Abstract
Targeted alteration of the genome lies at the heart of the exploitation of S. pombe as a model system. The rate of analysis is often determined by the efficiency with which a target locus can be manipulated. For most loci this is not a problem, however for some loci, such as fin1 +, rates of gene targeting below 5% can limit the scope and scale of manipulations that are feasible within a reasonable time frame. We now describe a simple modification of transformation procedure for directing integration of genomic sequences that leads to a 5-fold increase in the transformation efficiency when antibiotic based dominant selection markers are used. We also show that removal of the pku70 + and pku80 + genes, which encode DNA end binding proteins required for the non-homologous end joining DNA repair pathway, increases the efficiency of gene targeting at fin1 + to around 75–80% (a 16-fold increase). We describe how a natMX6/rpl42 + cassette can be used for positive and negative selection for integration at a targeted locus. To facilitate the evaluation of the impact of a series of mutations on the function of a gene of interest we have generated three vector series that rely upon different selectable markers to direct the expression of tagged/untagged molecules from distinct genomic integration sites. pINTL and pINTK vectors use ura4 + selection to direct disruptive integration of leu1 + and lys1 + respectively, while pINTH vectors exploit nourseothricin resistance to detect the targeted disruption of a hygromycin B resistance conferring hphMX6 cassette that has been integrated on chromosome III. Finally, we have generated a series of multi-copy expression vectors that use resistance to nourseothricin or kanamycin/G418 to select for propagation in prototrophic hosts. Collectively these protocol modifications and vectors extend the versatility of this key model system.
Introduction
The genetic malleability of the fission yeast S. pombe has helped it to maintain a prominent position alongside the more extensively exploited budding yeast Saccharomyces cerevisiae, as a powerful model system for the characterisation of the basic facets of eukaryotic cell and molecular biology. This malleability is based upon an extensive repertoire of classical and molecular genetic techniques [1], [2], [3]. As in budding yeast these techniques were initially based upon the exploitation of key auxotrophic markers.
Classical genetic analysis the adenine biosynthesis pathway in S. pombe highlighted the utility of the colony-colour change resulting from the accumulation of P-ribosylaminoimidazole in ade6 mutants that is then oxidised to a red pigment [4]. The ability to use this red pigmentation as a reporter for Ade6 function made this locus a major focus for studies of core genetic principles. These studies led to the development of a number of useful genetic tools including ade6.M210/ade6.M216 hetero-allelic complementation for the selection and maintenance of diploid strains [5] and the use of the sup3.5 opal suppressor tRNAser mutation as a marker for selection in an ade6.704 mutant background [6], [7], [8]. Cross species complementation of S. pombe leu1 mutations with the S. cereviaisae LEU2 + gene was initially used to apply existing budding yeast technology to fission yeast [9], but remains a widely used selectable marker to this day because the lack of homology to sequences in the S. pombe genome means that it does not direct integration into a specific genomic site. However, when used as a marker to select for site specific integration, multiple integration events can occur [10], suggesting either that the heterologous expression of the LEU2 + gene is barely sufficient for growth at low copy number or that the budding yeast enzyme is less attuned to fission yeast physiology than the native 3-isopropyl malate dehydrogenase enzyme, Leu1. Transposition of the lessons learnt from the exploitation of the budding yeast ornithine decarboxylase URA3 + gene for positive and negative selection [11] led to the deletion of the ura4 + gene from S. pombe to generate the ura4.d18 allele that is so widely used in the field today [12] with many ura4 + based vectors [13], [14], [15], [16]. Continued developments are considerably expanding the array of available auxotrophy-complementing markers to include: ade7, his1, his2, his3, his5, arg3, arg12, lys1, lys2 and tyr1 [17], [18], [19], [20], [21], [22], [23], [24]. However, his3 +, LEU2 +and ura4 + remain the most widely-used markers for selection of multi-copy vectors in common use. Integration vectors that target a particular heterologous locus have been less extensively developed, however the pDUAL series and pJK148 vectors are used widely as they exploit recombination to convert the leucine auxotrophy of leu1.32 to leucine prototrophy to select integration at the leu1 locus [25], [26], [27]. The pJK210 uses a similar rescue of ura4.294 to target integration at the ura4 locus [25].
While these auxotrophic selection markers offer powerful tools, they also create the need to introduce an increasingly complex array of background markers into a strain of interest. Not only is this time consuming but many combinations of deficiencies in amino acid provision compromise a host strain’s fitness on certain media, which may complicate the interpretation of the phenotype arising from the mutation of interest. Furthermore, the sensitivity of the broadly acting TOR signalling network to addition of leucine to the medium [28] indicates that provision of amino acids demanded by the use of auxotrophic markers and perhaps the auxotrophic markers themselves are not merely passive players in cellular homeostasis, but can influence the control networks that impinge upon diverse processes from metabolism, through cell cycle control, sexual differentiation, and the actin cytoskeleton. Thus, controlling the genetic context within which the consequences of particular mutations are studied in prototrophs not only accelerates the rate of analysis, but avoids both anticipated and unforeseen complications arising from interplay between pathways.
Following the highly successful exploitation of antibiotic resistance genes as dominant selectable markers for PCR based tagging and deletion approaches in the budding yeast S. cerevisiae [29], [30], [31], [32], [33], [34], the technology has been adapted for use in a variety of fungi including S. pombe. Genes conferring resistance to kanamycin/G418, hygromycin B, phleomycin/bleomycin and nourseothricin/ClonNat are highly effective dominant markers in fission yeast [35], [36], [37], [38]. They have been extensively exploited in an increasing array of “PCR tagging vectors” in which oligo-nucleotides, that fuse vector sequences to short stretches of homology to the target locus, are used to amplify cassettes that will place “tags” and markers of choice in particular genomic contexts following targeted recombination into the host genome [29], [30], [38], [39], [40], [41], [42], [43], [44], [45]. While this approach is very powerful, it still faces the challenge that the number of manipulations is limited by the range of markers available. However, this problem can be circumvented by flanking the marker with loxP sites so that it can be excised from the genome following integration by the induction of Cre recombinase [46]. Host strains can then be sequentially modified with the same selectable marker, irrespective of previous manipulations.
Although attempts to define the extent of the problem have proved challenging [25], [47], [48], many anecdotal accounts suggest that the efficiency of targeting different loci by PCR tagging is variable; some loci can be targeted with very high efficiency while others only poorly, or not at all. It has been suggested that illegitimate recombination poses a major issue in these cases. In such circumstances the problem may be resolved by extending the region of homology with the genome can enhance the efficiency of targeting. In many other fungi deletion of the genes encoding the DNA end recognition proteins that are required for non-homologous end joining (NHEJ), Ku70 and Ku80 [49] greatly increases targeting efficiencies [50], [51], [52], [53], [54], [55], [56].
We now show that the removal of either the Ku70 or Ku80 homologues from S. pombe (Pku70 and Pku80 respectively) increases targeting efficiency at the fin1 + locus from 5% to 80% (16-fold increase). A modification to transformation procedures enhances transformation frequencies by a further 5–8 fold when selecting for antibiotic resistance markers. We show that a PCR cassette that combines the cycloheximide sensitivity of rpl42 + in an rpl42.sP56Q background [57] with the nourseothricin resistance conferred by natMX6 [58] offers a robust and cheaper alternative to positive and negative selection cycles with ura4 + and 5 fluoro-orotic acid (FOA) [12]. We describe two complementary integration vector series that exploit disruption of an auxotrophic marker with a second auxotrophic marker to direct the regulated expression of tagged or untagged molecules from a reproducible genome context. A further set of integration vectors exploits antibiotic resistance markers to direct the integration of both tagged and untagged expression cassettes into a site on chromosome III. We have also switched the markers in our multi-copy pREP41 based tagging/expression series [16] to generate vectors that exploit natMX6 or kanMX6 as a selectable marker.
Materials and Methods
Strain Growth, Selection and Maintenance
E. coli strain DH5α was used to propagate plasmids in standard LB medium. The S. pombe leu1.32 his2 h+(IH147) and 972h- (IH5974) strains were grown by standard procedures [2]. The following stock solutions of 100 mg ml−1 Geneticin (MP Biomedicals, 158782), Hygromycin B (Calbiochem, 400050), Nourseothricin/ClonNat (Werner BioAgents, 96736-11-7), Phleomycin (Sigma, P9564), and Cyclohexamide (Sigma, C1988) were added to generate final concentrations of 100 µg/ml in the growth medium where appropriate.
Transformation of S. pombe
Cells were grown to mid-log phase in YES (4×106 cells/ml). After harvesting cells were washed with H2O, 0.1 M Lithium Acetate (pH 4.9) and re-suspended in 0.1 M Lithium Acetate (pH 4.9) at 109 cells ml−1. After 1 h incubation at 25°C 1–5 µg DNA and 290 µl of 50% PEG4000 (freshly made in sterile 0.1 M Lithium Acetate, pH 4.9 for each transformation) was added to 100 µl cell-suspension. After 1h incubation at 25°C and 15 min heat shock at 43°C, cells were harvested, washed with H2O and re-suspended in MSL-N or spread to YES.
Molecular Genetics
Generating pINTL vectors
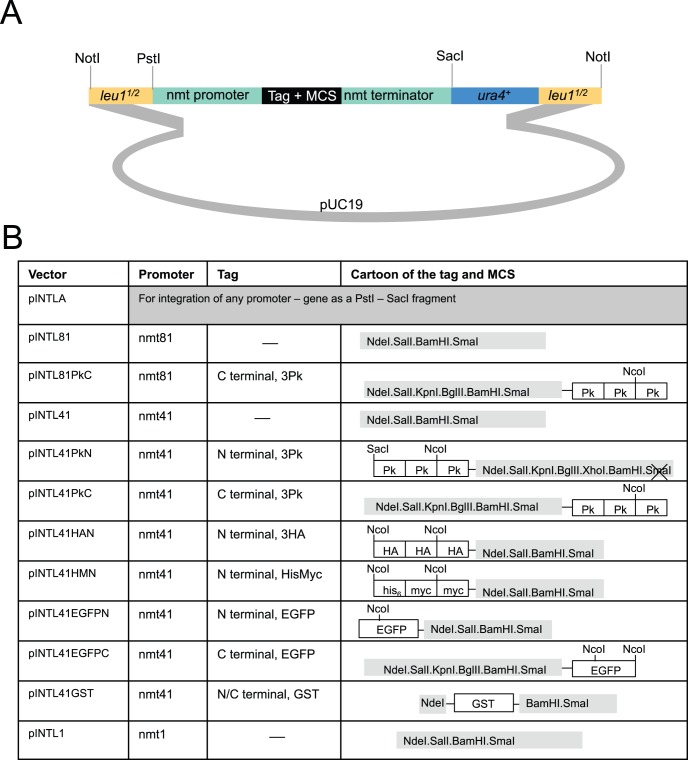
A 686 bp fragment of pUC19, including the MCS and NdeI site, were removed by Phusion mediated deletion (New England Biolabs) using oligo nucleotides BH1 and BH2, creating a NotI site. A 2.32 kb fragment extending from −456 to +752 of the S. pombe leu1 + gene was amplified as a NotI fragment using oligo-nucleotides BH3 and BH4 and cloned into the modified pUC19 vector to create pINT1. An 1.78 kb fragment extending from –516 to +1186 of the S. pombe ura4 + gene was amplified from pURA4 [12] using oligonucleotides BH5 and BH6 to flank the ura4 + sequences with 5′ PstI site and 3′ SacI sites. Oligos BH7 and BH8 were used to amplify pINT1, which was then used as recipient for the amplified ura4 + fragment within the leu1 + open reading frame by Gibson-mediated integration (New England Biolabs) to generate pINTLA that can act as recipient for any PstI/SacI fragment containing promoter-insert-terminator (Figure S1). The remaining plasmids in the pINTL series were generated by cloning the appropriate PstI–SacI fragment from a relevant pREP tagging vector [16]. pINTL41PkN was generated by insertion of the PstI/SacI fragment of pREP41PkN into pINTLA followed by the SacI/SacI fragment from pREP41PkN. Full sequences of the pINTL vectors are presented in Figure S1.
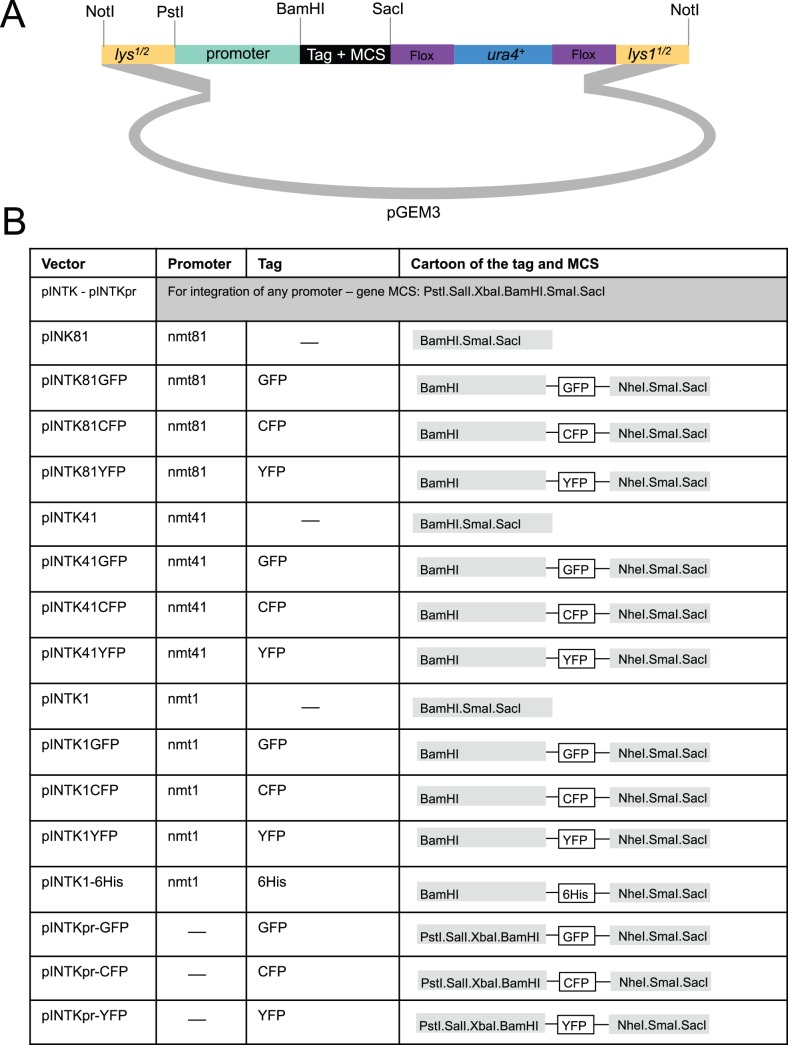
Generating pINTK vectors
The lys1 5′ region was amplified (VS642/VS644) to introduce HindIII NotI sites at one end and PstI site at the other end. The lys1 3′region was amplified (VS645/VS646) to introduce a KpnI site at one end and EcoRI NotI sites at the other end. Both fragments were cloned HindIII - PstI and KpnI – EcoRI, respectively into pGEM3. The loxP–ura4 cassette was generated by PCR amplification of the ura4+ gene (VS647/VS648) to introduce KpnI site and a LoxP site on one end and SmaI SacI sites with the LoxP site on the other end. This fragment was then cloned as a KpnI – SmaI fragment into the lys1+ containing vector to generate pINTK, pINTK81, pINTK41 and pINTK1 by cloning the nmt promoters from pREP81, pREP41 and pREP1 respectively as PstI – BamHI fragments into pINTK. GFP, CFP and YFP tags were amplified to introduce a BamHI site at the 5′end and NheI SmaI SacI sites at the 3′end. The tags were then cloned BamHI – SacI into the pINTK81/41/1. The 6His tag was cloned as a BamHI – NheI fragment generated after annealing of complementary oligonucleotides (VS1482/VS1483) into the pINTK1GFP to generate pINTK1-6His. The sequence of the pINTK vector is presented in Figure S2.
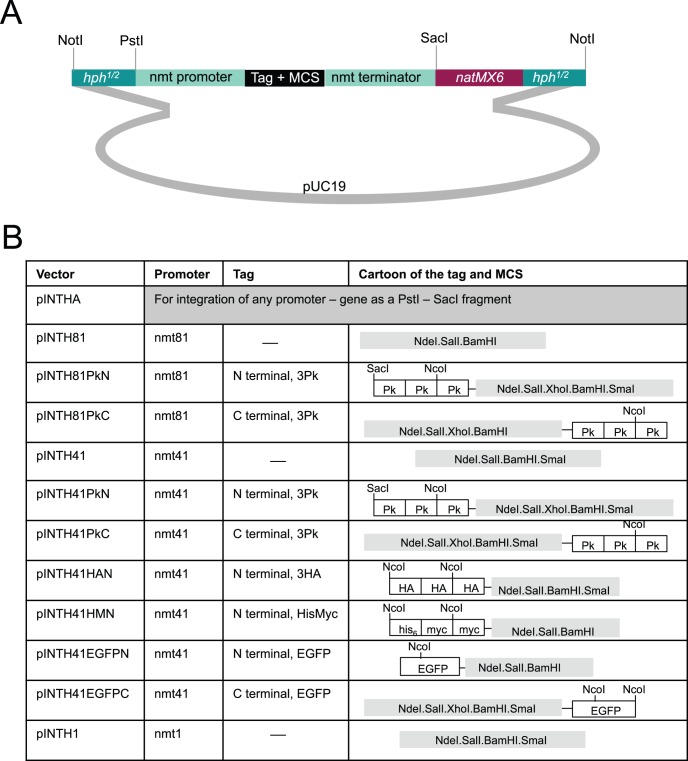
Generating pINTH vectors
The first step in the generation of the pINTH vector was Quickchange (Stragene) silent mutagenesis to remove the PstI and NdeI sites from hphMX6 (CTGCAG>CTGCAA and CATATG>CATTTG, respectively) and a XmaI/SmaI site from natMX6 (CCCGGG>CCCAGG). The following fragments were amplified using the indicated primers to introduce restriction sites at their termini before being cloned into the vector ZeroBluntTOPO (Invitrogen): one 0.85 kb half of hphMX6 flanked with PstI and NotI HindIII (primers DF1 and DF2); the remaining 0.85 kb fragment flanked by EcoRI and EcoRI NotI (primers DF3 and DF4); natMX6 flanked with SacI and EcoRI (primers DF5 and DF6). The SacI – EcoRI natMX6 fragment was inserted into pUC19 followed by the EcoRI and Pst1-HindIII hphMX6 fragments to generate pINT*. pINT* was digested PfoI and KpnI, end filled with Klenow polymerase (New England Biolabs) to remove a 189 bp fragment and re-ligated to remove the NdeI site of pUC19 to generate pINT**. PstI SacI sequences containing the nmt promoter - cloning site/tag - nmt teminator cassettes from pREP1, pREP41 and pREP81 based plasmids were then inserted between PstI SacI sites in pINT** to generate the vector series. Because the multi-cloning site of pREP41PkN and pREP81PkN contains a SacI site, the pINTH41PkN and pINTH81PkN were generated by sequential insertion of the appropriate SacI and PstI-SacI fragments from pREP41PkN and pREP81PkN respectively. pINT* was digested with NdeI, end filled with Klenow polymerase (New England Biolabs) and re-ligated to generate pINTHA. Full sequences of the pINTH vectors are presented in Figure S3.
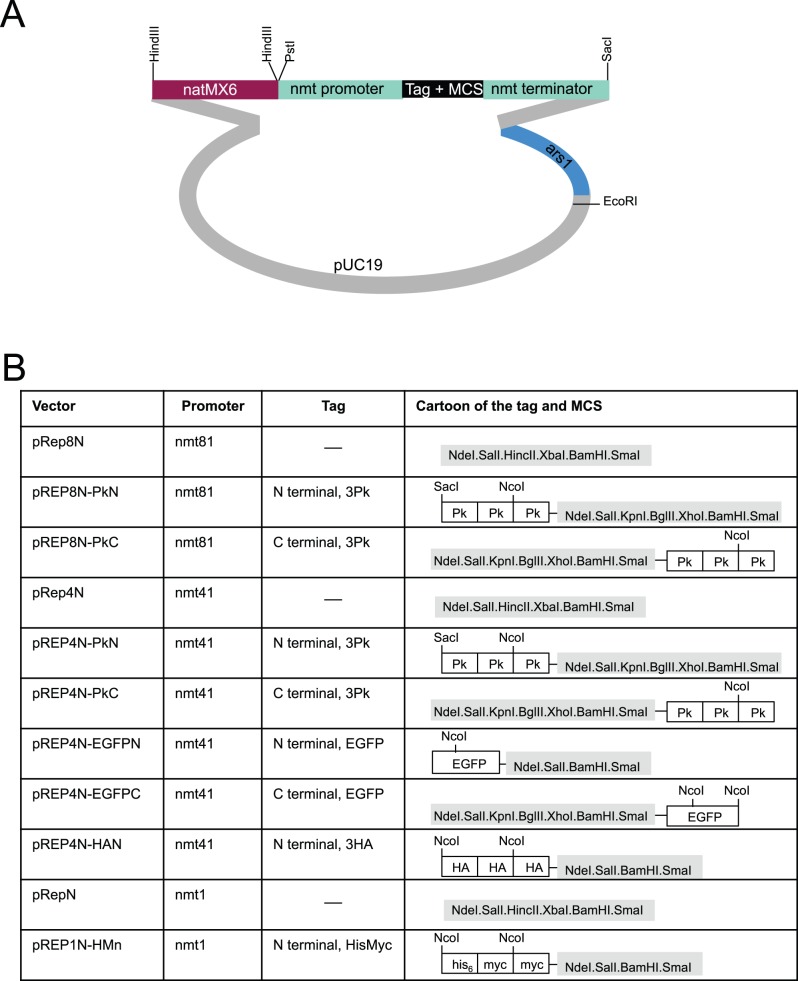
Generating pREPN and pREPK vectors
To generate the pREPN vectors, the SmaI sites of the natMX6 gene in natMX6 cassette were destroyed (the NdeI site in pFA6anatMX6 is outside of the cassette). The natMX6 gene was amplified with the oligonucleotides AG1and AG2 that had 20 nucleotides homology to the ends of the natMX6 cassette and 80 bp homology with the sequences adjacent to the LEU2 + integration site of pREP1. 5 µg of the natMX6 fragment was transformed alongside 1 µg of the appropriate LEU2 + based pREP vector that had been linearised by digestion with KpnI that cut inside the LEU2 + marker gene of the relevant vector in host IH147. Plasmids were isolated from two antibiotic resistant leucine auxotrophic colonies with the DNA isolation kit (Flowgen), before 5 µl was transformed into DH5α. Restriction mapping and sequencing of DNA from two transformants identified the desired vector. The pREPK series were made in the same way as pREPN, but the co-transformation used a KanMX6 fragment amplified from pFA6a-kanMX6 [40].
Biochemistry
Generation of cell extracts and western blotting of these extracts was as described previously [59].
Results
A natMX6/rpl42 Cassette for Positive and Negative Selection
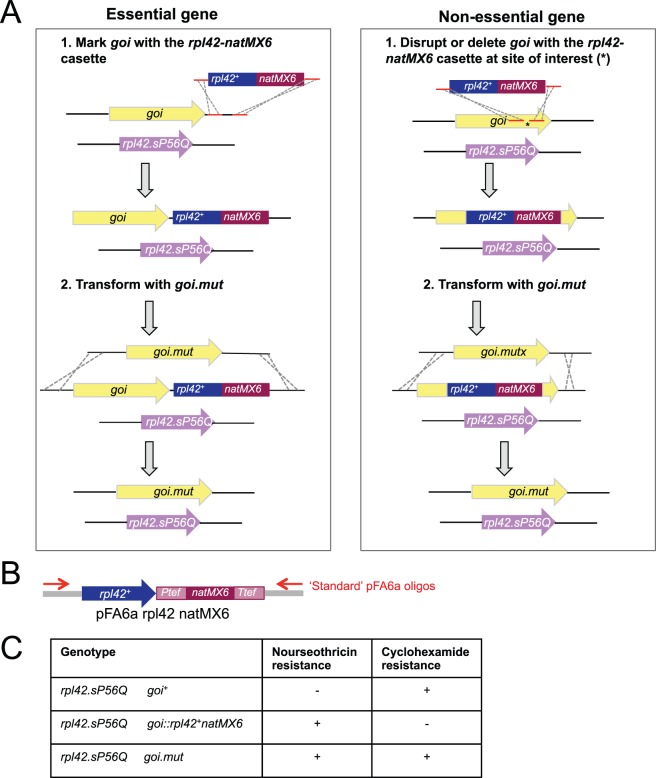
To study the significance of phosphorylation events in the timing and execution of cell division we mutate candidate sites at endogenous loci [60], [61], [62]. We first integrate a marker at the gene of interest (goi +) before transforming this new host strain with a fragment whose homology to the genome extends beyond either side of the integration site. As this fragment harbours a goi mutation, positively selecting for marker loss and screening by DNA sequencing identifies the candidate with the desired mutation (Figure 1A). Although the ability to apply both positive and negative selection for ura4 + make it an ideal marker for this purpose [12], the cost of FOA can become limiting in a programme that targets multiple mutants to multiple loci. Similarly, the need to express human equilibrative nucleoside transporter, hENT1, to use thymidine analogues for positive selection [63] limits the appeal of this alternative approach. To generate a rapid and cheap alternative to these two options we have combined the strong positive selection of natMX6 [58] with the rpl42 recessive cycloheximide resistance marker system developed by Krogan and colleagues [57] in a natMX6/rpl42 + double cassette in pFA6arpl42natMX6 (Figure 1B). Resistance to nourseothricin selects for integration of this cassette in an rpl42.sP56Q host strain. Subsequent replacement of the cassette by an overlapping sequence is selected for by placing transformants onto plates containing cycloheximide (Figure 1C).
Figure 1. Manipulating native loci with an rpl42+/natMX6 cassette.
A) Approaches used for targeted mutagenesis. B) The structure of the pFA6arpl42natMX6 plasmid. C) The phenotype switches arising from the progression through the indicated genotypes.
Overnight MSL-N Incubation Enhances Transformation Efficiency
Our ability to manipulate native loci has been confounded by varying efficiencies of targeted integration. For some loci, such as fin1 +, targeted integration was uncommon, with around a 5% chance that a transformant was the desired integration event, prompting us to seek strategies that may improve transformation and targeting efficiencies.
When using antibiotic selection for integrative transformation, transformants are incubated in non-selective conditions for 18 hours before applying selection to enable them to accumulate sufficient enzyme from the newly generated expression cassette to survive the otherwise lethal impact of the antibiotic [40]. Traditionally this “recovery phase” has been applied by spreading cells on non-selective plates before replica plating the ensuing lawn of cells onto antibiotic containing plates 18 hours later [40]. While highly effective, it is inevitable that the transfer efficiency during replica plating is less than 100%. Furthermore, if the targeting event compromises fitness, vigorous growth of the non-transformed host clones may out compete the less fit transformant clones. We therefore sought recovery conditions in which cell division would be blocked and yet the antibiotic metabolising enzymes could accumulate in all transformants before exposure of the entire mix of transformants and untransformed neighbours to selection pressure.
Cells are unable to divide in the absence of a nitrogen source [64]. We therefore asked whether we could simply substitute the overnight incubation on solid medium with incubation in a liquid minimal medium that lacked a nitrogen source. The MSL medium that was developed by Richard Egel [65], is ideal for this goal because it efficiently invokes a nitrogen starvation response. Amino acid supplements were omitted from this medium in our tests to limit provision of nitrogen from in vivo amino acid catabolism.
We used the integration of a marker at the pku80+ and leu1+ loci to assess the impact of MSL-N recovery phase of differing durations upon the transformation efficiency. Cells were grown to mid log phase (4×106 cells ml−1) in rich YES medium before standard procedures were used to make the cells competent to receive DNA. The DNA fragments that were added to these competent cells were generated by PCR amplification of pFA6a antibiotic resistance deletion vector series templates with the same oligonucleotides being used with each template [40], [58]. For each marker tested the transformed cell mix was split into 6. One portion was immediately spread onto non-selective YES plates at 25°C, while the others were re-suspended in 1 ml of MSL-N and incubated with agitation at 25°C. The MSL-N transformation mixes were spread onto selective plates 2, 4, 8, 16 and 24 hours later. Cell counts confirmed that no cell division occurred during the 24 hours of incubation in MSL-N medium (data not shown). The lawn of cells on the YES plates that had received the transformation mix immediately were replica plated 20 hours after the initial spreading of the transformation mix. There were no major differences in the number of transformants between any of the protocols when uracil prototrophy was used as the selectable marker to detect ura4 + integration (Table 1). In contrast, when antibiotic resistance formed the basis for the selection for the integration event between 5 to 8 fold more transformants were obtained in the samples that received a 24 hour MSL-N recovery period than when the aliquot had been replica plated aliquot (Table 1). PCR analysis revealed a similar rate of integrative transformation in either the replica plated or liquid recovery samples (data not shown).
Table 1. Transformation efficiencies.
| 108 cells transformed with 1 µg DNA for the indicated transformation | |||||
| ura4 + into leu1+ | kanMX6 into pku80+ | natMX6 into pku80+ | hphMX6 into pku80+ | bleMX6 into pku80+ | |
| Spread directly to selective media | 950 | 0 | 0 | 0 | 0 |
| Spread to YES, replica plate to selective media after 20 h | 1000 | 150 | 140 | 120 | 100 |
| Incubate in MSL for 2 h, spread to selective media | 900 | 0 | 0 | 0 | 0 |
| Incubate in MSL for 4 h, spread to selective media | 950 | 3 | 2 | 0 | 1 |
| Incubate in MSL for 8 h, spread to selective media | 1000 | 4 | 5 | 3 | 3 |
| Incubate in MSL for 16 h, spread to selective media | 1100 | 650 | 700 | 600 | 650 |
| Incubate in MSL for 20 h, spread to selective media | 1100 | 850 | 800 | 750 | 800 |
The table shows the number of transformants obtained when 1 µg of the indicated DNA fragments was transformed into identical numbers of competent cells of the indicated strains. Each transformation mix was split into seven equal aliquots that were treated as indicated in the column on the left.
Enhanced Efficiency of Integrative Targeting in pku70.Δ or pku80.Δ Backgrounds
Our attempts to target integration at a range of loci concur with the anecdotal experiences of the S. pombe community that the efficiency of targeting different loci varies widely. In many fungi removal of the Ku70 and Ku80 end recognition proteins blocks non-homologous end joining DNA repair pathway [49] to greatly enhances the frequency of gene targeting [50], [51], [52], [53], [54], [55], [56]. We therefore asked whether deletion of either molecule might enhance the 5% efficiency of integration at the S. pombe fin1 + locus.
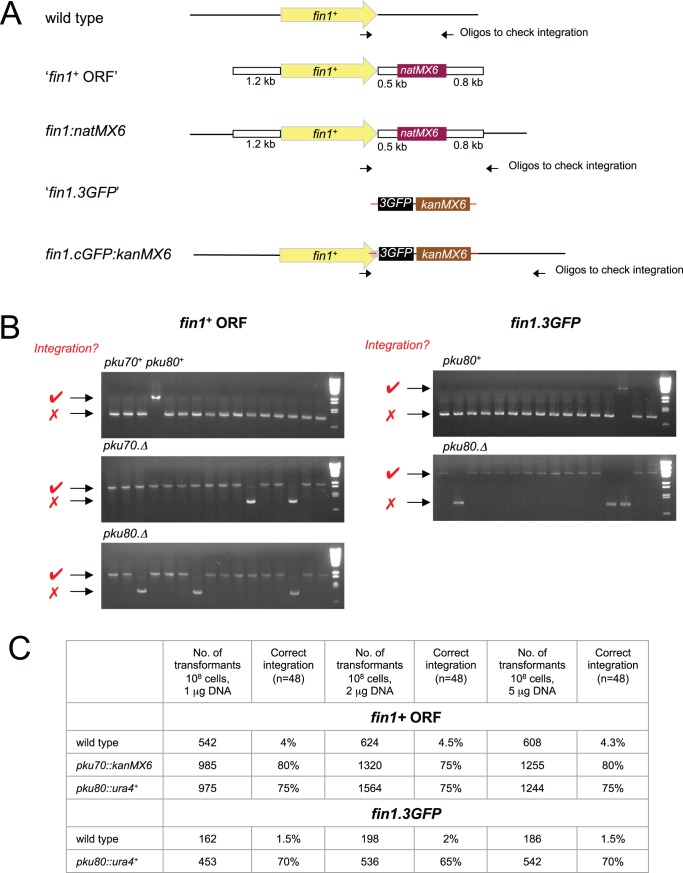
Two types of DNA fragment were used for transformation: a large fragment excised from a plasmid in which the natMX6 marker was flanked by extensive regions of homology (1.2 kb 5′ and 0.8 kb 3′) to the fin1 + locus (Figure 2A, upper “fin1 + ORF”) and a short fragment with 80 bp regions of homology either side of the stop codon that generated a fin1+.3GFP fusion sequence by standard PCR amplification [40] from the pSM1023 template [66], [67] (Figure 2A, lower “fin1+.3GFP”). For the “fin1 + ORF” DNA fragment, a single sample of donor DNA was split into four. One quarter was transformed into a pku70::kanMX6 strain, another into an otherwise isogenic pku70 + strain and the remaining two aliquots into pku80::ura4+ and isogenic pku80 + hosts. As the selectable marker for the “fin1+.3GFP” fragment was the same geneticin/G418 resistance marker that had been used to delete pku70 + with kanMX6, this fin1+.3GFP fragment was only transformed into pku80::ura4+ and isogenic hosts. Diagnostic PCR analysis of transformants from each comparison revealed that the efficiency of gene targeting was elevated to between 75 and 80% (at least 16 fold increase) by the removal of either Pku70 or Pku80 (Figure 2B, C). Such a marked improvement in transformation efficiency upon removal of these end recognition factors prompted us to generate strains in which pku70 + and pku80 + have been replaced with the kanMX6, hphMX6 and natMX6 cassettes. These strains have been deposited in the Yeast Genome Resource Centre Japan (http://yeast.lab.nig.ac.jp/nig/index_en.html, for YGRC strain numbers see Table 2).
Figure 2. Inclusion of pku70.Δ and pku80.Δ in host strain radically enhances targeting at the fin1 + locus.
A) Cartoons depicting the structure of the DNA fragments used to direct the integration of a natMX6 cassette 3′ to the Fin1 coding sequences at the fin1 + locus and the integration of sequences encoding three GFP molecules, a stop codon and the kanRMX6 marker at the end of the fin1 + locus. B) PCR amplification reactions with the oligonucleotides indicated by arrows in panel A to monitor the structure of the genomic regions at the fin1 + locus. For the “fin1 ORF” transformation amplification gives an 850 bp fragment (red cross next to each panel), whereas with successful integration generates an 2050 bp fragment (red tick next to each panel). For the “fin1.3GFP” transformation amplification with the same primers used to screen “ fin1 ORF” transformants generated an 850 bp fragment in the recipient host (red cross next to each panel) and an 4650 bp fragment in the correct transformant (red tick next to each panel). C) A table showing the frequency of correct integration events in the indicated strains with the indicated concentrations of each DNA fragment as determined by PCR analysis of 48 candidate transformants in each case.
Table 2. Strains used in this study.
| Lab Strain number | Genotype | YGRC strain number | Source |
| IH5974 | 972 h− | Lab stock | |
| IH1308 | ura4.D18 h− | Lab stock | |
| IH8794 | rpl42.sP56Q leu1.32 ura4.D18 h+ | Roguev et al. 2007 | |
| IH5221 | pku70::his3 leu1.32 his3.d1 ade6.M216 ura4.d18 h− | FY23684 | Lab stock |
| IH6067 | pku70::kanMX6 leu1.32 ura4.D18 his2 h+ | FY23686 | Manolis et al. 2001 |
| IH12994 | pku70::natMX6 ura4.D18 | FY23687 | This study |
| IH12959 | pku70::hphMX6+ leu1.32 ura4.D18 his2 h+ | FY23685 | This study |
| IH6114 | pku80::ura4+ leu1.32 ura4.D18 his2 h+ | FY23691 | Manolis et al. 2001 |
| IH13006 | pku80::kanMX6 | FY23689 | This study |
| IH12958 | pku80::natMX6+ leu1.32 ura4.D18 | FY23690 | This study |
| IH12960 | pku80::hphMX6 ura4.D18 his2 h+ | FY23688 | This study |
| IH5869 | hph171k h− | FY23692 | This study |
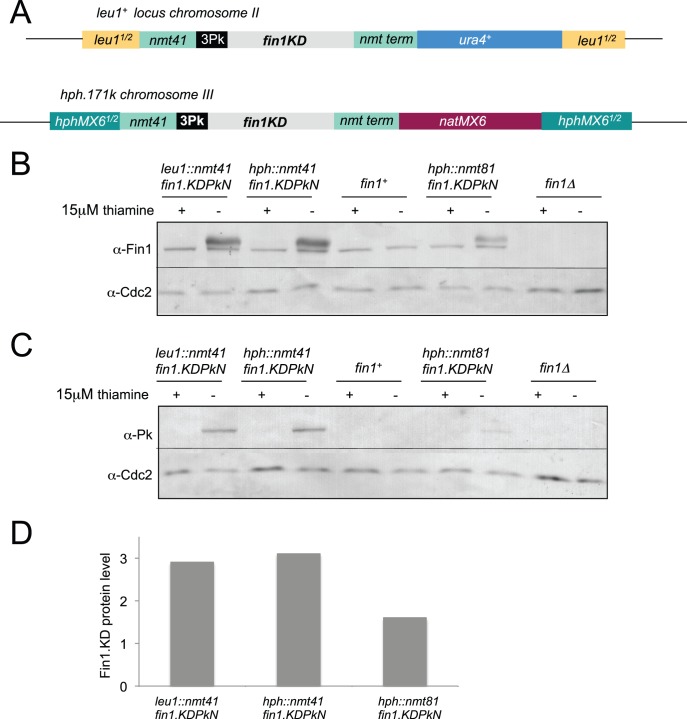
| IH6365 | leu1::nmt41fin1.KD-pkn:ura4+ ura4.D18 | This study | |
| IH6366 | leu1::nmt81fin1.KD-pkn:ura4+ ura4.D18 | This study | |
| IH6364 | hph171k::nmt41fin1.KD-pkn:natMX6 ura4.D18 | This study | |
| IH6409 | hph171k::nmt81fin1.KD-pkn:natMX6 ura4.D18 | This study |
Integration Vectors
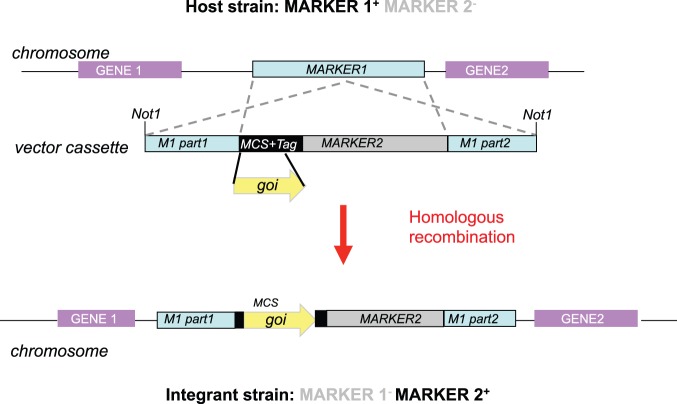
Although the expression of molecules from multi-copy vectors can be highly informative, the highly variable stoichiometry of protein levels between neighbouring cells can make it difficult to derive concrete conclusions from a particular manipulation. In contrast, direct comparisons can be made between the consequences of expressing different mutant alleles when integrated in the same vector context into the same genomic location. We have therefore developed three different vector series that each direct integration of an expression cassette into distinct, defined locations in the fission yeast genome. The same principle is employed in each case; the correct integration event is identified through the simultaneous gain of one marker and the disruption, and therefore loss, of another (Figure 3). For two systems the markers are classic amino acid auxotrophies, while the third exploits dominant antibiotic markers. Each system exploits the nmt1 based thiamine repressible promoter series: nmt1, nmt1* and nmt1** derived from the plasmids pREP1, pREP41 and pREP81 respectively [14], [15], [68]. The “A” plasmid in each series has no insert but can receive the entire promoter – gene – terminator expression cassette from existing pREP plasmids as a Pst1-Sac1 fragment. It is also the easiest recipient vector for integration of any sequence of choice.
Figure 3. A cartoon indicating the approach used by all three integration vector systems.
Auxotrophic pINTL and pINTK Integration Vectors
In the INTL vectors the leu1 + gene has been disrupted by a cloning module (expression or expression + tag) and ura4 + (Figure 4). The entire cassette is flanked by Not1 restriction sites. Once the desired sequences have been inserted, the Not1 fragment is excised and transformed into an ura4.d18 host. In correct transformants the disruption of leu1 + by the vector sequences flips auxotrophy from leu+ ura− to leu− ura+. INTK vectors direct the disruption of lys1 + with a similar ura4 + expression/tagging module to switch auxotrophy from lys+ ura− to lys− ura+ (Figure 5). The use of ura4 + as a positive selection in both the INTL and INTK systems excluding subsequent use of ura4+ based multi-copy vectors in either case. Consequently the ura4 + sequences within the INTK cassette have been flanked with loxP sites to facilitate marker excision upon expression of Cre recombinase (Figure 5).
Figure 4. The pINTL series of vectors for the expression of a gene of interest from the leu1 locus.
Cartoons depicting the structure of the indicated pINTL vectors.
Figure 5. The pINTK series of vectors for the expression of a gene of interest from the lys1 locus.
Cartoons depicting the structure of the indicated pINTK vectors.
Nourseothricin Resistance Based pINTH Integration Vectors
Modification of the amino acid requirements and amino acid content of the medium can significantly change flux through the TOR signalling pathway to impact upon diverse aspects of cell physiology [28]. We therefore generated an integrative vector system that can be used in prototrophs because it relies upon switching resistance to antibiotics rather than amino acid requirements. To achieve this goal we needed to select a site at which to integrate the recipient antibiotic marker that would later act as a target site for vector integration. As the smallest chromosome, chromosome III, harbours the smallest proportion of the genome of the three chromosomes, inserting a marker on this chromosome would lend itself to easier manipulation in subsequent crosses to introduce an expression cassette into a particular background. We therefore scanned chromosome III using the dataset of Wihelm et al. to find regions with low or no transcriptional activity [69]. Because no transcription was detected around position 171385 we integrated the Hygromycin B resistance cassette, hphMX6, at this site to generate an integration target locus that we refer to as the “hph.171k” locus (available from Yeast Genome Resource Centre Japan (http://yeast.lab.nig.ac.jp/nig/index_en.html). YGRC strain number listed in Table 2). The growth rate and fitness of hph.171k cells was indistinguishable from wild type at all temperatures tested (20°C, 25°C, 30°C, 32°C, 36°C) in both rich YES and minimal EMM2 medium. We then generated the pINTH series of vectors shown in Figure 6B that can be used to target any integration to any hphMX6 sequence in the genome (Figure 6B). In our case we use it to target hph.171k.
Figure 6. The pINTH series of vectors for the expression of a gene of interest from the hph.171k locus on chromosome III.
Cartoons depicting the structure of the indicated pINTH vectors.
To assess the level of expression obtained following integration of the pINTH expression cassettes at hph.171k we cloned a fin1 allele that encodes a catalytically inactive kinase, fin1.KD [62], into the pINTH41PkN vector. The Not1 restriction enzyme digested fragment was transformed into a hph.171k host (Figure 7A). 135 of the 141 transformants obtained were hygromycin resistance negative and nourseothricin resistance positive (i.e. a targeting efficiency of 96% in this hph.171k pku70+ pku80+ host). After backcrossing, protein samples were prepared from mid-log phase cultures 15 hours after expression from the nmt41 promoter was de-repressed by the removal of thiamine and processed for western blotting. To compare the expression level of the pINTH vectors with that obtained with the pINTL series vectors, the same fin1.KD insert had been cloned into the pINTL41PkN vector before integration into the genome, backcrossing and the production of protein extracts from mid-log phase cultures 15 hours after thiamine removal. The levels of Fin1.KD protein attained following induction of expression from either the leu1 targeted pINTL41PkN or the hph.171k targeted pINTH41PkN construct (Figure 7B, C) were indistinguishable from one another (3 fold higher than that of the native Fin1 kinase (Figure 7B)). As expected, the expression levels from the nmt81 based cassette was lower than from the nmt41 cassettes. We note that the 2 fold differential in protein levels between the two strength promoters (Figure 7D) is less than the 10 fold difference reported for the production of RNA levels from the nmt81 and nmt41 promoters on multi-copy vectors [70], however, Fin1 is subject to proteolytic control [62] making it impossible to draw solid conclusions about transcription rates when integrated into the leu1 + locus.
Figure 7. Integration at either the hph.171k or leu1 loci gave identical levels of protein expression.
A) Cartoons showing the structure of the two nmt41 integrated cassettes from which catalytically inactive Fin1.KD fusion proteins (three “Pk” SV5 epitopes fused, in frame, to their amino termini) are expressed upon removal of thiamine. B) Cells were grown to early log phase in EMM2+15 µM thiamine at 25°C before being washed three times in thiamine free EMM2 medium and re-suspended in EMM2 at a density of 1.8×105. Protein extracts were prepared from the mid-log phase cultures and processed for Western Blots after a further 15 hours culture at 25°C. Blots were cut in two; high molecular weight regions were probed with Fin1 antibodies while the loading control, While Cdc2 was detected on the lower molecular weight portion of the same blot. C) The same samples as shown in B probed with Cdc2 and mAb336 antibodies that recognised the Pk tags on the Fin1.KD3Pk fusion protein. D) A plot of the intensity ratios between the Fin1 and Cdc2 bands in each lane of the blots in B setting the ratio seen in wild type cells as 1 and that detected in fin1.Δ control as 0.
pREPN and pREPK Vectors
Multi-copy plasmids that can be selected for in prototrophs to drive the expression of tagged molecules remain popular. To generate a series of vectors we used in vivo gap repair in fission yeast [24], [71] to switch the markers in pREP81 and pREP41/42 based vector backbones [68]. We have previously reported the construction of these donor vectors [16]. Of the four antibiotic resistance markers in use in S. pombe, only nourseothricin/ClonNat and Kanamycin/G418 resistance can be used in the minimal medium in which the nmt1 based promoters of the pREP series vectors can be de-repressed, making these the only markers that would be of utility for a pREP based series of vectors. The LEU2 + marker of pREP81 and pREP41 derived plasmids was removed by restriction digestion and the linear vector sequences were co-transformed with a DNA fragment in which the natMX6 or the kanMX6 cassette had been amplified with primers that had 80 bp of homology with either end of the opened vector sequences. Plasmids were re-isolated from two nat + or kan + transformants and the new vector sequenced. As reported previously for this approach [24], [71], recombination had faithfully created the desired vectors in each case (Figure 8, 9). The transformation efficiency and stability of the pREP1N plasmid was indistinguishable from that of pREP1 (data not shown).
Figure 8. The pREPN series of vectors for the expression of a gene of interest from an ectopic plasmid.
Cartoons depicting the structure of the indicated pREPN vectors.
Figure 9. The pREPK series of vectors for the expression of a gene of interest from an ectopic plasmid.
Cartoons depicting the structure of the indicated pREPK vectors.
Discussion
We describe a number of the tools that we have developed to assist our efforts that exploit the molecular genetics of S. pombe to understand the signal transduction pathways that control cell division. The tools and methods presented in this paper make a significant contribution to resolving the problem of locus-dependent gene targetting efficiency. Manipulation of all loci has now become routine with the enhancements of transformation efficiency after switching the recovery incubation to an overnight incubation in un-supplemented MSL and the removal of the NHEJ response by deleting either pku70 + or pku80 + (the choice of which deletion to use depends upon the genomic location of the gene of interest to be targeted). We have generated pkuX0::kanMX6, pkuX0::hygMX6, pkuX0::natMX6 strains for greatest flexibility in designing a particular knockout strategy (available from the Yeast Genome Resource Centre Japan (http://yeast.lab.nig.ac.jp/nig/index_en.html)). We note that ura4 + and LEU2 + deleted alleles have been generated in other studies [72], [73].
While removal of the NHEJ pathway radically enhances the frequency of gene targeting in our work on cell cycle control, our experience with genes in the TOR signalling pathway has been different as targeting can be less efficient in pku70.Δ and pku80.Δ strains than in wild type strains (data not shown). Why this should be is unclear, however we suggest that pku70 + and pku80 + deletions be used as host strains for manipulations of genes in processes other than TOR signalling, but reverting to targeting in a wild type prototrophic strain should targeting efficiencies prove to be poor.
We found that the efficiency of integration was radically enhanced by altering the nature of the recovery period that is used to enable the expression of antibiotic resistance markers before they are challenged with selective conditions. Switching a from recovery phase on solid medium to a liquid recovery phase in nitrogen free medium increased transformation efficiencies 5–8 fold with antibiotic resistance cassettes. Although the greatest enhancement of transformation efficiencies arose at the 24 hour time point, an overnight incubation is normally sufficient and avoids delays in strain construction. While the MSL-N recovery period was a great benefit when the integrated expression cassette directed the expression of antibiotic resistance markers to counteract the otherwise lethal impact of antibiotics, it had only a modest impact when the selection relied upon ura4 + complementation of ura4.d18 in media lacking uracil. We assume that this is due to the inherent differences in the nature of the selection pressure in the two cases. Expression of antibiotic resistance molecules is required to prevent an apparently immediate death from a lethal assault by the antibiotic, whereas the ornithine decarboxylase is required to permit the generation of uracil. Cells will simply remain in a stationary phase until ornithine decarboxylase levels reach the critical threshold to allow them to resume growth and division.
The generation of three series of integration vectors that allow the expression of wild type, mutant, tagged or un-tagged native molecules from three different loci greatly facilitates the analysis of the impact of mutations on individual molecules or compound interactions of mutant molecules in a protein complex. The pINTXX.A vectors can accept the entire promoter–gene–terminator cassette as a Pst1-Sac1 restriction fragment from any existing pREP based vector [74]. Furthermore, the cloning approaches can be adapted to express full length non-coding RNAs from these sites of integration [75]. While we describe the full vector series here, we have used some members of each series in a number of studies that validate the application of these vectors in molecular cell biology in fission yeast [62], [74], [76], [77], [78], [79], [80], [81], [82], [83], [84].
Supporting Information
DNA sequences of the pINTL series.
(DOCX)
DNA sequence of pINTK.
(DOCX)
DNA sequences of the pINTH series.
(DOCX)
Acknowledgments
We are indebted to: Michael Knop, Peter Wagner and Berl Oakley for the motivation to conduct the work, Marc Sohrmann (University of Lausanne, Switzerland) and Christian Fankhauser (University of Lausanne, Switzerland) for technical contributions and Elmar Schiebel (ZMBH Heidelberg, Germany), Nevan Krogan (UCSF, USA) and Antony Carr (MRC GDSC, Sussex University, UK) for plasmids and Antony Carr for strains and Simon Whitehall for encouragement to compile this manuscript.
Funding Statement
Swiss National Science Foundation (www.snf.ch). École polytechnique fédérale de Lausanne (www.epfl.ch). Cancer Research UK [CRUK] C147/A6058. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Data Availability: The authors confirm that all data underlying the findings are fully available without restriction. Data is in the manuscript, and new strains are available from the Yeast Genetic Resource Center Japan (YGRC/NBRP): http://yeast.lab.nig.ac.jp/nig/index_en.html. All plasmids are available upon request from Iain.Hagan@CRUK.manchester.ac.uk.
References
- 1.Gutz H, Heslot H, Leupold U, Loprieno N (1974) Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. New York: Plenum press. 395–445.
- 2. Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods In Enzymology 194: 795–823. [DOI] [PubMed] [Google Scholar]
- 3. Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reaume SE, Tatum EL (1949) Spontaneous and nitrogen mustard-induced nutritional deficiencies in Saccharomyces cerevisiae . Arch Biochem 22: 331–338. [PubMed] [Google Scholar]
- 5.Leupold U (1970) Genetical methods for Schizosaccharomyces pombe. In: Prescott D, editor. Methods in cell physiology. New York: Academic Press. 169–177.
- 6. Carr AM, MacNeill SA, Hayles J, Nurse P (1989) Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 kinase gene: implications for cdc2 protein structure and function. Molecular and General Genetics 218: 41–49. [DOI] [PubMed] [Google Scholar]
- 7. Hottinger H, Pearson D, Yamao F, Gamulin V, Cooley L, et al. (1982) Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet 188: 219–224. [DOI] [PubMed] [Google Scholar]
- 8. Hofer F, Hollerstein H, Janner F, Minet M, Thuriaux P, et al. (1979) The genetic fine structure of nonsense suppressors in Schizosaccharomyces pombe. . Current Genetics 1: 45–61. [DOI] [PubMed] [Google Scholar]
- 9. Beach D, Nurse P (1981) High-Frequency Transformation Of the Fission Yeast Schizosaccharomyces-Pombe. Nature 290: 140–142. [DOI] [PubMed] [Google Scholar]
- 10. Russell P, Nurse P (1987) Negative regulation of mitosis by wee1 &, a gene encoding a protein- kinase homolog. Cell 49: 559–567. [DOI] [PubMed] [Google Scholar]
- 11. Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Molecular & general genetics : MGG 197: 345–346. [DOI] [PubMed] [Google Scholar]
- 12. Grimm C, Kohli J, Murray J, Maundrell K (1988) Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 + gene as a selectable marker. Molecular and General Genetics 215: 81–86. [DOI] [PubMed] [Google Scholar]
- 13. Barbet N, Muriel WJ, Carr AM (1992) Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe . Gene 114: 59–66. [DOI] [PubMed] [Google Scholar]
- 14. Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- 15. Maundrell K (1990) Nmt1 of Fission Yeast - a Highly Transcribed Gene Completely Repressed By Thiamine. Journal of Biological Chemistry 265: 10857–10864. [PubMed] [Google Scholar]
- 16. Craven RA, Griffiths DJ, Sheldrick KS, Randall RE, Hagan IM, et al. (1998) Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221: 59–68. [DOI] [PubMed] [Google Scholar]
- 17. Kikuchi Y, Kitazawa Y, Shimatake H, Yamamoto M (1988) The primary structure of the leu1+ gene of Schizosaccharomyces pombe. Curr Genet 14: 375–379. [DOI] [PubMed] [Google Scholar]
- 18. Apolinario E, Nocero M, Jin M, Hoffman CS (1993) Cloning and manipulation of the Schizosaccharomyces pombe his7+ gene as a new selectable marker for molecular genetic studies. Curr Genet 24: 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burke JD, Gould KL (1994) Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet 242: 169–176. [DOI] [PubMed] [Google Scholar]
- 20. Fujita Y, Giga-Hama Y, Takegawa K (2005) Development of a genetic transformation system using new selectable markers for fission yeast Schizosaccharomyces pombe. Yeast 22: 193–202. [DOI] [PubMed] [Google Scholar]
- 21. Hoffman RL, Hoffman CS (2006) Cloning the Schizosaccharomyces pombe lys2+ gene and construction of new molecular genetic tools. Curr Genet 49: 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waddell S, Jenkins JR (1995) arg3+, a new selection marker system for Schizosaccharomyces pombe: application of ura4+ as a removable integration marker. Nucleic Acids Res 23: 1836–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, et al. (1995) The chromodomain protein Swi6 - a key component at fission yeast centromeres. Science 269: 1429–1431. [DOI] [PubMed] [Google Scholar]
- 24. Chino A, Watanabe K, Moriya H (2010) Plasmid construction using recombination activity in the fission yeast Schizosaccharomyces pombe. PloS one 5: e9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keeney JB, Boeke JD (1994) Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136: 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, et al. (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nature biotechnology 24: 841–847. [DOI] [PubMed] [Google Scholar]
- 27. Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, et al. (2004) pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21: 1289–1305. [DOI] [PubMed] [Google Scholar]
- 28. Hartmuth S, Petersen J (2009) Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J Cell Sci 122: 1737–1746. [DOI] [PubMed] [Google Scholar]
- 29. Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- 30. Longtine M, McKenzie A, Demarini DJ, Shah NG, Wach A, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. . Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- 31. Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- 32. Wach A (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12: 259–265. [DOI] [PubMed] [Google Scholar]
- 33. Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- 35. Bähler J, Steever AB, Wheatley S, Wang YL, Pringle JR, et al. (1998) Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. Journal of Cell Biology 143: 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 37. Burland TG, Pallotta D, Tardif MC, Lemieux G, Dove WF (1991) Fission yeast promoter-probe vectors based on hygromycin resistance. Gene 100: 241–245. [DOI] [PubMed] [Google Scholar]
- 38. Sato M, Dhut S, Toda T (2005) New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22: 583–591. [DOI] [PubMed] [Google Scholar]
- 39. Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- 40. Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, et al. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- 41. Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- 42. Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670. [DOI] [PubMed] [Google Scholar]
- 43. Germino M, Sohail H, Germino E, Germino J (2006) A vector for double epitope tagging with a recyclable marker. Yeast 23: 763–769. [DOI] [PubMed] [Google Scholar]
- 44. Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, et al. (2006) HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast 23: 623–632. [DOI] [PubMed] [Google Scholar]
- 45. Snaith HA, Anders A, Samejima I, Sawin KE (2010) New and old reagents for fluorescent protein tagging of microtubules in fission yeast; experimental and critical evaluation. Methods in cell biology 97: 147–172. [DOI] [PubMed] [Google Scholar]
- 46. Erler A, Maresca M, Fu J, Stewart AF (2006) Recombineering reagents for improved inducible expression and selection marker re-use in Schizosaccharomyces pombe. Yeast 23: 813–823. [DOI] [PubMed] [Google Scholar]
- 47. Grimm C, Kohli J (1988) Observations on integrative transformation in Schizosaccharomyces pombe. Molecular & general genetics : MGG 215: 87–93. [DOI] [PubMed] [Google Scholar]
- 48. Grallert B, Nurse P, Patterson TE (1993) A study of integrative transformation in Schizosaccharomyces pombe. Molecular & general genetics : MGG 238: 26–32. [DOI] [PubMed] [Google Scholar]
- 49. Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends in biochemical sciences 23: 394–398. [DOI] [PubMed] [Google Scholar]
- 50. Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America 101: 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kooistra R, Hooykaas PJ, Steensma HY (2004) Efficient gene targeting in Kluyveromyces lactis. Yeast 21: 781–792. [DOI] [PubMed] [Google Scholar]
- 52. Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, et al. (2006) A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poggeler S, Kuck U (2006) Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378: 1–10. [DOI] [PubMed] [Google Scholar]
- 54. Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, et al. (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal genetics and biology : FG & B 45: 68–75. [DOI] [PubMed] [Google Scholar]
- 55. Naatsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, et al. (2012) Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PloS one 7: e39720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li ZH, Du CM, Zhong YH, Wang TH (2010) Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Applied microbiology and biotechnology 87: 1065–1076. [DOI] [PubMed] [Google Scholar]
- 57. Roguev A, Wiren M, Weissman JS, Krogan NJ (2007) High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nature methods 4: 861–866. [DOI] [PubMed] [Google Scholar]
- 58. Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe . Yeast 15: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 59. Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, et al. (2006) S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev 20: 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petersen J, Hagan IM (2005) Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature 435: 507–512. [DOI] [PubMed] [Google Scholar]
- 61. Grallert A, Chan KY, Alonso-Nunez ML, Madrid M, Biswas A, et al. (2013) Removal of Centrosomal PP1 by NIMA Kinase Unlocks the MPF Feedback Loop to Promote Mitotic Commitment in S. pombe. Current biology : CB 23: 213–222. [DOI] [PubMed] [Google Scholar]
- 62. Grallert A, Connolly Y, Smith DL, Simanis V, Hagan IM (2012) The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nature cell biology 14: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sivakumar S, Porter-Goff M, Patel PK, Benoit K, Rhind N (2004) In vivo labeling of fission yeast DNA with thymidine and thymidine analogs. Methods 33: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Egel R (1971) Physiological aspects of conjugation in fission yeast. Planta 98: 89–96. [DOI] [PubMed] [Google Scholar]
- 65. Egel R, Willer M, Kjaerulff S, Davey J, Nielsen O (1994) Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast 10: 1347–1354. [DOI] [PubMed] [Google Scholar]
- 66. Maekawa H, Usui T, Knop M, Schiebel E (2003) Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. Embo J 22: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM (2004) Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Basi G, Schmid E, Maundrell K (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136. [DOI] [PubMed] [Google Scholar]
- 69. Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, et al. (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- 70. Forsberg SL (1993) Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Research 21: 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelly DA, Hoffman CS (2002) Gap repair transformation in fission yeast to exchange plasmid-selectable markers. BioTechniques 33: 978, 980, 982. [DOI] [PMC free article] [PubMed]
- 72. Miyoshi T, Sadaie M, Kanoh J, Ishikawa F (2003) Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. The Journal of biological chemistry 278: 1924–1931. [DOI] [PubMed] [Google Scholar]
- 73. Kibe T, Tomita K, Matsuura A, Izawa D, Kodaira T, et al. (2003) Fission yeast Rhp51 is required for the maintenance of telomere structure in the absence of the Ku heterodimer. Nucleic acids research 31: 5054–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Petersen J, Hagan IM (2003) S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol 13: 590–597. [DOI] [PubMed] [Google Scholar]
- 75. Bitton DA, Grallert A, Scutt PJ, Yates T, Li Y, et al. (2011) Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Molecular systems biology 7: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fankhauser C, Simanis V (1994) The Cdc7 protein-kinase is a dosage-dependent regulator of septum formation in fission yeast. Embo Journal 13: 3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, et al. (2001) The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. . EMBO Journal 20: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Grallert A, Patel A, Tallada VA, Chan KY, Bagley S, et al. (2013) Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nature cell biology 15: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Drummond DR, Hagan IM (1998) Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. Journal of Cell Science 111: 853–865. [DOI] [PubMed] [Google Scholar]
- 80. Tay YD, Patel A, Kaemena DF, Hagan IM (2013) Mutation of a conserved residue enhances the sensitivity of analogue-sensitised kinases to generate a novel approach to the study of mitosis in fission yeast. Journal of cell science 126: 5052–5061. [DOI] [PubMed] [Google Scholar]
- 81. Murone M, Simanis V (1996) The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. Embo Journal 15: 6605–6616. [PMC free article] [PubMed] [Google Scholar]
- 82. Schmidt S, Sohrmann M, Hofmann K, Woolard A, Simanis V (1997) The Spg1 GTPase is an essential dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. . Genes and Development 11: 1519–1534. [DOI] [PubMed] [Google Scholar]
- 83. Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V (2008) Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J Cell Sci 121: 843–853. [DOI] [PubMed] [Google Scholar]
- 84. Krapp A, Collin P, Cokoja A, Dischinger S, Cano E, et al. (2006) The Schizosaccharomyces pombe septation initiation network (SIN) is required for spore formation in meiosis. J Cell Sci 119: 2882–2891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences of the pINTL series.
(DOCX)
DNA sequence of pINTK.
(DOCX)
DNA sequences of the pINTH series.
(DOCX)