Abstract
Type 2 diabetes (T2DM) is an important risk factor for cardiovascular disease (CVD)—the leading cause of death in the US. Yet not all subjects with T2DM are at equal risk for CVD complications; the challenge lies in identifying those at greatest risk. Therapies directed towards treating conventional risk factors have failed to significantly reduce this residual risk in T2DM patients. Thus newer targets and markers are needed for the development and testing of novel therapies. Herein we review two complementary mass spectrometry-based approaches—Mass Spectrometric Immunoassay (MSIA) and tandem mass spectrometry as multiple reaction monitoring (MRM)—for the analysis of plasma proteins and post translational modifications (PTMs) of relevance to T2DM and CVD. Together, these complementary approaches allow for high-throughput monitoring of many PTMs and the absolute quantification of proteins near the low picomolar range. In this review article, we discuss the clinical relevance of the HDL proteome and Apolipoprotein A-I PTMs to T2DM and CVD as well as provide illustrative MSIA and MRM data on high density lipoprotein (HDL) proteins from T2DM patients to provide examples of how these mass spectrometry approaches can be applied to gain new insight regarding cardiovascular risk factors. Also discussed are the reproducibility, interpretation and limitations of each technique with an emphasis on their capacities to facilitate the translation of new biomarkers into clinical practice.
Keywords: Proteomics, HDL, Apolipoprotein A-I, Diabetes, Cardiovascular Disease
1 Introduction
1.1 An Unmet Clinical Need in Diabetes and Cardiovascular Disease
Nearly 70% of the deaths attributed to type 2 diabetes mellitus (T2DM) are a result of cardiovascular events with the majority due to coronary artery disease [1]. Life expectancy for people with type 2 diabetes decreased by 5–10 years and adults with T2DM have an annual mortality of about 5.4%, which is double the rate for non-diabetic adults [2]. The mechanism(s) behind the association between T2DM and cardiovascular events is not clearly defined. As such, a significant need exists for the identification of causal molecular risk factors as well as assays to predict, characterize and monitor T2DM and associated comorbidities and outcomes. This underscores the value of early and accurate detection, and the importance of accelerating the rate at which new candidate therapies are entering clinical trials. Recent trials on the treatment of T2DM and the risk of cardiovascular disease reveal the need for a better molecular-level understanding of the complexity of these conjoined diseases. In particular, reports from several major studies over the past few years have found that intensive glucose lowering alone does not reduce cardiovascular disease (CVD) outcomes in diabetic populations [3–6]—indicating that therapeutic measures beyond glucose control are needed. Likewise, more recent findings by the ACCORD Study group showed that combined aggressive improvements in glucose, lipid and blood pressure levels did not demonstrate reduction of CVD outcomes [7, 8].
For the drug development industry, these important findings have precipitated a paradigm-shifting FDA Guidance to Industry regarding the development of antidiabetic therapies [9]. Now, in addition to demonstrating efficacious lowering and maintaining of blood glucose levels [using conventional glycated hemoglobin (HbA1c) as a marker], new antidiabetic drugs must be concurrently evaluated (during and after development) for CVD risk. Accordingly, there is an immediate and urgent need for the development of prognostic biomarkers that better predict CVD risk and provide early and accurate indications as to the efficacy of new T2DM treatments to modify CVD risk.
1.2 Clinical Relevance of High Density Lipoprotein as a Biomarker for CVD in Diabetes
Lipoproteins are lipid rich particles responsible for transporting lipids, fats, cholesterol and other insoluble biomolecules such as fat-soluble vitamins. These particles can be classified into five major groups as (from smallest-to-largest) high density lipoprotein (HDL), low density lipoprotein (LDL), intermediate density lipoprotein (IDL), very low density lipoprotein (VLDL), and chylomicrons. In terms of cardiovascular disease, the most studied lipoproteins are HDL and LDL, commonly referred to as “good” and “bad” cholesterol, respectively. Low HDL and high LDL have been linked to increases in CVD events. However, as Navab et al point out [10], as many as 40% of individuals experiencing a cardiovascular event have HDL and LDL cholesterol levels that fall within the normal range. Notably, Kontush and Chapman [11] pointed out that the functions of HDL can be impaired under the hyperglycemic and oxidative stress conditions frequently found in diabetic patients.
Functionally, HDL plays several roles in CVD. The major atheroprotective role involves the removal of free cholesterol from peripheral tissues and its transport to the liver for excretion through the reverse cholesterol transport (RCT) pathway [12]. HDL also has anti-inflammatory and anti-oxidant effects that may impact the progression of atherosclerosis in the context of diabetes [13]. The ability of HDL to exert some of its anti-inflammatory functions may be related to the proteomic composition of certain HDL particles. For example, HDL protects against LDL oxidation—a function that is possibly related to its Apolipoprotein A-I (apoA1) content [14, 15]. Additionally, Green et al have shown that Apolipoprotein J (apoJ, a.k.a. clusterin) in HDL is enriched after niacin therapy [16], a medicine associated with decreased progression of atherosclerosis. Yet, as highlighted by Navab, HDL from subjects with CVD is believed to be inflammatory, increasing monocyte chemotactic activity [17]. Some HDL particles also carry serum amyloid A (SAA – in the forms of SAA1, SAA2 and SAA4), proteins known to elicit an inflammatory cascade by facilitating the binding of HDL to vascular proteoglycans [18]. Given that HDL can mediate both pro and anti-inflammatory effects, the HDL proteome appears to be a promising target for the discovery of biomarkers related to CVD in the context of T2DM, and is discussed as an example of how proteomic approaches can further elucidate the complexities of CVD risk factor discovery.
2 The Application of Proteomics to Apolipoprotein A-I and HDL
In this review article, we focus on an emerging two-pronged approach for the discovery and development of new prognostic biomarkers of CVD risk. The first approach focuses on the relative abundance of posttranslational modifications (PTMs) of targeted proteins. The second approach concentrates on discovery followed by multiplexed absolute quantification of well-defined protein and peptide targets vis-à-vis tandem mass spectrometry-based multiple reaction monitoring (MRM). Focusing on apolipoprotein A-I (apoA1) and HDL constituents as examples, we highlight mass spectrometric immunoassay (MSIA) and MRM as synergistic mass spectrometry-based approaches to top-down (intact protein-level) and bottom-up (peptide surrogate-based) proteomics, respectively, that are poised to establish a pathway for translating biomarkers of T2DM and CVD into clinical practice. Prior to a discussion on HDL proteomics, a brief overview of existing methods for HDL isolation is warranted:
2.1 Isolation of HDL for Proteomic Studies
There are four major ways to isolate HDL for proteomic experiments—all of which have been described [19] or reviewed [20] elsewhere. The most established method is by density which confers the name of high density lipoprotein. This is accomplished by the use of sequential ultracentrifugation. HDL lies between a density of 1.06 and 1.21 g/mL. In this density range, two subfractions exist, the denser HDL3 (1.12 - 1.21 g/mL) and the less the dense HDL2 (1.06 - 1.12 g/mL). Most of the HDL proteins reside in the HDL3 subfraction. A second common approach is to separate HDL by size (6–15 nm). This can be achieved by gel filtration, native PAGE and NMR (non-physical separation). A third approach is to fractionate HDL by charge, into preβ, pre α and α. This can be done using agarose electrophoresis, isoelectric focusing or anion exchange. Notably, the preβ HDL fraction serves as the initial acceptor of cholesterol in the efflux process. Finally, HDL can be isolated by its protein content—which is predominately apoA1 and apoA2. Isolation is accomplished through antibody affinity [20, 21] or, for apoA2, sulfhydryl covalent chromatography.
There is no real concordance between the different HDL techniques. For example, HDL3 contains apoA1 and apoA2 as well as preβ, and α particles. Thus, these separation techniques are based on physico-chemical properties and not HDL function. These techniques and the concerns of using one particle method are well addressed by Gordon et al [19].
Among the different methods to isolate HDL, we focus this review on two high throughout, reproducible techniques. These are the isolation of HDL proteins by ultracentrifugation for MRM, and MSIA (i.e., affinity pipette-tip based extraction) of individual HDL proteins. MSIA can be applied to any HDL protein, but we focus our discussion here on HDL’s major protein apoA1. Given longer sample preparation times and reproducibility challenges, the other methods described above for HDL isolation are more suitable for discovery proteomics rather than population studies.
2.2 Prong 1: Expanded Molecular Definition of HDL Proteins
Conventionally, HDL has been described as a composite of a limited number of apolipoproteins, with apoA1 (a ~28 kDa, monomeric protein) accounting for approximately 70% of the protein composition of the lipoprotein particle [15, 22]. Several studies in the past decade, such as those by Shao et al and Zheng et al [23, 24], have demonstrated that PTMs of apoA1 are of functional significance. In particular, Shao et al demonstrated that Met oxidation is associated with reduced cholesterol efflux activity [25], which is believed to be a main athero-protective mechanism of HDL function. But apoA1 also acts as a cofactor for lecithin cholesterol acyltransferase (LCAT), a critical early enzyme in the RCT pathway which catalyzes the conversion of free cholesterol to cholesterol ester, making it sufficiently hydrophobic to enter the HDL particle [26]. As shown by Nobecourt et al and Park et al [27–29], modifications of HDL, such as glycation of apoA1 inhibits the ability of apoA1 to act as a cofactor for LCAT, reducing the RCT pathway’s ability to function properly.
Additionally, enhanced oxidative stress in T2DM increases lipid peroxides (from fatty acid oxidation) in lipoproteins. In 1998 two reports by Garner et al [30, 31] showed that apoA1 directly reduces lipid peroxides with the concomitant oxidation of its Met residues. At low levels of oxidative stress, the oxidative damage to apoA1 in this process does not impede the protein-protein interaction with LCAT. However, Shao and co-workers [32] showed that apoA1 ceases to be a cofactor for LCAT, reducing the activity of LCAT to 20% of normal activity in conditions of high levels of oxidative stress in vitro. Of equal interest, when exogenously administered, the genetic variant apoA1 Milano (which codes for an Arg173Cys protein mutation) is protective against CVD [33]. Thus, the study of microheterogeneity present in a single constituent of HDL (i.e., different isoforms of apoA1) presents a significant (and biologically well-founded) opportunity for investigating changes in HDL related to T2DM and CVD. Moreover, similar phenomena may hold true for other HDL proteins and other lipoproteins.
In T2DM, the attenuated anti-oxidant capacity of apoA1 and the loss of its ability to act as a cofactor for LCAT in vivo may be due to physical damage to the apoA1 protein molecule. This may be one explanation as to why a significant portion of patients experiencing cardiovascular events have a “healthy” concentration of HDL cholesterol [10]. As such, there is a significant interest in the lipoprotein research community to evaluate and develop markers of CVD based on qualitative changes in the apoA1 protein molecule such as oxidation, glycation, truncation and other PTMs.
2.2.1 Mass Spectrometric Immunoassay and PTMs
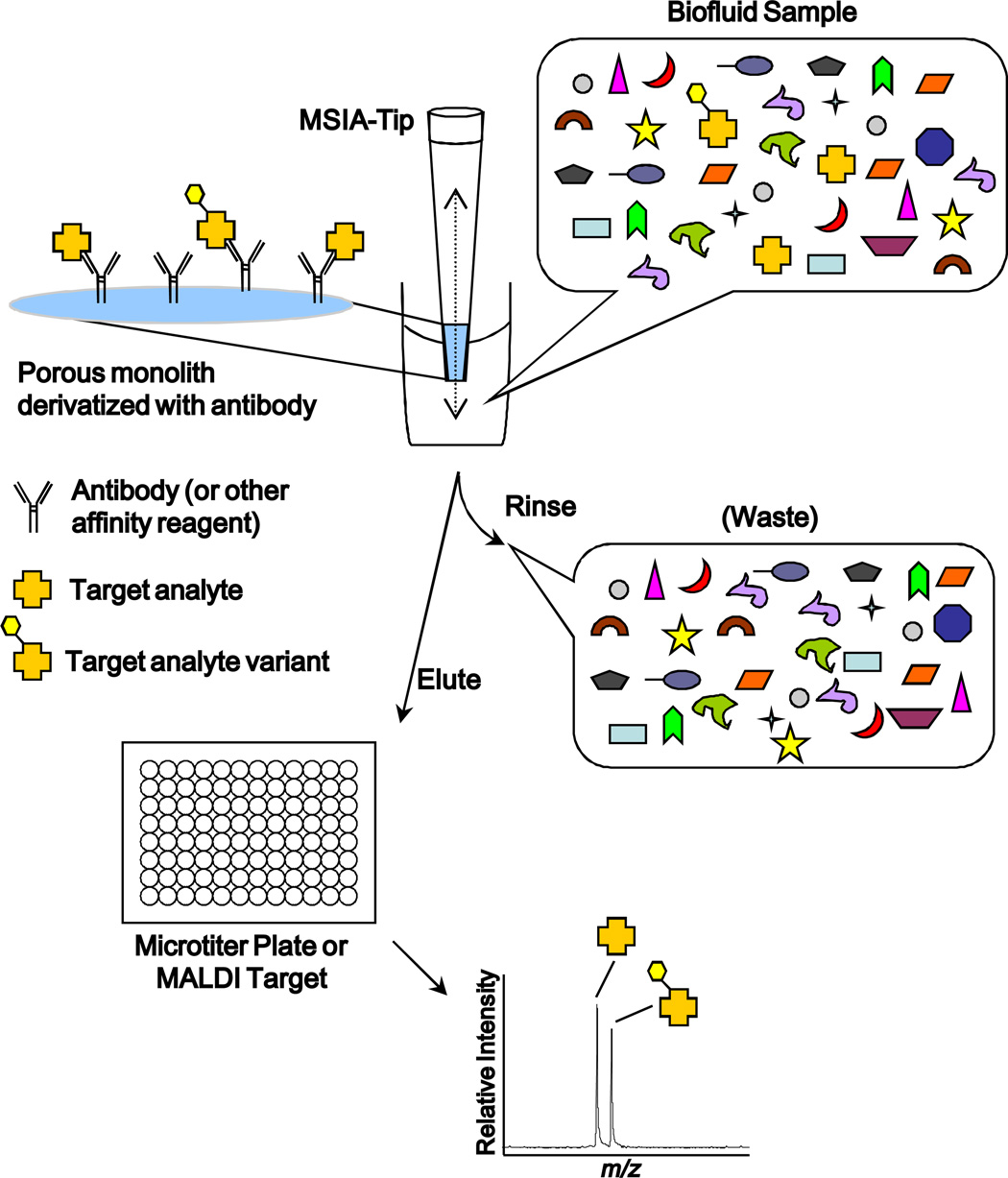
Mass spectrometric immunoassay (MSIA) [21, 34–64] represents a high throughput means by which to identify and quantify such molecular variants. MSIA is based on the isolation of analytes from biological milieu by immobilized antibodies, followed by analysis using mass spectrometry—i.e., immunoaffinity capture in front of mass spectrometry (Figure 1). This general approach to protein analysis was pioneered by Nelson and colleagues in the mid 1990’s and has been modified and adapted by others into various differential forms, e.g., SISCAPA [65] and iMALDI [66]. The specificity of this approach comes from a combination of antibody selectivity paired with medium-to-high resolution mass spectrometry-based protein identification and quantification. The high-value information content provided by the mass accuracy of modern mass spectrometers (i.e., single Dalton-level mass accuracy on intact proteins) is hard to overstate and is best explained in practical terms: When immunoaffinity approaches are employed to isolate intact proteins prior to mass spectral analysis (i.e., using a top-down approach), most single amino acid- and post-translationally modified variants can readily be determined and quantified [21, 34–38, 42,44–64]—(PTMs with a relative abundance under 1% notwithstanding.) In fact, as illustrated below with apoA1, in some cases it is possible to discern both genotype and protein phenotype in a single analysis [21, 55, 56, 58] (Figure 2).
Figure 1.
The MSIA process. A biological sample is repetitively passed through the MSIA-Tip, performing immunoaffinity capture of the targeted analytes. After nonspecific components within the biological fluid are rinsed away, the concentrated target analyte is eluted for analysis by mass spectrometry.
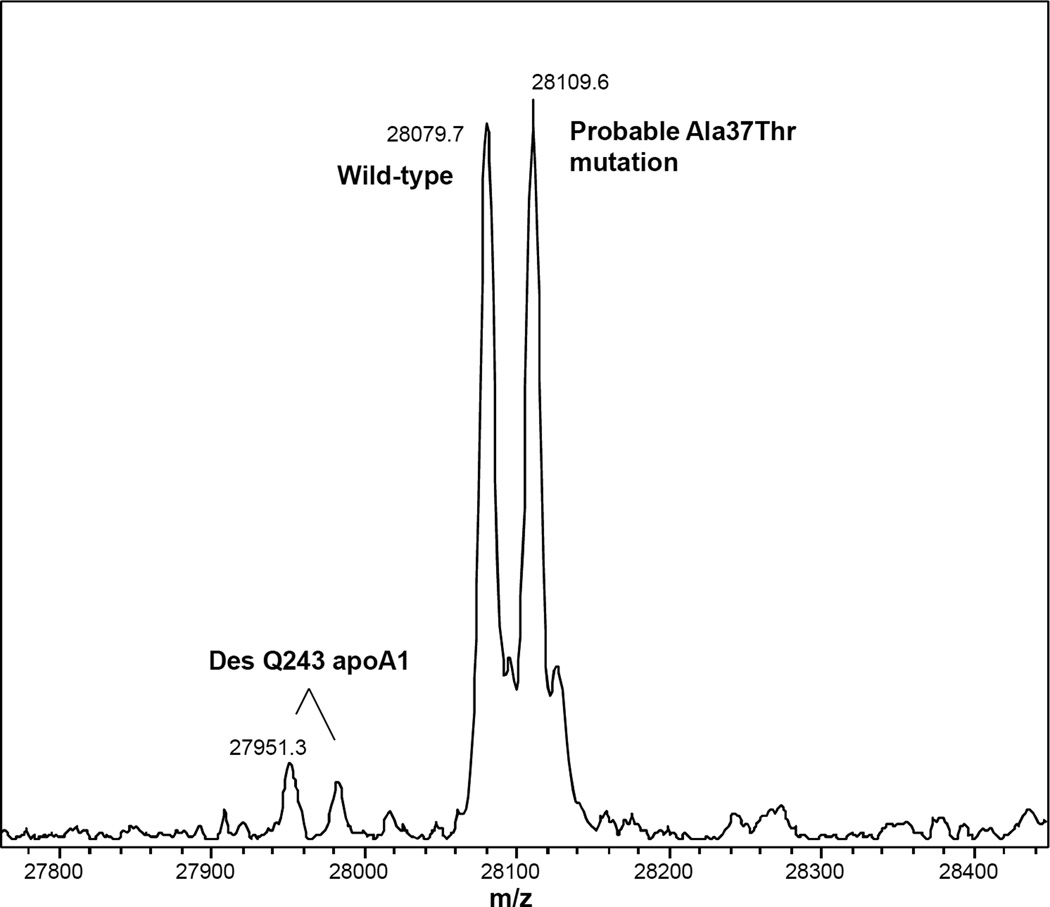
Figure 2.
Charge deconvoluted apoA1 MSIA spectrum from an individual heterozygous for a rare genetic form of apoA1. The observed mass shift of +30 Da suggests that the mutation codes for the previously observed Ala37Thr form of apoA1 [84]. The specificity of antibody capture and observed mass that corresponds with a known human mutation, combined with prior population studies in which similar peak “splitting” has not been observed [21, 36] support the assertion that approximately half of the apoA1 represented in this spectrum carries a sequence-altering mutation.
The benefit of using mass spectrometry as a detection modality as compared to ELISA, RIA and other immuno-recognition based assays is that the output data are qualitative in nature—i.e., able to detect known variants, and discover new variants with similar immuno-reactivity simultaneously—and do not indiscriminately quantify all immuno-reactive protein isoforms captured by the antibody under the heading of a single, nominal protein. (Approaches that lack this degree of specificity miss molecular variants that can possess unique biological function). For example, in Figure 1, the intensity of the first mass spectral peak relates directly to the quantity of native/unmodified target protein present. But, in contrast to ELISA, any qualitatively distinct forms of the analyte protein (at least in terms of molecular weight) are quantified in a separate register (i.e., different m/z value). Notably, no specialized antibodies are required as part of this process; in practice, off-the-shelf monoclonal and/or polyclonal antibodies with the highest available binding affinity (lowest empirical equilibrium dissociation constant, Kd) are employed. Overall, the process may be envisioned as an ultra-high resolution western blot that can be run in high throughput.
The reproducibility of MSIA and MRM-based MSIA for both absolute quantification as well as semi-quantification (in which the relative percent abundance of variant forms is determined) is in line with mass spectrometry-based assays of small molecules that are routinely employed in the clinic—i.e., less than 20 % CV and often much better [59–61, 67–69] (see also Supporting Information Figure 1).
In practice the primary limitation of immunoaffinity capture-based techniques such as MSIA lies in the availability of high quality antibodies. Antibodies must not only possess a low Kd, but must be highly purified. For polyclonal antibodies this means immunogen-based affinity purification and, for monoclonal antibodies, this means at least Protein A or Protein G-based affinity purification. Also, formulation buffers must not contain functional groups that would compete with the covalent binding of antibodies to the immobilization surface so, for example, antibodies formulated in buffers containing non-antibody amino groups (e.g., Tris or carrier proteins) cannot be used when coupling antibodies to carboxyl-functionalized MSIA tips using carbonyldiimidazole or carbodiimide chemistry.
When these pre-requisite conditions are met, it is possible to successfully analyze proteins over a wide range of concentrations by MSIA and related techniques: from albumin at ~40 mg/mL to numerous medium abundance proteins (µg/mL range) [70], to angiotensin I and angiotensin II (low ng/mL) [71, 72], all the way down to parathyroid hormone (PTH) [68] (~30 pg/mL) and B-type natriuretic peptide (BNP) [42] (~10 pg/mL), with individual variant forms of PTH and BNP quantifiable in the single-digit pg/mL range. (Notably, however, quantification of the relative abundance of intact protein variants is limited by the overall signal-to-noise ratio of the mass spectrum. In this case, the limits of detection (LODs) typically range from 0.5 to 1%. This means that some PTMs may not be detected due to low relative abundance.)
The cases of PTH and BNP indicate the future direction of MSIA-related development: identification of individual variant forms of clinically relevant proteins followed by development of variant-specific quantitative assays for those forms with potentially distinctive clinical impact.
2.2.2 MSIA Applied to ApoA1
MSIA targets specific protein molecules—e.g., apoA1—it does not target HDL or the HDL complex per se as HDL is a molecularly ill-defined biological entity. If the target protein exists as part of a larger non-covalent protein complex, the rinses that take place following affinity capture are designed to ensure that binding partners are removed prior to mass spectral analysis. Niederkofler et al first utilized MSIA to investigate apoA1 (and other HDL apolipoproteins) using MALDI-MS [73]. At 28 kDa, however, the 16 Da mass shifts imparted to the apoA1 protein molecule by sulfoxidation of Met residues cannot be resolved by most commercially available MALDI instruments. For technical reasons outside the scope of this article, the electrospray ionization (ESI) process overcomes this loss of resolution, allowing for detection of small mass changes (e.g., due to oxidation) to intact proteins above ~25 kDa in mass.
The major methodological techniques for linking antibodies to pipette tips and performing MALDI- [21, 34–54, 59–64] and ESI-based [55–58, 69] MSIA on human plasma samples have been described in detail in the literature. The specific details of the methodology that was employed to generate the additional illustrative MSIA data presented herein can be found online in Supporting Information.
ApoA1 contains three Met residues; as such, each molecule of apoA1 may contain 0, 1, 2, or 3 oxidized Met residues (Figure 3). The relative distribution of apoA1 in each of these 4 oxidation states can readily be determined with an apoA1 ESI-MSIA. This degree of analytical clarity affords the expression of a unique measurement of apoA1 oxidation which we call total [Met] oxidation capacity. This term refers to the observed percentage of maximum Met oxidation—i.e. the case in which all three Met residues of the apoA1 are oxidized to sulfoxides. To calculate percent total oxidation capacity, the mass spectral peak areas of native (unoxidized), singly, doubly, and triply oxidized apoA1 are multiplied by 0, 0.33, 0.66, and 1, respectively, before normalizing for total apoA1 peak area. We view this as a useful metric by which to quantify apoA1 oxidation in future studies of samples from clinical diabetes trials.
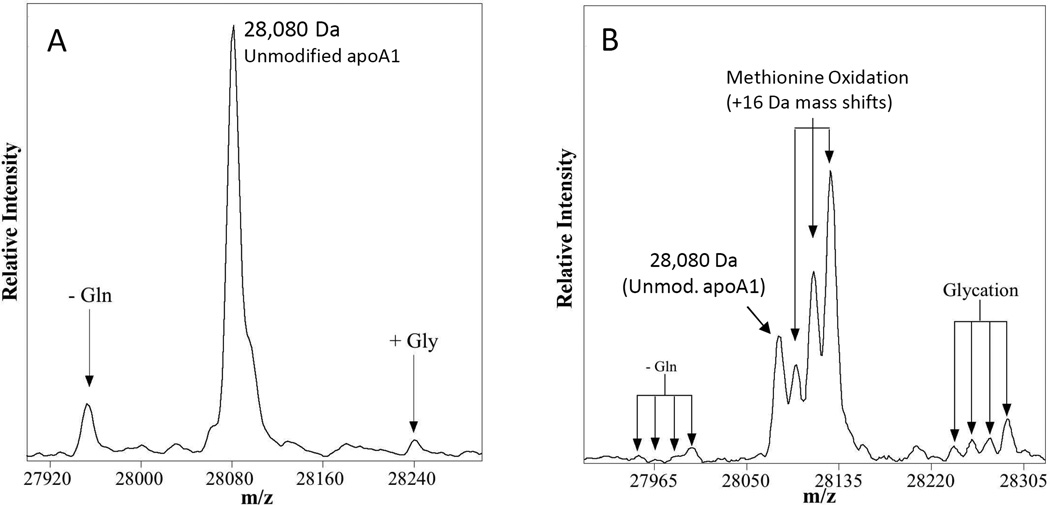
Figure 3.
Representative charge deconvoluted ESI-MSIA-mass spectra of apoA1 from the human popoulation. A) The three observed peaks at 27,951 Da, 28,080 Da and 28,242 Da represent C-terminal truncation of Gln243, unmodified apoA1 and non-enzymatically glycated apoA1, respectively. This spectrum was obtained from a nominally healthy individual. B) Charge deconvoluted spectrum of apoA1 from a patient with T2DM. Sixteen-dalton mass shifts (up to 3 per apoA1 molecule) are caused by oxidation of Met residues. The facts that 1) only three 16-Da oxidation events can occur per apoA1 molecule (one per Met residue; see also Supporting Information Figure 1) and 2) the oxidized forms of apoA1 also occur on truncated and glycated apoA1 support the assignment of these 16-Da mass-shifted peaks as oxidized forms of apoA1.
A portion of the data that we have collected to date using this assay were published by the Nelson group in 2011 as part of a multivariate study of protein heterogeneity in patients ranging from nominally healthy to T2DM to T2DM with heart disease (congestive heart failure, CHF) [57]. In general, the charge deconvoluted mass spectra observed for apoA1 reveal a base peak (most intense peak) at 28,079.8 ± 0.5 Da (SE) that corresponds well to the calculated protonated mass of unmodified apoA1 at 28,079.6 Da (Figure 3). In addition to this main peak, two other peaks at 27,951.8 ± 0.8 Da and 28,242.1 ± 0.7 Da are identified on the basis of mass as C-terminally truncated des-Gln243-apoA1 [73] (calc. MH+avg = 27,951.4 Da) and non-enzymatically glycated apoA1 (calc. MH+avg = 28,241.7 Da). Consistent with these assignments, oxidation of apoA1 was found superimposed on all 3 forms of apoA1 as a series of zero to three +16 Da mass shifts (Figure 3).
Rare heterozygous genetic variants in which a change in amino acid sequence introduces a +30 Da mass shift into half of the mass spectral signal (Figure 2) constitute ~1% of the samples from the general population that we have analyzed to date. Table 1 summarizes a cross-section of the apoA1 ESI-MSIA data published as part of a multivariate dataset in 2011 [57] and includes additional data on apoA1 glycation. Though the clinical relevance of these variant forms of apoA1 has yet to be determined, it is clear that the data provided by ESI-MSIA generate unique insights into the inner workings of this important HDL protein.
Table 1.
Relative Percent Abundance ± SD of apoA1 protein variants observed by ESI-MSIA in human plasma samples whose donors ranged from nominally healthy to T2DM with congestive heart failure (CHF). No statistically significant differences between groups are evident at these n-values (ANOVA followed by Tukey's HSD).
| Unmodified ApoA1 | Oxidized ApoA1a | Glycated ApoA1 | C-terminally Truncated ApoA1b | |
|---|---|---|---|---|
| Mass Shift (Da) | 0 Da | +16, +32, +48 Da | +162 Da | −128 Da |
| Controls (n = 7) | 84 ± 13 | 4.0 ± 5.9 | 2.7 ± 1.2 | 5.9 ± 1.6 |
| CHF (n = 7) | 82 ± 9.4 | 6.8 ± 5.6 | 3.1 ± 1.2 | 3.7 ± 1.8 |
| T2DM (n = 9) | 76 ± 9.6 | 7.3 ± 7.2 | 5.0 ± 4.2 | 6.7 ± 3.9 |
| T2DM & CHF (n = 12) | 77 ± 12 | 8.4 ± 7.2 | 4.4 ± 0.9 | 6.5 ± 5.5 |
Data presented as percent of total methionine oxidation capacity (see text)
I.e., Native apoA1 des Q243
2.3 Prong 2: Expanded Definition of HDL Protein Composition and Subsequent Quantification
Recent proteomic studies have increased the compositional description of the HDL proteome significantly beyond the traditional concept of an HDL particle. For example, Vaisar et al [74] not only demonstrated both a remarkable diversity of proteins and peptides identified within HDL, but also reported a high representation of proteins involved in non–lipid transport functions, including acute phase response, complement regulation, proteolysis, and coagulation. Using peptide spectral counts and extracted ion chromatograms, they noted a trend for an increase in Apolipoprotein E (apoE), paraoxonase 1 (PON1), Complement C3, Apolipoprotein L-I (apoL1) and Apolipoprotein C-IV (apoC-IV) in the HDL particles of subjects with CVD; and HDL enriched in apoJ in healthy controls.
As a follow-on to these discovery studies, we have explored the protein composition of HDL using Multiple Dimensional Protein Identification Technology “MudPIT” (LC-LC-MS/MS). Using this approach, we isolated HDL from a control subject, proteolytically digested one microgram, and fractionated it with separate strong cation exchange (SCX) and reversed phase (RP) columns as part of an exploratory LC-LC-MS/MS run (details on methodology are described online in the Supporting Information). In excess of 60 proteins were identified as part of the HDL complex (Table 2), which is in good agreement with Vaisar et al [74]. However, after such discovery-oriented analyses, the subsequent challenge for conventional proteomics lies in studying these HDL-complexed proteins in large population cohorts.
Table 2.
HDL proteins identified using MudPIT
| Lipid Metabolism | Complement Regulation | Acute Phase Response |
|---|---|---|
| apoA-I | Complement component C4B | VDBP |
| apoA-II | Complement component C4A | Transthyretin |
| apoA-IV | Complement component C9 | Plasminogen |
| apoE | Vitronectin | Others |
| apoC-I | Complement component C3 | Albumin |
| apoC-II | Proteinase Inhibitor | Apo B 100 |
| apoC-III | alpha-2-antiplasmin | Platelet factor 4* |
| apoC-IV* | Angiotensinogen | α-1antichymotrypsin |
| apoL-I | α-2-HS-glycoprotein | Fibronectin* |
| apoM | Kininogen-1 | Fibrocystin L* |
| apoF | haptoglobin-related protein | Titin* |
| apoD | AMP | Teneurin-3* |
| apoH | SERF1 | Tolloid like protein 1* |
| Clusterin | AHSG | Bloom Syndrome* protein |
| LCAT | Acute Phase Response | |
| CETP* | Fibrinogen | Uncharacterized* Proteins |
| PLTP* | PRBP | |
| PON1 | IAHC | |
| SAA | Hemopexin | |
| SAA 1* | transferrin | |
| SAA 2* | A1G1 | |
| SAA 4* | A1M* |
Abbrevations
PON1: Paraoxonase 1
A1GI: alpha 1 glycoprotein 1
LCAT: Lecithin-cholesterol acyltransferase
PLTP:Phospholipid transfer protein
IAHC:inter-α-trypsin inhibitor heavy chain H4
PRBP: Plasma retinol–binding protein
A1M: α-1-microglobulin/bikunin
AMP,bikunin
SERF1, serpin peptidase inhibitor
AHSG, Alpha-2-HS-glycoprotein
SAA, Serum Amyloid Protein
Clusterin: ApoJ
VDBP: Vitamin D Binding Proteins
Not verified by MRM
2.3.1 MRM in Protein Quantification
By design, mass spectrometry-based MRM approaches are naturally suited to such high-throughput, high specificity analyses—and have been used in this mode for over 15 years to quantify small molecules. Additionally, MRM offers high dynamic range and the ability to quantify up to 100 targets (peptides) in a single run. As demonstrated by Kuzyk, Yocum and others [75, 76], MRM has emerged at the forefront of quantitative proteomics as a viable approach for quantifying proteins of moderate to low concentration in plasma.
A prerequisite of the approach is that the peptide targets of interest and their optimal mass-specific gas-phase molecular fragmentation transitions must be known. These targets can be determined empirically using discovery-based shot-gun proteomic data, and/or computationally using algorithms designed around optimal digest and (electrospray) ionization probabilities (e.g., as demonstrated by Lopez et al [68]). Moreover, site-specific PTM’s may be included in the quantitative profile—either having been directly observed during previous analyses (e.g., via MSIA or other efforts directed toward qualitative discovery), or anticipated through causal biological pathways. Notably, as described by Abbatiello et al [77], there is ongoing effort across different laboratories to define comprehensive lists of MRM peptide transitions for specific applications.
2.3.2 MRM Data Qualification and Analysis
Quantitative MRM experiments may be conducted after the transition list has been optimized, selecting at least three transitions per peptide target with the corresponding transition-specific instrument settings. In practice, chromatographic co-elution of the three peptide-specific transitions provides verification that observed signals are indeed genuine. The most sensitive and/or specific transition is typically employed for quantification with the other two transitions serving as “qualifier” transitions. Analyte quantities are determined by dividing the chromatographic MRM peak area of the target peptide by that of an exogenously added stable isotope standard (SIS) peptide analog. Absolute quantification is made possible by creating standard curves to accompany each batch of samples.
The sensitivity of MRM-based analysis for ~ 50 unenriched, medium to high abundance plasma proteins (many of which may be carried on HDL) with limits of quantitation, recovery and linearity has been published by Kuzyk et al [78] and by Anderson and Hunter [70]. In this approach a standard sample is diluted several fold to generate a range of analyte concentrations. These concentrations are combined with SIS peptide mixtures added at the same concentrations anticipated for their naturally occurring in vivo counterparts. The limit of quantitation is defined as the lowest protein concentration that can be detected with CV < 20% on replicate runs. For these unenriched plasma proteins, sensitivity in actual plasma samples down to the low µg/mL range could be achieved. As pointed out by Kuzyk and co-workers [78], however, the on-column raw sensitivity of MRM-based quantitative approaches (in the absence of matrix interferences) can be in the attomole range.
2.3.3 MRM Applied to HDL Proteins
HDL is a good candidate for MRM-based analysis for two reasons: First, HDL carries numerous proteins; second, the most abundant HDL proteins (apoA1 and apoA2) are susceptible to modifications that are of functional significance. The study of HDL by shotgun-based LC-MS/MS in the last few years has resulted in increasing numbers of proteins identified in HDL particles—from 13 [79] up to 75 in our most recent experiments (Table 2). There is a strong interest in the lipid community to identify whether proteins found in HDL are a part of the functional HDL complex or are a result of HDL isolation techniques (e.g., centrifugation and immunoseparation). Emerging evidence by Davidson et al [80] indicates that the proteins identified in HDL can be clustered based on areas of functionality. These areas are metabolism and transport, inflammation, immune system and complement factors, growth factors, hormone-binding proteins, and proteins involved in hemostasis. The HDL surface monolayer can also serve as a binding matrix not only for intact apolipoproteins and associated proteins but also for a wide range of small peptides that were originally thought to be associated mostly with plasma albumin. Davidson’s group discovered more than 100 peptides in the range of 1000 to 5000 Da attached to HDL. The circulating life of these peptides is possibly extended by binding HDL, and other lipoproteins.
Quantitative proteomic profiles have the promise of assigning specific roles to subclasses of HDL in health and disease. One of the major barriers to progress in this field lies in translating discovery experiments (e.g., those by Vaiser et al [74] and Davidson et al [80]) into the study of large clinical cohorts. These translational studies require a high throughput assay that is reproducible across different laboratories. MRM has this capacity to translate discovery proteomics into practice through the analysis of a large number targets in a relatively high throughput manner. Figure 4 illustrates the relative abundance and LC-MRM performance of 25 HDL proteins identified by MudPIT and verified by MRM in patient samples. Most of the proteins identified (e.g., apoA1, apoAII, apoE, apoCIII, PON1, LCAT, Complement C3, Serum Amyloid Protein) are involved in CVD pathogenesis. Using the absolute intensity of extracted ion chromatograms from shotgun LC-MS/MS runs Green et al reported that apoE and apoCII are increased in CVD while, apoJ, phospholipid transfer protein, and apoF are decreased in subjects with CAD [16]. We believe the application of MRM-based proteomics to HDL holds the key to robust validation of results such as these and promises to significantly enhance the quantitative reproducibility and overall performance of proteomics as a discipline.
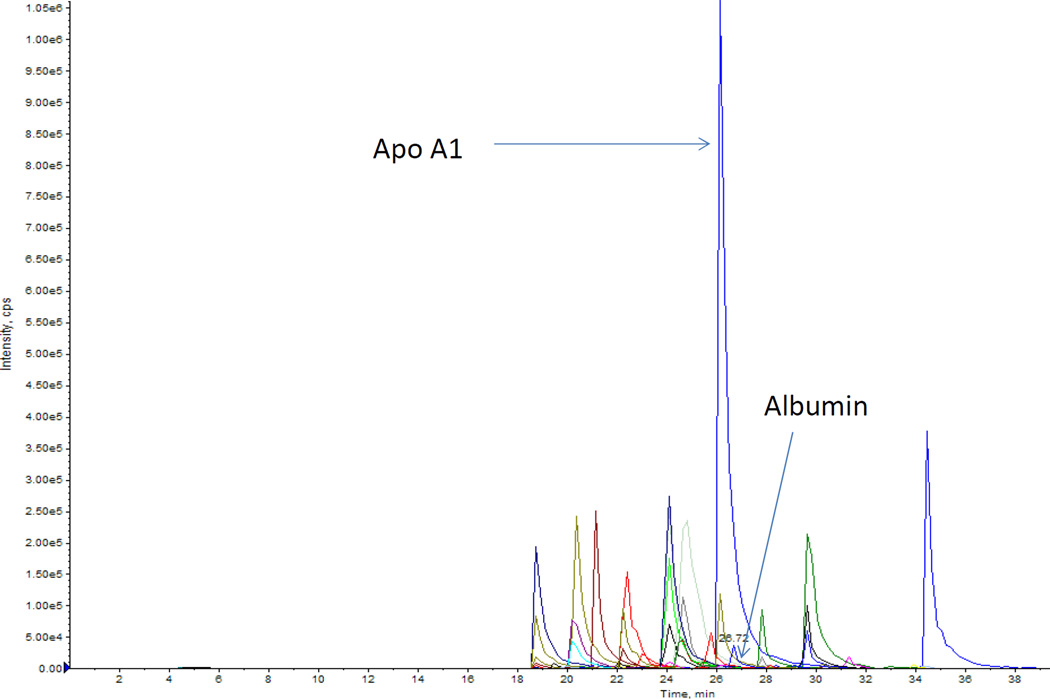
Figure 4.
Twenty five HDL proteins identified by MRM. HDL fractions isolated by ultracentrifugation were subjected to MRM after appropriate sample preparation. A graphical workflow description of MRM can be found in a 2009 paper by Kuzyk et al [75]. The purpose of MRM experiments is to provide a multiplexed platform for monitoring several HDL proteins simultaneously in a high throughput manner. The high abundance of apoA1 and low abundance of albumin reflects the success of HDL isolation by ultracentrifugation. Each peptide is represented by 3 transitions with identical retention times. These proteins are listed in Table 2.
2.3.4 MRM Applied to PTMs of HDL Proteins
Met oxidation (+16 Da) of apoA1 can serve as a practical example of how one might apply MRM to study modifications of HDL proteins. First, the MRM targets or transitions must be identified. These can be obtained for post-translationally modified peptides using initial discovery/mapping samples into which the PTM of interest has been fortified. To accomplish this for Met oxidation of apoA1, we have found it useful to add 5% hydrogen peroxide to pure HDL that is isolated by differential centrifugation [81]. In this case, shotgun proteomics using MudPIT analysis of the sample reveals several oxidized peptides that can serve as targets for MRM. After reduction, alkylation, digestion, and purification, samples are screened for potential modifications by examining the initial MS spectra. Figure 5 illustrates the informatics-mediated sequence coverage and subsequent “discovery” of oxidized Met148. After confirming the sequence specificity of the modification, a list of all abundant ions from the MS/MS spectra is generated for each modified peptide. Each peptide (modified or unmodified) may generate tens of putative transitions that must be aligned with calculated b- or y-ions, instrumentally optimized (e.g., for declustering potential, collision energy, etc.), then tested empirically for sensitivity and specificity. Useful working details on MRM methodologies are provided in Supporting Information.
Figure 5.
The scaffold output of oxidized apolipoprotein A1 after the addition of 5% H2O2 after MudPIT experiment. We highlight peptide LSPLGEEMR (M148) and the relative abundance of the induced M148 oxidation (+16Da). Yellow highlighted peptides are the ones found in our sample. Green W (tryptophans) and M (Mets) are oxidized. The box on the left indicates that 30 peptides with +16 Da modifications were identified with 95% confidence based on the scaffold algorithm.
2.3.5 MRM Limitations
One of the main limitations of MRM is the fact that not all proteins have functionally useful surrogate peptides—i.e., peptides that possess a set of transitions that provide consistently strong signal-to-noise ratios in patient samples. Efficiency of tryptic digestion and robustness of nanoflow liquid chromatography are also common challenges. Overall, the sensitivity of MRM in human samples is limited by sample interferences. When albumin is depleted, for example, MRM-based analyses can be used to quantify lower ng/ml range proteins of otherwise unmodified whole blood plasma. Additionally, some PTMs are unstable (e.g. carbonyl adducts) and, as such, can present problems for SIS synthesis. Despite the challenges presented by difficulties such as these, the future of MRM-based proteomics looks bright and, as described by Abbatiello et al [77], there are ongoing efforts to standardize MRM-based methodologies across different laboratories. Under such efforts, creation of a master list of peptide transitions with correspondingly optimized sample preparation techniques will help minimize variability in results across different laboratories.
3 Combining MSIA and MRM: Absolute and Relative Quantification of Proteins
Proteins are quantified in terms of concentration—e.g., ng of protein-of-interest per mL blood plasma (ng/mL). As we point out above, however, a single nominal protein is not necessarily a singular, unique molecular species. Thus the question arises, “How much of each unique variant form of a protein of interest is present in a sample?” The answer to this question may be expressed in either absolute terms (ng/mL) and/or in relative terms—e.g., as a percentage of total nominal protein—the units in which HbA1c measurements are expressed.
Quantitative MRM-based proteomics focuses on the absolute quantification of singular molecular species (peptides) as surrogates for a larger protein and, as such, outside of the development of multiple assays for each qualitatively unique form of the target protein, this approach cannot provide an assessment of the relative abundances of different post-translationally modified forms of proteins that may be present in a sample. MSIA naturally excels at relative quantification but, without specific efforts directed toward making MSIA fully quantitative [39, 45, 50,62–64], MSIA does not provide information on absolute protein concentration. We propose that a straightforward mathematical merger of independently validated data provided by quantitative MRM-based proteomics and MSIA can be used to provide both absolute and relative quantification of all unique variant forms of a target protein—opening up new, unexplored territory in the hunt for novel markers of disease.
The proposed merger of the two approaches will be particularly useful in cases such as the study of HDL proteins in which just one of the proteins, apoA1, exists as at least a dozen unique molecular forms (Figure 3). Using MRM, dozens of targets can be monitored simultaneously and samples can be run in tandem in a semi-automated manner. MSIA assays are grouped by nominal protein(s) and are generally run on up to 96 samples in a batch. The automated, medium-to-high throughput nature of both of these techniques makes them well suited to biomarker studies in which dozens of protein assays must be run on hundreds to thousands of samples.
The cost of MRM-based analyses largely reflects the combined expense of synthesizing new stable isotope-labeled standards (SIS) and instrument time. The development of SIS libraries, and dedicated mass spectrometry instruments to particular assays will likely reduce its costs and perhaps eventually replace the need to use poorly characterized antibodies in ELISAs. The cost of MSIA lies in the affinity extraction tips, antibodies and instrument time. On a per-assay basis it is competitive with many clinical diagnostics currently on the market.
The proposed strategy of merging MRM and MSIA data fits well with the fact that there is a national move underway to standardize the use of MRM assays across different labs by building an MRM transition atlas with SIS libraries [82]. Use of appropriate standards has, in many cases, been able to improve MRM reproducibility to the point where CVs in MRM runs are less than 20% per the FDA required standard. Ultimately, quantitative MRM assays will likely replace antibody based non-mass spectrometric assays for most protein targets—as they have for several small molecule analytes (e.g. testosterone) where head to head comparisons to immune recognition-based assays have revealed the superior analytical capabilities of the MRM approach [83].
4 Conclusions and a Look Toward the Future
Targeted mass spectrometry-based methods such as MSIA and MRM provide a technical platform for the identification of disease-related protein variants along with reproducible, sensitive quantification of numerous proteins and their posttranslationally modified variant forms—all at unprecedented qualitative specificity using minimal blood volume. As such, the prospects of discovery and validation of new biomarkers can be quite enticing to those of us who are technically oriented analytical scientists. But we cannot traverse the translation gauntlet alone: New biomarkers must be validated and ultimately translated in the context of specific clinical questions. Thus the verification and validation of biomarkers using mass spectrometry-based technologies will be dependent on tight-knit collaborations of analytical scientists with biostatisticians and clinician-scientists who have ready access to patients and are able to collect well characterized samples in a consistent manner. Toward the end of the translation pathway active collaborations with industries in need of new markers will be critical to validating marker utility in “real-life” samples—i.e., existing archived samples from large observational cohorts and randomized trials. We anticipate that such large-scale collaborative efforts will also help ease administrative and regulatory burdens for commercialization of new markers.
For example, in order to develop and bring to market new medications for patients with diabetes, the FDA now requires older patients at higher risk for CVD outcomes to be enrolled in Phase III trials that run for years instead of three-to-six months. In tandem, we anticipate that MSIA and MRM will improve our understanding of candidate biomarkers’ diagnostic and prognostic capacities. An industry-validated protein / protein-PTM panel that is more comprehensive than the industry standard of HbA1c alone and correlates better with CVD phenotype outcomes—allowing, for example, reclassification of patient subsets for more efficient clinical trial design vis-à-vis enrollment of fewer people while maintaining event rates—has significant potential for influencing industry-wide T2DM drug development decisions and holding significant sway with the FDA. Once validated in prospective cohorts, such markers have the potential to reduce the cost of developing newer therapies, reclassify patients’ risk for CVD and translate into clinical practice.
Supplementary Material
Acknowledgements
This work was supported by Award Number R24DK090958 from the National Institute of Diabetes And Digestive And Kidney Diseases. Dr. Yassine was supported by K23HL107389 and American Heart Association grant 12CRP11750017. MudPIT and MRM mass spectrometric data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant P30ES06694 to the Southwest Environmental Health Sciences Center (SWEHSC to Dr. Lau), NIH/NCI grant P30CA023074 to the Arizona Cancer Center (AZCC) and by the BIO5 Institute of the University of Arizona. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes And Digestive And Kidney Diseases or the National Institutes of Health.
Abbreviations
- HDL
High Density Lipoprotein
- CVD
Cardiovascular Disease
- T2DM
Type 2 Diabetes
- RCT
Reverse Cholesterol Transport
- MudPIT
Multidimensional Protein Information Technology
- MRM
Multiple Reaction Monitoring
- MSIA
Mass Spectrometric Immunoassay
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Coccheri S. Approaches to prevention of cardiovascular complications and events in diabetes mellitus. Drugs. 2007;67:997–1026. doi: 10.2165/00003495-200767070-00005. [DOI] [PubMed] [Google Scholar]
- 2.Tuomilehto J, Lindstrom J, Qiao Q. Strategies for the prevention of type 2 diabetes and cardiovascular disease. Eur Heart J Suppl. 2005;7:D18–D22. [Google Scholar]
- 3.Brunner EJ, Shipley MJ, Witte DR, Fuller JH, Marmot MG. Relation between blood glucose and coronary mortality over 33 years in the Whitehall Study. Diabetes Care. 2006;29:26–31. doi: 10.2337/diacare.29.01.06.dc05-1405. [DOI] [PubMed] [Google Scholar]
- 4.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England journal of medicine. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 5.Gerstein HC, Miller ME, Byington RP, Goff DC, Jr, et al. Effects of intensive glucose lowering in type 2 diabetes. The New England journal of medicine. 2008;358:2545–2559. doi: 10.1056/NEJMoa0802743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duckworth W, Abraira C, Moritz T, Reda D, et al. Glucose control and vascular complications in veterans with type 2 diabetes. The New England journal of medicine. 2009;360:129–139. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 7.Group AS, Ginsberg HN, Elam MB, Lovato LC, et al. Effects of combination lipid therapy in type 2 diabetes mellitus. The New England journal of medicine. 2010;362:1563–1574. doi: 10.1056/NEJMoa1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Group AS, Cushman WC, Evans GW, Byington RP, et al. Effects of intensive blood-pressure control in type 2 diabetes mellitus. The New England journal of medicine. 2010;362:1575–1585. doi: 10.1056/NEJMoa1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FDA Guidance for Industry: Diabetes Mellitus — Evaluating Cardiovascular Risk in New Antidiabetic Therapies to Treat Type 2 Diabetes. U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER); 2008. [Google Scholar]
- 10.Navab M, Ananthramaiah GM, Reddy ST, Van Lenten BJ, et al. The double jeopardy of HDL. Ann Med. 2005;37:173–178. doi: 10.1080/07853890510007322. [DOI] [PubMed] [Google Scholar]
- 11.Kontush A, Chapman MJ. Why is HDL functionally deficient in type 2 diabetes? Curr Diab Rep. 2008;8:51–59. doi: 10.1007/s11892-008-0010-5. [DOI] [PubMed] [Google Scholar]
- 12.Rothblat GH, Phillips MC. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Current Opinion in Lipidology. 2010;21:229–238. doi: 10.1097/mol.0b013e328338472d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowri MS, Van der Westhuyzen DR, Bridges SR, Anderson JW. Decreased protection by HDL from poorly controlled type 2 diabetic subjects against LDL oxidation may Be due to the abnormal composition of HDL. Arterioscler Thromb Vasc Biol. 1999;19:2226–2233. doi: 10.1161/01.atv.19.9.2226. [DOI] [PubMed] [Google Scholar]
- 14.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, et al. Antiinflammatory properties of HDL. Circ Res. 2004;95:764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 15.Panzenbock U, Stocker R. Formation of methionine sulfoxide-containing specific forms of oxidized high-density lipoproteins. Biochim Biophys Acta. 2005;1703:171–181. doi: 10.1016/j.bbapap.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Green PS, Vaisar T, Pennathur S, Kulstad JJ, et al. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation. 2008;118:1259–1267. doi: 10.1161/CIRCULATIONAHA.108.770669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navab M, Hama SY, Hough GP, Subbanagounder G, et al. A cell-free assay for detecting HDL that is dysfunctional in preventing the formation of or inactivating oxidized phospholipids. J Lipid Res. 2001;42:1308–1317. [PubMed] [Google Scholar]
- 18.Chiba T, Chang MY, Wang S, Wight TN, et al. Serum amyloid A facilitates the binding of high-density lipoprotein from mice injected with lipopolysaccharide to vascular proteoglycans. Arterioscler Thromb Vasc Biol. 2011;31:1326–1332. doi: 10.1161/ATVBAHA.111.226159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon SM, Deng JY, Lu LJ, Davidson WS. Proteomic Characterization of Human Plasma High Density Lipoprotein Fractionated by Gel Filtration Chromatography. Journal of proteome research. 2010;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenson RS, Brewer HB, Chapman MJ, Fazio S, et al. HDL Measures, Particle Heterogeneity, Proposed Nomenclature, and Relation to Atherosclerotic Cardiovascular Events. Clinical Chemistry. 2011;57:392–410. doi: 10.1373/clinchem.2010.155333. [DOI] [PubMed] [Google Scholar]
- 21.Niederkofler EE, Tubbs KA, Kiernan UA, Nedelkov D, Nelson RW. Novel mass spectrometric immunoassays for the rapid structural characterization of plasma apolipoproteins. J Lipid Res. 2003;44:630–639. doi: 10.1194/jlr.D200034-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- 23.Shao B, Oda MN, Bergt C, Fu X, et al. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J Biol Chem. 2006;281:9001–9004. doi: 10.1074/jbc.C600011200. [DOI] [PubMed] [Google Scholar]
- 24.Zheng L, Nukuna B, Brennan ML, Sun M, et al. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J Clin Invest. 2004;114:529–541. doi: 10.1172/JCI21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc Natl Acad Sci U S A. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rader DJ. Lecithin: cholesterol acyltransferase and atherosclerosis: another high-density lipoprotein story that doesn't quite follow the script. Circulation. 2009;120:549–552. doi: 10.1161/CIRCULATIONAHA.109.881979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobecourt E, Davies MJ, Brown BE, Curtiss LK, et al. The impact of glycation on apolipoprotein A-I structure and its ability to activate lecithin : cholesterol acyltransferase. Diabetologia. 2007;50:643–653. doi: 10.1007/s00125-006-0574-z. [DOI] [PubMed] [Google Scholar]
- 28.Nobecourt E, Tabet F, Lambert G, Puranik R, et al. Nonenzymatic Glycation Impairs the Antiinflammatory Properties of Apolipoprotein A-I. Arteriosclerosis Thrombosis and Vascular Biology. 2010;30:766-U279. doi: 10.1161/ATVBAHA.109.201715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park KH, Jang W, Kim KY, Kim JR, Cho KH. Fructated apolipoprotein A-I showed severe structural modification and loss of beneficial functions in lipid-free and lipid-bound state with acceleration of atherosclerosis and senescence. Biochemical and Biophysical Research Communications. 2010;392:295–300. doi: 10.1016/j.bbrc.2009.12.179. [DOI] [PubMed] [Google Scholar]
- 30.Garner B, Waldeck AR, Witting PK, Rye KA, Stocker R. Oxidation of high density lipoproteins. II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem. 1998;273:6088–6095. doi: 10.1074/jbc.273.11.6088. [DOI] [PubMed] [Google Scholar]
- 31.Garner B, Witting PK, Waldeck AR, Christison JK, et al. Oxidation of high density lipoproteins. I. Formation of methionine sulfoxide in apolipoproteins AI and AII is an early event that accompanies lipid peroxidation and can be enhanced by alpha-tocopherol. J Biol Chem. 1998;273:6080–6087. doi: 10.1074/jbc.273.11.6080. [DOI] [PubMed] [Google Scholar]
- 32.Shao BH, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, et al. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. Jama. 2003;290:2292–2300. doi: 10.1001/jama.290.17.2292. [DOI] [PubMed] [Google Scholar]
- 34.Nelson RW, Krone JR, Bieber AL, Williams P. Mass spectrometric immunoassay. Analytical chemistry. 1995;67:1153–1158. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 35.Nedelkov D. Population proteomics: investigation of protein diversity in human populations. Proteomics. 2008;8:779–786. doi: 10.1002/pmic.200700501. [DOI] [PubMed] [Google Scholar]
- 36.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci U S A. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedelkov D, Tubbs KA, Niederkofler EE, Kiernan UA, Nelson RW. High-throughput comprehensive analysis of human plasma proteins: a step toward population proteomics. Analytical chemistry. 2004;76:1733–1737. doi: 10.1021/ac035105+. [DOI] [PubMed] [Google Scholar]
- 38.Nelson RW, Borges CR. Mass spectrometric immunoassay revisited. Journal of the American Society for Mass Spectrometry. 2011;22:960–968. doi: 10.1007/s13361-011-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson RW, Nedelkov D, Tubbs KA, Kiernan UA. Quantitative mass spectrometric immunoassay of insulin like growth factor 1. Journal of proteome research. 2004;3:851–855. doi: 10.1021/pr0499388. [DOI] [PubMed] [Google Scholar]
- 40.Nelson RW, Williams P, Krone JR. USPTO (Ed.), USA. 2005. [Google Scholar]
- 41.Nelson RW, Williams P, Krone JR. USPTO (Ed.), USA. 2007. [Google Scholar]
- 42.Niederkofler EE, Kiernan UA, O'Rear J, Menon S, et al. Detection of Endogenous B-Type Natriuretic Peptide at Very Low Concentrations in Patients with Heart Failure. Circulation: Heart Failure. 2008;1:258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 43.Niederkofler EE, Tubbs KA, Gruber K, Nedelkov D, et al. Determination of beta-2 microglobulin levels in plasma using a high-throughput mass spectrometric immunoassay system. Analytical chemistry. 2001;73:3294–3299. doi: 10.1021/ac010143j. [DOI] [PubMed] [Google Scholar]
- 44.Tubbs KA, Kiernan UA, Niederkofler EE, Nedelkov D, et al. Development of recombinant-based mass spectrometric immunoassay with application to resistin expression profiling. Analytical chemistry. 2006;78:3271–3276. doi: 10.1021/ac060013g. [DOI] [PubMed] [Google Scholar]
- 45.Kiernan UA, Addobbati R, Nedelkov D, Nelson RW. Quantitative multiplexed C-reactive protein mass spectrometric immunoassay. Journal of proteome research. 2006;5:1682–1687. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- 46.Kiernan UA, Nedelkov D, Nelson RW. Multiplexed mass spectrometric immunoassay in biomarker research: a novel approach to the determination of a myocardial infarct. Journal of proteome research. 2006;5:2928–2934. doi: 10.1021/pr060062+. [DOI] [PubMed] [Google Scholar]
- 47.Kiernan UA, Nedelkov D, Niederkofler EE, Tubbs KA, Nelson RW. High-throughput affinity mass spectrometry. Methods Mol Biol. 2006;328:141–150. doi: 10.1385/1-59745-026-X:141. [DOI] [PubMed] [Google Scholar]
- 48.Kiernan UA, Nedelkov D, Tubbs KA, Niederkofler EE, Nelson RW. Proteomic characterization of novel serum amyloid P component variants from human plasma and urine. Proteomics. 2004;4:1825–1829. doi: 10.1002/pmic.200300690. [DOI] [PubMed] [Google Scholar]
- 49.Kiernan UA, Nedelkov D, Tubbs KA, Niederkofler EE, Nelson RW. Selected expression profiling of full-length proteins and their variants in human plasma. Clin Proteomics J. 2004;1:7–16. [Google Scholar]
- 50.Kiernan UA, Phillips DA, Trenchevska O, Nedelkov D. Quantitative mass spectrometry evaluation of human retinol binding protein 4 and related variants. PLoS One. 2011;6:e17282. doi: 10.1371/journal.pone.0017282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiernan UA, Tubbs KA, Gruber K, Nedelkov D, et al. High-throughput protein characterization using mass spectrometric immunoassay. Analytical biochemistry. 2002;301:49–56. doi: 10.1006/abio.2001.5478. [DOI] [PubMed] [Google Scholar]
- 52.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, et al. Comparative urine protein phenotyping using mass spectrometric immunoassay. Journal of proteome research. 2003;2:191–197. doi: 10.1021/pr025574c. [DOI] [PubMed] [Google Scholar]
- 53.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Comparative phenotypic analyses of human plasma and urinary retinol binding protein using mass spectrometric immunoassay. Biochem Biophys Res Commun. 2002;297:401–405. doi: 10.1016/s0006-291x(02)02212-x. [DOI] [PubMed] [Google Scholar]
- 54.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Detection of novel truncated forms of human serum amyloid A protein in human plasma. FEBS Lett. 2003;537:166–170. doi: 10.1016/s0014-5793(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 55.Borges CR, Jarvis JW, Oran PE, Nelson RW. Population studies of vitamin d binding protein microheterogeneity by mass spectrometry lead to characterization of its genotype-dependent o-glycosylation patterns. Journal of proteome research. 2008;7:4143–4153. doi: 10.1021/pr8002936. [DOI] [PubMed] [Google Scholar]
- 56.Borges CR, Jarvis JW, Oran PE, Rogers SP, Nelson RW. Population studies of intact vitamin D binding protein by affinity capture ESI-TOF-MS. J Biomol Tech. 2008;19:167–176. [PMC free article] [PubMed] [Google Scholar]
- 57.Borges CR, Oran PE, Buddi S, Jarvis JW, et al. Building multidimensional biomarker views of type 2 diabetes on the basis of protein microheterogeneity. Clin Chem. 2011;57:719–728. doi: 10.1373/clinchem.2010.156976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borges CR, Rehder DS, Jarvis JW, Schaab MR, et al. Full-length characterization of proteins in human populations. Clin Chem. 2010;56:202–211. doi: 10.1373/clinchem.2009.134858. [DOI] [PubMed] [Google Scholar]
- 59.Oran PE, Jarvis JW, Borges CR, Nelson RW. C-peptide Microheterogeneity in Type 2 Diabetes Populations. Proteomics. Clinical applications. 2010;4:1–6. doi: 10.1002/prca.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oran PE, Jarvis JW, Borges CR, Sherma ND, Nelson RW. Mass spectrometric immunoassay of intact insulin and related variants for population proteomics studies. Proteomics. Clinical applications. 2011;5:454–459. doi: 10.1002/prca.201000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oran PE, Sherma ND, Borges CR, Jarvis JW, Nelson RW. Intrapersonal and Populational Heterogeneity of the Chemokine RANTES. Clin Chem. 2010;56:1432–1441. doi: 10.1373/clinchem.2010.147884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trenchevska O, Kamcheva E, Nedelkov D. Mass spectrometric immunoassay for quantitative determination of protein biomarker isoforms. Journal of proteome research. 2010;9:5969–5973. doi: 10.1021/pr1007587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trenchevska O, Kamcheva E, Nedelkov D. Mass spectrometric immunoassay for quantitative determination of transthyretin and its variants. Proteomics. 2011 doi: 10.1002/pmic.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trenchevska O, Nedelkov D. Targeted quantitative mass spectrometric immunoassay for human protein variants. Proteome Sci. 2011;9:19. doi: 10.1186/1477-5956-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson NL, Anderson NG, Haines LR, Hardie DB, et al. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA) J. Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 66.Warren EN, Elms PJ, Parker CE, Borchers CH. Development of a protein chip: a MS-based method for quantitation of protein expression and modification levels using an immunoaffinity approach. Analytical chemistry. 2004;76:4082–4092. doi: 10.1021/ac049880g. [DOI] [PubMed] [Google Scholar]
- 67.Krastins B, Prakash A, Sarracino DA, Nedelkov D, et al. Rapid development of sensitive, high-throughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clinical biochemistry. 2013 doi: 10.1016/j.clinbiochem.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez MF, Rezai T, Sarracino DA, Prakash A, et al. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin Chem. 2010;56:281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 69.Rehder DS, Borges CR. Possibilities and pitfalls in quantifying the extent of cysteine sulfenic acid modification of specific proteins within complex biofluids. BMC Biochem. 2010;11:25. doi: 10.1186/1471-2091-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Molecular & cellular proteomics : MCP. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Reid JD, Holmes DT, Mason DR, Shah B, Borchers CH. Towards the Development of an Immuno MALDI (iMALDI) Mass Spectrometry Assay for the Diagnosis of Hypertension. Journal of the American Society for Mass Spectrometry. 2010 doi: 10.1016/j.jasms.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 72.Mason DR, Reid JD, Camenzind AG, Holmes DT, Borchers CH. Duplexed iMALDI for the detection of angiotensin I and angiotensin II. Methods. 2012;56:213–222. doi: 10.1016/j.ymeth.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Niederkofler EE, Tubbs KA, Kiernan UA, Nedelkov D, Nelson RW. Novel mass spectrometric immunoassays for the rapid structural characterization of plasma apolipoproteins. Journal of Lipid Research. 2003;44:630–639. doi: 10.1194/jlr.D200034-JLR200. [DOI] [PubMed] [Google Scholar]
- 74.Vaisar T, Pennathur S, Green PS, Gharib SA, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuzyk MA, Smith D, Yang J, Cross TJ, et al. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Molecular & cellular proteomics : MCP. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yocum AK, Chinnaiyan AM. Current affairs in quantitative targeted proteomics: multiple reaction monitoring-mass spectrometry. Brief Funct Genomic Proteomic. 2009;8:145–157. doi: 10.1093/bfgp/eln056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abbatiello S, Schilling B, Mani D, Feng X, et al. Development and application of a system suitability standard and protocol to assess data quality in LC-MRM-MS across multiple MS platforms. http://proteomics.cancer.gov/programs/CPTAC. 2010
- 78.Kuzyk MA, Smith D, Yang J, Cross TJ, et al. MRM-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Molecular & cellular proteomics : MCP. 2009 doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davidsson P, Hulthe J, Fagerberg B, Camejo G. Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins. Arterioscler Thromb Vasc Biol. 2010;30:156–163. doi: 10.1161/ATVBAHA.108.179317. [DOI] [PubMed] [Google Scholar]
- 80.Davidson WS, Silva RA, Chantepie S, Lagor WR, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McPherson PA, Young IS, McKibben B, McEneny J. High density lipoprotein subfractions: isolation, composition, and their duplicitous role in oxidation. J Lipid Res. 2007;48:86–95. doi: 10.1194/jlr.M600094-JLR200. [DOI] [PubMed] [Google Scholar]
- 82.Parker CE, Pearson TW, Anderson NL, Borchers CH. Mass-spectrometry-based clinical proteomics--a review and prospective. Analyst. 2010;135:1830–1838. doi: 10.1039/c0an00105h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sacks SS. Are routine testosterone assays good enough? Clin Biochem Rev. 2005;26:43–45. [PMC free article] [PubMed] [Google Scholar]
- 84.Fullerton SM, Buchanan AV, Sonpar VA, Taylor SL, et al. The effects of scale: variation in the APOA1/C3/A4/A5 gene cluster. Hum Genet. 2004;115:36–56. doi: 10.1007/s00439-004-1106-x. [DOI] [PubMed] [Google Scholar]
- 85.Boys BL, Kuprowski MC, Noel JJ, Konermann L. Protein oxidative modifications during electrospray ionization: solution phase electrochemistry or corona discharge-induced radical attack? Analytical chemistry. 2009;81:4027–4034. doi: 10.1021/ac900243p. [DOI] [PubMed] [Google Scholar]
- 86.Lantz RC, Lynch BJ, Boitano S, Poplin GS, et al. Pulmonary biomarkers based on alterations in protein expression after exposure to arsenic. Environ Health Perspect. 2007;115:586–591. doi: 10.1289/ehp.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Breci L, Hattrup E, Keeler M, Letarte J, et al. Comprehensive proteornics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis, and isoelectric focusing. Proteomics. 2005;5:2018–2028. doi: 10.1002/pmic.200401103. [DOI] [PubMed] [Google Scholar]
- 88.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.