Abstract
Background
A need exists to expand the characterization of tetrahydrobiopterin (BH4) responsiveness in patients with phenylketonuria (PKU), beyond simply evaluating change in blood phenylalanine concentrations. The clinical interpretation of BH4 responsiveness should be evaluated within the context of phenylalanine hydroxylase (PAH) genotype.
Aim
This investigation seeks to use a modified version of a previously developed PAH genotype severity tool, the assigned value (AV) sum, to assess the molecular basis of responsiveness in a clinical cohort and to explore the tool’s ability to differentiate BH4 responsive groups.
Methods
BH4 response was previously clinically classified in 58 patients with PKU, with three response groups emerging: definitive responders, provisional responders, and non-responders. Provisional responders represented a clinically ambiguous group, with an initial decrease in plasma phenylalanine concentrations, but limited ability to improve dietary phenylalanine tolerance. In this retrospective analysis, mutations in the PAH gene were identified in each patient. PAH genotype was characterized through the AV sum approach, in which each mutation is given an AV of 1, 2, 4, or 8; the sum of both mutations’ AV corresponds to genotype severity, with a lower number representing a more severe phenotype. An AV sum cutoff of 2 (indicative of the most severe genotypes) was used to dichotomize patients and predict BH4 responsiveness. Provisional responders were classified with the definitive responders then the non-responders to see with which group they best aligned.
Results
In 17/19 definitive responders, at least one mutation was mild or moderate in severity (AV sum>2). In contrast, 7/9 provisional responders carried two severe or null mutations (AV sum=2), suggesting little molecular basis for responsiveness. Non-responders represent a heterogeneous group with 15/25 patients carrying two severe mutations (AV sum=2), 5/25 patients carrying one moderate or mild mutation in combination with a severe or null mutation (AV sum>2), and the remaining five patients carrying an uncharacterized mutation in combination with a severe mutation. Predictive sensitivity of the AV sum was maximized (89.5% vs. 67.9%) with limited detriment to specificity (79.4% vs. 80.0%), by classifying provisional responders with the non-responders rather than with the definitive responders.
Conclusions
In our clinical cohort, the AV sum tool was able to identify definitive responders with a high degree of sensitivity. As demonstrated by both the provisional responder group and the substantial number of non-responders with AV sums>2, a potential exists for misclassification when BH4 response is determined by relying solely on change in plasma phenylalanine concentrations. PAH genotype should be incorporated in the clinical evaluation of BH4 responsiveness.
Keywords: Phenylketonuria, Tetrahydrobiopterin, Sapropterin dihydrochloride, BH4, Phenylalanine hydroxylase, Genotype
1. Introduction
Phenylalanine hydroxylase (PAH; EC 1.14.16.1) genotype is playing an increasingly important role in the management of patients with phenylketonuria (PKU; OMIM 261600), especially with the emergence of tetrahydrobiopterin (BH4) therapy. Found to lower blood phenylalanine concentrations in a subset of patients with PKU [1], BH4 is believed to improve the activity of certain dysfunctional PAH enzymes by optimizing cellular BH4 concentrations, acting as a pharmacological chaperone, and/or overcoming kinetic variants[2-4]. BH4’s modes of action are contingent on the enzymes produced from the mutated gene. As such, PAH genotype should play a pivotal role in defining BH4 responsiveness.
PAH genotype is currently not a standard criterion for BH4 response classification. Patients are typically categorized as either “responders” or “non-responders” based only on percent change in blood phenylalanine concentrations after being administered BH4 [5,6]. Protocols assessing responsiveness are highly divergent with respect to variables that can affect circulating phenylalanine concentrations, such as diet prescription compliance, length of evaluation, dose of BH4, and use of a pre-BH4 phenylalanine load [6,7]. Not surprisingly, inconsistencies in the relationship between PAH genotype and response classification have emerged [6,8,9]. Discordant categorization is rarely attributed to response misclassification, despite some “responsive” patients having severe PAH genotypes [10,11] or limited to no long-term clinical benefits with continued use [12-17]. Thus, a need exists to expand the scope of BH4 response classification.
We recently described a novel clinical algorithm for assessing BH4 responsiveness which includes both change in plasma phenylalanine concentrations and ability to modify dietary restrictions as criteria [18]. This approach allowed us to identify a subgroup of patients which experienced an initial marked decrease in plasma phenylalanine concentrations, but had only marginal improvements in dietary phenylalanine tolerance. Similar patients have been reported in protocols different from ours [13,14]. It is unclear if these patients represent a truly responsive group or are merely artifacts of the protocols assessing responsiveness. PAH genotype may help to shed light on the nature of this subgroup.
From a clinical perspective, PAH genotypes are often difficult to interpret. The severity of a mutation or genotype can be explored through open-access databases like the Phenylalanine HydroxylaseLocus Knowledgebase ([19]. While not intended for BH4 response classification, this tool may serve as a starting point for incorporating PAH genotype into the clinical definition of BH4 responsiveness. The goals of this investigation are to use this tool to assess the molecular basis of responsiveness in our clinical cohort and to explore the utility of using a genotype severity tool to differentiate BH4 responsive groups.
2. Patients and methods
2.1. Patients and clinical BH4 response classification
Patients at least 4 years of age, diagnosed PAH-deficient hyper-phenylalaninemia were enrolled in a single-center, clinical trial assessing BH4 responsiveness. Response was classified using a multi-criteria approach outlined in Table 1 and detailed elsewhere [18]. Briefly, patients were first categorized based on change in plasma phenylalanine concentrations after one month of 20 mg/kg/day BH4 therapy (sapropterin dihydrochloride; Kuvan®, BioMarin Pharmaceutical Inc., Novato, CA). Patients with ≥15% decrease in plasma phenylalanine concentrations continued BH4 therapy and were further segregated based on subsequent ability to increase dietary phenylalanine tolerance and decrease medical food needs while maintaining plasma phenylalanine concentrations ≤360 μmol/L. Three BH4 response groups emerged: definitive responders, provisional responders, and non-responders. Provisional responders represent a clinically ambiguous group, experiencing an initial decrease in plasma phenylalanine concentrations but being unable to substantially change their dietary phenylalanine tolerance or medical food needs. Noncompliant patients or those lost to follow-up remain unclassified. Informed consent was received for all patients. This study was approved by the Emory University Institutional Review Board.
Table 1.
Clinical BH4 response classification of patients with PKU using a novel, multi-criteria algorithm.
| Response classification |
Classification criteria |
|---|---|
| Definitive responder |
|
| Provisional responder |
|
| Non-responder Unclassified |
|
Change in plasma phenylalanine concentrations assessed after one month of BH4 therapy (20 mg/kg/day).
Dietary criteria contingent on maintaining plasma phenylalanine concentrations ≤ 360 μmol/L.
2.2. PAH mutation identification
PAH genotypes were assessed retrospectively, and were not evaluated as part of the clinical BH4 response classification. When available, PAH genotypes were taken from participants’ medical records. These PAH mutations were identified using polymerase chain reaction and DNA sequencing of the 13 coding exons and flanking regions. If only one mutation was identified, a second sample was analyzed using a PAH gene-specific comparative genomic hybridization array [20]. For patient who had not been clinically genotyped, a filter paper blood spot was collected which provided DNA that was analyzed using high-resolution melt profiling, as previously described [21]. Mutations were characterized by location (i.e. exon, intron, untranslated region) and by type (missense, mRNA processing, nonsense, or deletion).
2.3. Assessing PAH genotype severity using assigned value (AV) sum
PAH genotype severity was assessed using the assigned valued (AV) sum approach developed by Guldberg et al. [19]. The method was created by evaluating nearly 300 functionally hemizygous patients with PKU and using the patients’ phenotypic severity to classifying a total of 105 different mutations. Each mutation was given an AV of 1, 2, 4, or 8. A lower mutation AV corresponds to a more severe phenotype. Mutations with an AV of 1 are considered particularly severe in nature, with many classified as putative null mutations. Mutations with AV>1 are associated with moderate or mild phenotypes, suggesting that the mutation retains some functionality. To assess the severity of a patient’s genotype, both mutations’ AVs are added together (the “AV sum”). AV sums range from 2 to 16, again with a lower number indicating a more severe phenotype.
Some minor modifications to the AV sum approach were necessary for our analysis. First, there were certain mutations which had been assigned to multiple AVs due to a wide range of clinical phenotypes observed in the original analysis. In those instances, we only used the mutation AV most frequently designated by Guldberg et al. (see Appendix in Ref. [19]). To expand our ability to give a patient an AV sum, decidedly severe mutations not previously evaluated in the AV sum analysis—such as large deletions, frame shift mutations, and disruptions of canonical splice site motifs—were given a mutation AV of 1. Finally, since not all mutations identified in our clinic population had a designated mutation AV, some patients were given an “indefinite AV sum” (e.g. ≥2, ≥3, ≥4, etc). The indefinite AV sum is, at minimum, one greater than the AV for the characterized mutation.
2.4. Assessment of classification approaches and statistics
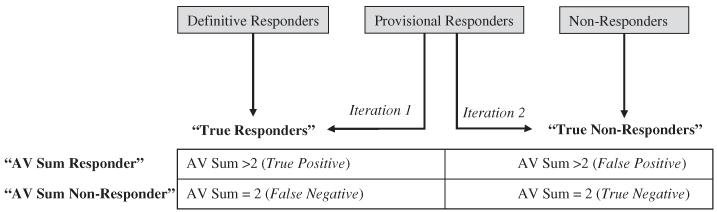
The ability of the AV sum to differentiate the clinically designated BH4 responses and the genetic basis of responsiveness were simultaneously assessed, as outlined in Fig. 1. Patients were first dichotomized into “true responder” and “true non-responder” groups based on the clinical response classification described in Section 2.1 and Table 1. Due to the clinical ambiguity of the provisional responder group, two iterations were evaluated: (1) provisional responders were classified with the definitive responders in a single “true responder” group and (2) provisional responders were classified with the non-responders in a single “true non-responder” group. Patients were then classified by their AV sum. Patients with an AV sum>2 were classified “AV sum responders” those with an AV sum=2 were classified as “AV sum non-responders.” This threshold was selected, as an AV sum of 2 represents a severe genotype with limited to no molecular basis for responsiveness. Patients with the indefinite AV sum of ≥2 were obligate “AV sum non-responders,” since an AV sum above 2 could not definitively be assigned. Since obligate AV sum non-responders have the potential to bias the analysis, results are presented both with and without these patients. To quantify the ability of AV sum to classify BH4 response, sensitivity, specificity, positive predictive value, and negative predictive value were calculated. Patients with an unclassified BH4 response, while presented in the descriptive and summary statistics, were excluded from this portion of the analysis.
Fig. 1.
Classification of clinical BH4 response and assigned value (AV) sum to evaluate the utility of a PAH genotype severity tool.
3. Results
3.1. Summary of identified mutations
A total of 58 patients were genotyped: 19 definitive responders, 9 provisional responders, 25 non-responders, and 5 unclassified patients. Of the expected 116 alleles, 114 mutations were identified (98.3% detection rate). In two patients, only one mutation could be identified, although their clinical and biochemical profiles indicated PAH-deficient hyperphenylalaninemia. There were 47 different mutations identified within our clinical cohort. Mutations affected all 13 exons, 7 introns (intron 1, 4, 5, 6, 8, 10 and 12), and the 3′ untranslated region. As Table 2 shows, missense mutations comprise the majority of the 47 distinct mutations and the majority of 116 alleles.
Table 2.
Frequency of PAH mutation types in patients with PKU evaluated for BH4 responsiveness (N = 58).
| Mutation type | Of the 47 distinct mutations, n (%) |
Of the 116 alleles, n (%) |
|---|---|---|
| Missense mutations | 30 (63.8%) | 78 (67.2%) |
| mRNA processing mutations | 8 (17.0%) | 24 (20.7%) |
| Nonsense mutations | 4 (8.5%) | 7 (6.0%) |
| Deletions | 5 (10.6%) | 5 (4.3%) |
| Mutation not identified | – | 2 (1.7%) |
3.2. PAH genotype AV sum by BH4 response classification
Table 3 presents the PAH genotypes and AV sums of all patients, separated into their respective BH4 response groups. The majority of definitive responders (17/19 patients) had an AV sum>2, indicating that at least one mutation is moderate or mild in severity. The remaining two definitive responders carried a severe mutation (AV=1) in combination with an uncharacterized mutation, and were given an indefinite AV sum of ≥2. In contrast, 7/9 provisional responders had a severe PAH genotype (AV sum=2). The two remaining provisional responders had AV sums of 5.
Table 3.
PAH genotypes and AV sums of 58 patients evaluated for BH4 responsiveness.
| PtID | Mutation 1 | Mutation 2 AV |
Mutation 1 AV |
Mutation 2 sum |
AV |
|---|---|---|---|---|---|
| Definitive responders (n=19) | |||||
| 148 | p.I65T | p.A403V | 2 | 8 | 10 |
| 128 | p.V190A | p.X453_453 + 2del | 8 | - | ≥9 |
| 155 | c.441 +1G>A | p.V190A | 1 | 8 | 9 |
| 135 | p.V245A | p.F299C | 8 | 1 | 9 |
| 104 | p.A104D | p.Y414C | 4 | 4 | 8 |
| 102 | p.L48S | p.I65T | 4 | 2 | 6 |
| 110 | p.V245L | p.Y414C | - | 4 | ≥5 |
| 158 | p.R68S | c.1065 + 3A>G | 4 | 1 | 5 |
| 111 | p.R68S | c.509 + 1G>A | 4 | 1 | 5 |
| 122 | p.N133_Q134>Rfs p.Y414C | 1 | 4 | 5 | |
| 113 | p.L348V | p.L348V | 2 | 2 | 4 |
| 114 | p.L348V | p.L348V | 2 | 2 | 4 |
| 132 | p.I65T | p.E205Da | 2 | - | ≥3 |
| 107 | p.I65T | - | 2 | - | ≥3 |
| 134 | p.F39L | p.G272X | 2 | 1 | 3 |
| 131 | p.I65T | c.1066-3C>T | 2 | 1 | 3 |
| 105 | p.I65T | p.F299C | 2 | 1 | 3 |
| 106 | c.1066-11 G>A | p.P366H | 1 | ≥2 | |
| 136 | p.P275S | EX9_EX13del | - | 1 | ≥2 |
| Provisional responders (n=9) | |||||
| 144 | p.A104D | p.R408W | 4 | 1 | 5 |
| 100 | p.L48S | c.1315+1G>A | 4 | 1 | 5 |
| 112 | p.R408W | EX6_IVS6del | 1 | 1 | 2 |
| 153 | c.509 + 1G>A | p.R408W | 1 | 1 | 2 |
| 117 | p.Q20X | p.R408W | 1 | 1 | 2 |
| 109 | p.R408W | c.1315+1G>A | 1 | 1 | 2 |
| 138 | p.P281L | c.1315+1G>A | 1 | 1 | 2 |
| 115b | p.R408W | p.R408W | 1 | 1 | 2 |
| 126c | c.912 + 1G>A | p.R408W | 1 | 1 | 2 |
| Non-responders (n=25) | |||||
| 129 | p.R241C | c.912 + 1G>A | 8 | 1 | 9 |
| 154 | p.A403V | p.R408W | 8 | 1 | 9 |
| 141 | p.R68S | p.R408W | 4 | 1 | 5 |
| 121 | p.L348V | p.R408W | 2 | 1 | 3 |
| 123 | p.I65T | p.R111X | 2 | 1 | 3 |
| 101 | c.1315+1G>A | - | 1 | ≥2 | |
| 108 | p.L41P | p.E280K | 1 | ≥2 | |
| 120 | p.R252W | p.Q304R | 1 | ≥2 | |
| 124 | p.I283F | p.R408W | 1 | ≥2 | |
| 145 | p.L311P | p.Y386C | 1 | ≥2 | |
| 103 | c.1315+1G>A | c.1315+1G>A | 1 | 1 | 2 |
| 116b | p.R408W | p.R408W | 1 | 1 | 2 |
| 119d | p.R261X | c.1066-11G>A | 1 | 1 | 2 |
| 125 | p.R252Q | c.1315+1G>A | 1 | 1 | 2 |
| 151 | p.R252Q | c.1315+1G>A | 1 | 1 | 2 |
| 127 | p.R158Q | c.1315+1G>A | 1 | 1 | 2 |
| 133 | p.R408W | c.1315+1G>A | 1 | 1 | 2 |
| 137 | p.R252W | p.R408W | 1 | 1 | 2 |
| 139 | p.P281L | c.1315+1G>A | 1 | 1 | 2 |
| 140 | p.A395P | p.R408W | 1 | 1 | 2 |
| 142e | p.E280K | p.F299C | 1 | 1 | 2 |
| 147e | p.E280K | p.F299C | 1 | 1 | 2 |
| 149f | c.60 + 5G>T | p.G272X | 1 | 1 | 2 |
| 150f | c.60 + 5G>T | p.G272X | 1 | 1 | 2 |
| 152c | c.912 + 1G>A | p.R408W | 1 | 1 | 2 |
| Unclassified patients (n=5) | |||||
| 130 | p.I65T | p.Y414C | 2 | 4 | 6 |
| 146 | p.G218V | p.S349P | - | 1 | ≥2 |
| 143 | p.E280K | EX6_IVS6del | 1 | 1 | 2 |
| 118d | p.R261X | c.1066-11 G>A | 1 | 1 | 2 |
| 157b | p.R408W | p.R408W | 1 | 1 | 2 |
Variant of unknown pathogenesis (c.615G>C).
Patient 115 and patient 116 are siblings; patient 157 is unrelated.
Patient 126 and patient 152 are siblings.
Patient 118 and patient 119 are siblings.
Patient 142 and patient 147 are siblings.
Patient 149 and patient 150 are siblings.
Non-responders represented a particularly heterogeneous group. The majority of non-responders (15/25 patients) had an AV sum of 2, indicating a severe PAH genotype. However, 5/25 non-responders had an AV sum>2, carrying a mild or moderate mutation in combination with a severe mutation. The remaining 5 non-responders had a severe mutation (AV=1) in combination with an uncharacterized mutation, and were assigned an indefinite AV sum≥2. The unclassified patients’ AV sums indicate their genotypes are primarily severe. One unclassified patient, who was lost to follow-up, has an AV sum of 6.
3.3. Discordant BH4 response classification of matching PAH genotypes
Several patients had a PAH genotype matching one or more enrolled patient, including five pairs of siblings, four pairs of unrelated patients, and one unrelated patient matching a sibling pair. Of these, two sibling sets and two unrelated sets had discordant clinical BH4 response classification. In these four instances, one patient was classified as a non-responder and the other patient was classified as a provisional responder. The PAH genotype AV sum in each instance was 2, indicating that both mutations were severe in nature. Interestingly, none of these discordant classifications included a patient being classified as a definitive responder.
3.4. Ability of AV sum to predict BH4 response
Table 4 shows the ability of the AV sum to predict clinical BH4 response classification. Categorizing provisional responders with the non-responder group improved sensitivity and negative predictive value with little detriment to specificity and positive predictive value. As expected, excluding patients with an indefinite AV sum of ≥2 improved the sensitivity of using AV sum to classify BH4 responsiveness.
Table 4.
Sensitivity, specificity, positive predictive value, and negative predictive value using genotype AV sum cutoff of > 2 to predict clinical BH4 response classification
| “True responder” citeriar |
“True non-responder” criteria |
Sensitivity | Specificity | Positive predictive value |
Negative predictive value |
|---|---|---|---|---|---|
| Iteration 1: analysis of all classified patients (n=53) | |||||
| Definitive responders, provisional responders |
Non-responders | 67.9% | 80.0% | 79.2% | 69.0% |
| Definitive responders |
Non-responders, provisional responders |
89.5% | 79.4% | 70.8% | 93.1% |
| Iteration 2: analysis excluding patients with indefinite av sums of ≥2 (n=46) | |||||
| Definitive responders, provisional responders |
Non-responders | 73.1% | 75.0% | 79.2% | 68.2% |
| Definitive responders |
Non-responders, provisional responders |
100% | 75.9% | 70.8% | 100% |
4. Discussion
PAH genotype severity has important implications for clinical classification of BH4 responsiveness. With seven of nine of our provisional responders carrying two severe or null mutations, there is strong evidence to suggest they do not represent a truly responsive group. The discordant classification of four sets of patients with matching PAH genotypes—with one patient being classified as a non-responder and one patient being classified as a provisional responder—further suggests that the initial change in plasma phenylalanine concentrations in the provisional responders cannot necessarily be attributed to a drug effect. These findings highlight the potential for patient misclassification in extended protocols relying solely on change in plasma phenylalanine concentrations. As BH4 response classification continues to evolve, it is essential that the definition becomes more comprehensive to encompass change in plasma phenylalanine concentrations, change in dietary phenylalanine tolerance and medical food needs, and PAH genotype. Identification of misclassified patients must also become a crucial element of BH4 response assessment.
In our clinical cohort, one mild or moderate mutation was necessary but not sufficient for BH4 responsiveness. In two instances, a definitive responder carried an uncharacterized mutation in combination with a severe mutation. The literature, while sparse, indicates that the uncharacterized mutation in each of these patients—p.P275S [13,22] and p.P366H [23,24], respectively—does not produce a severe phenotype, even when coupled with a severe or null mutation. Thus, it appears that all of our definitive responders have an AV sum>2, including these two patients. Surprisingly, the ability of AV sum to differentiate our non-responder group is less straightforward. Assuming our clinical classification of BH4 responsiveness is accurate, relying solely on AV sum to predict response classification led to a substantial number of false-positive cases. These genotypic inconsistencies, however, may potentially expose inherent limitations of current BH4 response protocols, especially those spanning days or weeks. The lack of demonstrated decrease in plasma phenylalanine concentrate may have been affected by numerous factors, including: overall metabolic state of the patient, change in health status, non-compliance with BH4, or alteration of dietary intake [2,25-27]. Extensive evaluation of these potentially misclassified patients may elucidate limitations of the AV sum approach or clinical BH4 response protocols.
The concept of evaluating PAH genotype for BH4 responsiveness is not a novel one. Efforts have been made to identify “responsive” alleles from the clinical results of various BH4 response protocols[27,28]. This approach, however, is limited in that it is reliant on divergent protocols which do not assess patient misclassification, and ambiguity has arisen. A simple, BH4-specific clinical tool has yet to be developed. In contrast, PAH genotype AV sum is an easy tool, developed independent of BH4 response classification. While our data may be preliminary in nature, the AV sum approach appears to provide a high degree of sensitivity for identifying patients who have both biochemical and dietary benefits from BH4 therapy. AV sum, in its current state, may serve as a tool for screening patients who should be evaluated for responsiveness. In retrospective analyses, the AV sum may help identify potentially misclassified patients.
While our data are promising, some limitations of our study should be noted. Although a group of 58 patients with PKU assessed at a single clinic is substantial, the external validity of our findings needs to be assessed. Moreover, we could not confirm that the two mutations are in trans in each patient due to incomplete parental studies. There is a potential that some patients’ mutations are in cis and that these patients may harbor an additional unidentified mutation; however, these cases are relatively atypical and rare [29]. Furthermore, some adjustments to the AV sum approach should be considered before widespread implementation. For example, the mutation c.1066-3C>T is classified as a severe mutation (AV=1), but is known to maintain some normal splicing properties and can result in a mild phenotype [30,31]. An expansion of the number of mutations with an AV score would also be necessary. The AV sum tool, should be considered a starting point for the clinical utilization of PAH genotype for response classification.
In conclusion, AV sum appears to be a useful clinical tool for identifying potential candidates for BH4 therapy and retrospectively evaluating BH4 response misclassification. As our provisional responder group exemplifies, a change in phenylalanine concentrations does not always indicate BH4 responsiveness. Our findings underscore the importance of factors such as genotype and dietary phenylalanine tolerance when assessing a patient’s response to BH4.
Acknowledgments
We would like to acknowledge the contributions of the medical and research staff who aided in the development and implementation of this study. We would also like to thank the participants and their families for their contributions.
Role of funding source
The data presented are part of an investigator-initiated trial funded by sponsor-id=“gs1” id=“gts0005”>BioMarin Pharmaceutical Inc. This study was also supported in part by PHS Grant UL1 RR025008 from the sponsor-id=“gs2” id=“gts0010”>Clinical and Translational Science Award program, National Institutes of Health, National Center for Research Resources.
Abbreviations
- AV
assigned value
- BH4
tetrahydrobiopterin
- PAH
phenylalanine hydroxylase
- PKU
phenylketonuria
Footnotes
Conflict of interest statement
This investigator-initiated protocol was supported in part by BioMarin Pharmaceutical Inc. Rani H. Singh and Meghan E. Quirk currently have an investigator-initiated protocol with a material supply agreement with BioMarin Pharmaceutical Inc. Additionally, Rani H. Singh is involved in four sponsor-initiated protocols in collaboration with BioMarin Pharmaceutical Inc.
References
- [1].Kure S, Hou DC, Ohura T, Iwamoto H, Suzuki S, Sugiyama N, Sakamoto O, Fujii K, Matsubara Y, Narisawa K. Tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. J. Pediatr. 1999;135:375–378. doi: 10.1016/s0022-3476(99)70138-1. [DOI] [PubMed] [Google Scholar]
- [2].Staudigl M, Gersting SW, Danecka MK, Messing DD, Woidy M, Pinkas D, Kemter KF, Blau N, Muntau AC. The interplay between genotype, metabolic state and cofactor treatment governs phenylalanine hydroxylase function and drug response. Hum. Mol. Genet. 2011;20:2628–2641. doi: 10.1093/hmg/ddr165. [DOI] [PubMed] [Google Scholar]
- [3].Erlandsen H, Pey AL, Gamez A, Perez B, Desviat LR, Aguado C, Koch R, Surendran S, Tyring S, Matalon R, Scriver CR, Ugarte M, Martinez A, Stevens RC. Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16903–16908. doi: 10.1073/pnas.0407256101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pey AL, Perez B, Desviat LR, Martinez MA, Aguado C, Erlandsen H, Gamez A, Stevens RC, Thorolfsson M, Ugarte M, Martinez A. Mechanisms underlying responsiveness to tetrahydrobiopterin in mild phenylketonuria mutations. Hum. Mutat. 2004;24:388–399. doi: 10.1002/humu.20097. [DOI] [PubMed] [Google Scholar]
- [5].Bernegger C, Blau N. High frequency of tetrahydrobiopterin-responsiveness among hyperphenylalaninemias: a study of 1,919 patients observed from 1988 to 2002. Mol. Genet. Metab. 2002;77:304–313. doi: 10.1016/s1096-7192(02)00171-3. [DOI] [PubMed] [Google Scholar]
- [6].Blau N, Hennermann JB, Langenbeck U, Lichter-Konecki U. Diagnosis, classification, and genetics of phenylketonuria and tetrahydrobiopterin (BH4) deficiencies. Mol. Genet. Metab. 2011;104(Supplement):S2–S9. doi: 10.1016/j.ymgme.2011.08.017. [DOI] [PubMed] [Google Scholar]
- [7].Levy H, Burton B, Cederbaum S, Scriver C. Recommendations for evaluation of responsiveness to tetrahydrobiopterin (BH(4)) in phenylketonuria and its use in treatment. Mol. Genet. Metab. 2007;92:287–291. doi: 10.1016/j.ymgme.2007.09.017. [DOI] [PubMed] [Google Scholar]
- [8].Belanger-Quintana A, Garcia MJ, Castro M, Desviat LR, Perez B, Mejia B, Ugarte M, Martinez-Pardo M. Spanish BH4-responsive phenylalanine hydroxylase-deficient patients: evolution of seven patients on long-term treatment with tetrahydrobiopterin. Mol. Genet. Metab. 2005;86(Suppl. 1):S61–S66. doi: 10.1016/j.ymgme.2005.07.024. [DOI] [PubMed] [Google Scholar]
- [9].Trefz FK, Scheible D, Gotz H, Frauendienst-Egger G. Significance of genotype in tetrahydrobiopterin-responsive phenylketonuria. J. Inherit. Metab. Dis. 2009;32:22–26. doi: 10.1007/s10545-008-0940-8. [DOI] [PubMed] [Google Scholar]
- [10].Elsas LJ, Greto J, Wierenga A. The effect of blood phenylalanine concentration on Kuvan response in phenylketonuria. Mol. Genet. Metab. 2011;102:407–412. doi: 10.1016/j.ymgme.2010.12.003. [DOI] [PubMed] [Google Scholar]
- [11].Leuzzi V, Carducci C, Chiarotti F, Artiola C, Giovanniello T, Antonozzi I. The spectrum of phenylalanine variations under tetrahydrobiopterin load in subjects affected by phenylalanine hydroxylase deficiency. J. Inherit. Metab. Dis. 2006;29:38–46. doi: 10.1007/s10545-006-0096-3. [DOI] [PubMed] [Google Scholar]
- [12].Levy HL, Milanowski A, Chakrapani A, Cleary M, Lee P, Trefz FK, Whitley CB, Feillet F, Feigenbaum AS, Bebchuk JD, Christ-Schmidt H, Dorenbaum A. Efficacy of sapropterin dihydrochloride (tetrahydrobiopterin, 6R-BH4) for reduction of phenylalanine concentration in patients with phenylketonuria: a phase III randomised placebo-controlled study. Lancet. 2007;370:504–510. doi: 10.1016/S0140-6736(07)61234-3. [DOI] [PubMed] [Google Scholar]
- [13].Lambruschini N, Perez-Duenas B, Vilaseca MA, Mas A, Artuch R, Gassio R, Gomez L, Gutierrez A, Campistol J. Clinical and nutritional evaluation of phenylketonuric patients on tetrahydrobiopterin monotherapy. Mol. Genet. Metab. 2005;86(Suppl. 1):S54–S60. doi: 10.1016/j.ymgme.2005.05.014. [DOI] [PubMed] [Google Scholar]
- [14].Trefz FK, Scheible D, Frauendienst-Egger G. Long-term follow-up of patients with phenylketonuria receiving tetrahydrobiopterin treatment. J. Inherit. Metab. Dis. 2010 doi: 10.1007/s10545-010-9058-x. SpringerLink, Online. [DOI] [PubMed] [Google Scholar]
- [15].Trefz FK, Burton BK, Longo N, Casanova MM, Gruskin DJ, Dorenbaum A, Kakkis ED, Crombez EA, Grange DK, Harmatz P, Lipson MH, Milanowski A, Randolph LM, Vockley J, Whitley CB, Wolff JA, Bebchuk J, Christ-Schmidt H, Hennermann JB. Efficacy of sapropterin dihydrochloride in increasing phenylalanine tolerance in children with phenylketonuria: a phase III, randomized, double-blind, placebo-controlled study. J. Pediatr. 2009;154:700–707. doi: 10.1016/j.jpeds.2008.11.040. [DOI] [PubMed] [Google Scholar]
- [16].Vernon HJ, Koerner CB, Johnson MR, Bergner A, Hamosh A. Introduction of sapropterin dihydrochloride as standard of care in patients with phenylketonuria. Mol. Genet. Metab. 2010;100:229–233. doi: 10.1016/j.ymgme.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Burton BK, Adams DJ, Grange DK, Malone JI, Jurecki E, Bausell H, Marra KD, Sprietsma L, Swan KT. Tetrahydrobiopterin therapy for phenylketonuria in infants and young children. J. Pediatr. 2011;158:410–415. doi: 10.1016/j.jpeds.2010.08.016. [DOI] [PubMed] [Google Scholar]
- [18].Singh RH, Quirk ME. Using change in plasma phenylalanine concentrations and ability to liberalize diet to classify responsiveness to tetrahydrobiopterin therapy in patients with phenylketonuria. Mol. Genet. Metab. 2011;104:485–491. doi: 10.1016/j.ymgme.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guldberg P, Rey F, Zschocke J, Romano V, Francois B, Michiels L, Ullrich K, Hoffmann GF, Burgard P, Schmidt H, Meli C, Riva E, Dianzani I, Ponzone A, Rey J, Guttler F. A European multicenter study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am. J. Hum. Genet. 1998;63:71–79. doi: 10.1086/301920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tayeh MK, Chin EL, Miller VR, Bean LJ, Coffee B, Hegde M. Targeted comparative genomic hybridization array for the detection of single- and multiexon gene deletions and duplications. Genet. Med. 2009;11:232–240. doi: 10.1097/GIM.0b013e318195e191. [DOI] [PubMed] [Google Scholar]
- [21].Dobrowolski SF, Ellingson C, Coyne T, Grey J, Martin R, Naylor EW, Koch R, Levy HL. Mutations in the phenylalanine hydroxylase gene identified in 95 patients with phenylketonuria using novel systems of mutation scanning and specific genotyping based upon thermal melt profiles. Mol. Genet. Metab. 2007;91:218–227. doi: 10.1016/j.ymgme.2007.03.010. [DOI] [PubMed] [Google Scholar]
- [22].Mallolas J, Vilaseca MA, Campistol J, Lambruschini N, Cambra FJ, Estivill X, Mila M. Mutational spectrum of phenylalanine hydroxylase deficiency in the population resident in Catalonia: genotype-phenotype correlation. Hum. Genet. 1999;105:468–473. doi: 10.1007/s004390051132. [DOI] [PubMed] [Google Scholar]
- [23].Langenbeck U, Burgard P, Wendel U, Lindner M, Zschocke J. Metabolic phenotypes of phenylketonuria. Kinetic and molecular evaluation of the Blaskovics protein loading test. J. Inherit. Metab. Dis. 2009;32:506–513. doi: 10.1007/s10545-009-1152-6. [DOI] [PubMed] [Google Scholar]
- [24].Fiori L, Fiege B, Riva E, Giovannini M. Incidence of BH4-responsiveness in phenylalanine-hydroxylase-deficient Italian patients. Mol. Genet. Metab. 2005;86(Suppl. 1):S67–S74. doi: 10.1016/j.ymgme.2005.06.017. [DOI] [PubMed] [Google Scholar]
- [25].Treacy E, Pitt JJ, Seller K, Thompson GN, Ramus S, Cotton RG. In vivo disposal of phenylalanine in phenylketonuria: a study of two siblings. J. Inherit. Metab. Dis. 1996;19:595–602. doi: 10.1007/BF01799832. [DOI] [PubMed] [Google Scholar]
- [26].Pey AL, Desviat LR, Gamez A, Ugarte M, Perez B. Phenylketonuria: genotypephenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum. Mutat. 2003;21:370–378. doi: 10.1002/humu.10198. [DOI] [PubMed] [Google Scholar]
- [27].Zurfluh MR, Zschocke J, Lindner M, Feillet F, Chery C, Burlina A, Stevens RC, Thony B, Blau N. Molecular genetics of tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. Hum. Mutat. 2008;29:167–175. doi: 10.1002/humu.20637. [DOI] [PubMed] [Google Scholar]
- [28].Blau N, Erlandsen H. The metabolic and molecular bases of tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. Mol. Genet. Metab. 2004;82:101–111. doi: 10.1016/j.ymgme.2004.03.006. [DOI] [PubMed] [Google Scholar]
- [29].Dobrowolski SF, Heintz C, Miller T, Ellingson C, Ozer I, Gokcay G, Baykal T, Thony B, Demirkol M, Blau N. Molecular genetics and impact of residual in vitro phenylalanine hydroxylase activity on tetrahydrobiopterin responsiveness in Turkish PKU population. Mol. Genet. Metab. 2011;102:116–121. doi: 10.1016/j.ymgme.2010.11.158. [DOI] [PubMed] [Google Scholar]
- [30].Desviat LR, Perez B, Belanger-Quintana A, Castro M, Aguado C, Sanchez A, Garcia MJ, Martinez-Pardo M, Ugarte M. Tetrahydrobiopterin responsiveness: results of the BH4 loading test in 31 Spanish PKU patients and correlation with their genotype. Mol. Genet. Metab. 2004;83:157–162. doi: 10.1016/j.ymgme.2004.06.007. [DOI] [PubMed] [Google Scholar]
- [31].Jaruzelska J, Abadie V, d’Aubenton-Carafa Y, Brody E, Munnich A, Marie J. In vitro splicing deficiency induced by a C to T mutation at position —3 in the intron 10 acceptor site of the phenylalanine hydroxylase gene in a patient with phenylketonuria. J. Biol. Chem. 1995;270:20370–20375. doi: 10.1074/jbc.270.35.20370. [DOI] [PubMed] [Google Scholar]