Abstract
Picornaviruses infecting pigs, described for many years as ‘porcine enteroviruses’, have recently been recognized as distinct viruses within three distinct genera (Teschovirus, Sapelovirus and Enterovirus). To better characterize the epidemiology and genetic diversity of members of the Enterovirus genus, faecal samples from pigs from four provinces in Vietnam were screened by PCR using conserved enterovirus (EV)-specific primers from the 5′ untranslated region (5′ UTR). High rates of infection were recorded in pigs on all farms, with detection frequencies of approximately 90 % in recently weaned pigs but declining to 40 % in those aged over 1 year. No differences in EV detection rates were observed between pigs with and without diarrhoea [74 % (n = 70) compared with 72 % (n = 128)]. Genetic analysis of consensus VP4/VP2 and VP1 sequences amplified from a subset of EV-infected pigs identified species G EVs in all samples. Among these, VP1 sequence comparisons identified six type 1 and seven type 6 variants, while four further VP1 sequences failed to group with any previously identified EV-G types. These have now been formally assigned as EV-G types 8–11 by the Picornavirus Study Group. Comparison of VP1, VP4/VP2, 3Dpol and 5′ UTRs of study samples and those available on public databases showed frequent, bootstrap-supported differences in their phylogenies indicative of extensive within-species recombination between genome regions. In summary, we identified extremely high frequencies of infection with EV-G in pigs in Vietnam, substantial genetic diversity and recombination within the species, and evidence for a much larger number of circulating EV-G types than currently described.
Introduction
A wide variety of viruses infect the domestic pig (Sus scrofa) and many, such as rotaviruses and caliciviruses, are associated with severe enteric disease. A large group of viruses, collectively termed ‘porcine enteroviruses’, members of the virus family Picornaviridae, have also been implicated in a wide range of disease presentations in pigs (Knowles et al., 1979, 2006). A better understanding of their epidemiology and pathogenicity was achieved once it was recognized that the group incorporated members of three different picornavirus genera, Teschovirus, Sapelovirus and Enterovirus, with distinct infection profiles and disease associations (Kaku et al., 2001; Krumbholz et al., 2002; Knowles, 2006). Among these, teschoviruses have been identified as the cause of a frequently severe epidemic form of encephalomyelitis, as well as a range of other systemic disease manifestations including diarrhoea, respiratory disease and myocarditis. Sapeloviruses have also been linked with enteric and respiratory disease presentations and more recently, with polioencephalomyelitis (Lan et al., 2011).
The third group, now classified as members of the genus Enterovirus, were originally isolated from skin lesions of pigs (Knowles, 1988) but have not to date been clearly linked to enteric or other disease presentations (Knowles, 2006). Enteroviruses (EVs) infecting pigs are genetically distinct from other EVs and have recently been assigned as members of species G (EV-G) (Knowles et al., 2012), a separate species from those infecting humans (species A–D), cows (E and F) and non-human primates (A, B, D, H and J). The original isolates of EV-G (PEV-9 and PEV-10) were serologically distinct from each other and are now recognized and reassigned as separate EV-G types, EV-G1 and -G2. Very recently, molecular methods have identified further genetic variants, now classified as EV-G3 and EV-G4 from pigs and wild boars in Hungary (Boros et al., 2011, 2012a), EV-G5 from a sheep (Boros et al., 2012b) and EV-G6 from a pig in Korea (Moon et al., 2012). These type assignments were based on sequence divergence of the VP1 gene, where types display >25 % nucleotide sequence divergence from each other (http://www.picornaviridae.com/); these are a substitute for demonstrating serological relationships, which remain as yet uncharacterized for these newly assigned types.
In the current study, we investigated the infection frequency, disease associations and genetic diversity of EVs in pigs from Vietnam. This study was part of a larger longitudinal cohort research programme within farming communities in Vietnam, providing a baseline survey of enteric pathogens on pig farms in the study province, Dong Thap, located in the Mekong delta.
Results
Infection frequency of EV
Detection frequencies of EV RNA were high within all four surveyed districts of Dong Thap province, with 92 of 102 (90 %) farms testing EV positive for at least one pig. Rates of infection were highest among weaners (93 %) aged 7–14 weeks, and declined moderately with the age of the pigs, with 42 % of those older than 1 year of age continuing to shed detectable levels of EV RNA (Table 1). Frequencies of EV detection in pigs with and without diarrhoea were similar in all age ranges. There was no significant association between viral load as determined by EV threshold cycle (Ct) values and clinical status (data not shown).
Table 1. Detection of EV RNA in pig faecal samples.
| Group | Age range (weeks)* | With diarrhoea | Without diarrhoea | P-value† | ||
| Pigs tested (N) | EV frequency (%) | Pigs tested (N) | EV frequency (%) | |||
| Suckler | <7 | 9 | 89 | 33 | 85 | 1.0 |
| Weaner | 7–14 | 29 | 97 | 29 | 90 | 0.66 |
| Grower | 15–23 | 10 | 70 | 28 | 71 | 1.0 |
| Gilts | 24–52 | 0 | – | 12 | 58 | – |
| Sows, boars | >52 | 22 | 41 | 26 | 42 | 1.0 |
| Total | – | 70 | 74 | 128 | 72 | 0.74 |
Typical age range for group.
Two-tailed Fisher’s exact test.
Genetic diversity of EV variants
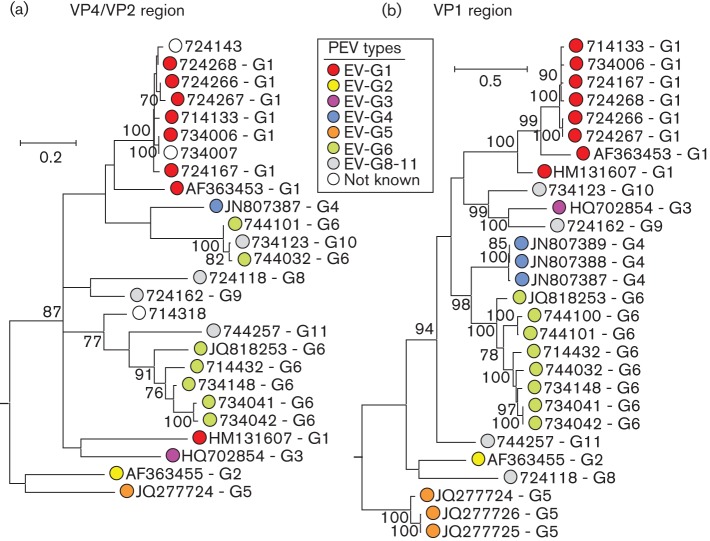
The genetic diversity of EVs detected in a subsample of 20 positive faecal specimens was determined by sequence comparisons in the VP4/VP2 and VP1 regions (Fig. 1). All but one sample could be amplified in the VP4/VP2 region, while 17 of 20 were positive in VP1. Consensus sequences from these two regions identified species G EV variants in all infected pigs. The latter region was additionally used for type assignments; phylogenetic analysis of this region identified six type 1 (EV-G1) variants, seven EV-G6 and four further variants that did not cluster with sequences from this study or with those previously published and available in public sequence databases. These latter variants showed >25 % nucleotide sequence divergence from other EV-G types over the VP1 coding sequences and on consultation with the ICTV Picornavirus Study group, they have been assigned as new types EV-G8–EV-G11 (http://www.picornaviridae.com/enterovirus/ev-g/ev-g.htm).
Fig. 1.
Phylogenetic comparisons of sequences from VP4/VP2 region (nt 810–1250) (a) and VP1 (nt 2469–3317) (b) from study samples and available sequences of other EV-G variants from GenBank. Maximum-likelihood trees were reconstructed using 500 bootstrap resamples to demonstrate the robustness of groupings; values of ≥70 % are shown. The tree was rooted by inclusion of the more divergent EV (bovine) species E variant, GenBank accession no. AF123432 (not shown). The tree was drawn to scale; bar shows indicated evolutionary distance.
Pigs infected with type 1 were typically younger (median age 4 months, range 3–9) than those infected with type 6 (median 7 months, range 5–56) and other types (median 104 months, range 52–165), differences that were significantly different despite the small sample size (P = 0.018; Kruskal–Wallace non-parametric test; Table 2). In contrast, no differences in frequency of diarrhoea or sampling location were observed between EV-G types (Table 2, P>0.05).
Table 2. Clinical characteristics of EV-infected pigs.
| Sample | Location | Age* | Diarrhoea | Ct (s) | EV-G type |
| 714133 | Cao Lanh | 3 | No | 25.5 | 1 |
| 724267 | Chau Thanh | 4 | Yes | 24.6 | 1 |
| 724268 | Chau Thanh | 4 | Yes | 28.1 | 1 |
| 724266 | Chau Thanh | 4 | Yes | 30.1 | Unk/ND |
| 734041 | Thanh Binh | 7 | Yes | 27.3 | 6 |
| 734042 | Thanh Binh | 7 | No | 30.0 | 6 |
| 734148 | Thanh Binh | 7 | No | 30.7 | 6 |
| 714318 | Cao Lanh | 7 | No | 29.9 | Unk/ND |
| 714432 | Cao Lanh | 7 | No | 31.5 | 6 |
| 734006 | Thanh Binh | 9 | Yes | 29.6 | 1 |
| 744100 | Hong Ngu | 9 | Yes | 31.1 | 6 |
| 744101 | Hong Ngu | 9 | No | 28.0 | 6 |
| 734007 | Thanh Binh | 9 | Yes | 29.2 | Unk/ND |
| 734123 | Thanh Binh | 52 | Yes | 25.9 | 10 |
| 744032 | Hong Ngu | 56 | No | 29.9 | 6 |
| 744257 | Hong Ngu | 104 | No | 28.5 | 11 |
| 724118 | Chau Thanh | 165 | No | 29.6 | 8 |
| 724167 | Chau Thanh | Unk | No | 30.5 | 1 |
| 724162 | Chau Thanh | Unk | Yes | 30.3 | 9 |
| 724143 | Chau Thanh | Unk | No | 28.9 | Unk/ND |
ND, Not determined; Unk, Unknown.
Age in months; samples ranged by age.
Recombination in the EV-G genome
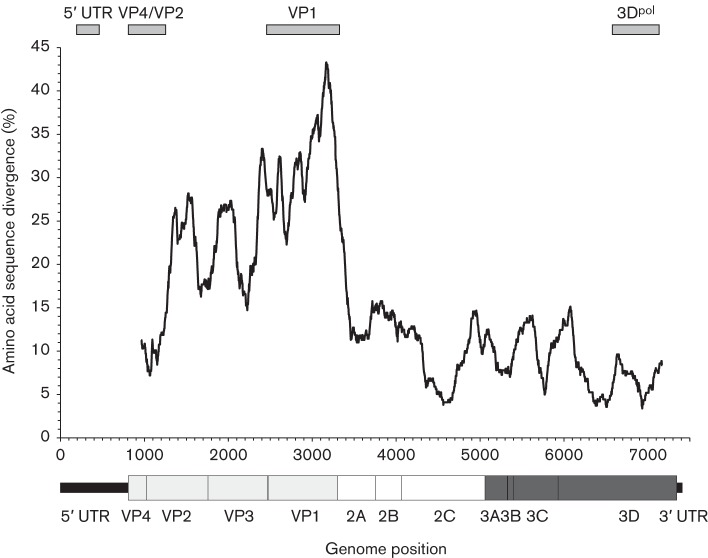
The occurrence of recombination in picornavirus genomes is variable, even between closely related viruses in the same genus. We previously hypothesized that the generation of recombinants was favoured in genome regions showing restricted genetic variability and with consequent greater likelihood of creating biologically compatible genome combinations (Simmonds, 2006). Through a sliding window comparison of complete genome sequences of EV-G, amino acid sequences divergence was greatest in the structural gene (VP4–VP1)-encoding region and much more restricted in the non-structural gene block and in the 5′ untranslated region (5′ UTR, Fig. 2).
Fig. 2.
Amino acid sequence divergence across the genome of EV-G types 1–6. Divergence values were calculated using a window size of 300 bases, incrementing by a codon between windows. The genome diagram beneath is plotted to scale with the x-axis. Genome regions sequenced for recombination analysis (5′ UTR, VP4/VP2, VP1 and 3Dpol) are shown above the graph.
Distinct patterns of divergence and codon usage have been observed between picornavirus groups that undergo frequent recombination between structural and non-structural gene regions and those where recombination is infrequent or undetected (Simmonds, 2006). EV-G sequences showed characteristics of the former, with mean pairwise distances between structural gene sequences of EV-G substantially higher than non-structural gene regions (0.23 compared with 0.09, 2.5 : 1). The structural gene region additionally showed a higher frequency of non-synonymous to synonymous substitutions (dN/dS = 0.20) and more random codon usage (effective number of codons (ENc) of 55; Fares et al., 2002) than non-structural genes (dN/dS = 0.09; ENc of 51). Indeed, the divergence scan (Fig. 2) was highly comparable to that observed in EV species A–C, each of which is documented to undergo frequent recombination between non-structural regions and the 5′ UTR, and in species B, additionally with VP4 (Oberste et al., 2004; Simmonds, 2006).
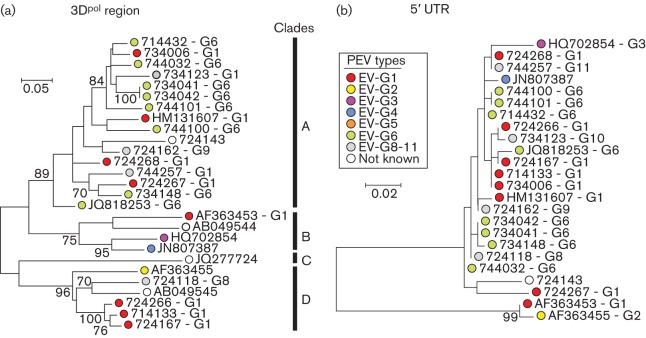
Sequences from the 3Dpol region fell into four main bootstrap-supported clades on phylogenetic analysis of nucleotide sequences (Fig. 3a). Membership of the four clades (designated A–D) bore little relationship to type assignments based on VP1 sequences; members of different types were found within each of the four clades, and conversely, sequences of the same type were also found in different clades. Pairwise distances within and between 3Dpol groups fell into two, largely distinct, ranges (Fig. S1, available in the online Supplementary Material) with a distance threshold matching clade membership of approximately 14 %, comparable to that dividing recombinant forms in other EVs (McWilliam Leitch et al., 2010, 2012). However, variants that were closely similar in VP1, such as the subcluster of three sequences 724167, 714133 and 734006, remained grouped together in 3Dpol (and in VP4/VP2), consistent with a degree of retained genetic linkage between the two regions over short periods of sequence divergence.
Fig. 3.
Phylogenetic comparisons of sequences from 3Dpol (nt 6585–7121) (a) and 5′ UTR (nt 196–470) (b) from the study samples and available sequences of other EV-G variants from GenBank. Maximum-likelihood trees were constructed from nucleotide sequences with 500 bootstrap resamplings to demonstrate robustness of groupings; values of ≥70 % are shown. The tree was drawn to scale; bar shows indicated evolutionary distance.
The phylogeny of VP1 and groupings of EV-G variants into types was mirrored in part in the VP4/VP2 regions but with some exceptions that were indicative of recombination between the two genome regions. The GenBank sequence HM131607 was assigned as EV-G1 based on its grouping with other type 1 variants in VP1 but it adopted a different tree position in VP4/VP2. A more complex change in phylogeny occurs for the type 6 variants obtained in the current study; two of these (744032 and 744101) that cluster with type 6 in the VP1, cluster to form a new clade in VP4 that also includes the newly assigned EV-G10 variant, 734123.
The phylogeny of the 5′ UTR was even less congruent with VP1 than VP4/VP2 (Fig. 3b). All but three variants possessed 5′ UTR sequences that were nearly identical to each other and fell into a separate bootstrap-supported clade that grouped separately from the prototype type 1 and type 2 sequences, GenBank accession nos AF363453 and AF363455. The most divergent 5′ UTR sequence was JQ277724, a type 5 variant isolated from a sheep and proposed to be an interspecies recombinant virus with a 5′ UTR most similar to those of EV-E and -F (Boros et al., 2012b).
Discussion
This study provides evidence for a broad ecology of EV-G and highlights substantial genetic diversity and a propensity for intra-species recombination. High rates of EV-G infection were detected in young pigs; detection frequencies of over 90 % in young pigs and 40 % in adult pigs are consistent with 92 and 20 % positivity rates in the equivalent age groups reported from the Czech Republic (Prodělalová, 2012) although substantially higher than in China (10 and 5 %, respectively (Yang et al., 2013), Hungary (Boros et al., 2011), Italy (Sozzi et al., 2010) and Spain (Buitrago et al., 2010). High rates of infection in young pigs likely reflect their exposure to environmental sources of infection and infection with potentially multiple types of circulating EV-G strains. Although numbers were relatively small, type 1 infections were specifically found in younger pigs (sucklers and weaners; 3–9 weeks) and type 6 infections were primarily detected in the age range 7–9 months, while all but one of the older pigs (age range 52–165 months) were infected with types 8–11. The observations are consistent with successive rounds of infection of pigs in Vietnam, starting with EV-G1, followed by EV-G6 and thereafter more sporadic infections with rarer types. This perhaps mirrors age-associated differences in infection frequencies of different EV serotypes in young children (Khetsuriani et al., 2006). However, it is also possible that observed associations were the result of different sampling times in different farms and districts; this age association requires confirmation with larger numbers.
We found no association between PEV detection and enteric disease (diarrhoea) in this cross-sectional study (Table 1). These findings are consistent with a previous study that clearly differentiated EV-G from more pathogenic porcine picornaviruses (Prodělalová, 2012). In this respect, EV-G infections resemble those of EV species infecting humans that are largely non-pathogenic, appear to be ubiquitous and frequently detected in the enteric and respiratory tracts of healthy people, but for which infections with particular variants may be associated with neurological syndromes. There is limited evidence for systemic infections of pigs with EV-G; one experimental infection study of specific-pathogen-free pigs showed detectable viral RNA in plasma, spinal cord and brain (Yang et al., 2013), and three of 12 inoculated pigs developed flaccid paralysis of the hind limbs. These intriguing results suggest the possibility that certain EV-G types may be pathogenic. Unfortunately, the study did not provide the necessary sequence data to support this conclusion (the cited GenBank accession numbers were incorrect in the manuscript) and, indeed, evidence for pathogenic variants of EV-G has not been reported elsewhere in the literature, either from field epidemiology studies or from laboratory infection trials.
The genetic diversity of EV-G is incompletely characterized. Both prototype strains [EV-G1, originally described as PEV-9, and EV-G2 (PEV-10)] have been detected in recent surveys of domestic pig populations (Sozzi et al., 2010; Ren et al., 2012) and several further examples of EV-G1 were detected in the current study. A substantial proportion of our study group were infected with EV-G6, to date described as a single variant from Korea (Moon et al., 2012). Reports of EV-G3, EV-G4 and EV-G5 are similarly sparse, in each case restricted to the single publications in which they were first described (Boros et al., 2011, 2012a, b). The likelihood that viruses in species G are actually highly diverse genetically (and likely serologically) but incompletely sampled is supported by observations in the current study in which genetic analysis of a relatively small sample identified four new EV types (G8–G11). Further evidence for the currently uncharted diversity of this EV species is provided by analysis of partial VP1 sequences published from previous surveys deposited in GenBank (La Rosa et al., 2006; Sozzi et al., 2010; Nix et al., 2013). Although sequences in these studies were too short for formal type assignments and were not suitable for uncovering robust phylogenetic relationships, two surveys of infected pigs in Italy (La Rosa et al., 2006; Sozzi et al., 2010) generated sequences potentially representing six new types, while a further potential new type was observed in Bolivia (Nix et al., 2013). Larger scale and more systematic surveying of variants infecting pigs in different countries will undoubtedly reveal the existence of much larger numbers of EV-G types in the future.
Several differences in phylogeny relationships were observed between VP1 (used for type assignment) and other genome regions (3Dpol, VP4/VP2 and 5′ UTR). Changes in phylogeny are, by analogy with the better documented human EVs, strongly indicative of recombination events occurring during EV-G diversification. EV-G shares with other EV species a number of genome properties, such as restricted amino acid sequence diversity in NS gene regions and a modularly functioning 5′ UTR, that favour recombination by increasing the likelihood that a chimeric sequence will be biologically viable, as discussed previously in reviews by Simmonds (2006) and Lukashev (2005). Previous descriptions of recombination in EV-G are restricted to those involving the 5′ UTR; Boros et al. (2012b) identified a highly divergent 5′ UTR sequence on an EV-G variant (EV-G5) that grouped with those from bovine EVs (EV species E, F). This probably represents an interspecies recombination event of the type documented previously for example between rhinovirus species A and C (Huang et al., 2009; McIntyre et al., 2010) and between EV species A and B (Santti et al., 1999). Bovine EV 5′ UTR sequences in species E and F are similarly interspersed phylogenetically. In a second study, the EV-G1 variant Ch-ah-f1 (GenBank accession no. HM131607) showed a change in phylogeny relationships in the 5′ UTR with the prototype EV-G1 and G2 strains (GenBank accession nos AF363453 and AF363455) and the type 3 sequence (GenBank accession no. HQ702854). On analysis of the expanded dataset in the current study (Fig. 3b), this finding is actually a manifestation of the separate clustering of the older AF363453 and AF363455 variants from the more recently detected EV-G variants, and not a recombination event specific to Ch-ah-f1. Remarkably, it appears that all existing types circulating in Europe and Asia, including modern versions of EV-G1 and -G2, share a common 5′ UTR sequence acquired in the period following the original isolation dates of the EV-G prototype strains (1973 for EV-G1 and 1975 for EV-G2 respectively; N. J. Knowles, personal communication).
Through the identification of additional phylogenetic incompatibilities in other genome regions, the current study additionally identifies extensive recombination between structural (VP1) and non-structural gene regions (3Dpol) and within the capsid (between VP1 and VP4/VP2). Membership of the four bootstrap-supported clades in 3Dpol indeed bears little relationship to their type assignments based on VP1 sequences (Fig. 3a), while further, different, recombination events probably underlie the altered position of the type 1 variant, HM131607, and splitting of sequences from study samples assigned as EV-G6 into two clades in the VP4/VP2 region (Fig. 1a). Clades in 3Dpol show a degree of divergence from each other comparable to those previously characterized in species A and B human EVs, and which were used to subdivide members of the same serotype into a series of recombinant forms (McWilliam Leitch et al., 2010, 2012). In EV-G, however, these groups are not numerous, and individual 3Dpol clades contain examples of several different types; this is rarely observed in species A (EV71) or species B (echoviruses 9, 11 and 30) EVs. These differences reflect probable differences in transmission dynamics and reservoir sizes, and potentially differences in recombination frequency between species. Larger datasets are required to compare timescales of recombination events and recombinant form half-lives with those previously established for human EVs and parechoviruses (McWilliam Leitch et al., 2009, 2010, 2012; Calvert et al., 2010).
In conclusion, this study reveals the high frequency of infection with species G EVs in pigs in Vietnam, with almost universal infection among post-weaning pigs. EV-G variants were highly genetically diverse and, although a relatively small number were analysed, suggest much greater genetic diversity within species G than has been characterized to date.
Methods
Study location and design.
The survey was carried out between February and May 2012 in Dong Thap province in southern Vietnam. The study included four of 12 districts (Cao Lanh, Chau Thanh, Hong Ngu and Thanh Binh) from which a total of 102 farms were randomly selected and sampled. From each farm, freshly voided individual faecal samples (~5 g) were randomly collected from 10 pigs, plus up to four samples per farm from any pigs with frank diarrhoea. Samples were recorded as diarrhoeic or not based on faecal consistency. Farmer survey questionnaires were used to collect information on animal and farm characteristics as well as farming practices. The study was approved and implemented by the Subdepartment of Animal Health Dong Thap province and veterinary students from Nong Lam University. A total of 198 faecal samples were subsampled from the total sample set, representing one to three randomly selected pigs per farm, including all of the 70 diarrhoeic samples.
Porcine EV screening.
RNA was extracted from 200 µl of 10 % (w/v) faecal suspensions using a MagNA Pure 96 Viral NA small volume kit (Roche) and an automated extractor (Roche). The presence of PCR inhibitors and RNA quality control were assessed by spiking samples with an RNA internal extraction control (Equine Arterivirus) prior to extraction. The total RNA recovered (60 µl in nuclease-free water) was stored at −80 °C until use. cDNA was screened for EVs by real-time reverse transcription (RT)-PCR using primers from the 5′ UTR as described previously (Beld et al., 2004). A total of 20 samples that were representative of the geographical range and ages of infected pigs were selected for further genetic analysis (type identification, recombination detection). These were re-extracted from the original specimens using Qiagen RNA extraction kits.
Amplification of VP4/VP2, VP1, 3Dpol and 5′ UTR sequences.
These four genome regions were amplified by nested RT-PCR using primers listed in Table S1. For each PCR, 6 µl extracted RNA was used for cDNA synthesis followed by nested PCR. The reverse transcription step was combined with first-round PCR using Superscript III One-Step RT-PCR system with Platinum Taq High Fidelity according to the manufacturer’s instructions (Invitrogen). The RT-PCR conditions were, sequentially, 43 °C for 1 h and 20 cycles of 53 °C for 1 min and 55 °C for 1 min, followed by 70 °C for 15 min and 94 °C for 2 min. PCR comprised 40 cycles of 94 °C (30 s), 50 °C (30 s), and 68 °C (105 s) and a final extension at 68 °C for 5 min. After that, 1 µl of the first-round reaction was used for the nested PCR with second-round primers and GoTaq DNA polymerase (Promega). PCR amplification included 30 cycles of denaturation (at 94 °C for 30 s), annealing (50 °C, 30 s) and elongation (72 °C, 90 s) in a thermal cycler. Products of the second-round PCR were visualized by agarose gel electrophoresis and sequenced.
Amplicons were sequenced directly using ABI 7200 BigDye capillary sequencing (Applied Biosystems) in a total volume of 10 µl containing 7 µl DNA/RNA-free water, 1 µl BigDye, 1 µl inner sense primer or antisense primer and 1 µl DNA amplicon. Reactions were carried out with 25 cycles of 30 s at 96 °C, 20 s at 50 °C and 4 min at 60 °C.
Sequences were imported and aligned using the SSE sequence editor (Simmonds, 2012) and phylogenetic trees reconstructed using maximum-likelihood methods as implemented in the mega 5.2 software package (Tamura et al., 2011). The optimum maximum-likelihood model (lowest Bayesian information criterion score and typically greatest maximum-likelihood value) for each sequence dataset was first determined and used for phylogenetic reconstruction. Different models were selected for different datasets: Tamura–Nei gamma distribution (five rates selected) with invariant sites for VP4; general time-reversible gamma distribution (five rates) with invariant sites for VP1; Tamura–Nei, gamma distribution (five rates) for 3Dpol and Kimura two-parameter gamma distribution for the 5′ UTR. However, running the datasets with second- or third-choice models created trees with identical topologies and with similar branch lengths and bootstrap values (data not shown). Phylogenetic analysis of each dataset used bootstrap resampling to determine the robustness of grouping.
Codon usage and pairwise distances were computed using built-in functions in the SSE sequence editor. All nucleotide positions were numbered using the annotation provided for the EV-G3 sequence, GenBank accession no. HQ702854 (swine/K23/2008/HUN).
Acknowledgements
We would like to thank the Subdepartment of Animal Health of Dong Thap and all the farmers who participated in the survey for their support. This work has been funded by the Vietnam Initiative on Zoonotic Infections (VIZIONS), part of the Wellcome Trust Major Overseas Programme (UK) (WT/093724/Z/10/Z).
Footnotes
One supplementary table and one supplementary figure are available with the online version of this paper.
References
- Beld M., Minnaar R., Weel J., Sol C., Damen M., van der Avoort H., Wertheim-van Dillen P., van Breda A., Boom R. (2004). Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J Clin Microbiol 42, 3059–3064 10.1128/JCM.42.7.3059-3064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á., Pankovics P., Reuter G. (2011). Characterization of a novel porcine enterovirus in domestic pig in Hungary. Infect Genet Evol 11, 1096–1102 10.1016/j.meegid.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Boros A., Nemes C., Pankovics P., Bíró H., Kapusinszky B., Delwart E., Reuter G. (2012a). Characterization of a novel porcine enterovirus in wild boars in Hungary. Arch Virol 157, 981–986 10.1007/s00705-012-1255-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A., Pankovics P., Knowles N. J., Reuter G. (2012b). Natural interspecies recombinant bovine/porcine enterovirus in sheep. J Gen Virol 93, 1941–1951 10.1099/vir.0.041335-0 [DOI] [PubMed] [Google Scholar]
- Buitrago D., Cano-Gómez C., Agüero M., Fernandez-Pacheco P., Gómez-Tejedor C., Jiménez-Clavero M. A. (2010). A survey of porcine picornaviruses and adenoviruses in fecal samples in Spain. J Vet Diagn Invest 22, 763–766 10.1177/104063871002200519 [DOI] [PubMed] [Google Scholar]
- Calvert J., Chieochansin T., Benschop K. S., McWilliam Leitch E. C., Drexler J. F., Grywna K., da Costa Ribeiro H., Jr, Drosten C., Harvala H. & other authors (2010). Recombination dynamics of human parechoviruses: investigation of type-specific differences in frequency and epidemiological correlates. J Gen Virol 91, 1229–1238 10.1099/vir.0.018747-0 [DOI] [PubMed] [Google Scholar]
- Fares M. A., Elena S. F., Ortiz J., Moya A., Barrio E. (2002). A sliding window-based method to detect selective constraints in protein-coding genes and its application to RNA viruses. J Mol Evol 55, 509–521 10.1007/s00239-002-2346-9 [DOI] [PubMed] [Google Scholar]
- Huang T., Wang W., Bessaud M., Ren P., Sheng J., Yan H., Zhang J., Lin X., Wang Y. & other authors (2009). Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS ONE 4, e6355 10.1371/journal.pone.0006355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku Y., Sarai A., Murakami Y. (2001). Genetic reclassification of porcine enteroviruses. J Gen Virol 82, 417–424 [DOI] [PubMed] [Google Scholar]
- Khetsuriani N., Lamonte A., Oberste M. S., Pallansch M. (2006). Neonatal enterovirus infections reported to the national enterovirus surveillance system in the United States, 1983–2003. Pediatr Infect Dis J 25, 889–893 10.1097/01.inf.0000237798.07462.32 [DOI] [PubMed] [Google Scholar]
- Knowles N. J. (1988). The association of group III porcine enteroviruses with epithelial tissue. Vet Rec 122, 441–442 10.1136/vr.122.18.441 [DOI] [PubMed] [Google Scholar]
- Knowles N. J. (2006). Porcine enteric picornaviruses. In Diseases of Swine, pp. 337–345 Edited by Straw B. E., Zimmermann J. J., D’Allaine S. D., Taylor D. J. Ames: Blackwell [Google Scholar]
- Knowles N. J., Buckley L. S., Pereira H. G. (1979). Classification of porcine enteroviruses by antigenic analysis and cytopathic effects in tissue culture: description of 3 new serotypes. Arch Virol 62, 201–208 10.1007/BF01317552 [DOI] [PubMed] [Google Scholar]
- Knowles N. J., Hovi T., Hyypia T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., Skern T., Stanway G., Yamashita T., Zell R. (2012) Picornaviridae. In Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses, pp. 855–880 Edited by King A. M. Q., Lefkowitz E., Adams M. J., Carstens E. B. London: Academic Press [Google Scholar]
- Krumbholz A., Dauber M., Henke A., Birch-Hirschfeld E., Knowles N. J., Stelzner A., Zell R. (2002). Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J Virol 76, 5813–5821 10.1128/JVI.76.11.5813-5821.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Muscillo M., Di Grazia A., Fontana S., Iaconelli M., Tollis M. (2006). Validation of rt-PCR assays for molecular characterization of porcine teschoviruses and enteroviruses. J Vet Med B Infect Dis Vet Public Health 53, 257–265 10.1111/j.1439-0450.2006.00955.x [DOI] [PubMed] [Google Scholar]
- Lan D., Ji W., Yang S., Cui L., Yang Z., Yuan C., Hua X. (2011). Isolation and characterization of the first Chinese porcine sapelovirus strain. Arch Virol 156, 1567–1574 10.1007/s00705-011-1035-7 [DOI] [PubMed] [Google Scholar]
- Lukashev A. N. (2005). Role of recombination in evolution of enteroviruses. Rev Med Virol 15, 157–167 10.1002/rmv.457 [DOI] [PubMed] [Google Scholar]
- McIntyre C. L., McWilliam Leitch E. C., Savolainen-Kopra C., Hovi T., Simmonds P. (2010). Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J Virol 84, 10297–10310 10.1128/JVI.00962-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam Leitch E. C., Bendig J., Cabrerizo M., Cardosa J., Hyypiä T., Ivanova O. E., Kelly A., Kroes A. C., Lukashev A. & other authors (2009). Transmission networks and population turnover of echovirus 30. J Virol 83, 2109–2118 10.1128/JVI.02109-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam Leitch E. C., Cabrerizo M., Cardosa J., Harvala H., Ivanova O. E., Kroes A. C., Lukashev A., Muir P., Odoom J. & other authors (2010). Evolutionary dynamics and temporal/geographical correlates of recombination in the human enterovirus echovirus types 9, 11, and 30. J Virol 84, 9292–9300 10.1128/JVI.00783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam Leitch E. C., Cabrerizo M., Cardosa J., Harvala H., Ivanova O. E., Koike S., Kroes A. C., Lukashev A., Perera D. & other authors (2012). The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol 86, 2676–2685 10.1128/JVI.06065-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H. J., Song D., Seon B. H., Kim H. K., Park S. J., An D. J., Kim J. M., Kang B. K., Park B. K. (2012). Complete genome analysis of porcine enterovirus B isolated in Korea. J Virol 86, 10250 10.1128/JVI.01548-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix W. A., Khetsuriani N., Peñaranda S., Maher K., Venczel L., Cselkó Z., Freire M. C., Cisterna D., Lema C. L. & other authors (2013). Diversity of picornaviruses in rural Bolivia. J Gen Virol 94, 2017–2028 10.1099/vir.0.053827-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Peñaranda S., Pallansch M. A. (2004). RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J Virol 78, 2948–2955 10.1128/JVI.78.6.2948-2955.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodělalová J. (2012). The survey of porcine teschoviruses, sapeloviruses and enteroviruses B infecting domestic pigs and wild boars in the Czech Republic between 2005 and 2011. Infect Genet Evol 12, 1447–1451 10.1016/j.meegid.2012.04.025 [DOI] [PubMed] [Google Scholar]
- Ren L., Zhang W., Yang S., Shen Q., Fan K., Hua X. (2012). Sequencing of a porcine enterovirus strain prevalent in swine groups in China and recombination analysis. Vet Microbiol 159, 265–268 10.1016/j.vetmic.2012.03.036 [DOI] [PubMed] [Google Scholar]
- Santti J., Hyypiä T., Kinnunen L., Salminen M. (1999). Evidence of recombination among enteroviruses. J Virol 73, 8741–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. (2006). Recombination and selection in the evolution of picornaviruses and other Mammalian positive-stranded RNA viruses. J Virol 80, 11124–11140 10.1128/JVI.01076-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. (2012). SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes 5, 50 10.1186/1756-0500-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi E., Barbieri I., Lavazza A., Lelli D., Moreno A., Canelli E., Bugnetti M., Cordioli P. (2010). Molecular characterization and phylogenetic analysis of VP1 of porcine enteric picornaviruses isolates in Italy. Transbound Emerg Dis 57, 434–442 10.1111/j.1865-1682.2010.01170.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang Y., Shen Q., Zhang W., Hua X. (2013). Prevalence of porcine enterovirus 9 in pigs in middle and eastern China. Virol J 10, 99 10.1186/1743-422X-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]