Abstract
Beyond the genome, epigenetics has become a promising approach in understanding the interactions between the gene and the environment. Epigenetic regulation includes DNA methylation, histone modifications, and non-coding RNAs. Among these, DNA methylation, which is the addition of a methyl group to the fifth base of cytosine to produce 5-methylcytosine (5-mC), is most commonly studied. Epigenetic regulation has changed given the discovery of 5-hydroxymethylcytosine (5-hmC), considered the “sixth base”, and the nature of TET proteins to catalyze 5-mC oxidation to 5-hmC. 5-hydroxymethylation has been proposed to be a stable intermediate between methylation and demethylation and has raised questions about the functions of 5-hmC in gene regulation in cells, tissues, and organs in response to environmental exposure. Herein, we have provided an introduction to the chemistry of 5-hydroxymethylation, and the techniques for detection of 5-hydroxymethylation. In addition, we have reviewed current reports describing how 5-hmC responds to environmental factors, leading to the development of disease. And finally, we have discussed the potential use of 5-hmC in the study of disease development. All in all, it is our goal to provide innovative and convincing epigenetic studies for understanding the etiology of environmentally-related human disease, and translate these epigenetic findings into lifestyle recommendations and clinical practices to prevent and cure disease.
Keywords: 5-hydroxymethylcytosine, 5-methylcytosine, 5-formylmethylcytosine, 5-carboxylmethylcytosine, DNA methylation, DNA hydroxymethylation, demethylation, base excision repair, oxidation, glucosylation, deamination, ten-eleven translocation proteins, environmental health, epigenetics, environmental biosensor
I. Introduction
Each cell of the body has the same genome, yet very different forms and functions. Carefully orchestrated epigenetic mechanisms play a role in the cell/tissue-specific gene expression regulating cell differentiation, maintenance, and proliferation [1]. There are three distinct epigenetic mechanisms, including histone modifications, non-coding RNAs, and DNA methylation, that work singularly or conjointly to epigenetically regulate the chromatin structure [1, 2]. These mechanisms modify downstream gene transcription and function in order to fine-tune biological mechanisms at the cellular level. There is increasing evidence that 5-hydroxymethycytosine (5-hmC), considered to be the “sixth base” [3], may also play a role in epigenetic regulation. Given the fact that epigenetic changes are sensitive to environmental stimuli, in this review, we focus on the role of DNA hydroxymethylation in gene regulation and how it interacts with environmental factors, leading to disease development.
DNA methylation and hydroxymethylation are proposed as epigenetic modifications of gene regulation. Chemistry of DNA methylation and hydroxymethylation is summarized in Figure 1. DNA methyltransferases (DNMTs) catalyze the covalent addition of a methyl group from S-adenosyl-methionine (SAM) to the C5 position of cytosine in CpG dinucleotides (Figure 1), which predominantly cluster in densely populated CpG islands (CGI), often located in gene promoters and the first intron and exon [4–7]. Initially, 5-methylcytosine (5-mC) is established by de novo DNMT3A and DNMT3B [8], and during semi-conservative DNA replication, DNMT1 maintains methylation by using the hemi-methylated DNA template to regenerate the symmetrical methylation site on the new DNA strand [5]. 5-mC is found in 1–4% of all cytosines (or ~ 80% of CpG dinucleotides) in the mammalian genome [6, 9, 10]. CGI methylation occurs mainly in the promoter regions, where ~70% of the CGIs are unmethylated except for those on the inactive X chromosome and some associated with imprinted genes [11]. DNA methylation in critical regulatory regions, such as gene promoters, demonstrates the influential role of 5-mC in gene repression. 5-mC has a high affinity for methyl-CpG binding proteins (MBPs) and their interaction causes the recruitment of chromatin-modifying enzymes at the promoter, leading to gene repression by sterically hindering the binding of transcription factors and basal transcriptional machinery [5]. Although the mechanisms of DNA methylation are well defined, the opposing role of DNA demethylation has not yet been fully understood. During zygote formation, there is rapid demethylation during zygote cleavage, and rapid methylation after implantation, independent of cell division, which indicates that active DNA demethylation pathways are responsible [1, 4]. The discovery of ten-eleven translocation (TET) proteins brought an incredible interest to the 5-hmC mediated DNA demethylation pathways and the biological role of 5-hmC in development [12]. TET enzymes are responsible for catalyzing the oxidation of 5-mC to 5-hmC and the iterative oxidation of 5-hmC to 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC) DNA derivatives, believed to be the intermediates in the DNA demethylation pathways [4, 6, 13] (Figure 1).
Figure 1.
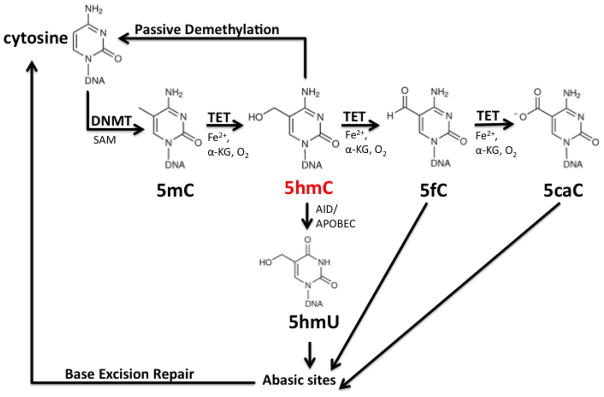
Chemistry of DNA methylation and hydroxymethylation. 5methylcytosine (5-mC) is produced from the addition of S-adenosylmethionine (SAM) onto the 5-carbon of Cytosine by DNA methyltransferases (DNMT). Ten-eleven-translocation (TET) proteins then catalyze the interactive oxidation of 5mC to 5-hydroxymethylcytosine (5-hmC), 5-formlycytosine (5-fC) and 5-carboxycytosine (5-caC), with required cofactors alpha-ketoglutarate (a-KG), Iron (Fe2+) and oxygen. 5-hmC, 5-fC or 5-caC could act as an intermediate in both passive and active DNA demethylation pathways involving DNA repair enzymes like AID or APOBEC (activation-induced cytidine deaminase).
TET Proteins
TET expression levels vary between cells and organs. TET1 and TET2 are highly expressed in mouse ES cells while TET3 is seen predominantly in oocytes and one-cell zygotes [4]. TET1 and TET3 are expressed in several adult tissues but most abundant in brain while TET2 is enriched in hematopoietic cells [3]. There is limited number of studies about how TET proteins are targeted to specific genes in distinct cell types and developmental stages. Whether TET expression profiles correlate to 5-hmC product formation is also very complicated and controversial. The brain (~0.80%) and spinal cord (~0.45%) are the most 5-hmC enriched cells/organs among different tissues [6], [16]. Studies showed global 5-hmC levels in human tissues are not associated with 5-mC content or TET gene expression [14]. However, additional studies are required to evaluate this finding in other tissue/cell types and in a large-scale human population studies. All TET proteins have a catalytic domain that contains both a cysteine rich region and 2-oxoglutarate-Fe(II) dioxygenase activity [6, 9, 15]. In mammals, TET proteins (TET1, TET2, TET3) catalyze the transfer of a hydroxyl group to 5-mC [9, 13, 15] to form 5-hmC/5-fC/5-caC. TET1 can oxidize both fully and hemi-methylated DNA, and does not require CpG dinucleotides [4]. In the N-terminal, the CXXC domain of the TET proteins (TET1 and TET3 only) is a DNA binding domain, with a high affinity for clustered unmethylated DNA [4, 15]. Subsequent genome-wide mapping of mouse and human embryonic stem cells determined that the CXXC domain of TET1 overlaps with the unmethylated DNA generally located in the CGIs of promoter regions [4], signaling many chromatin associated proteins. TET1 has been shown to activate gene transcription by reducing the 5-mC mediated recruitment of the polycomb repression complex 2 (PRC2) to chromatin [4]. In contrast, TET proteins have been shown to recruit the Sin3a (directly) and the Ezh2 complex (indirectly), to deacetylate and trimethylate histone H3, and repress target genes [4, 6]. To recap, TET proteins may both repress and activate gene transcription by the recruitment of chromatin remodeling enzymes through the CXXC domain, and/or as the intermediate of gene demethylation through the catalytic 5-mC oxidation to 5-hmC/5-fC/5-caC.
Non-enzymatic Regulation by 5-hmC
The substitution of 5-mC for 5-hmC can non-catalytically regulate gene transcription by interfering with DNA-protein interactions. Once established, these 5-hmC residues can influence gene regulation. 5-hmC has a large, polar, hydroxymethyl group, which extends into the major groove of DNA, pushing the attached cytosine further away from the duplex, and creating a polar cavity that increases solvation dynamics (the interaction of DNA and water molecules) [17]. The solvation effects of 5-hmC causes local geometric changes in intra-base pair fluctuations (shear, stretch, stagger, and buckle), which decreases rigidity around the local helical axis conformation, and increases the ability of the duplex to propeller twist and open [17]. 5-hmC generally functions to activate gene expression by destabilizing the DNA structure to allow transcriptional machinery to access the transcriptional start site [3]. Using synthetic oligonucleotides, the replacement of 5-mC with 5-hmC diminishes the binding affinity of MeCP2 (methyl-CpG binding protein 2), suggesting that 5-hmC residues are functionally equivalent to cytosine residues [5]. The degree of 5-hmC content can reduce methylation binding domain (MBD) interactions to re-organize the chromatin structure, and reverse the repressive effects of 5-mC upon transcriptional regulation. 5-hmC modifications generally are transcriptionally activating, but exceptions always arise. In contrast, 5-hmC residues have been demonstrated to interact with MBDs or methyl-CpG binding proteins (MBPs) to recruit many chromatin remodeling enzymes that maintain the repressive transcriptional regulation. For example, in mouse embryonic stem cells (ESCs), MBD3 (methyl-CpG binding protein 3) directly binds 5-hmC modified DNA (but not 5-mC) and recruits NuRD (nucleosome remodeling and deacetylase) complexes to repress gene transcription [18]. In addition, evidence is emerging that 5-hmC might act as a new landmark that recognizes and recruits specific DNA binding proteins [6, 15], which can further complicate the role of 5-hmC transcriptional regulation.
Enzymatic Regulation by 5-hmC
TET proteins catalyze the generation of 5-hmC residues that act as intermediates for DNA demethylation, to affect gene transcription. Proposed mechanisms indicate that 5-hmC is a stable intermediate for both “passive” and “active” DNA demethylation [3] (Figure 1). “Passive” (replication-dependent) DNA demethylation occurs as a result of the inability of DNMT1 to maintain DNA methylation, resulting in the passive loss of DNA methylation at a specific CpG site [3, 5, 6, 14]. Less defined are the mechanisms of “active” (or replication-independent) DNA demethylation, which suggest that 5-hmC is either spontaneously or enzymatically converted to cytosine through DNA repair pathways [9]. Possible mechanisms of “active” DNA demethylation include: the recognition of 5-hmC by activation-induced cytidine deaminase (AID/APOBEC), which mediates deamination to 5-hydroxymethlyuracil (5-hmU), generating an abasic site, which is recognized and subsequently removed by DNA glycosylases (SMUG1 (single-strand-selective monofunctional uracil-DNA glycosylase 1) and TDG (thymine DNA glycosylase); and base excision repair (BER) machinery, to restore an unmodified cytosine (Figure 1) [4, 6]. Regardless of the route of TET mediated demethylation, 5-hmC is an essential intermediate in the removal of 5-mC resides in the DNA demethylation pathway, which can drastically change global methylation patterns and biological mechanisms. Therefore, further investigations are needed to fully understand the DNA demethylation pathway.
Recent studies have demonstrated that TET enzymatic activity is affected by many factors. Isocitrate dehydrogenase enzymes (IDH1 and IDH2) commonly undergo gain-of-function mutations in human gliomas (~75%) and acute myeloid leukemia (~20%) patients that catalyze the production of α-ketoglutarate (α-KG) and 2-hydroxyglutarate (2-HG) respectively [19, 20]. 2-HG is a proposed oncometabolite that competitively inhibits α-KG dioxygenases, such as TET [20]. In TET1 overexpressing HEK293 cells, the co-expression of tumor derived mutant IDH1 and IDH2 substantially decreased the 5-hmC content, compared to the wild-type [19]. In vitro, 2-HG was shown to inhibit TET activity, and clinically, glioma samples subjected to immunohistochemical (IHC) staining demonstrated that 5-hmC was significantly decreased in IDH1-mutatated-gliomas, compared to the control, despite consistent 5-mC levels [19]. Oxidative stress has also been proposed to affect TET activity. Under high oxidative stress conditions, when homeostasis cannot be maintained, increased NAD+ levels in the mitochondria activate sirtuin NAD+ dependent deacetylases (Sirt) [20]. In mammals, increased Sirt3 results in the deacetylation of IDH2, and consequent activation of metabolic activity [20]. Therefore, activated IDH2 increases the conversion of isocitrate to α-KG, and increases TET enzymatic activity, potentially changing global methylation patterns. Research continues to associate epigenetic mechanisms as responses to environmental sensors that can potentially disrupt cellular and biological processes, resulting in disease development.
Measurement of DNA Hydroxymethylation
Tools that exist for the detection of methylated DNA, such as bisulfite conversion and methylated-DNA-specific-antibodies, have failed to distinguish between 5-mC and 5-hmC DNA, so it is critical to develop techniques that can differentiate between these two modifications. Currently, the most accessible technique developed for hydroxymethylated DNA enrichment and detection is the use of enzymatic and antibody approaches. 5-hmC DNA quantitation, by techniques such as PCR, high-performance liquid chromatography (HPLC), thin layer chromatography (TLC), or liquid chromatography-mass spectrometry (LC-MS) can detect 5-hmC at single CpG dinucleotides. We summarized several common techniques to reveal (1) global hydroxymethylation, (2) loci-specific hydroxymethylation, and (3) whole genome profiling for hydroxymethylated DNA loci (Table 1). All the techniques described could be used on distinct tissues, cell types or biological specimens.
Table 1.
Common Methods for 5-hmC detection
| Type | Detection Level | Detection Principle | Pre-modification of DNA1 | Processing Time2 | Specialized Instrument3 |
|---|---|---|---|---|---|
| Global | Whole Sample | ELISA4 | Required | 6–8 hrs | Not Required, |
| Loci Specific | Loci | GSRE, TET1- oxidation, PCR and sequencing | Required | 1–3 days | Not Required |
| Whole Genome | Loci | Massive Parallel Sequencing | Required | Variable | Required5 |
Antibody binding (anti-5-hydroxymethylcytosine) or enzyme modifications (i.e. GT, GSRE, TET1)
Time for completion of ELISA is usually less than one working day and depends on the commercial vendor’s manual. For loci 5-hmC detection, it requires 1–3 days for pretreatment of DNA, PCR and/or sequencing. Whole genome massive parallel sequencing requires library construction, massive sequencing, data analysis, data mining and gene validation. Time for completion varies between facilities.
Instruments other than the thermocycler and plate reader.
Techniques using chromatography and mass spectroscopy to detect global hydroxymethylation are available but not commonly used.
Massive parallel sequencing on different types of sequencers is available in core facilities or commercial vendors.
Quantitation for Global Hydroxymethylation
The content of 5-hmC across the genome can be measured using enzyme-linked immunosorbent assay (ELISA), with capture and detection antibodies followed by colorimetric quantification. The capture antibody for 5-hmC has no or negligible cross-reactivity to both methylated and unmethylated cytosines. Additionally, there are other protocols to examine the 5-hmC content across the genome, using enzymatic digestion coupled with HPLC, TLC, or LC-MS. Input DNA are first glucosylated by 5-hmC glucosyltransferase (GT), which transfers a glucose moiety from uridine diphosphoglucose (UDPG) onto preexisting 5-hmC’s within DNA [21, 22]. The difference in the chemical structure (mass) between glucosylated 5-hmC and unglucosylated 5-mC or cytosine generates a distinct profile for 5-hmC in the chromatography [23–27]. To locate the 5-hmC distribution in the tissues, immunohistochemistry or immunofluorescence staining using an antibody against 5-hmC is employed to examine the distribution of 5-hmC among different cell types.
Specific Hydroxymethylated DNA/Loci/Gene DNA Quantitation
Current bisulfite sequencing technology cannot distinguish 5-mC and 5-hmC at specific CpG sites; therefore, realtime PCR (RTPCR), coupled with a glucosyl-5-hmC sensitive restriction endonuclease (GSRE) digestion [28], is used to measure the percentage of 5-hmC of specific CpG sites. First, the hydroxy group of 5-hmC is protected with a glucosylated moiety by glucosyltransferase prior to PCR. Glucosylated 5-hmC is next cut by a GSRE, like Taq1a, and according to the RTPCR assay, the cleaved glucosylated 5-hmC has a smaller threshold value (Ct) than the unglucosylated cytosine/5-mC. The percentage 5-hmC at specific CpG sites is quantified using the Ct difference between the glucosylated and unglucosylated samples. It is used for detection of PCR products smaller than 150 base pairs. For longer fragments, we suggest the new TET-1 assisted bisulfite sequencing. It involves the UDPG-mediated protection of 5-hmC, and recombinant mouse TET1 (mTET1)-mediated oxidation of 5-mC to 5-caC. After the subsequent bisulfite treatment and PCR amplification, both cytosine and 5-caC (derived from 5-mC) are converted to thymine (T), whereas 5-hmC reads as cytosine (C). The treated genomic DNA is suitable for locus-specific sequencing [9, 29, 30].
Whole Genome DNA Quantitation
Researchers that are interested in quantifying the whole genome profiling hydroxymethylation loci could use the anti-5-hmC antibody to immunoprecipitate and enrich the sonicated DNA-protein complexes. The enriched DNA products are subjected to a methylation oligonucleotide array (hMeDIP-chip) or next generation sequencing (hMeDIP-Seq) [31, 32].
II. Environmental Exposure Linked to 5-hmC Perturbations
Cytosine modifications, such as 5-mC and 5-hmC, are epigenetic mechanisms that enable genes to respond to external environmental cues. Environmental exposure has been shown to disrupt epigenetic mechanisms and interfere with 5-mC and 5-hmC profiles in many diseases, as summarized in Tables 2 and 3. TET proteins are also apt to environmental perturbations. Therefore, researchers have been investigating 5-hmC content as a novel epigenetic marker to understand the link between epigenetic signatures and disease states.
Table 2.
Examples of environmental exposures contributed to TET regulation and 5-hydroxymethylation changes.
| Environmental Exposures | Observed changes | References |
|---|---|---|
| Ascorbic Acid (Vitamin C) | Increased TET1 and TET2 activity and increased 5hmC in mouse embryonic stem cells | [33], [34]. [35] |
| Phenobarbital | Increased 5hmC in mature mouse liver tissue | [36] |
| Diethylstilbestrol | Decreased 5hmC in mouse adult uterine tissue | [37] |
| Hydroquinone | Increased TET1 and 5hmC in human embryonic kidney cell culture | [38] |
| House dust mite | Increased 5hmC in mouse lung tissue | [40] |
Table 3.
Examples of diseases associated with changes in 5-hydroxymethylation or TET regulation.
| Diseases | Observed changes | References |
|---|---|---|
| Cancer | ||
| Gliomas | Decreased 5hmC | [42] |
| Hepatocellular Carcinoma | Decreased 5hmC in both human and rodent HCC | [43] |
| Melanomas | Decreased TET2 and 5hmC | [44], [45] |
| Acute myeloid leukemia | Increased TET1 and 5hmC in bone marrow samples | [47] |
| Myelodysplastic syndrome | Decreased TET2 and 5hmC in bone marrow samples | [50] |
| Late-onset diseases | ||
| Huntington’s disease | Decreased 5hmC in mouse brain | [51] |
| Alzheimer’s disease | Increased 5hmC in mouse brain of females offspring experiencing prenatal stress | [54] |
| Psychosis | Increased TET1 and 5hmC in brain cortex tissue of psychotic and depressed patients | [55] |
Ascorbate Acid
Vitamin C, or ascorbic acid (AA), stimulates the drastic erasure of 5-mC by promoting TET activity and generating 5-hmC mediated DNA demethylation [33–35]. TET enzymes require both Fe2+ and α-KG co-factors [3, 9], and AA is another co-factor required for full catalytic activity of TET proteins [34]. AA directly interacts with the catalytic domains of TET1 and TET2 to reduce Fe3+ to Fe2+ and enhance TET mediated oxidation [33]. AA not only increases 5-hmC, but also the iterative oxidation products 5-fC and 5-caC in mouse ESCs [33]. The reducing effects of AA are specific, since other antioxidant enzymes (glutathione, selenite, vitamin B1, vitamin E, L-carnitine, lipoic acid) do not induce the enzymatic activity of TET [35]. Mouse ESCs treated with AA for 12 and 72 hours displayed a progressive increase in 5-hmC, and decrease in 5-mC, at the gene promoters, but it was not influenced by the gene expression of TET or DNMT [35]. In addition, the global induction of 5-hmC in response to AA treatment was reversible; after three days of AA removal, both the 5-hmC and 5-mC profiles returned to the baseline levels. These findings confirmed the dynamic interplay between 5-mC and 5-hmC from exogenous exposures mediated by TET oxidation. Additionally, these investigations suggest the nutritional value of AA, especially during embryonic development, where drastic global changes in DNA methylation occur at the fetus genome.
Phenobarbital
Phenobarbital, a widely used barbiturate medication to control seizures, is classified as a non-genotoxic carcinogen. In the mouse liver, PB perturbs 5-mC and 5-hmC profiles associated with tumor formation [16, 36]. Mice administered PB through drinking water displayed significant differential hydroxymethylation in juvenile (30–33 days of phenobarbital exposure) and mature (91 days of phenobarbital exposure) liver tissue. In mature liver tissue, phenobarbital induced proximal promoter enrichment of 5-hmC, coupled with a decrease in 5-mC, to increase the gene transcription of a collection of candidate genes with a potential role in liver tumorigenesis [36]. Additionally, phenobarbital has provoked the rapid and prolonged gene expression of cytochrome P450 2b10 (Cyp2b10), a member of the Cyp family of genes, critical for xenobiotic metabolism in the liver [36]. Therefore, by epigenetically inducing the expression of Cyp2b10 in the liver, the increased rate of phenobarbital metabolites produced, including carcinogens, could promote liver tumor formation. Wisp1, a Wnt signaling pathway gene, was over-expressed and associated with increased 5-hmC levels following phenobarbital exposure [16]. It supports the previous findings in both rodents and humans of Wisp1 gene expression increasing during the disease progression of hepatocellular carcinoma. These findings exemplify the use of 5-hmC as a sensitive biosensor, which can be used to distinguish control versus phenobarbital exposed rodents.
Diethylstilbestrol
Neonatal exposure to diethylstilbestrol, an endocrine disrupting chemical, resulted in the adult onset of uterine and vaginal cancer in women. Mice postnatally exposed to DES showed global 5-hmC reduction in uterine tissues of diethylstilbestrol treated mice (8 weeks old), when compared to the control [37]. Furthermore, diethylstilbestrol mediated the reduction of TET1, TET2, Dnmt1, and Dnmt3a genes, which remained repressed in adult uterine tissue. The decreased 5-hmC profile observed in adult mice in response to diethylstilbestrol treatment could be due to the impaired TET oxidation of 5-mC, and/or declining 5-mC preservation due to DNMTs reduction. Results suggest that neonatal exposure to endocrine disruption chemicals may result in aberrant 5-mC/5-hmC mediated DNA methylation, altering gene expression patterns throughout adulthood, and possibly increasing disease risk.
Hydroquinone
Hydroquinone, a copious metabolite of benzene (a carcinogen found in car exhaust, industrial emissions, and cigarette smoke), caused an increase in TET1 activity and global 5-hmC in human embryonic kidney cell cultures [38]. Hydroquinone exposure increased the global 5-hmC content and decreased 5-mC profiles through a reactive oxygen species (ROS) associated mechanism, since the hydroquinone effects were annulled upon N-acetyl-cysteine (NAC) rescue. In addition, the targeted siRNA mediated knockdown of TET1 caused a reversal of 5-hmC levels back to baseline. It suggests that numerous environmental exposures that induce reactive oxygen species/oxidative stress may influence 5-hmC patterns and increase the disease risk.
Environmental allergens
Asthma is a significant public health concern, and its prevalence has increased dramatically in past 30 years among both children and adults [39]. Since the prevalence of asthma cannot be explained by traditional genetic inheritance, much consideration has been given to lifestyle and environmental exposures being related to increased disease risk. House dust mite, a common environmental allergen, induces airway hyperresponsiveness and inflammation in mouse models through epigenetic alterations in the lung [40]. Exposure to dust mite has resulted in global increases in both 5-mC and 5-hmC in the lung, compared to saline controls. In addition, gene specific methylation of Pde4d, Pom121l2 and Ncx3 has been linked to dysregulated gene expression and cell functions of the lungs. How 5-mC and 5-hmC mediate gene regulation towards environmental allergens requires further investigation.
III. 5-hmC in Disease Development
Cancer
Epigenetic dysregulation is well described in cancer pathogenesis. Using human lung, brain, kidney, liver, skin, and small intestine tissues, the levels of 5-hmC are remarkably depleted in malignant tissues, compared to the benign tissues [41–43]. One explanation is that tumor formation involves rapid cell proliferation, which could lead to the passive loss of 5-hmC [41]. In support of this theory, proliferating cells stained with Ki67 did not express 5-hmC [3, 41]. However, more research is needed to understand the mechanisms linking the loss of 5-hmC in proliferating cells, which leads to increased cancer risks. If this theory is proved, genome wide 5-hmC and 5-mC profiles in conjunction with Ki67 staining could be valuable biomarkers for cancer diagnosis and management.
Gliomas exhibit epigenetic changes in response to disease progression. Immunohistochemistry (IHC) detection of 5-hmC in formalin fixed, paraffin-embedded, human glioblastomas revealed that 5-hmC decreased significantly with the progression of the tumor grade, regardless of fetal or adult origin [42]. Upon stratifying the 5-hmC IHC stains into high and low content tissues, 5-hmC reduction became a significant prognostic indicator of decreased life expectancy in fetal and adult glioblastomas. This indicates the critical role of 5-hmC in disease state progression and disease management.
In hepatocellular carcinoma, clinical and animal data indicate that 5-hmC may serve as a prognostic marker for disease outcomes. IHC detection of 5-hmC on hepatocellular carcinoma samples showed a drastic decrease in 5-hmC content in the tumor tissue, compared to the benign normal liver tissue [43]. Subsequently, lower levels of 5-hmC were associated with decreased overall survival in hepatocellular carcinoma. Male rats exposed to diethylnitrosamine, a potent carcinogen for hepatocellular carcinoma, exhibited a gradual dose-dependent decrease in 5-hmC IHC detection up through 24 weeks of treatment, and showed remarkably reduced 5-hmC in tumor versus non-tumor tissue [43]. These data further confirm the role of 5-hmC depletion as a biomarker of cancer development.
Melanomas are aggressive cancers that are capable of transforming from benign nevi into lethal metastases in a small surface area. Using tissue sections from patients in different stages of melanoma progression, IHC staining exhibited significant decreases in global 5-hmC levels, depending on the degree of disease [44, 45], even though the 5-mC profiles among the samples were constant, regardless of the disease state. IHC staining of TET2 was robust in nevi samples and reduced in advanced melanomas [45]. Therefore, a reduction in TET2 and 5-hmC indicate the epigenetic hallmark representing the disease progression of melanoma. These findings also further implicate the dangers of UV exposure in melanoma disease pathogenesis [44].
Myeloid malignancies are cancers originating in blood stem cells in the bone marrow and include acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) [46]. In bone marrow samples from mixed lineage leukemia (MLL)-rearranged AML patients (TET1 is a fusion partner of the MLL gene), TET1 is overexpressed [47]. Using small hairpin RNA (shRNA) against TET1 in mouse MLL-rearranged leukemic cells, it led to decreased 5-hmC in a dose-dependent manner, and induced apoptosis [47]. In vivo, TET1-knockout mice exhibited similar results to TET1-shRNA models, with TET1 reduction correlating to significant decreases in 5-hmC, compared to the controls [47]. Results suggested that TET1 plays a role in the pathogenesis of MLL-rearranged leukemia, which is marked by a global increase in 5-hmC and the transcriptional activation of many targets promoting cell proliferation and inhibiting apoptosis, resulting in cell transformation and AML disease pathogenesis. In MDS, TET2 and 5-hmC expression appear to be necessary for the normal differentiation of hematopoietic stem cells [48, 49]. TET2 mutations in bone marrow mononuclear cells were associated with ~16% of MDS patients, and the level of 5-hmC was significantly lower in MDS patients with mutant TET2 [50]. Upon risk-stratifying the 5-hmC patients, high 5-hmC levels had increased MDS survival rates when compared to those with low 5-hmC level [50]. It suggested that the depletion of 5-hmC can be used as diagnostic and prognostic tools for MDS patients.
Developmental Syndromes
In normal human tissues, average 5-hmC values vary depending on tissue type. Brain tissues have the highest 5-hmC content, up to 10 to 20-fold more than peripheral tissues and ESCs [14, 51]. Many neurodegenerative disorders are associated with aberrant methylation, once the epigenetic signature is impaired. Since neuronal cells in the adult brain do not divide mitotically, high levels of 5-hmC are maintained. High 5-hmC content indicates that active demethylation pathways may be upregulated [51, 52]. Under normal conditions in the central nervous system, abundant MeCP2 have a higher affinity for 5-hmC resides, compared to 5-mC, facilitating active gene transcription [52]. In classical Rett Syndrome, a neurodevelopmental disorder that affects primarily girls and results in the loss of language and motor skills, the loss-of-function MeCP2 mutation disrupts the neuronal maturation through the epigenetic dysregulation of MeCP2 function [53]. Given the abundant levels of 5-hmC in the central nervous system, decoding the role of 5-hmC in neuronal gene regulation and neurodevelopmental disorders is a growing interest among researchers.
Late-onset Diseases
Huntington’s disease is a fatal genetic disorder characterized by chorea, and cognitive and psychiatric decline. In a mouse model of Huntington’s disease, 5-hmC was significantly reduced in the striatum and cortex of Huntington’s disease mouse brains, compared to the control, and the IHC stain co-localized with NeuN, a marker of mature neurons [51]. Upon deep sequencing and genomic mapping, 436 striatal and 199 cortical genes were differentially hydroxymethylated [51]. Therefore, 5-hmC reduction in Huntington’s disease mouse brains further complements previous findings on aging and the decrease in 5-hmC in neurodegenerative disorders.
Alzheimer’s disease is the most common form of dementia among the aging population, and results from the aberrant production and cleavage of amyloid-β (Aβ) plaques and tau protein tangles. The etiology of Alzheimer’s disease is unknown, since disease onset occurs after decades of exposure at different stages of life. One factor investigated in Alzheimer’s disease mouse models is the role of stress during prenatal development, which has been shown to negatively affect cognition and hippocampal functioning, predominantly in females [54]. In female offspring experiencing prenatal stress, compared to the males and un-stressed control, the dorsal and ventral subregions of the hippocampus showed increased 5-hmC. This suggests that in utero prenatal stress can impact epigenetic mechanisms via 5-hydroxymethylation that increase Alzheimer’s disease risk.
Increasing evidence is implicating the role of epigenetic modifications in neuropsychiatric disorders. Using post-mortem brain cortex tissue from psychotic (bipolar disorder, schizophrenia) and depressed patients for examining the TET1 mRNA and protein expression, it was shown TET1 expression levels were increased in both the psychotic and depressed patients when compared to the controls [55]. In another study, the global 5-hmC levels were significantly increased in the psychotic patients, compared to controls [56]. In addition, decreased 5-hydroxymethylation was correlated to the decreased glutamic acid decarboxylase67 (GAD67) mRNA expression in psychotic subjects [55, 56]. Hence, 5-hmC can be used to investigate the pathogenesis of psychosis.
IV. Conclusion and Future Perspective
Given the nature of 5-hmC in the epigenetic regulation of cell functions, and its possible link to disease development, there is the possibility of employing global 5-hmC or specific hydroxymethylated genes as environmental biosensors. 5-hmC, the “sixth base”, is as stable as an epigenetic marker as 5-mC in the genome. The discovery of TET proteins, which was a breakthrough in the field of understanding gene regulation by DNA hydroxymethylation, allows us to track the development of 5-hmC via TET proteins in response to environmental exposure. Similar to other epigenetic studies, there are several challenges in performing 5-hmC studies. First, the exact mechanism by which TET proteins or 5-hmC regulate gene transcription is unclear. What are the other epigenetic factors that modulate the TET proteins and process the 5-hmC-mediated transcription signal? Second, the distribution of 5-hmC varies among the cell types and cell tissues. Are there any cell type-specific 5-hmC signatures? Most of the 5-hmC studies are focused on prenatal development, especially the stem cell differentiation and lineage. There are limited studies on the biological roles of 5-hmC in adult tissues. If 5-hmC markers could be validated in various biological samples, the 5-hmC signature could be applied in clinical research, such as in the development of non-invasive diagnostic and prognostic approaches, by testing for epigenetic markers of body fluids (i.e. serum and urine). Third, is there any critical window for the establishment of 5-hmC marks under the exposure of environmental factors? Not many studies have demonstrated the establishment of 5-hmC from the prenatal to the aging or diseased stages. No doubt, it could be assayed in the rodent model first to understand its role in disease development, and manipulate the studies on human tissues.
With the advance of “omic” profiling techniques and bioinformatics database searches, 5-hmC signatures can be discovered at every stage of cell development, or in response to any environmental exposure. Proper bioinformatics and statistical analyses on high-throughput epigenomic technologies, such as microarray or deep sequencing, are required, due to the huge amount of data that these tests provide. A set of follow-up studies, in vitro or in vivo, are also needed to validate and characterize a new set of molecular participants in disease development. We expect to identify markers that could allow us to reset the disruptive hydroxymethylome in disease development. Additionally, techniques for detecting single CpG hydroxymethylation have been established and applied for detecting gene specific hydroxymethylation. It helps to understand the epigenetic modulation of cell functions via modification at a specific gene promoter, leading to new designs for therapeutic drugs. In order to apply 5-hmC as a promising environmental biosensor, we must track epigenetic changes in different tissues and cell types, and the time course in a number of subjects. Applying 5-hmC studies to human population studies would speed up the understanding of the use of 5-hmC in predicting exposure and disease risk. However, methods of statistical analysis, such as data modeling and multiple testing, must be carefully chosen when we apply genetic and epigenetic analyses in environmental diseases. All in all, the studies of 5-hmC and its interactions with environmental exposure are exciting; however, many questions remain unanswered. It is believed that with the new basic knowledge of 5-hmC biology, advanced sequencing technology, and proper statistical and epidemiological strategies, we could expect the findings from 5-hmC studies to be applied for lifestyle recommendations and/or changes in clinical practices, which can lead to an improvement in disease management and, ultimately, in public health.
Acknowledgments
The authors acknowledge the following support: NIEHS (ES016887-WY Tang, P30ES03819) and NIH (HL10342-W Mitzner).
Footnotes
Conflict of Interest
T. Dao, R.Y.S. Cheng, M.P. Revelo, W. Mitzner, and WY Tang declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003 Mar;33(3):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004 May;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics & Chromatin. 2013 May;6(10):1–9. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & Development. 2011 Dec;25(23):2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Research. 2004 Aug;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delatte B, Fuks F. TET proteins: on the frenetic hunt for new cytosine modifications. Briefings in Functional Genomics. 2013 May;12(3):191–204. doi: 10.1093/bfgp/elt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristensen LS, Treppendahl MB, Grønbæk K. Analysis of Epigenetic Modifications of DNA in Human Cells. Current Protocols in Human Genetics. 2013 Apr;77:20.2.1–20.2.22. doi: 10.1002/0471142905.hg2002s77. [DOI] [PubMed] [Google Scholar]
- 8.Costello JF, Plass C. Methylation matters. J Med Genet. 2001 May;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009 May;324(5929):930–935. doi: 10.1126/science.1170116. This study demonstrated the discovery of the TET proteins which catalyze the oxidation of 5-methylcytosine to 5-hydroxymethylation in mammalian DNA. This breakthrough suggested a novel mechanism of regulation of DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson JP, Moggs JG, Wolf CR, Meehan RR. Epigenetic profiles as defined signatures of xenobiotic exposure. Mutation Research - Genetic Toxicology and Environmental Mutagenesis. 2013 Aug;:1–22. doi: 10.1016/j.mrgentox.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011 Aug;10(15):2428–2436. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Chen T. Tet family of 5-methylcytosine dioxygenases in mammalian development. J Human Genet. 2013 Jul;58(7):421–427. doi: 10.1038/jhg.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011 Dec;13(1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Research. 2012 Mar;22(3):467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012 Jun;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson JP, Hunter JM, Lempiainen H, Muller A, Terranova R, Moggs JG, Meehan RR. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Research. 2013 Jun;41(11):5639–5654. doi: 10.1093/nar/gkt232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of Methylcytosine from Hydroxymethylcytosine in DNA Molecules. J Am Chem Soc. 2011 Jan;133(3):486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011 Dec;147(7):1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-Hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011 Jan;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chia N, Wang L, Lu X, Senut M, Brenner C, Ruden DM. Environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics. 2011 Jul;6(7):853–856. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- 21.Morera S, Imberty A, Aschke-Sonnenborn U, Ruger W, Freemont PS. T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism. Journal of Molecular Biology. 1999 Sep;292(3):717–730. doi: 10.1006/jmbi.1999.3094. [DOI] [PubMed] [Google Scholar]
- 22.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011 Jan;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharyya S, Yu Y, Suzuki M, Campbell N, Mazdo J, Vasanthakumar A, Bhagat TD, Nischal S, Christopeit M, Parekh S, Steidl U, Godley L, Maitra A, Greally JM, Verma A. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Research. 2013 Sep;41(16):e157. doi: 10.1093/nar/gkt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Zhang L, Zhang L, Cai C. Analysis of global deoxyribonucleic acid 5-hydroxymethylcytosine in tissue by liquid chromatography-tandem mass spectrometry. Se Pu. 2012 May;30(5):533–537. doi: 10.3724/sp.j.1123.2011.12015. [DOI] [PubMed] [Google Scholar]
- 25.Gomez C, Pozo OJ, Diaz R, Sancho JV, Vilaroca E, Salvador JP, Marco MP, Hernandez F, Segura J, Ventura R. Mass spectrometric characterization of urinary toremifene metabolites for doping control analyses. J Chromatogr A. 2011 Jul;1218(29):4727–4737. doi: 10.1016/j.chroma.2011.05.073. [DOI] [PubMed] [Google Scholar]
- 26.Mao W, Hu J, Hong T, Xing X, Wang S, Chen X, Zhou X. A convenient method for selective detection of 5-hydroxymethylcytosine and 5-formylcytosine sites in DNA sequences. Org Biomol Chem. 2013 Jun;11(21):3568–3572. doi: 10.1039/c3ob40447a. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Zhang L, Zhou K, Ye X, Zhang J, Xie A, Chen L, Kang JX, Cai C. Simultaneous determination of global DNA methylation and hydroxymethylation levels by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J Biomol Screen. 2012 Aug;17(7):877–884. doi: 10.1177/1087057112447946. [DOI] [PubMed] [Google Scholar]
- 28.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010 Oct;38(19):e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012 Dec;7(12):2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min J-H, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012 Jun;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. This technique could differentiate 5-mC and 5-hmC at a single-base resolution and could be applied for detection of either locus-specific or genome-wide 5-hydroxymethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olkhov-Mitsel E, Bapat B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 2012 Oct;1(2):237–260. doi: 10.1002/cam4.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan L, Xiong L, Xu W, Wu F, Huang N, Xu Y, Kong L, Zheng L, Schwartz L, Shi Y, Shi YG. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Research. 2013 Apr;41(7):e84. doi: 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, Jiao Y, Li C, Liu S, Wu D, Gu W, Yang YG, Xu GL, Wang H. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. J Am Chem Soc. 2013 Jul;135(28):10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 34.Minor EA, Court BL, Young JI, Wang G. Ascorbate Induces Ten-Eleven Translocation (Tet) Methylcytosine Dioxygenase-mediated Generation of 5-Hydroxymethylcytosine. Journal of Biological Chemistry. 2013 May;288(19):13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, Lorincz MC, Ramalho-Santos M. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013 Jul;500(7461):222–226. doi: 10.1038/nature12362. This study investigates the role of Vitamin C in regulating TET activity and DNA methylation profiles in mouse embryonic stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson JP, Lempiäinen H, Tefferi A, Nestor CE, Müller A, Bolognani F, Oakeley EJ, Schübeler D, Terranova R, Reinhardt D, Moggs JG, Meehan RR. Non-genotoxic carcinogen exposure induces defined changes in the 5-hydroxymethylome. Genome Biology. 2012 Oct;13(10):R93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jefferson WN, Chevalier DM, Phelps JY, Cantor AM, Padilla-Banks E, Newbold RR, Archer TK, Kinyamu HK, Williams CJ. Persistently Altered Epigenetic Marks in the Mouse Uterus After Neonatal Estrogen Exposure. Mol Endocrinol. 2013 Oct;27(10):1666–1677. doi: 10.1210/me.2013-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coulter JB, O’Driscoll CM, Bressler JP. Hydroquinone increases 5-hydroxymethylcytosine formation through Ten Eleven Translocation 1 (Tet1) 5-methylcytosine dioxygenase. J Biol Chem. 2013 Oct;288(40):28792–28800. doi: 10.1074/jbc.M113.491365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selgrade MK, Blain RB, Fedak KM, Cawley MA. Potential risk of asthma associated with in utero exposure to xenobiotics. Birth Defect Res C Embryo Today. 2013 Mar;99(1):1–13. doi: 10.1002/bdrc.21028. [DOI] [PubMed] [Google Scholar]
- 40.Shang Y, Das S, Rabold R, Sham JS, Mitzner W, Tang WY. Epigenetic Alterations by DNA Methylation in House Dust Mite–Induced Airway Hyperresponsiveness. Am J Respir Cell Mol Biol. 2013 Aug;49(2):279–287. doi: 10.1165/rcmb.2012-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine Is Strongly Depleted in Human Cancers but Its Levels Do Not Correlate with IDH1 Mutations. Cancer Res. 2011 Dec;71(24):7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-Hydroxymethylcytosine Is Associated with Neural Progenitor Phenotype in Normal Brain and Shorter Survival in Malignant Glioma. PLoS ONE. 2012 Jul;7(7):e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, Yang Z, Wu L, Xia F, Bie P, Cui Y, Bian XW, Qian C. Decrease of 5-Hydroxymethylcytosine Is Associated with Progression of Hepatocellular Carcinoma through Downregulation of TET1. PLoS ONE. 2013 May;8(5):e62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee C-W, Hu Di, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-Hydroxymethylcytosine is an Epigenetic Hallmark of Melanoma. Cell. 2012 Sep;150(6):1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambichler T, Sand M, Skrygan M. Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma Res. 2013 Jun;23(3):218–220. doi: 10.1097/CMR.0b013e32835f9bd4. [DOI] [PubMed] [Google Scholar]
- 46.Tefferi A, Thiele J, Vardiman JW. The 2008 World Health Organization classification system for myeloproliferative neoplasms: order out of chaos. Cancer. 2009 Sep;115(17):3842–3847. doi: 10.1002/cncr.24440. [DOI] [PubMed] [Google Scholar]
- 47.Huang H, Jiang X, Li Z, Li Y, Song CX, He C, Sun M, Chen P, Gurbunxani S, Wang J, Hong GM, Elkahloun AG, Arnovitz S, Wang J, Szulwach K, Lin L, Street C, Wunderlich M, Dawlaty M, Neilly MB, Jaenisch R, Yang FC, Mulloy JC, Jin P, Liu PP, Rowley JD, Xu M, He C, Chen J. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013 Jul;110(29):11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011 Nov;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 49.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010 Dec;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Guangsen Z, Yi Y, Xiao L, Pei M, Liu S, Luo Y, Zhong H, Xu Y, Zheng W, Shen J. Decreased 5-hydroxymethylcytosine levels are associated with TET2mutation and unfavorable overall survival in myelodysplastic syndromes. Leuk Lymphoma. 2013 Nov;54(11):2466–2473. doi: 10.3109/10428194.2013.778408. [DOI] [PubMed] [Google Scholar]
- 51.Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Luo HR, Guo C, Han C, Tang TS. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington’s disease. Hum Mol Genet. 2013 Sep;22(18):3641–3653. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- 52.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012 Dec;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Della Sala G, Pizzorusso T. Synaptic plasticity and signaling in Rett syndrome. Dev Neurobiol. 2013 Aug; doi: 10.1002/dneu.22114. [DOI] [PubMed] [Google Scholar]
- 54.Sierksma ASR, Prickaerts J, Chouliaras L, Rostamian S, Delbroek L, Rutten BPF, Steinbusch HWM, van den Hove DLA. Behavioral and neurobiological effects of prenatal stress exposure in male and female APPswe/PS1dE9 mice. Neurobiol Aging. 2013 Jan;34(1):319–337. doi: 10.1016/j.neurobiolaging.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012 Sep;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grayson DR, Guidotti A. The Dynamics of DNA Methylation in Schizophrenia and Related Psychiatric Disorders. Neuropsychopharmacology. 2013 Jan;38(1):138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]


