Abstract
The brain-specific angiogenesis inhibitors 1-3 (BAI1-3) comprise a subfamily of adhesion G protein-coupled receptors (GPCRs). These receptors are highly expressed in the brain and were first studied for their ability to inhibit angiogenesis and tumor formation. Subsequently, BAI1 was found to play roles in apoptotic cell phagocytosis and myoblast fusion. Until recently, however, little was known about the physiological importance of the BAI subfamily in the context of normal brain function. Recent work has provided evidence for key roles of BAI1-3 in the regulation of synaptogenesis and dendritic spine formation. In this review, we summarize the current understanding of the BAI subfamily with regard to the receptors’ downstream signaling pathways, physiological actions and potential importance as novel drug targets in the treatment of psychiatric and neurological diseases.
Keywords: brain, angiogenesis, inhibitor, G protein-coupled receptor, synapse, spine
Adhesion G protein-coupled receptors
G protein-coupled receptors (GPCRs) are a superfamily of seven-transmembrane (7TM) receptors that constitute one of the largest gene families in the human genome [1]. GPCRs recognize a diverse array of extracellular stimuli and transduce intracellular signaling cascades via heterotrimeric G proteins and other signaling intermediates to control cellular physiology [2]. GPCRs are important targets for therapeutics and thus have been intensively studied, but nonetheless there are still more than 100 GPCRs that are considered orphan receptors because they lack identified ligands. Many of these orphan receptors are of interest with regard to human disease either because their tissue distributions suggest they might be tractable drug targets or because genetic studies have linked them to human diseases [3].
The largest family of orphan GPCRs is the adhesion GPCRs [4-6]. These receptors are characterized by extremely long N-termini and a conserved GPCR proteolysis site (GPS) motif that results in autoproteolytic cleavage of the N-terminus, separating it from the rest of the 7TM region [7]. The GPS motif of adhesion GPCRs has recently been shown via X-ray crystallography to be a part of a much larger GPCR autoproteolysis-inducing (GAIN) domain [8, 9]. Although the N-termini of most adhesion GPCRs are cleaved from the 7TM regions at some point during receptor processing, the two receptor fragments can remain non-covalently associated for some period of time [4, 10, 11]. The N-termini of adhesion GPCRs are heavily glycosylated and contain a wide variety of domains known to play roles in cellular adhesion and other biological processes.
An intriguing subfamily of adhesion GPCRs is the trio of brain-specific angiogenesis inhibitors: BAI1, BAI2 and BAI3 [12, 13]. Progress in the understanding of these receptors has seen a dramatic step forward recently with the elucidation of various signaling pathways, regulatory mechanisms, and physiological roles. In this review, we will outline these newly revealed facets of BAI biology, their relevance to neurological and psychiatric disorders, and the various ways in which these receptors might be targeted therapeutically.
Brain-Specific Angiogenesis Inhibitors
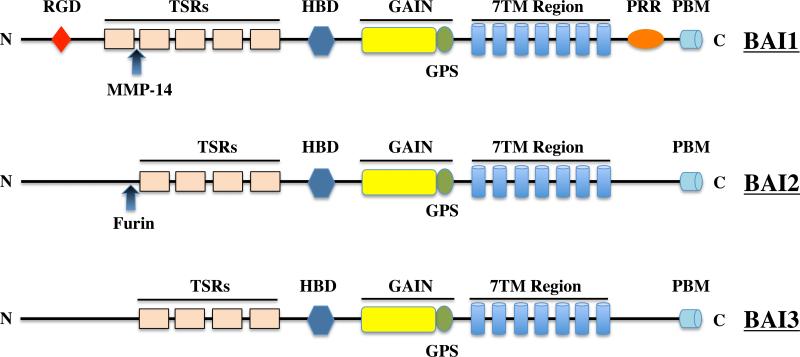
The members of the BAI subfamily contain a variety of conserved domains on both their N-terminal and C-terminal regions (Figure 1). For example, the N-termini of the BAI subtypes each contain multiple thrombospondin type 1 repeats (TSRs), one hormone-binding domain (HBD) each and one GAIN domain. The BAI1 N-terminus also features an integrin-binding RGD (Arg-Gly-Asp) motif in addition to its five TSRs, whereas the BAI2 and BAI3 N-termini do not possess RGD motifs and have only four TSRs. BAI1, BAI2 and BAI3 each share 45% identity to each other at the amino acid level from the N-terminus to first TM domain. The C-terminus of each BAI subtype ends with a PDZ-binding motif, QTEV (Gln-Thr-Glu-Val). The C-terminus of BAI1 also features a proline-rich region (PRR), which can bind to Src homology 3 (SH3) domains and WW domains [14].
Figure 1. Schematic representation of the three members of the BAI subfamily.
The major domains found in each receptor are shown, and known proteolytic events are also indicted. Abbreviations: PBD, PDZ-binding motif; PRR, proline-rich region; 7TM, seven-transmembrane regions; GPS, GPCR proteolytic side; GAIN, GPCR autoproteolysis-inducing domain; HBD, hormone-binding domain; TSR, thrombospondin type 1 repeats; RGD, Arg-Gly-Asp integrin-binding motif
The BAI subtypes are widely expressed in both fetal and adult brain tissue [15]. BAI1 mRNA is found at high levels in the cerebral cortex, the hippocampus, olfactory bulb, thalamic nuclei and basal ganglia [16-18]. During development, transcripts for murine BAI1 and BAI2 mRNA peak at postnatal day 10, whereas BAI3 mRNA peaks 1 day after birth [19]. BAI2 is more ubiquitously expressed than BAI1 during development, exhibiting expression in brain, skin, kidney, skeletal muscle and thymus, but is largely limited to the brain after birth. In contrast, BAI3 is more limited to the central nervous system (CNS) at all developmental stages [19, 20]. BAI1 protein is found in neurons, astrocytes, microglia, and macrophages, with the most robust expression observed in neurons and astrocytes [17, 18, 21, 22]. BAI3 is known to be expressed in hippocampal neurons [23], but the expression profile for BAI2 and BAI3 across different cell types has not yet been characterized as fully as for BAI1. The fact that the BAI subfamily members are expressed in multiple cell types in various tissues suggests distinct functions for these receptors depending on cellular context. Indeed, as discussed below, a variety of divergent physiological roles have been uncovered for BAI1-3.
BAI Autoproteolysis
One of the defining characteristics of adhesion GPCRs is the presence of a GAIN domain, which is an evolutionarily-conserved region that contains an integral GPS motif [9, 10]. The GAIN domains functions as a site of autoproteolysis within the adhesion GPCRs, separating the N-termini of the receptors from their 7TM regions. The GPS motif is a stretch of approximately 40 amino acids within the much larger GAIN domain, which encompasses approximately 320 residues. The GPS motif includes a conserved catalytic serine/threonine residue, which performs a nucleophilic attack on the carbonyl carbon of the peptide backbone, resulting in autoproteolytic cleavage [9, 24, 25]. Following cleavage, the receptors’ N-termini can remain non-covalently associated with the 7TM regions, as has been demonstrated for CD97 [26], CIRL/latrophilin-1 [27, 28], GPR56 [29, 30], BAI1 [11] and BAI2 [31]. The ability of the BAI subfamily to undergo autoproteolysis appears to be cell-specific, as BAI1 and BAI3 do not readily undergo proteolytic cleavage in HEK293T cells [10, 11, 13]. A similar lack of cleavage has been seen in other adhesion GPCRs, such as GPR111 and GPR115, upon expression in HEK-293 cells [32]. Interestingly, BAI1 has been shown to undergo cleavage at the GPS motif in human malignant glioma cells [33, 34] and all three BAI family members are autoproteolytically cleaved in mouse brain lysates [10, 31]. These findings indicate a possible role for regulatory factors, present in neurons and glia but absent in HEK-293T cells, which modulate the BAI subtypes such that autoproteolytic cleavage can occur. It should be noted that although BAI1 and BAI3 do not readily undergo autoproteolysis in HEK293T cells, these receptors are still efficiently trafficked to the plasma membrane [10, 11], indicating that proteolysis at the GPS motif is not required for proper surface trafficking. These observations are intriguing because mutations in the GAIN domains of other adhesion GPCRs can cause protein misfolding and improper trafficking, which in some cases results in human disease [27, 30, 35-37].
Upon proteolysis of the GAIN domain, the N-terminus of BAI1 becomes a 120-kDa protein termed Vasculostatin-120, which has been extensively studied with regard to its ability to inhibit angiogenesis and tumor formation [12, 33, 34]. Further cleavage occurs upstream of the GAIN domain by matrix metalloproteinase 14 to produce a 40-kDa fragment, Vasculostatin-40, which also possesses anti-angiogenic properties [38]. BAI1 was initially identified in a screen for targets of the p53 tumor suppressor [39], but was subsequently shown to be down-regulated via epigenetic regulation independently of p53 expression [40, 41]. The anti-angiogenic effects of the BAI1 N-terminus appear to be primarily mediated through interactions of the TSRs with the scavenger receptor CD36, which induces pro-apoptotic signaling upon binding of the BAI1 TSRs [34]. Thus, autoproteolytic cleavage of the GAIN domain is important because it liberates the BAI1 N-terminus to exert various physiological actions and also because it may modulate the signaling activity of the receptor's seven-transmembrane region, as discussed in the next section. Future studies aimed at understanding how autoproteolysis is regulated in diverse cell types will shed light on BAI subfamily signaling and also potentially shed light on distinct physiological roles for these receptors in different tissues.
BAI Signaling
Although the adhesion GPCRs belong to the GPCR superfamily, only a few of these receptors have actually been shown to couple to G proteins. Both GPR56 [29, 42, 43] and CD97 [44] have been shown to activate the Rho pathway via coupling to Gα12/13, and CIRL-1/latrophilin-1 has been demonstrated to couple to both Gαq and Gαo [45, 46]. GPR133 [47] and GPR114 [48] can couple to Gαs to increase cyclic AMP, and GPR97 can couple to Gαo [48]. However, the G protein coupling preferences of the vast majority of adhesion GPCRs remain to be characterized.
As for the BAI subfamily, BAI2 was found to promote activation of nuclear factor of activated T-cells (NFAT, a transcription factor important for immune responses) via a promiscuous G protein, Gα16, revealing an ability of this receptor to couple to G proteins [31]. BAI1 has also been found to signal through G proteins, specifically Gα12/13 to activate the Rho pathway in HEK293T cells [11]. Interestingly, these studies revealed that removal of the BAI1 N-terminus resulted in enhanced downstream Rho activation as well as increased association with β-arrestins and stimulation of ERK [11]. Increases in constitutive activity following removal of N-terminal regions have also been observed for other adhesion GPCRs, including GPR56 [29, 49], CD97 [44] and BAI2 [31]. These observations may provide fundamental insight into the mechanism of activation for adhesion GPCRs, as it is possible that conformational changes induced by the binding of large extracellular ligands to adhesion GPCR N-termini may relieve inhibitory constraints that are imposed by the N-termini on the receptors’ 7TM regions, thereby allowing for the activation of receptor signaling.
In addition to the G protein-dependent signaling that has been demonstrated for BAI1 and BAI2, several G protein-independent pathways have also been established for the BAI family (Figure 2). The ability of BAI1 to exert effects on intracellular signaling pathways was first shown in studies that identified an interaction between the C-terminus of BAI1 and the intracellular adaptor protein ELMO [21]. ELMO can interact with Dock180 to form a functional guanine nucleotide exchange factor (GEF), which activates the small GTPase Rac by facilitating the exchange of GDP for GTP [50, 51]. BAI3 can also interact with ELMO-Dock180 to activate the Rac pathway [23]. More recently, BAI1 was shown to stimulate Rac via a mechanism independent of either classical G proteins or the ELMO-Dock180 complex, as the PDZ binding motif of BAI1 was found to interact with the PDZ domain of the Rac-GEF Tiam1 and the polarity protein Par3 [22]. BAI1 was shown to enhance the synaptic localization of the Tiam1/Par3 complex, thereby leading to increased Rac activation in cultured hippocampal neurons. A comprehensive list of known binding partners of the BAI subtypes is presented in Table 1.
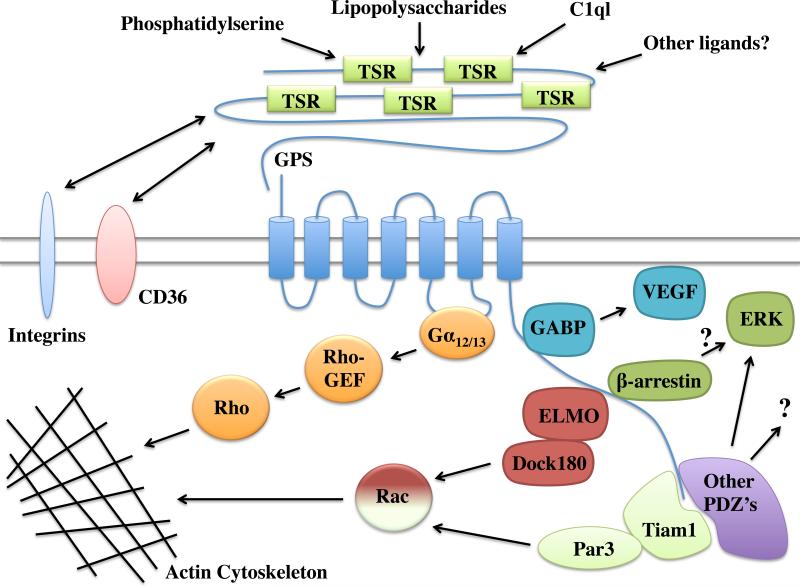
Figure 2. Signaling pathways and binding partners of the BAI subfamily of adhesion GPCRs.
BAI1 activates Rac via an ELMO/Dock180 C-terminal interaction upon binding to exposed phosphatidylserine on apoptotic cells [21] and lipopolysaccharides [64]. BAI1 can also activate Rac via Tiam1/Par3 complex association with the C-terminal PDZ-binding motif [22]. BAI1 can activate Rho via Gα12/13 activation of p115RhoGEF and also stimulate downstream phosphorylation of ERK, possibly through association with β-arrestins [11]. BAI1 has also been shown to bind to a variety of PDZ domains that may differentially regulate signaling and localization in a cell-specific manner [11]. BAI2 suppresses vascular endothelial growth factor (VEGF) expression via interaction with GA-binding protein gamma (GABP) [82]. BAI3 can activate Rac via association with ELMO [23] and also can bind to C1ql proteins via its N-terminal TSRs [84]. Furthermore, the N-terminus of BAI1 can interact with CD36 [34] and integrins [100]. For most of the interactions shown here, it is not yet known whether the interactions are general for all three members of the BAI subfamily or unique to only one subtype. However, the full spectrum of currently-known BAI interactions are represented here in order to provide a comprehensive view of the range of potential signaling pathways for the various BAI subtypes.
Table 1. Comprehensive list of interacting partners identified for BAI1-3.
The region of interaction and functional significance are indicated for each partner.
| NT Interaction | Region | Function | Refs | |
|---|---|---|---|---|
| BAI1 | CD36 | TSRs | Inhibits angiogenesis | [34] |
| αvβ5 Integrin | TSRs | Blocks proliferation of endothelial cells | [100] | |
| Phosphatidylserine | TSRs | Engulfment of apoptotic cells | [21] | |
| MMP-14 | AAs S326 L327 | Cleaves BAI1 NT | [38] | |
| Lipopolysaccharides | TSRs | Bacterial internalization | [64] | |
| BAI2 | Furin | AAs R296 S297 | Cleaves BAI1 NT | [31] |
| BAI3 | C1q-like Proteins | TSRs | Regulation of synaptic density | [84] |
| CT Interaction | Region | Function | Refs | |
|---|---|---|---|---|
| BAI1 | ELMO | α-helix region | Rac GEF | [21] |
| TIAM1 | PDZ-Binding Motif | Rac GEF | [22] | |
| Par3 | PDZ-Binding Motif | Cellular polarity | [22] | |
| MAGI-3 | PDZ-Binding Motif | Potentiates ERK signaling | [11] | |
| MAGI-1, MAGI-2, PSD95, INADL, SAP97, Chapsyn-110, MALS-1, Densin-180, PAPIN, Syntrophins | PDZ-Binding Motif | Unknown | [71],[11],[101] | |
| IRSp53 | Proline rich region | Unknown | [74] | |
| BAP-3 | C-terminus | Unknown | [73] | |
| BAP-4 | C-terminus | Unknown | [16] | |
| β-arrestin2 | C-terminus | Signal regulation | [11] | |
| BAI2 | Glutaminase interacting protein | C-terminus | Unknown | [102] |
| GA-binding protein gamma | C-terminus | Transcriptional regulation of VEGF | [82] | |
| BAI3 | ELMO | α-helix region | Rac GEF | [23] |
Functional Roles of the BAI Subtypes
As mentioned earlier, the BAI subtypes possess multiple TSRs on their N-termini. TSRs were first identified as regions of thrombospondin-1 that mediate the anti-angiogenic activity of this secreted protein [52]. Thus, much of the early research on the BAI subtypes revolved around the ability of these receptors to inhibit experimental angiogenesis and tumor formation [39, 53, 54]. Restoration of BAI1 can inhibit the growth of tumors derived from gliomas and renal cell carcinomas [34, 55-58]. Moreover, expression of the isolated BAI1 N-terminus (Vasculostatin-120) and the association of its TSRs with CD36 can mimic the anti-angiogenic effects of full-length BAI1 [12, 33, 34]. Thus, the anti-angiogenic actions of BAI1 parallel the classical anti-angiogenic actions of thrombospondin-1, which interacts with CD36 to induce apoptosis in endothelial cells and thereby inhibit angiogenesis [59, 60]. In addition to the TSRs on the BAI N-termini, there is also a putative hormone-binding domain located between the TSRs and the GAIN domain. Nothing is known at present about the role of this domain with regards to BAI subfamily biology, including whether this domain actually binds to hormones, but this could be an interesting area of future research.
In addition to its role in regulating angiogenesis, a novel role for BAI1 was uncovered following identification of an interaction between the BAI1 C-terminus and the Rac-GEF ELMO-Dock180, a conserved signaling complex known to be important in promoting the internalization of apoptotic cells [21]. BAI1 was identified as a receptor upstream of this signaling module and a key player in the engulfment of cells that have undergone apoptosis. One classical feature of apoptotic cells is the exposure of phosphatidylserine on the outer leaflet of the cell membrane [61]. The TSRs on the BAI1 N-terminus were found to interact with exposed phosphatidylserine on apoptotic debris, thereby eliciting the activation of the Rac signaling pathway [21]. Activated Rac is known to promote cytoskeletal rearrangement via actin polymerization, allowing for the engulfment and internalization of apoptotic cell debris [62]. The ability of BAI1 to bind to externalized phosphatidylserine on apoptotic cells has also been shown to be important for myoblast fusion, as genetic deletion of BAI1 was observed to reduce the size of myofibers and impair muscle regeneration in vivo [63]. Additionally, the BAI1 TSRs have been shown to bind to lipopolysaccharides on Gram-negative bacteria to mediate bacterial phagocytosis [64]. Interestingly, a key feature shared by the processes of phagocytosis and angiogenesis is that they are both known to be highly regulated by thrombospondin interactions with CD36 [65]. Thus, although it has not yet been determined if CD36 plays a role in BAI1-mediated regulation of phagocytosis, it is possible that BAI1 regulates both angiogenesis and phagocytosis via the action of the five TSRs on the BAI1-NT in a manner that parallels the regulation of angiogenesis and phagocytosis by thrombospondin-1.
BAIs at the Synapse
Beyond the regulation of angiogenesis and phagocytosis, a third major action of thrombospondins and TSRs in general is control of synaptogenesis [66]. Thrombospondins are known to promote excitatory synaptogenesis [67, 68], and other TSR-containing proteins, such as semaphorin-5A [69] and UNC-5 [70], are best known for the roles they play in synaptic development. An accumulating body of evidence suggests an important role for BAI1 as a synaptic protein, including the interaction of the PDZ-binding motif of BAI1 with PSD-95 [11, 71], a scaffold protein that regulates spine formation and shape [72]. BAI1 has also been reported to bind to PDZ domains from the synaptic scaffold protein MAGI-1/BAP1 [73]. The observation that BAI1 is capable of binding to PDZ domains led to proteomic analyses revealing that the C-terminus of BAI1 can robustly associate with PDZ domains from a number of distinct scaffold proteins, including SAP97 (DLG1), Densin-180, MAGI-2 and MAGI-3 [11]. Co-expression with MAGI-3 was found to augment BAI1 constitutive activity in HEK-293T cells, but only if the receptor's PDZ-binding motif was intact, thereby providing an example as to how PDZ interactions can modulate BAI1-mediated signaling [11].
The BAI1-interacting PDZ proteins mentioned above are all known to be concentrated in the post-synaptic density (PSD), a macromolecular signaling assembly found in the post-synaptic regions of excitatory CNS synapses. Interestingly, BAI1 itself has also recently been shown to be highly concentrated in PSD fractions from brain tissue [11, 22]. Moreover, another protein that associates with the BAI1 C-terminus, insulin receptor substrate 53 (IRSp53; also known as ‘BAI1-associated protein 2” or BAIAP2) [74], is enriched in the PSD [75, 76]. When the BAI1/IRSp53 interaction was identified, little was known about the cellular functions of IRSp53 and no physiological significance was established for this interaction. Over the past decade, however, IRSp53 has been demonstrated to be a key regulator of dendritic spines [77] and suggested to play a role in autism spectrum disorder (ASD) [78].
Evidence that BAI1 can regulate synaptic function and dendritic spine morphology has come from recent studies identifying the PDZ protein Tiam1 as a BAI1-interacting protein [22]. Tiam1 is best known as a Rac-GEF that can induce cytoskeletal changes in dendritic spines [79]. Duman et al. found that BAI1 signaling to Rac in cultured hippocampal neurons was dependent on BAI1 binding to Tiam1; in contrast, mutations blocking the ability of BAI1 to bind ELMO/Dock180 had no effect on BAI1 signaling to Rac in this system [22]. BAI1 was found in these studies to be localized to dendritic spines, consistent with the aforementioned biochemical evidence that BAI1 in highly enriched in the PSD [11, 22]. The studies by Duman et al. revealed that knockdown of BAI1 resulted in a reduction in spine density and a less mature phenotype in the remaining spines. These findings suggest that BAI1 may play a key role in dendritic spine maturation and synaptogenesis.
Important roles in neurogenesis and synaptogenesis have also recently been suggested for the other BAI family members. Studies on BAI2-deficient mice revealed that loss of BAI2 induces a depression-resistant phenotype [80]. BAI2 knockout mice were found to be resistant to social defeat and less prone to immobility in the tail suspension test, two well-established rodent assays of depressive behavior. Importantly, these differences could not be attributed to any deficits in motor activity or spatial learning. The BAI2-deficient mice were also found to exhibit increased neurogenesis in the dentate gyrus of the hippocampus [80], which may be related to the depression-resistant phenotype of these mice given that enhanced neurogenesis in the dentate gyrus often correlates with resistance to depression [81]. Regarding the mechanism by which BAI2 might regulate neurogenesis, BAI2 has previously been reported to suppress vascular endothelial growth factor (VEGF) expression via interaction with GA-binding protein gamma [82], and VEGF is known to stimulate neurogenesis in the adult hippocampus [83]. Thus, loss of BAI2 expression might plausibly lead to increased VEGF expression and increased neurogenesis. Further work will test this model and also elucidate additional phenotypes of mice deficient in the various BAI subtypes.
BAI3 was recently shown to control dendritic arborization and branching in cultured neurons, at least partly via interactions with ELMO1 [23]. BAI3 knockdown in hippocampal neurons both in vitro and in vivo resulted in longer and thinner dendrites, indicating a deficit in maturation and thereby suggesting a role for BAI3 in regulating dendritic arbor formation [23]. Interestingly, BAI3 has also recently been found to interact via its TSRs with the members of C1ql family of secreted complement-like proteins [84]. The C1ql proteins are highly expressed in the CNS and capable of forming both homomeric and heteromeric complexes [85, 86]. Bolliger et al. found that incubating primary hippocampal neurons with low concentrations of C1ql3 decreased the density of excitatory synapses in a manner that was blocked by the addition of a TSR-containing fragment of BAI3 [84]. Further work in this area will shed light on how C1ql interactions with BAI3 might regulate cellular physiology and whether C1ql proteins interact with all BAI subtypes or only with BAI3.
Relevance to Human Disease
The aforementioned effects of the members of the BAI subfamily on dendritic spines may be of clinical interest because abnormalities in spine density and morphology are associated with a range of human diseases [87, 88]. For example, Fragile X syndrome is the most common inherited form of mental retardation and is characterized by the development of abnormally long and thin dendritic spines in the cerebral cortex and hippocampus [89]. Furthermore, one of the key pathological features of Parkinson's disease is the loss of dendritic spines in the striatum [89, 90], and alterations in spine morphology have been linked to schizophrenia and other psychiatric disorders [91]. Chronic drug abuse has also been shown to result in dramatic changes to spine density and morphology in brain regions associated with reward and addiction [92]. Given this large body of evidence linking spine dysregulation with disease, it is of critical importance to elucidate the underlying elements that regulate dendritic spine development and morphology in order to identify novel targets for therapeutic interventions for these disorders. Thus, the recent studies demonstrating regulation of dendritic spine development and morphology by the members of the BAI subfamily mark these receptors as potentially exciting new therapeutic targets for the treatment of psychiatric and neurological disorders associated with dendritic spine pathology.
Clinical interest in the BAI subtypes has also arisen from genetic studies linking these receptors to various psychiatric and neurological disorders. For example, BAI1 may possibly be implicated in ASD, as the BAI1 gene was localized to a hot spot for germline mutations in patients with autism [93]. Moreover, BAI1 interacts with PSD-95 [11, 71], and there is evidence that regulation of PSD-95 may be a common downstream feature of multiple genes associated with autism [94]. Additionally, single-nucleotide polymorphisms in BAI3 [95] as well as changes in BAI3 copy number [96] have been linked to the development of schizophrenia, and BAI3 expression has been found to be regulated by treatment with lithium, a drug used to treat psychiatric conditions such as bipolar disorder and certain types of schizophrenia [97, 98].
The BAI Subtypes as Pharmacological Targets
Given the converging lines of evidence from in vitro, in vivo and genetic studies suggesting that the members of the BAI subfamily may play roles in a variety of psychiatric and neurological disorders, these receptors have emerged as potentially important new therapeutic targets. Important future steps toward the targeting of the BAI subtypes for therapeutic purposes will include identification of the receptors’ ligands and further elucidation of the receptors’ downstream signaling pathways (Box 1). Such advances will facilitate high-throughput screening approaches aimed at finding small molecule agonists, antagonists and modulators for the BAI subtypes. High-throughput screening to find small-molecule ligands for adhesion GPCRs is indeed tractable, as recent screens of the adhesion GPCR GPR97 identified beclomethasone dipropionate as a small molecule agonist for GPR97 [48]. It can be envisioned that small molecule BAI agonists could be developed to potentially promote dendritic spine stabilization and rescue spines from the immature phenotypes associated with certain psychiatric and neurological disorders.
One of the most important conceptual advances in the field of pharmacology in the past 15 years has been the concept of biased ligands, which are ligands that preferentially activate certain pathways downstream of a given receptor [99]. Since BAI1 can signal through a multitude of distinct pathways, it may be possible to develop biased agonists that specifically activate one of these pathways without activating others in order to fine-tune cellular responses and maximize clinical benefit. Moreover, small molecules could be developed to regulate the association of the N-terminus with the 7TM region of these receptors, as it has been shown that this interaction modulates receptor-signaling activity. In a related vein, small molecules that bind to the GPS motif to accelerate or inhibit GPS cleavage might also be predicted to alter receptor signaling activity, and thus the GPS motifs of the BAI subtypes and other adhesion GPCRs could provide unique therapeutic targets not found in other GPCRs. Based on the recent advances that have been made in understanding the signaling activity of the BAI subtypes, it is evident that there are a variety of distinct approaches that might be taken in targeting the members of this receptor subfamily for the potential treatment of psychiatric and neurological disorders.
Concluding Remarks
The regulation of dendritic spines and synaptic plasticity is extremely complex and involves a multitude of signaling pathways and regulatory proteins. Achieving an understanding of the key signaling molecules involved is of critical importance not only to understanding normal physiology, but also to better understanding and hopefully treating neurological disorders that have a basis in aberrant synapse formation and morphology. This review provides an overview of the BAI subfamily of adhesion GPCRs, with an emphasis on recent studies that have uncovered critical roles for the BAI subfamily members at the synapse. These receptors can regulate neuronal function and synaptogenesis through several different signaling pathways, and recent genetic studies have linked this subfamily to multiple psychiatric and neurological disorders. Further elucidation of BAI1-3 signaling, regulation and activation by ligands will likely lead to substantial new insights in the area of synaptic biology.
Highlights for “The BAI Subfamily of Adhesion GPCRs: Synaptic Regulation and Beyond” by Stephenson et al.
BAI1, BAI2 & BAI3 are receptors that can regulate neuronal function & synaptogenesis.
BAI1 has been shown to signal via both G protein- and -independent pathways.
Genetic studies link BAI1-3 to a variety of psychiatric & neurological disorders.
The BAI subtypes may prove to be beneficial therapeutic targets for these disorders.
Box 1. Outstanding questions.
How are the various signaling pathways downstream of the BAI subtypes activated and regulated?
Can these signaling pathways be modulated by the various N-terminal ligands of the BAI subtypes?
Are the synaptic functions of the BAI subtypes redundant or unique?
Are the actions of BAI1 as a mediator of engulfment related in some way to the actions of this receptor at the synapse?
What roles do the BAI subtypes play in neurological disorders?
What aspects of BAI functionality could potentially be targeted for modulation by small molecule ligands?
Can the members of the BAI subfamily be exploited pharmacologically for therapeutic benefit in neurological and psychiatric disorders?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierce KL, et al. Seven-transmembrane receptors. Nature reviews. Molecular cell biology. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum DM, et al. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung S, et al. Orphan GPCR research. British journal of pharmacology. 2008;153(Suppl 1):S339–346. doi: 10.1038/sj.bjp.0707606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paavola KJ, Hall RA. Adhesion G Protein-Coupled Receptors: Signaling, Pharmacology & Mechanisms of Activation. Mol Pharmacol. 2012 doi: 10.1124/mol.112.080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjarnadottir TK, et al. The adhesion GPCRs: a unique family of G protein-coupled receptors with important roles in both central and peripheral tissues. Cell Mol Life Sci. 2007;64:2104–2119. doi: 10.1007/s00018-007-7067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yona S, et al. Adhesion-GPCRs: emerging roles for novel receptors. Trends in biochemical sciences. 2008;33:491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Langenhan T, et al. Sticky signaling--adhesion class g protein-coupled receptors take the stage. Science signaling. 2013;6:re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- 8.Arac D, et al. Dissecting signaling and functions of adhesion G protein-coupled receptors. Annals of the New York Academy of Sciences. 2012;1276:1–25. doi: 10.1111/j.1749-6632.2012.06820.x. [DOI] [PubMed] [Google Scholar]
- 9.Promel S, et al. Matching structure with function: the GAIN domain of Adhesion-GPCR and PKD1-like proteins. Trends in pharmacological sciences. 2013;34:470–478. doi: 10.1016/j.tips.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Arac D, et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephenson JR, et al. Brain-specific Angiogenesis Inhibitor-1 Signaling, Regulation, and Enrichment in the Postsynaptic Density. The Journal of biological chemistry. 2013;288:22248–22256. doi: 10.1074/jbc.M113.489757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cork SM, Van Meir EG. Emerging roles for the BAI1 protein family in the regulation of phagocytosis, synaptogenesis, neurovasculature, and tumor development. J Mol Med (Berl) 2011;89:743–752. doi: 10.1007/s00109-011-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park D, Ravichandran KS. Emerging roles of brain-specific angiogenesis inhibitor 1. Advances in experimental medicine and biology. 2010;706:167–178. doi: 10.1007/978-1-4419-7913-1_15. [DOI] [PubMed] [Google Scholar]
- 14.Kay BK, et al. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 15.Shiratsuchi T, et al. Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1). Cytogenetics and cell genetics. 1997;79:103–108. doi: 10.1159/000134693. [DOI] [PubMed] [Google Scholar]
- 16.Koh JT, et al. Characterization of mouse brain-specific angiogenesis inhibitor 1 (BAI1) and phytanoyl-CoA alpha-hydroxylase-associated protein 1, a novel BAI1-binding protein. Brain research. Molecular brain research. 2001;87:223–237. doi: 10.1016/s0169-328x(01)00004-3. [DOI] [PubMed] [Google Scholar]
- 17.Sokolowski JD, et al. Brain-specific angiogenesis inhibitor-1 expression in astrocytes and neurons: implications for its dual function as an apoptotic engulfment receptor. Brain, behavior, and immunity. 2011;25:915–921. doi: 10.1016/j.bbi.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori K, et al. Brain-specific angiogenesis inhibitor 1 (BAI1) is expressed in human cerebral neuronal cells. Neuroscience research. 2002;43:69–74. doi: 10.1016/s0168-0102(02)00018-4. [DOI] [PubMed] [Google Scholar]
- 19.Kee HJ, et al. Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS letters. 2004;569:307–316. doi: 10.1016/j.febslet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Kee HJ, et al. Expression of brain-specific angiogenesis inhibitor 2 (BAI2) in normal and ischemic brain: involvement of BAI2 in the ischemia-induced brain angiogenesis. Journal of cerebral blood flow and metabolism. 2002;22:1054–1067. doi: 10.1097/00004647-200209000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 22.Duman JG, et al. The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. The Journal of neuroscience. 2013;33:6964–6978. doi: 10.1523/JNEUROSCI.3978-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanoue V, et al. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei W, et al. Characterization of cis-autoproteolysis of polycystin-1, the product of human polycystic kidney disease 1 gene. J Biol Chem. 2007;282:21729–21737. doi: 10.1074/jbc.M703218200. [DOI] [PubMed] [Google Scholar]
- 25.Deyev IE, Petrenko AG. Regulation of CIRL-1 proteolysis and trafficking. Biochimie. 2010;92:418–422. doi: 10.1016/j.biochi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Gray JX, et al. CD97 is a processed, seven-transmembrane, heterodimeric receptor associated with inflammation. J Immunol. 1996;157:5438–5447. [PubMed] [Google Scholar]
- 27.Krasnoperov V, et al. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. The Journal of biological chemistry. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 28.Volynski KE, et al. Latrophilin fragments behave as independent proteins that associate and signal on binding of LTXN4C. Embo Journal. 2004;23:4423–4433. doi: 10.1038/sj.emboj.7600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paavola KJ, et al. The N Terminus of the Adhesion G Protein-coupled Receptor GPR56 Controls Receptor Signaling Activity. Journal of Biological Chemistry. 2011;286:28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Z, et al. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 31.Okajima D, et al. Brain-specific angiogenesis inhibitor 2 (BAI2) may be activated by proteolytic processing. Journal of receptor and signal transduction research. 2010;30:143–153. doi: 10.3109/10799891003671139. [DOI] [PubMed] [Google Scholar]
- 32.Promel S, et al. Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Developmental dynamics. 2012;241:1591–1602. doi: 10.1002/dvdy.23841. [DOI] [PubMed] [Google Scholar]
- 33.Kaur B, et al. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24:3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 34.Kaur B, et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer research. 2009;69:1212–1220. doi: 10.1158/0008-5472.CAN-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piao X, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 36.Ke N, et al. Biochemical characterization of genetic mutations of GPR56 in patients with bilateral frontoparietal polymicrogyria (BFPP). Biochem Biophys Res Commun. 2008;366:314–320. doi: 10.1016/j.bbrc.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 37.Chiang NY, et al. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem. 2011;286:14215–14225. doi: 10.1074/jbc.M110.183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cork SM, et al. A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene. 2012 doi: 10.1038/onc.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimori H, et al. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15:2145–2150. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- 40.Kaur B, et al. Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. The American journal of pathology. 2003;162:19–27. doi: 10.1016/S0002-9440(10)63794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu D, et al. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer research. 2011;71:5859–5870. doi: 10.1158/0008-5472.CAN-11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iguchi T, et al. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha(12/13) and rho pathway. Journal of Biological Chemistry. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 43.Luo R, et al. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward Y, et al. LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71:7301–7311. doi: 10.1158/0008-5472.CAN-11-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lelianova VG, et al. alpha-Latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. Journal of Biological Chemistry. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- 46.Rahman MA, et al. Norepinephrine exocytosis stimulated by alpha-latrotoxin requires both external and stored Ca2+ and is mediated by latrophilin, G proteins and phospholipase C. Philos T Roy Soc B. 1999;354:379–386. doi: 10.1098/rstb.1999.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohnekamp J, Schoneberg T. Cell Adhesion Receptor GPR133 Couples to G(s) Protein. Journal of Biological Chemistry. 2011;286:41912–41916. doi: 10.1074/jbc.C111.265934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupte J, et al. Signaling property study of adhesion G-protein-coupled receptors. Febs Letters. 2012;586:1214–1219. doi: 10.1016/j.febslet.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, et al. GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011;71:5558–5568. doi: 10.1158/0008-5472.CAN-10-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature cell biology. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 51.Lu M, Ravichandran KS. Dock180-ELMO cooperation in Rac activation. Methods in enzymology. 2006;406:388–402. doi: 10.1016/S0076-6879(06)06028-9. [DOI] [PubMed] [Google Scholar]
- 52.de Fraipont F, et al. Thrombospondins and tumor angiogenesis. Trends in molecular medicine. 2001;7:401–407. doi: 10.1016/s1471-4914(01)02102-5. [DOI] [PubMed] [Google Scholar]
- 53.Duda DG, et al. Overexpression of the p53-inducible brain-specific angiogenesis inhibitor 1 suppresses efficiently tumour angiogenesis. British journal of cancer. 2002;86:490–496. doi: 10.1038/sj.bjc.6600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang X, et al. Antiangiogenic activity of BAI1 in vivo: implications for gene therapy of human glioblastomas. Cancer gene therapy. 2006;13:385–392. doi: 10.1038/sj.cgt.7700898. [DOI] [PubMed] [Google Scholar]
- 55.Xiao XR, et al. [Therapeutic effect of brain-specific angiogenesis inhibitor 1 on glioblastoma: an animal experiment]. Zhonghua yi xue za zhi. 2006;86:1342–1346. [PubMed] [Google Scholar]
- 56.Kudo S, et al. Inhibition of tumor growth through suppression of angiogenesis by brain-specific angiogenesis inhibitor 1 gene transfer in murine renal cell carcinoma. Oncology reports. 2007;18:785–791. [PubMed] [Google Scholar]
- 57.Yoon KC, et al. Lipid-mediated delivery of brain-specific angiogenesis inhibitor 1 gene reduces corneal neovascularization in an in vivo rabbit model. Gene therapy. 2005;12:617–624. doi: 10.1038/sj.gt.3302442. [DOI] [PubMed] [Google Scholar]
- 58.Izutsu T, et al. Brain-specific angiogenesis inhibitor 1 is a putative factor for inhibition of neovascular formation in renal cell carcinoma. The Journal of urology. 2011;185:2353–2358. doi: 10.1016/j.juro.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 59.Dawson DW, et al. CD36 mediates the In vitro inhibitory effects of thrombospondin-1 on endothelial cells. J Cell Biol. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez B, et al. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat Med. 2000;6:41–48. doi: 10.1038/71517. [DOI] [PubMed] [Google Scholar]
- 61.Fadok VA, et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 62.Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 63.Hochreiter-Hufford AE, et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497:263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das S, et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2136–2141. doi: 10.1073/pnas.1014775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Science signaling. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams JC, Tucker RP. The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Developmental dynamics. 2000;218:280–299. doi: 10.1002/(SICI)1097-0177(200006)218:2<280::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 67.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 68.Eroglu C, et al. Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell. 2009;139:380–392. doi: 10.1016/j.cell.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kantor DB, et al. Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron. 2004;44:961–975. doi: 10.1016/j.neuron.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 70.Poon VY, et al. UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature. 2008;455:669–673. doi: 10.1038/nature07291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim IA, et al. Selectivity and promiscuity of the first and second PDZ domains of PSD-95 and synapse-associated protein 102. Journal of Biological Chemistry. 2002;277:21697–21711. doi: 10.1074/jbc.M112339200. [DOI] [PubMed] [Google Scholar]
- 72.El-Husseini AE, et al. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 73.Shiratsuchi T, et al. Cloning and characterization of BAP3 (BAI-associated protein 3), a C2 domain-containing protein that interacts with BAI1. Biochemical and biophysical research communications. 1998;251:158–165. doi: 10.1006/bbrc.1998.9408. [DOI] [PubMed] [Google Scholar]
- 74.Oda K, et al. Identification of BAIAP2 (BAI-associated protein 2), a novel human homologue of hamster IRSp53, whose SH3 domain interacts with the cytoplasmic domain of BAI1. Cytogenetics and cell genetics. 1999;84:75–82. doi: 10.1159/000015219. [DOI] [PubMed] [Google Scholar]
- 75.Abbott MA, et al. The insulin receptor tyrosine kinase substrate p58/53 and the insulin receptor are components of CNS synapses. J Neurosci. 1999;19:7300–7308. doi: 10.1523/JNEUROSCI.19-17-07300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bockmann J, et al. ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J Neurochem. 2002;83:1013–1017. doi: 10.1046/j.1471-4159.2002.01204.x. [DOI] [PubMed] [Google Scholar]
- 77.Choi J, et al. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. The Journal of neuroscience. 2005;25:869–879. doi: 10.1523/JNEUROSCI.3212-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toma C, et al. Association study of six candidate genes asymmetrically expressed in the two cerebral hemispheres suggests the involvement of BAIAP2 in autism. Journal of psychiatric research. 2011;45:280–282. doi: 10.1016/j.jpsychires.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nature cell biology. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 80.Okajima D, et al. Antidepressant-like behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J Physiol Sci. 2011;61:47–54. doi: 10.1007/s12576-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Samuels BA, Hen R. Neurogenesis and affective disorders. Eur J Neurosci. 2011;33:1152–1159. doi: 10.1111/j.1460-9568.2011.07614.x. [DOI] [PubMed] [Google Scholar]
- 82.Jeong BC, et al. Brain-specific angiogenesis inhibitor 2 regulates VEGF through GABP that acts as a transcriptional repressor. FEBS letters. 2006;580:669–676. doi: 10.1016/j.febslet.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 83.Jin K, et al. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2002;99:11946–11950. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bolliger MF, et al. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2534–2539. doi: 10.1073/pnas.1019577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuzaki M. Cbln and C1q family proteins: new transneuronal cytokines. Cellular and molecular life sciences. 2008;65:1698–1705. doi: 10.1007/s00018-008-7550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iijima T, et al. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. The European journal of neuroscience. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 87.Fiala JC, et al. Dendritic spine pathology: Cause or consequence of neurological. Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- 88.Blanpied TA, Ehlers MD. Microanatomy of dendritic spines: Emerging principles of synaptic pathology in psychiatric and neurological disease. Biol Psychiat. 2004;55:1121–1127. doi: 10.1016/j.biopsych.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 89.Irwin SA, et al. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 90.Villalba RM, Smith Y. Striatal spine plasticity in Parkinson's disease. Front Neuroanat. 2010;4:133. doi: 10.3389/fnana.2010.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costa E, et al. Dendritic spine hypoplasticity and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol Dis. 2001;8:723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- 92.Russo SJ, et al. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends in neurosciences. 2010;33:267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsai NP, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeRosse P, et al. The genetics of symptom-based phenotypes: toward a molecular classification of schizophrenia. Schizophrenia bulletin. 2008;34:1047–1053. doi: 10.1093/schbul/sbn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao HM, et al. Identification and characterization of three inherited genomic copy number variations associated with familial schizophrenia. Schizophrenia research. 2012;139:229–236. doi: 10.1016/j.schres.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 97.McCarthy MJ, et al. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS One. 2012;7:e32091. doi: 10.1371/journal.pone.0032091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McQuillin A, et al. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenetics and genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- 99.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 100.Koh JT, et al. Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Experimental cell research. 2004;294:172–184. doi: 10.1016/j.yexcr.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 101.Shiratsuchi T, et al. Cloning and characterization of BAI-associated protein 1: a PDZ domain-containing protein that interacts with BAI1. Biochem Biophys Res Commun. 1998;247:597–604. doi: 10.1006/bbrc.1998.8603. [DOI] [PubMed] [Google Scholar]
- 102.Zencir S, et al. Identification of brain-specific angiogenesis inhibitor 2 as an interaction partner of glutaminase interacting protein. Biochem Biophys Res Commun. 2011;411:792–797. doi: 10.1016/j.bbrc.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]