpressing the diguanylate cyclase PA4843. We hypothesized that such elevated intracellular c-di-GMP levels would achieve saturation of BrlR-c-di-GMP binding. Under the conditions tested, neither BrlR produced by PAO1/pJN-PA4843/pMJT-brlR nor by ΔsagS/pJN-PA4843/pMJT-brlR biofilms was able to bind biotinylated c-di-GMP (Fig. 4C). The finding indicated prebound c-di-GMP to interfere with binding to biotinylated c-di-GMP in our assay. Our findings furthermore suggested BrlR produced by ΔsagS biofilms to likely bind less or to be devoid of c-di-GMP in vivo.
Elevated c-di-GMP levels upon multicopy expression of PA4843 correlate with detectable brlR expression and BrlR production in exponential phase planktonic P. aeruginosa cells
Given the similarity between P. aeruginosa cells grown planktonically and ΔsagS biofilm cells with respect to low c-di-GMP levels, reduced/absent brlR transcript abundance, and increased susceptibility to antimicrobial agents compared to P. aeruginosa wild-type biofilm cells (Gupta et al., 2013, Liao et al., 2013, Liao & Sauer, 2012), and that elevating c-di-GMP levels increases brlR expression and drug susceptibility of ΔsagS biofilm cells, we asked whether modulating c-di-GMP levels in planktonic cells would equally result in increased brlR transcript abundance and thus heighten the resistance of planktonic cells to tobramycin and norfloxacin. We therefore first determined whether increasing c-di-GMP levels in P. aeruginosa under planktonic conditions would affect brlR transcript levels. In exponential-phase planktonic P. aeruginosa wild-type cells, the qRT-PCR threshold cycle (Ct) for brlR transcript levels was above 35 and similar to a no-RT control, indicating that brlR is either not expressed or transcribed at very low levels. However, multicopy overexpression of genes encoding the hypothetical diguanylate cyclases PA4929 and PA4843 (Winsor et al., 2009, Kulasekara et al., 2006, Petrova & Sauer, 2012) in P. aeruginosa grown to exponential phase resulted in decreased qRT-PCR Ct values, suggestive of 4- and 20-fold increases, respectively, in brlR transcript levels compared to the wild type harboring an empty vector (Fig. 5A). Moreover, the brlR transcript abundance in PAO1/pMJT-PA4843 was similar (a 1.7±0.4-fold increase was detected) to that detected in wild-type biofilms. It is of interest to note that mexA and mexE transcript levels were likewise found to be significantly increased in cells grown planktonically to exponential phase upon multicopy expression of PA4843 (Table 2).
Figure 5. Multicopy expression of PA4843 encoding a diguanylate cyclases correlates with increased brlR transcript levels, detectable BrlR, and elevated intracellular c-di-GMP levels in planktonic cells.
(A) qRT-PCR demonstrating increased brlR transcript levels under planktonic growth conditions in P. aeruginosa strains overexpressing the cyclases PA4843 (PAO1/pJN-PA4843) and PA4929 (PAO1/pMJT-PA4929). (B) Intracellular c-di-GMP levels present in planktonic cells of P. aeruginosa PAO1, PAO1 harboring the empty vector pJN105, and strains overexpressing PA4843, PA4929, and cells co-expressing PA4843 and dipA. c-di-GMP levels were quantitated per mg total protein using a HPLC-based approach. **, significantly different from wild-type PAO1 and vector control levels (p < 0.01). (C) Detection by immunoblot analysis of BrlR in cell extracts obtained from PAO1::PbrlR-brlR-His6V5 grown as biofilms and planktonic cells overexpressing genes encoding the diguanylate cyclases PA4843 and PA4929. PAO1::PbrlR-brlR-His6V5 harboring the empty pJN105 vector was used as control. PAO1::PbrlR-brlR-His6V5 harboring the empty pJN105 vector was used as control. Immunoblots were probed with anti-V5 antibody. Recombinant BrlR has an apparent molecular mass of 33 kDa. No band of that apparent mass was detected in total cell extract of biofilms harboring the chromosomally located promoter reporter construct (PAO1::P120-lacZ) that harbors the upstream region of brlR but is lacking the brlR structural gene. A total of 15 μg of total cell extracts were loaded. -, no arabinose was added; +, 1% arabinose was added to the growth medium to induce PA4843 expression from PBAD promoter. 1 and 2, refer to total cell extracts obtained from two separate set of biofilm cells by strain PAO1::PbrlR-brlR-His6V5. All experiments were done in triplicate and representative images are shown. Lower panel shows Coomassie-stained gels after immunoblotting demonstrating similar loading of all total cell extract (TCE) used for immunoblotting and probing for BrlR using anti-V5 antibodies. Error bars denote standard deviation.
brlR transcript abundance positively correlated with c-di-GMP levels. Under the conditions tested, a total of 8 pmol c-di-GMP per mg of total protein extract was detected for P. aeruginosa PAO1 grown planktonically to exponential phase. Similar results were obtained for the wild type harboring the empty vector pJN105 (Fig. 5B). Overexpression of PA4929 had little effect on the c-di-GMP levels. In contrast, however, overexpression of PA4843 resulted in c-di-GMP levels increasing by 5-fold to an average of 49.3 pmol/mg (Fig. 5B). The difference in c-di-GMP level detected in planktonic cells as a result of overexpressing genes encoding the putative diguanylate cyclases PA4843 and PA4929 further supports the notion of these cyclases differing in their specific activity to generate c-di-GMP.
We furthermore determined whether increased brlR expression correlated with BrlR protein production by using strain PAO1::PbrlR-brlR-His6V5 which enables the production of a recombinant BrlR protein at native levels. While BrlR was present in total cell extracts obtained from P. aeruginosa PAO1 biofilms as determined using immunoblot analysis, BrlR protein was not detectable in total cell extracts obtained from exponential phase P. aeruginosa PAO1 and PAO1/pNJT-PA4929 cells (Fig. 5C). In contrast, BrlR was detectable in total cell extracts obtained from planktonic cells overexpressing PA4843 (Fig. 5C).
High c-di-GMP levels contribute to resistance of exponential phase planktonic cells to tobramycin and norfloxacin
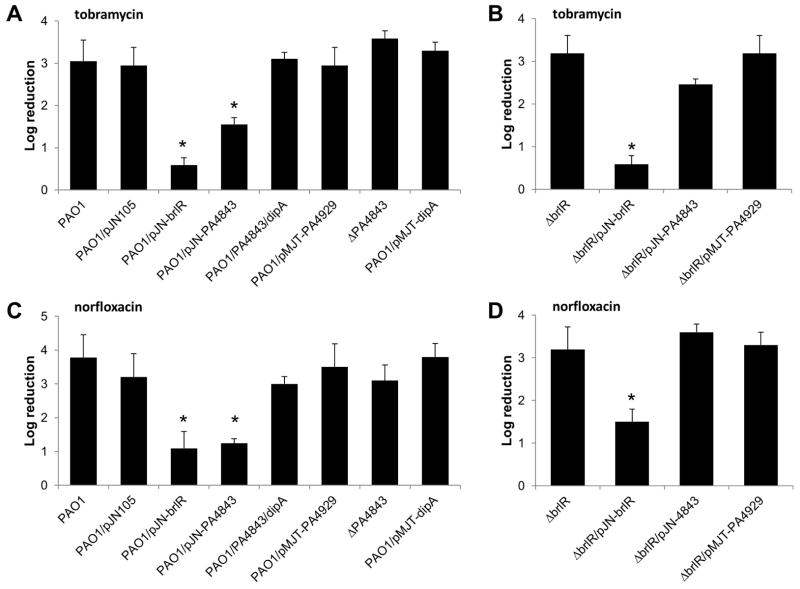
Considering that mexA, mexE, and brlR expression, as well as BrlR production, was increased in P. aeruginosa cells at high c-di-GMP levels and that BrlR was previously demonstrated to contribute to P. aeruginosa cells being more resistant to antimicrobial agents, we next asked whether modulating the level of c-di-GMP would not only affect the resistance to antimicrobial agents of P. aeruginosa cells grown as biofilms but also cells grown planktonically, by carrying out MIC assays. MICs were determined by 2-fold serial broth dilution in LB medium using 96-well microtiter plates and an inoculum of ~104 cells per well. MICs of planktonic cells of P. aeruginosa PAO1, PAO1/pMJT-PA4843, PAO1/pMJT-PA4929, and PAO1/pMJT1 vector control were compared. Under the conditions tested, the tobramycin-MIC for P. aeruginosa PAO1 was found to be 1 μg/ml. Compared to the wild type, no difference in the tobramycin-MIC was noted for PAO1/pMJT-PA4929 and vector control. In contrast, however, the MIC of a strain overexpressing PA4843 (PAO1/pMJT-PA4843) was 4–6-fold higher for tobramycin as indicated by an increase in the tobramycin-MIC for PAO1 from 1 μg/ml to 4–8 μg/ml upon overexpression of PA4843. It is of interest to note that the increase in tobramycin-MIC upon overexpression of PA4843 was comparable to the 4–6-fold increase in tobramycin-MIC observed upon overexpression of brlR (Liao & Sauer, 2012).
To further determine whether resistance of P. aeruginosa cells is heightened by increasing c-di-GMP levels, susceptibility assays were performed using planktonic cells overexpressing PA4843, which demonstrated elevated c-di-GMP levels compared to wild-type planktonic cells (Fig. 5B). Treatment of P. aeruginosa wild type grown planktonically to exponential phase with tobramycin and norfloxacin resulted in an overall decrease in the colony forming units (CFU/ml) from an average of 7.1×108 to 6.5×105 and 4.5×105, respectively. The decrease corresponded to a 3.0- and 3.2-log reduction in viability, respectively (Fig. 6A, C). In contrast, overexpression of PA4843 rendered P. aeruginosa planktonic cells significantly more resistant to tobramycin and norfloxacin compared to the wild type and vector control alone, as evidenced by a 1.6- and 1.2-log reduction (Fig. 6A, C). This corresponded to a reduction in CFU/ml from 4.5×108 to on average 9.0×106 and 2.5×106, respectively. Overall, overexpression of PA4843 not only resulted in elevated c-di-GMP levels (Fig. 5B), and detectable brlR transcript and BrlR protein levels in P. aeruginosa cells under planktonic growth conditions (Fig. 5A, C), but also rendered P. aeruginosa more resistant to tobramycin and norfloxacin in a manner similar to overexpressing brlR (Fig. 6B, D). Inactivation of PA4843 had no effect on susceptibility of planktonic cells regardless of the antibiotic used (Fig. 6A, C). Under the conditions tested, cells of the strain PAO1/pMJT-PA4929 were found to be as susceptible as wild type cells (Fig. 6A, C). The antibiotic resistance phenotype of PAO1/pMJT-PA4929 cells is in agreement with multicopy expression of PA4929 not significantly affecting c-di-GMP levels, tobramycin-MIC, and brlR transcript levels, nor resulting in detectable BrlR protein levels.
Figure 6. Multicopy expression of a gene encoding the diguanylate cyclase PA4843 renders exponential phase planktonic P. aeruginosa PAO1 but not ΔbrlR cells more resistant to antimicrobial agents.
All strains were grown planktonically to exponential phase. Susceptibility testing of indicated P. aeruginosa strains to (A–B) 50 μg/ml tobramycin and (C–D) 150 μg/ml norfloxacin. All planktonic cells were treated for 1 hr. Strain PAO1 harboring the empty plasmid pJN105 was used as vector control. Susceptibility is expressed as log reduction. Experiments were done at least in triplicate Error bars denote standard deviation. *, Significantly different from PAO1, p < 0.01 as determined by ANOVA and SigmaStat.
The link between c-di-GMP levels and resistance to antimicrobial agents was further supported by testing P. aeruginosa cells overexpressing PA4843 and dipA encoding the phosphodiesterase DipA (referred to as PAO1/pJN-PA4843/pMJT-dipA). Under planktonic growth conditions, the c-di-GMP levels in PAO1/pJN-PA4843/pMJT-dipA were comparable to those detected in P. aeruginosa wild-type cells (Fig. 5B), indicating DipA to counter the activity of the diguanylate cyclase PA4843. Moreover, PAO1/pJN-PA4843/pMJT-dipA cells grown planktonically were as susceptible to tobramycin and norfloxacin as wild-type planktonic cells (Fig. 6A, C). While our findings suggested resistance of P. aeruginosa to be linked to elevated c-di-GMP levels due to PA4843, the effect of c-di-GMP on resistance was BrlR-dependent, as overexpression of PA4843 in a ΔbrlR mutant did not result in significantly decreased susceptibility to tobramycin and norfloxacin (Fig. 6B, D).
High c-di-GMP levels, but not aggregative behavior, contribute to drug resistance of P. aeruginosa
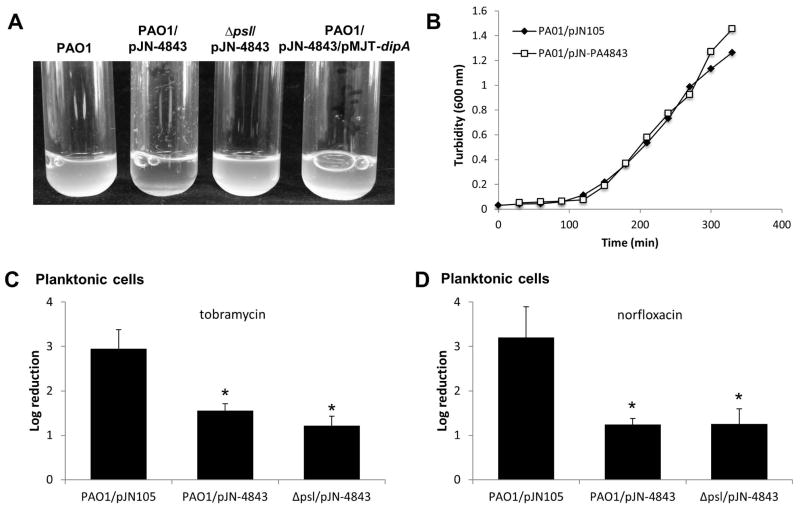
Overexpression of genes encoding diguanylate cyclases, resulting in elevated c-di-GMP levels, has been linked to the presence of an aggregative phenotype in liquid and the appearance of small colonies on agar plates (Fig. S1). Here, overexpression of PA4843 likewise resulted in an aggregative phenotype as evidenced by significant clumping of P. aeruginosa cells upon growth in liquid (Fig. 7A), with PAO1/pJN-PA4843 demonstrating emergence of small colony variants when grown on semi-solid medium. Small colony variance was not due to growth defects (Fig. 7B) but due to elevated c-di-GMP levels, as overexpression of dipA encoding the phosphodiesterase DipA in PAO1/pJN-PA4843 reduced or abrogated the aggregative phenotype of this strain and restored the colony morphology to wild-type levels (Figs. 7A, S1). Moreover, cells of the PAO1/pJN-PA4843/pMJT-dipA strain were as susceptible to norfloxacin and tobramycin as wild type cells (Fig. 6A, C).
Figure 7. Modulation of c-di-GMP levels, but not aggregative behavior, contributes to P. aeruginosa resistance to antimicrobial agents.
(A) Images demonstrate growth and aggregation of P. aeruginosa PAO1, PAO1/pJN-PA4843, Δpsl/pJN-PA4843, and PAO1/pJN-PA4843/pMJT-dipA in liquid and demonstrate that the hyperaggregative phenotype of PAO1/pJN-PA4843 depends on the presence of the Psl polysaccharide and elevated levels of c-di-GMP. (B) Overexpression of PA4843 does not affect growth in liquid. Due to differences in the aggregative phenotypes of the strains, bacterial cultures were homogenized prior to determining the turbidity at 600 nm. (C–D) Multicopy expression of PA4843 in P. aeruginosa PAO1 and Δpsl mutant renders P. aeruginosa more resistant to (C) tobramycin (50 μg/ml) and (D) norfloxacin (150 μg/ml). All planktonic cells were treated for 1 hr and experiments were done in triplicate. Error bars denote standard deviation. *, Significantly different from PAO1, p < 0.01 as determined by ANOVA and SigmaStat.
Aggregative behavior (clumping of cells) in liquid culture has previously been linked to the production of polysaccharide (Kirisits et al., 2005, Starkey et al., 2009, Colvin et al., 2011, Byrd et al., 2009). Recent evidence further suggests that Psl, a major biofilm matrix polysaccharide in P. aeruginosa PAO1, acts as a signal to stimulate two diguanylate cyclases, SiaD and SadC, to produce c-di-GMP (Irie et al., 2012). In turn, elevated intracellular concentrations of c-di-GMP lead to the increased production of Psl and other components of the biofilm (Irie et al., 2012). To determine whether brlR expression and increased resistance were dependent on Psl and hyperaggregation mimicking biofilm-like conditions, rather than on increased c-di-GMP levels, we determined whether brlR expression correlated with Psl abundance. No difference in brlR transcript levels was noted in cells lacking or overexpressing the Psl polysaccharide (Fig. S2). Likewise, no difference in brlR transcript levels compared to the wild type was noted upon differential expression of genes encoding the Pel polysaccharide biosynthetic operon (Fig. S2A). Moreover, no difference in susceptibility to tobramycin or norfloxacin was noted upon overexpression of the operons encoding the Psl or Pel polysaccharide biosynthetic machinery compared to P. aeruginosa PAO1 (Fig. S2B–C).
To further exclude the possibility that polysaccharide-dependent hyperaggregation mimicking biofilm-like conditions, rather than increased c-di-GMP levels, contributes to the resistance of P. aeruginosa to tobramycin and norfloxacin, we made use of a Δpsl mutant strain in which overexpression of PA4843 did not result in a hyperaggregative phenotype or the appearance of small colony variants (Fig. 7A). However, Δpsl/pJN-PA4843 grown planktonically to exponential phase was found to be as resistant to tobramycin and norfloxacin as the aggregative strain PAO1/pJN-PA4843 (Fig. 7C–D).
P. aeruginosa biofilm cells are rendered more susceptible by lowering intracellular c-di-GMP levels
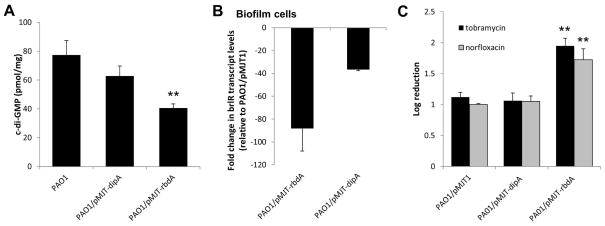
Our findings suggested a link between c-di-GMP levels and susceptibility to antimicrobial agents, with c-di-GMP levels comparable to those commonly detected in biofilm cells correlating with P. aeruginosa cells being more resistant to tobramycin and norfloxacin. We therefore reasoned that if elevated c-di-GMP levels render P. aeruginosa biofilm cells more resistant, reducing c-di-GMP levels in biofilm cells to levels commonly detected in planktonic cells, by overexpressing genes encoding phosphodiesterases involved in the degradation of c-di-GMP, would render biofilm cells more susceptible. To do so, we chose to overexpress dipA and rbdA encoding the previously characterized phopshodiesterases DipA and RbdA, respectively (Basu Roy et al., 2012, An et al., 2010). While 77 pmol/mg c-di-GMP was detected in wild-type biofilm cells, PAO1/pMJT-dipA biofilm cells harbored 62 pmol/mg and PAO1/pMJT-rbdA biofilm cells only harbored 40 pmol/mg c-di-GMP (Fig. 8A). Reduced c-di-GMP levels correlated with significantly reduced brlR expression in mutant biofilm cells, with P. aeruginosa biofilm cells overexpressing rbdA (PAO1/pMJT-rbdA) and harboring the lowest concentration of c-di-GMP demonstrating the lowest brlR transcript abundance compared to wild-type biofilms (Fig. 8B). PAO1/pMJT-dipA biofilm cells also demonstrated a reduction in brlR transcript levels relative to wild-type biofilms, but this effect was less dramatic than the one observed following rbdA overexpression. Despite reduced c-di-GMP levels and brlR transcript abundance, overexpression of dipA had very little effect on the susceptibility of biofilm cells to tobramycin and norfloxacin (Fig. 8C). In contrast, overexpressing rbdA rendered biofilm cells significantly more susceptible as evidenced by a 2-log reduction in viability compared to wild-type biofilms (Fig. 8C). These findings not only supported a correlation between brlR transcript abundance and c-di-GMP levels, but also indicated a link between susceptibility and the cellular level of c-di-GMP, with overexpression of rbdA but not dipA achieving sufficiently reduced c-di-GMP levels to render biofilm cells susceptible.
Figure 8. Multicopy expression of a gene encoding the phosphodiesterase RbdA renders P. aeruginosa biofilm cells more susceptible to antimicrobial agents.
P. aeruginosa wild-type biofilms and biofilms overexpressing rbdA and dipA were grown in tube reactors for 3 days. (A) Intracellular c-di-GMP levels present in biofilm cells of P. aeruginosa PAO1, PAO1 harboring the empty vector pJN105, and strains overexpressing rbdA and dipA. c-di-GMP levels were quantitated per mg total protein using a HPLC-based approach. (B) qRT-PCR demonstrating decreased brlR transcript levels in biofilm cells of P. aeruginosa strains overexpressing genes coding for the phosphodiesterases RbdA and DipA. Fold change in brlR transcript levels relative to biofilm cells formed by PAO1 harboring the empty vector pMJT1 (vector control) is shown. (C) Susceptibility of 3-day old P. aeruginosa wild-type biofilms and biofilms overexpressing rbdA and dipA. Biofilms were treated with tobramycin (150 μg/ml) or norfloxacin (450 μg/ml) for 1 hr under flowing conditions. P. aeruginosa harboring the empty vector pMJT1 was used as a control. Susceptibility is expressed as log reduction. **, significantly different from wild-type PAO1 and vector control levels (p < 0.01). Experiments were done in triplicate. Error bars denote standard deviation.
DISCUSSION
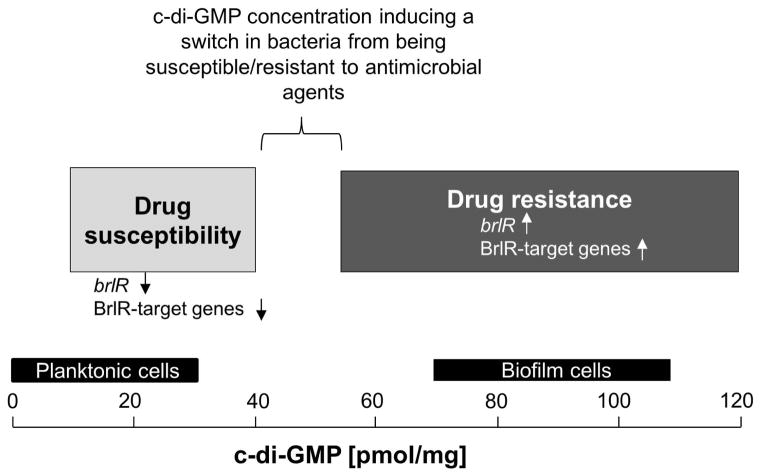
It has long been known that bacteria grown as biofilms are significantly less susceptible to various antimicrobial agents and components of the immune response than their planktonic counterparts. Moreover, while it is apparent that the mechanism(s) responsible for the observed reduced susceptibility of bacteria grown as biofilms (referred to as tolerance) differ from the resistance mechanisms commonly associated with multidrug resistance of bacteria (e.g. plasmid borne resistance, mutation), so far no single mechanism has been identified to contribute to the heightened resistance of bacteria grown as biofilms. Instead, the nature of the tolerance of biofilm cells has been deemed multifactorial with contributing factors believed to include slow growth, reduced metabolic rates, increased stress tolerance, and increased polysaccharide production. Additional differences between bacteria grown planktonically and as biofilms exist in the intracellular level of the secondary messenger molecule c-di-GMP. While planktonic cells have been described to contain less than 30 pmol/mg, mature P. aeruginosa biofilm cells contain on average 75–110 pmol c-di-GMP per mg total cell extract (Basu Roy et al., 2012, Barraud et al., 2009) (Figs. 2, 6B). Moreover, it is now widely accepted that c-di-GMP levels play an important role in the formation of biofilms, with high levels of this secondary messenger promoting biofilm formation contributing to biofilm-associated characteristics including increased polysaccharide production, increased adhesiveness, and autoaggregation, while suppressing motility (Kuchma et al., 2007, Merritt et al., 2007, Romling & Amikam, 2006, Simm et al., 2004, Thormann et al., 2006, Romling et al., 2005, Kirillina et al., 2004). By analyzing P. aeruginosa mutants previously demonstrated to be impaired at a similar biofilm developmental stage but differing in their susceptibility to antimicrobial agents, namely ΔsagS and ΔbfiS mutant biofilm cells (Gupta et al., 2013, Petrova & Sauer, 2009, Petrova & Sauer, 2010, Petrova & Sauer, 2011) (Fig. 1), we demonstrate a link between the level of c-di-GMP and drug resistance and recalcitrance to killing by antimicrobial agents (Fig. 9). We moreover demonstrate that biofilm cells can be rendered susceptible to antimicrobial agents by reducing c-di-GMP levels via overexpression of genes encoding phosphodiesterases. Likewise, we were able to demonstrate that planktonic cells are rendered more resistant to antimicrobial agents by increasing intracellular c-di-GMP levels to those more commonly found in biofilm cells. While our findings demonstrate that the concentration of the intracellular second messenger molecule c-di-GMP is a major contributing factor to drug tolerance, we were unable to determine the exact c-di-GMP concentration at which P. aeruginosa cells grown as biofilms were rendered susceptible and planktonic cells were rendered more resistant to antimicrobial agents. However, our findings indicate a range at which P. aeruginosa cells, regardless of growth conditions, experience a switch in susceptibility. Biofilm cells remain resistant to antimicrobial agents at c-di-GMP levels above 62 pmol/mg, but not at or below 40 pmol/mg (Fig. 5). In contrast, planktonic cells are rendered resistant when c-di-GMP levels reach or exceed 55 pmol/mg (Figs. 6–7). Our findings thus suggest a switch in susceptibility to occur when intracellular c-di-GMP levels exceed 40–55 pmol/mg, with the switch occurring in a growth mode independent manner (Fig. 9).
Figure 9. Model for the role of c-di-GMP levels rather than the mode of growth in the brlR expression, BrlR function, and resistance/susceptibility of P. aeruginosa cells to antimicrobial agents.
Based on our findings, the level of c-di-GMP is crucial for the susceptible-resistance switch by P. aeruginosa cells (see also “?” in Fig. 1). P. aeruginosa cells harboring ≤40 pmol/mg c-di-GMP are more susceptible to antimicrobial agents than P. aeruginosa cells harboring ≥55 pmol/mg c-di-GMP. Susceptibility was found to be independent of whether P. aeruginosa was grown planktonically or as biofilm. Light Grey bar, range of c-di-GMP levels determined here to correlate with P. aeruginosa cells being susceptible to antimicrobial agents. Low c-di-GMP levels correlated with significantly reduced or absent brlR expression and reduced DNA binding capability by BrlR. Dark Grey bar, range of c-di-GMP levels determined here to correlate with P. aeruginosa cells being resistant to antimicrobial agents. Elevated c-di-GMP levels correlated with significantly increased brlR expression and BrlR being able to bind to its target promoter DNA. BrlR target genes include mexAB-OprM and mexEF-OprN (Liao et al., 2013). Black bars indicate the range of intracellular c-di-GMP levels previously reported to be present in P. aeruginosa cells grown planktonically and as biofilm (Basu Roy et al., 2012, Barraud et al., 2009). BrlR target genes include the multidrug efflux pump genes mexAB-OprM and mexEF-OprN (Liao et al., 2013).
Aggregative phenotype (clumping of cells) in liquid and small colony phenotype on semi-solid surfaces are linked to elevated c-di-GMP levels and enhanced production of polysaccharide (Kirisits et al., 2005, Starkey et al., 2009, Colvin et al., 2011, Byrd et al., 2009). Here, we demonstrate a correlation between the aggregative phenotype due to the presence of high c-di-GMP levels and protection from antimicrobial agents. There are numerous reports linking the aggregative phenotype indicative of high c-di-GMP levels to heightened resistance and the long-term colonization of the cystic fibrosis (CF) lung. Haeussler et al. (Häußler et al., 1999, Häußler et al., 2003) demonstrated a correlation between P. aeruginosa small colony variant (SCV) recovery and poor lung function. Moreover, MICs of a broad range of antipseudomonas agents for SCVs were found to be two- to eight-fold higher than MIC values for revertants of SCVs (Häußler et al., 1999). Proctor et al. (Proctor et al., 2006, Proctor et al., 1995) linked the presence of SCVs to pathogenicity, as well as to reoccurring and persistent infection of the CF lung by Staphylococcus aureus. Likewise, Drenkard et al. (Drenkard & Ausubel, 2002) reported a link between phenotypic variants, persistent infection, and antibiotic resistance. The phenotypic variation observed by the authors was that of rough small colony variants (RSCV), which arose at a frequency of 10−6 to 10−7 when cultures were plated on Luria-Bertani (LB) agar containing kanamycin and that were detected in vivo in CF sputum samples of patients undergoing antibiotic treatment at the time of sampling. Moreover, RSCVs were found to be multi-drug resistant with the frequency of appearance of RSCVs correlating with increased MICs in liquid culture. Here, we likewise demonstrated a link between aggregative phenotypes, increased c-di-GMP levels, resistance and increased MIC.
The aggregative phenotype has been linked to enhanced production of biofilm matrix polysaccharides, Psl and Pel, by P. aeruginosa cells, with enhanced matrix production offering protection from environmental factors and antimicrobials including the aminoglycoside tobramycin (Colvin et al., 2011, Khan et al., 2010, Ma et al., 2012, Billings et al., 2013, Tseng et al., 2013). Here, we demonstrate that c-di-GMP levels, rather than polysaccharide production or the aggregative phenotype itself, contribute to drug resistance (Fig. 7, Fig. S2). Our findings are not necessarily contradicting previous reports attributing a protective role to polysaccharides, as the matrix polysaccharide Psl, has been shown to act as a signal in P. aeruginosa PAO1 to stimulate two diguanylate cyclases, SiaD and SadC, to produce c-di-GMP (Irie et al., 2012). It is thus likely that the link between polysaccharide production and susceptibility of P. aeruginosa cells to antimicrobial agents is based on c-di-GMP levels, with enhanced matrix production being an indicator of elevated c-di-GMP levels. However, while our findings indicate c-di-GMP to be a main contributor to drug resistance, c-di-GMP by itself does not confer resistance or recalcitrance to killing. Instead, our findings indicate c-di-GMP modulation to correlate with the modulation of brlR transcript abundance, BrlR production, modulation of BrlR-target gene expression, with high c-di-GMP levels correlating with enhanced BrlR production and BrlR-target gene expression, resulting in increased MIC and decreased susceptibility to antimicrobial agents. While it is unclear how c-di-GMP contributes to increased expression of brlR, our findings suggest c-di-GMP to be required for BrlR function, by enabling DNA binding to BrlR-target promoters PmexA and PmexE. Moreover, the c-di-GMP-dependent activation of BrlR requires the activity of the TCS hybrid SagS. Together with previous findings describing the function of SagS in promoting the initial steps of biofilm formation (Petrova & Sauer, 2011), these data suggest that SagS potentially functions to activate a specific diguanylate cyclase and thus, to elevate c-di-GMP levels following the transition to surface-associated growth (Fig. 1). In turn, this cyclase produces a tolerance- or protection-specific pool of c-di-GMP that activates BrlR to bind to its target DNA sites. However, the identity of this diguanylate cyclase remains unclear as neither of the diguanylate cyclases PA4843 or PA4929 function to produce the c-di-GMP pool that specifically activates BrlR.
Our findings indicate that in P. aeruginosa biofilms, resistance is a function of biofilm development and is regulated by the sensor hybrid SagS, which acts as a molecular switch in the transition of P. aeruginosa (i) from the motile to the sessile lifestyle, and (ii) from a susceptible to a highly resistant biofilm phenotype (Fig. 1). Thus, drug resistance and recalcitrance of biofilm cells to killing by antimicrobial agents is activated by a regulatory pathway, in which cues associated with transition to the surface are sensed by SagS and relayed to activate BrlR via elevation of c-di-GMP levels. Moreover, our findings indicate the presence of a tolerance- or protection-specific pool of c-di-GMP, which only contributes to resistance of P. aeruginosa cells when exceeding 42–55 pmol/mg (Fig. 9).
MATERIAL AND METHODS
Bacterial strains, plasmids, media, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in Lennox Broth (LB, BD Biosciences) in flasks at 220 rpm. Biofilms were grown as described below at 22°C in 20-fold diluted LB. Antibiotics for plasmid maintenance were used at the following concentrations: 50–75 μg/mL gentamicin, 60 μg/mL tetracycline and 200–250 μg/mL carbenicillin for P. aeruginosa; and 20 μg/mL gentamicin, 20 μg/mL tetracycline and 50 μg/mL ampicillin for E. coli.
Table 1.
Strains and plasmids used.
| Strains/Plasmids | Relevant genotype or description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 tonA | Invitrogen Corp. |
| S17-1 | λpir; hsdR pro recA; RP4 2-Tc : : Mu-Km : : Tn7, pro, res−, mod+, StrR, TrmR | (Simon et al., 1983) |
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type strain PAO1 | B.H. Holloway |
| ΔsagS | PAO1, ΔsagS (PA2824) | (Petrova & Sauer, 2011) |
| ΔbrlR | PAO1, ΔbrlR (PA4878) | (Liao & Sauer, 2012) |
| ΔPA4843 | PAO1, allelic gene replacement of PA4843 | This study |
| ΔPA4929 | PAO1, allelic gene replacement of PA4929 | This study |
| ΔbfiS | PAO1; PA4197::ISphoA; TetR | (Petrova & Sauer, 2009) |
| Δpsl | PAO1, WFPA60, ΔpslAB | (Ma et al., 2006) |
| Psl-OE | PAO1, WFPA801, pBAD-psl, arabinose inducible expression of psl operon with psl promoter region replaced by araC-PBAD promoter | (Ma et al., 2006) |
| Pel-OE | PAO1, pBAD-pel, arabinose inducible expression of pel operon with pel promoter region replaced by araC-PBAD promoter | (Ma et al., 2006) |
| Plasmids | ||
| pCR2.1-TOPO | TA cloning vector; KmR; AmpR | Invitrogen Corp. |
| pET101D | Vector for directional cloning and high level V5/6XHis fusion protein expression, AmpR | Invitrogen Corp. |
| pRK2013 | Helper plasmid for triparental mating; mob; tra; KmR | (Figurski & Helinski, 1979) |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD; GmR | (Newman & Fuqua, 1999) |
| pMJT1 | araC-PBAD cassette of pJN105 cloned into pUCP18, AmpR (CarbR) | (Kaneko et al., 2007) |
| pEX18Gm | Allelic replacement suicide vector; pUC18 MCS; oriT+; sacB+; GmR | (Hoang et al., 1998) |
| pMini-CTX-lacZ | attP site-specific integration vector, TetR | (Becher & Schweizer, 2000) |
| pEX18-PA4843 | Vector for allelic gene replacement of PA4843 | This study |
| pMJT-dipA | dipA (PA5017) cloned into pMJT1; AmpR (CarbR) | (Basu Roy et al., 2012) |
| pMJT-rbdA | rbdA (PA0861) cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-PA4843 | PA4843 cloned into pMJT1; AmpR (CarbR) | (Petrova & Sauer, 2012) |
| pMJT-PA4929 | PA4929 cloned into pMJT1; AmpR (CarbR) | This study |
| pMJT-brlR-His6V5 | C-terminal His6/V5-tagged brlR cloned into pMJT1; AmpR (CarbR) | (30) |
| pJN-PA4843 | PA4843 cloned into pJN105; GmR | (Petrova & Sauer, 2012) |
| pJN-brlR | brlR (PA4843) cloned into pJN105; GmR | (Liao & Sauer, 2012) |
| PbrlR-brlR-His6V5 | pMini-CTX harboring C-terminal His6/V5-tagged brlR under the control of the brlR promoter region (1–120 bp upstream of the brlR start codon) | This study |
Strain Construction
Isogenic PA4843 mutant was constructed by allelic replacement using sucrose-counter-selection as previously described (Schweizer & Hoang, 1995) with the gene replacement vector pEX18Gm (Hoang et al., 1998). Overexpression of PA4843, PA4929, rbdA, dipA, and brlR was accomplished by placing the respective genes under the control of an arabinose-inducible PBAD promoter in the pJN105 (Newman & Fuqua, 1999) or pMJT1 (Kaneko et al., 2007) vectors. C-terminal His6V5-tagging of BrlR was accomplished by subcloning brlR or brlR plus 1–120 bp upstream of the brlR start codon into pET101D (Invitrogen). C-terminal His6/V5-tagged brlR under control of its promoter was subsequently cloned into the mini-CTX vector (Hoang et al., 2000). The identity of vector inserts was confirmed by sequencing, and proper integration of the pMini-CTX vectors confirmed by PCR using Pser-up/-down PCR primers. Plasmids were introduced into P. aeruginosa via conjugation or electroporation. Primers used for strain construction are listed in Table S1.
Biofilm formation
For biofilm antibiotic susceptibility testing, biofilms were grown in a continuous flow tube reactor system (1 m long size 13 silicone tubing, Masterflex, Cole Parmer, Inc.) with an inner surface area of 25 cm2 at a flow rate of 0.1 ml/min, and in flow cell reactors (BioSurface Technologies) which also allowed for the analysis of biofilm architecture as previously described (Sauer et al., 2002, Sauer et al., 2004, Petrova & Sauer, 2009). Biofilms were grown in 20-fold diluted LB medium. The same growth conditions were used to cultivate biofilms to obtain protein and RNA. For plasmid maintenance, antibiotics were used as previously described (Liao & Sauer, 2012) in tube reactors at the following concentrations: 2 μg/mL gentamicin and 10 μg/mL carbenicillin.
In vivo quantification of c-di-GMP from P. aeruginosa
Cyclic di-GMP (c-di-GMP) was extracted in triplicate from wild-type and mutant strains using heat and ethanol precipitation (Morgan et al., 2006) and quantitated essentially as previously described (Petrova & Sauer, 2011, Basu Roy et al., 2013). Briefly, c-di-GMP was extracted in triplicate from wild-type and mutant strains grown planktonically to exponential phase or as biofilms for 3 days using heat and ethanol precipitation followed by centrifugation. Supernatants were combined, dried using a Speed-Vac and resuspended in 10 mM ammonium acetate. Samples (20 μl) were analyzed using an Agilent 1100 HPLC equipped with an autosampler, degasser, and Targa column (2.1 × 40 mm; 5 μm) at a detector set to 253 nm, and separated using a reverse-phase C18 flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B (buffer A, 10 mM ammonium acetate; buffer B, methanol plus 10 mM ammonium acetate). Commercially available cyclic di-GMP was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts.
Planktonic antibiotic susceptibility testing
To determine the role of c-di-GMP in antimicrobial susceptibility, P. aeruginosa strains grown planktonically in LB medium at 37°C to exponential phase were treated with tobramycin (50 μg/ml) or norfloxacin (150 μg/ml) for 1 hr, and subsequently homogenized, serially diluted and spread-plated onto LB agar. Viability was determined via CFU counts. Susceptibility is expressed as log reduction in CFU counts. Under the conditions tested, a total of 5×107±2×106 CFU/ml were detected on average for exponential phase planktonic cells.
Minimum inhibitory concentration (MIC) determination
The minimum inhibitory concentration (MIC) to tobramycin was determined by performing a series of 2-fold dilutions in LB medium, using 96-well microtiter plates. The method was adapted from Andrews (Andrews, 2001). Carbenicillin (200 μg/ml) was added for plasmid maintenance. The tobramycin concentrations used ranged from 0.08–40 μg/ml. The inoculum was ~104 cells per well, and growth inhibition was observed following overnight incubation at 37°C. MIC was defined as the lowest antibiotic concentration that yielded no visible growth.
Biofilm antibiotic susceptibility assays
For all susceptibility assays, biofilms were grown for 3 days. Under the conditions tested, a total of 3×109±1.5×108 CFU/biofilm were obtained on average following 3 days of biofilm growth. Following exposure of biofilms to tobramycin (150 μg/ml) and norfloxacin (450 μg/ml), biofilms were harvested by squeezing the tubing, followed by the extrusion of the cell paste as previously described (Sauer & Camper, 2001). To ensure complete disaggregation of cell aggregates, the resulting suspension was first homogenized using a tissue tearor, then serially diluted, and spread plated onto LB agar. Viability was determined via CFU counts. Susceptibility is expressed as log reduction.
The biofilm minimum bactericidal concentration (biofilm-MBC) has been defined as the concentration at which no further increase in log reduction is observed (Monzon et al., 2001, Villain-Guillot et al., 2007, Moriarty et al., 2007). To determine whether elevated levels of c-di-GMP affect the biofilm-MBC and resistance to killing, PAO1, ΔsagS, ΔsagS/pMJT-PA4929, and ΔsagS/pMJT-PA4843 cells were grown as biofilms for 3 days, after which time the medium was switched to the same medium containing tobramycin or norfloxacin at concentrations previously demonstrated to eradicate ΔsagS biofilm cells (Gupta et al., 2013). Following 24 hr of exposure to the antibiotic under continuous flow at 0.1 ml/min, biofilms were harvested and the surviving bacteria enumerated by viable counts.
Immunoblot analysis
Abundance of His6/V5-tagged BrlR present in exponential phase planktonic cells and biofilm cells was assessed by SDS-PAGE and immunoblot analysis. Briefly, total cell extracts from biofilms were obtained by sonication as previously described (Sauer et al., 2002) followed by centrifugation for 2 min at 21,200 × g to pellet unbroken cells. The protein concentrations were determined using a modified Lowry assay and bovine serum albumin as a standard (Peterson, 1977). The samples (3–15 μg) were resolved on an 11% polyacrylamide gel and subsequently transferred onto PVDF membrane using a TurboTransblot apparatus (Biorad). Western blots were probed with anti-V5 antibodies (Invitrogen) and developed using ImmunStar WesternC chemiluminescent reagents (Biorad). Following transfer, SDS/PAGE gels were Coomassie-stained to ensure equal loading.
Quantitative reverse transcriptase PCR (qRT-PCR)
Isolation of mRNA and cDNA synthesis was carried out as previously described (Allegrucci & Sauer, 2007, Allegrucci & Sauer, 2008, Southey-Pillig et al., 2005, Petrova & Sauer, 2009). qRT-PCR was performed using the BioRad CFX Connect Real- Time PCR Detection System and SsoAdvanced™ SYBR® Green Supermix (BioRad) with oligonucleotides listed in Table S1. mreB was used as a control. Relative transcript quantitation was accomplished using the CFX Manager Software (BioRad), by first normalizing transcript abundance (based on the threshold cycle value (Ct)) to mreB followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Streptavidin Magnetic Bead DNA and c-di-GMP Binding Assays
BrlR binding to the promoter regions of the mexAB-oprM and mexEF-oprN operons was determined using the streptavidin magnetic bead DNA binding assay as previously described (Petrova et al., 2011). Briefly, a total of 1 pmol of target DNA (biotinylated PmexA or PmexE, Table S1) was incubated for 30 min at room temperature with 15 μg of total cell extract containing the equivalent of 5 pmol of His6/V5-tagged BrlR (as indicated by immunoblot analysis using purified His6/V5-tagged BrlR) in 25 mM Tris-Cl, pH 8, 5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM EDTA, and 50 ng/uL poly(dI-dC) as nonspecific competitor DNA. Cells extracts were obtained from wild-type PAO1 and ΔsagS biofilms harboring pMJT-brlR-V5/His6 in the presence or absence of pJN-PA4843. For specific competition, non-biotinylated target DNA (up to 20-fold excess of target DNA) was used. c-di-GMP was added where indicated. Streptavidin magnetic beads (Thermo Scientific, 100 μg) were used to capture biotinylated DNA. Following four washes, the proteins co-purifying with the biotinylated DNA were separated by 11% SDS/PAGE and assessed by immunoblot analysis for the presence of BrlR using anti-V5 antibodies (Invitrogen). An aliquot prior to addition of streptavidin magnetic beads was used to determine total BrlR present in each DNA binding assay (loading control).
In order to assess the interaction between c-di-GMP and BrlR, the streptadivin magenitic bead assay was carried out essentially as described above with the following modifications: Instead of biotinylated promoter DNA, 25 pmol of biotinylated c-di-GMP was used; a 10-fold excess of non-biotinylated c-di-GMP was used as a specific competitor. Pulldowns were also carried out in the presence of non-biotinylated c-di-GMP (250 pmol) or by using biofilm cell extracts obtained from PAO1 and ΔsagS overexpressing PA4843. Detection of c-di-GMP binding by BrlR was done by immunoblot analysis as described above.
Statistical analysis
A Student’s t-test was performed for pair-wise comparisons of groups, and multivariant analyses were performed using a 1-way ANOVA followed by a posteriori test using Sigma Stat software.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 AI080710) and the Department of Defense (W81XWH-12-2-0063).
References
- Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M, Sauer K. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol. 2008;190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Wu Je, Zhang L-H. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing fomain. Appl Environ Microbiol. 2010;76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. Extraction and quantification of cyclic di-GMP from Pseudomonas aeruginosa. bio-protocol. 2013 doi: 10.21769/bioprotoc.828. http://www.bio-protocol.org/wenzhang.aspx?id=828. [DOI] [PMC free article] [PubMed]
- Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- Billings N, Ramirez Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. The extracellular matrix component Psl Provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013;9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua SL, Tan SYY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2013;57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, V, Gordon D, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- D’Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- Gupta K, Marques CNH, Petrova OE, Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol. 2013;195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häußler S, Tümmler B, Weißbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29:621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- Häußler S, Ziegler I, Löttel A, Götz Fv, Rohde M, Wehmhöhner D, Saravanamuthu S, Tümmler B, Steinmetz I. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc National Acad Sci. 2012;109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- Khan W, Bernier SP, Kuchma SL, Hammond JH, Hasan F, O’Toole GA. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int Microbiol. 2010;13:207–212. doi: 10.2436/20.1501.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Riddle of Biofilm Resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- Liao J, Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Schurr MJ, Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol. 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wang S, Wang D, Parsek MR, Wozniak DJ. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol Medical Microbiol. 2012;65:377–380. doi: 10.1111/j.1574-695X.2012.00934.x. [DOI] [PubMed] [Google Scholar]
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007:585–507. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon M, Oteiza C, Leiva J, Amorena B. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J Antimicrob Chemother. 2001;48:793–801. doi: 10.1093/jac/48.6.793. [DOI] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TF, Elborn JS, Tunney MM. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br J Biomed Sci. 2007;64:101–104. doi: 10.1080/09674845.2007.11732766. [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited Bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc National Acad Sci. 2012;109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RA, van Langevelde P, Kristjansson M, Maslow JN, Arbeit RD. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin Infect Dis. 1995;20:95–102. doi: 10.1093/clinids/20.1.95. [DOI] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Micro. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Romling R, Amikam D. Cyclic di-GMP as a second messenger. Curr Opinion Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP, Hoang TT. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotech. 1983;1:784–791. [Google Scholar]
- Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the Cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain-Guillot P, Gualtieri M, Bastide L, Leonetti JP. In vitro activities of different inhibitors of bacterial transcription against Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 2007;51:3117–3121. doi: 10.1128/AAC.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FSL. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucl Acids Res. 2009;37:D483–488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.