Abstract
Background
A good correlation between HCV core antigen (HCVAg) and different HCV-RNA assays has been described, but little data are available in HCV/HIV co-infection. We aimed to evaluate HCVAg in comparison with HCV-RNA and to determine their kinetics during antiviral treatment in selected HCV/HIV co-infected patients.
Methods
355 samples from 286 HCV/HIV co-infected subjects for whom HCV-RNA (Abbott RealTime) was requested were analysed also for HCVAg (Abbott ARCHITECT) in order to evaluate the correlation between the two parameters both in patients treated or untreated for chronic hepatitis C and according to different HCV genotypes. The differences between percentages were evaluated by chi square or Fisher’s exact test, while mean and median values were compared by Student’s t test or the Mann–Whitney test, respectively. All differences were considered significant for a p value <0.05.
Results
HCVAg was detectable on 288/315 sera (91.4%) positive for HCV-RNA and in 5 out of40 (12.5%) sera with undetectable HCV-RNA for a total concordance of 90.1%. The correlation was fair both in untreated (r = 0.742) and in treated (r = 0.881) patients and stronger for genotypes 1 and 4 than for genotype 3. Both HCV-RNA and HCVAg levels were significantly higher (p = 0.028 and p = 0.0098, respectively) in patients infected by genotype 1 than by genotype 3. The mean ratio of Log values between HCV-RNA (IU/mL) and HCVAg (fmol/liter) was 2.27 ± 1.09 in untreated and 2.20 ± 0.82 in treated patients (p = n.s.),consistent with a sensitivity of HCVAg corresponding to about 1,000 IU/mL of HCV-RNA, and ranged from 2.21 to 2.32 among HCV genotypes with no significant differences; five samples (1.4%; 2 genotype 1a or 1c, 3 genotype 3a) showed highly divergent values. The analysis of 18 monitoring profiles from patients treated with PEG-IFN and Ribavirin showed similar trends, except in one case in which relapse could be predicted by HCVAg and not by HCV-RNA.
Conclusion
These results suggest that HCVAg represents an adequate tool for determining an ongoing HCV infection also in HIV co-infected patients, with lower costs and faster turnaround time than HCV-RNA.
Background
The hepatitis C virus (HCV) was first identified in 1989 as the principal cause of post-transfusion non-A non-B hepatitis [1]. Worldwide an estimated 170 million people are infected with HCV and, due to common routes of transmission 4–5 million are co-infected with the human immunodeficiency virus (HIV) [2,3]. The current standard for diagnosing an active HCV infection is the detection and quantification of HCV-RNA, also recommended to monitor the antiviral treatment. Therapy guidelines for standard treatment with pegylated interferon (PEG-IFN) plus Ribavirin recommend the discontinuation of the therapy if HCV-RNA decline is <2 log at week 12 or when HCV RNA is still detectable at week 24 [4]. On therapy HCV-RNA evaluation is even more crucial to tailor new therapeutic regimens based on direct acting antivirals (DAA) drugs [5]. For the measurement of HCV-RNA, a number of qualitative and quantitative assays are commercially available offering a very low limit of detection and linear quantification over a broad dynamic range [6-9]. Although the available HCV-RNA testing systems are very sensitive and have a high-throughput performance, nucleic acid tests are expensive, labour intensive, and to avoid false positive results require technical skills, which limit their use [10].
A fully automated quantitative immunoassay for measuring the HCV core antigen (ARCHITECT HCVAg, Abbott Diagnostics, Wiesbaden, Germany) was registered within EC in 2009 [11]. The assay sensitivity across the different HCV genotypes ranges from 500 to 3,000 IU/mL of HCV-RNA [12-14]. Several studies have demonstrated a good correlation between HCVAg and different HCV-RNA assays [12,13,15-17] and suggested that HCVAg could be used as an alternative to HCV-RNA testing in some settings, for instance in hemodialysis patients [18]. Moreover HCVAg has been demonstrated to considerably reduce the window period in 97% of HCV-RNA positive/anti-HCV antibody negative specimens demonstrating that it represents a reliable marker of viral replication even in HCV pre-seroconversion phase [13,19,20]. To date, little data are available about HCVAg determination in HCV/HIV co-infected patients: studies carried out so far in this population included a small number of samples and did not give robust information on potential interference of HIV replicative activity on the specificity of HCVAg determination and on its sensitivity, particularly in the subjects with low HCV RNA level (HCV-RNA <104 IU/mL) [21].
The aims of the present study were to establish the clinical sensitivity and to evaluate the concordance and correlation of HCVAg with HCV-RNA in HCV/HIV co-infected patients, according to HCV genotypes, and in co-infected patients undergoing treatment for chronic hepatitis C.
Methods
Study population
Routine samples were selected from HCV/HIV positive patients admitted to the National Institute for Infectious Diseases (INMI) “L. Spallanzani” in Roma or to the Regional Hospital “Torrette” in Ancona for whom HCV-RNA and HIV-RNA testing had been required. The residual plasma samples were aliquoted, stored at -80°C and thawed only once for HCVAg testing. All samples come from the normal routine of our hospitals, therefore of course we did have access to the basic demographic information. However, after having collected all these data the patient list was completely anonymized, so that their ID recognition was no longer possible. In both Institutions a very large number of patients are followed for HIV and HCV infections every year, thus the age and gender information will not allow to identify them; in no way it is possible to link any study result with patient ID. The current Italian regulation on the protection of personal data have been strictly observed, in particular: “Codice in materia di protezione dei dati personali” (d.lg. 30 giugno 2003, n. 196) and “Provvedimento su amministratore di sistema”, issued by “Garante per la protezione dei dati personali” on November 27, 2008, and published in Gazzetta Ufficiale, December 24, 2008.
Samples were selected in order to obtain a balanced distribution of low, intermediate and high levels of HCV-RNA and to include sequential specimens from patients who had started PEG-IFN + RBV treatment for hepatitis C. Additionally, eleven routine specimens from HIV-positive individuals with high HIV viral load and negative for HBsAg, anti-HCV antibodies and HCV-RNA were collected in order to check the specificity of the HCVAg assay.
Serological and virological assays
HCVAg was quantified by ARCHITECT i2000SR (Abbott Diagnostics, Wiesbaden, Germany), a fully automated system, with a chemiluminescent immunoassay already described in detail [11,12]. The linear range of HCVAg quantification spans between 3.00 and 20,000 femtomoles/liter (fmol/L) (0.06 and 400 pg/ml), with an automated 1:9 dilution that extends the assay linearity up to 180,000 fmol/L (3,600 pg/mL). Samples with results greater than the cutoff value (>3 fmol/L) but ≤10 fmol/L were retested in duplicate and considered “true” positive if confirmed reactive.
Other serological markers (anti-HCV, HIV Ag/Ab) were evaluated using the respective assays on the Abbott ARCHITECT system.
HCV-RNA was quantified by means of a commercial real-time RT-PCR assay (RealTime™ HCV, Abbott Molecular Inc, Des Plaines, Il, USA) as specified by the manufacturer. The detection limit was 12 IU/mL. HCV genotypes were determined by VERSANT HCV Genotype 2.0 Assay (LiPa) (Innogenetics, Ghent, Belgium; distributed by Siemens Healthcare Diagnostics) [22] or by Abbott RealTime HCV Genotype II Assay (Abbott) [23].
Statistical analysis
The results were collected on a Microsoft Excel worksheet and analyzed for correlation (Spearman) and agreement (K statistic) by the Analyse-it software (Analyse Ltd, Birmingham, UK). The differences between percentages were evaluated by chi square or Fisher’s exact test, while mean and median values were compared by Student’s t test or the Mann–Whitney test, respectively. All differences were considered significant for a p value ≤0.05.
Results
Three hundred and fifty-five samples obtained from 292 HIV/HCV co-infected patients were included in this study. Of those, 106 were obtained from 43 patients under standard treatment with PEG-IFN + Ribavirin for chronic hepatitis C and 249 were taken from as many untreated patients.The mean and median ages did not differ significantly between genders and were 46.2 ± 7.9 and 47 (range: 20–84) years overall, respectively. As outlined in the study protocol, samples were stratified according to the HCV-RNA levels (Table 1). Sequential samples during treatment were available from 41 patients (3 or more draws from 18 patients, 2 draws from 23 patients).
Table 1.
Positivity and levels of HCVAg according to HCV-RNA levels in HIV/HCV co-infected patients
|
HCV-RNA IU/mL |
Treated |
Untreated |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N. | HCVAg+ | % | N. | HCVAg+ | % | N | HCVAg+ | % | Median | |
| <12 |
23 |
5 |
21.7 |
17 |
0 |
0 |
40 |
5 |
12.5 |
7.32 |
| 12-999 |
12 |
3 |
25.0 |
11 |
0 |
0 |
23 |
3 |
13.0 |
6.04 |
| 1000-9999 |
11 |
8 |
100 |
17 |
11 |
64.7 |
28 |
19 |
67.9 |
16.04 |
| 10000-99999 |
14 |
14 |
100 |
30 |
30 |
100 |
44 |
44 |
100 |
72.71 |
| 100000-999999 |
22 |
22 |
100 |
66 |
66 |
100 |
88 |
88 |
100 |
548.67 |
| ≥1000000 |
24 |
24 |
100 |
108 |
107 |
99.1 |
132 |
131 |
99.2 |
2968.20 |
| Total | 106 | 76 | 74.5 | 249 | 214 | 85.9 | 355 | 290 | 81.7 | 770.51 |
All the 11 samples from patients negative for HBV and HCV markers and anti-HIV antibody positive with HIV-RNA level >105 copies/mL were negative for HCVAg.
The overall concordance between HCVAg and HCV-RNA was 90.1% and theagreement was fair (kappa = 0.61). On samples from treated patients the qualitative agreement was significantly lower than on samples from untreated patients (84.0% vs. 92.8%; p = 0.01 by chi square). On the whole then, 285 samples were positive and 35 were negative (i.e. below the detection limit) for both markers, while 30 were positive for HCV-RNA (median level: 479 IU/ml; range 13–1,598,723) and negative for HCVAg and 5, all obtained from treated patients, were positive for HCVAg (median level: 7.32 fmol/L; range: 6.71-139.96) and negative for HCV-RNA. Furthermore, 3 out of 12 with RNA levels <1,000 UI/mL were positive for HCVAg among treated patients vs. none among untreated.
The positivity rates for HCVAg according to different levels of HCV-RNA and the median levels are reported in Table 1. Based on a polynomial regression analysis between HCV-RNA and HCVAg on the 22 samples positive for both and with a viral load between 100 and 10,000 IU/mL, the positivity threshold of the HCVAg assay corresponds to about 1,000-1,500 IU/mL of HCV-RNA, and indeed 282 out of 292 samples (96.6%) containing >1,000 IU/mL of HCV-RNA were positive for HCVAg. Five specimens (1.4%) showed a highly divergent quantitative result between the two assays, either because HCVAg levels were higher than HCV-RNA levels or because the Log ratio between the two parameters exceeded the mean plus 2 standard deviations measured on all samples. Details on the discrepant specimens, either qualitative or quantitative, are reported in Table 2.
Table 2.
Discrepant results between HCVAg and HCV-RNA
|
Case |
Genotype |
HCVAg |
HCV-RNA |
Log ratio |
Notes |
|---|---|---|---|---|---|
| n | Fmol/L | IU/mLI | RNA/Ag | ||
| 1 |
3a |
6.71 |
0 |
na |
Therapy 12 m, log ratio at T0 1.08, neg for both at 32 m |
| 2 |
1a |
11.21 |
0 |
na |
Therapy 8 m, log ratio at T0 2.46 |
| 2 |
1a |
139.96 |
0 |
na |
Therapy 16 m, log ratio at T0 2.46 |
| 3 |
3a |
7.32 |
<12 |
na |
Therapy 12 m, log ratio at T0 2.34, log ratio at 8 m 2.92 |
| 4 |
3a |
6.82 |
<12 |
na |
Therapy 4 m, log ratio at T0 2.43 |
| 5 |
na |
0.00 |
1142 |
na |
Untreated, single specimen |
| 6 |
3a |
0.20 |
1807 |
na |
Untreated, single specimen |
| 7 |
4 |
0.54 |
3468 |
na |
Untreated, single specimen |
| 8 |
3a |
1.89 |
4975 |
na |
Untreated, single specimen |
| 9 |
na |
1.78 |
7939 |
na |
Untreated, single specimen |
| 10 |
3a |
1.19 |
9206 |
na |
Untreated, single specimen |
| 11 |
1a |
2.69 |
1598723 |
14.44 |
Untreated, single specimen |
| 12 |
1c |
3,972.47 |
2097 |
0.92 |
Untreated, single specimen |
| 13 |
1a |
7.92 |
105551 |
5.59 |
Untreated, single specimen |
| 14 |
3a |
8.83 |
40643 |
4.87 |
Untreated, single specimen |
| 15 |
3a |
18.96 |
698555 |
4.57 |
Untreated, single specimen |
| 16 | 3a | 31.20 | 7338172 | 4.57 | Untreated, single specimen |
Five samples from four patients positive for HCVAg and negative for HCV-RNA, seven samples negative for HCVAg despite HCV-RNA levels higher than 1,000 UI/mL, five samples with a Log ratio between the two assays outside the expected range; na, not available; m, months.
The correlation between HCV-RNA and HCVAg was good in treated patients (Spearman r = 0.881; 95% confidence limits (CL) 0.821-0.921) and fair in untreated patients (Spearman r = 0.742; 95% CL 0.675-0.797; p = 0.05). Globally, the correlation was r = 0.815 with 95% CL: 0.774-0.849 (Figure 1). The ratio between HCV-RNA and HCVAg Log levels was also evaluated: the mean ± standard deviation and median ratio values were 2.20 ± 0.82 and 1.94 in treated patients vs. 2.27 ± 1.09 and 1.99 in untreated and (p = n.s.) and 2.25 ± 0.88 and 1.98 overall.
Figure 1.
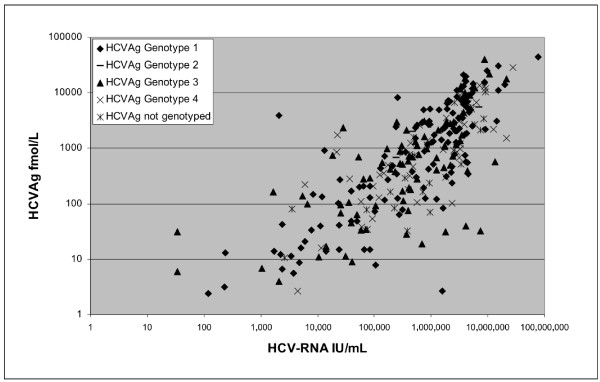
Correlation between HCVAg and HCV-RNA on 315 samples from patients with HCV RNA >12 IU/ml. Values are reported on a logarithmic scale and with different symbols according to genotype. The correlation coefficient (Spearman) was 0.869.
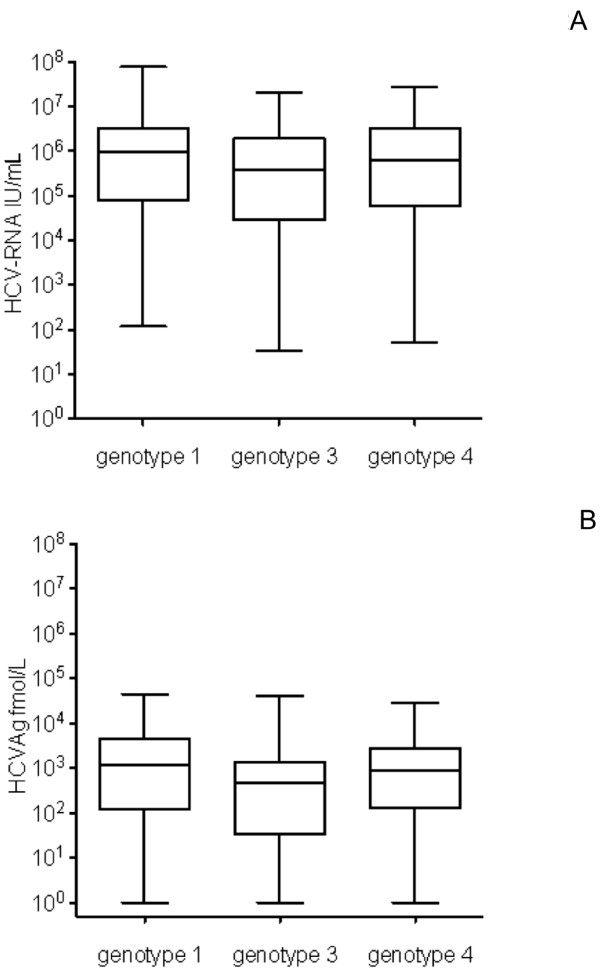
The HCV genotype was available for 245 of the 286 patients (86%), while in 41 cases it was not known due to low or undetectable HCV-RNA levels. Genotypes 1, 3 and 4 were the most represented as shown in Table 3. The correlation between HCV-RNA and HCVAg was fair (Spearman r = 0.816; 95% confidence limits (CL) 0.774-0.851; p < 0.0001) (Figure 1), and was greater for genotype 1 and 4 (Spearman r = 0.82 and 0.84) than for genotype 3 (Spearman r = 0.74). The Log ratio between HCV-RNA and HCVAg was not significantly different among genotype 1 (2.22 ± 0.80 and 1.94), genotype 3 (2.32 ± 0.88 and 2.00) and genotype 4 (2.21 ± 1.05 and 1.99). The distribution of HCV-RNA and HCVAg levels according to HCV genotypes 1, 3 and 4 are reported in Figure 2. Since HCV-RNA levels and HCVAg levels were not normally distributed, the difference among genotypes was assessed by a nonparametric test (Mann–Whitney). The difference was statistically significant for both parameters in genotype 1 vs genotype 3 (p = 0.028 for HCV-RNA and p = 0.0098 for HCVAg), while the difference between genotype 4 and 3 failed to reach the statistical significance (p = 0.122 and 0.114, respectively). No difference was observed between genotype 1 and genotype 4 (p = 0.902 and 0.578, respectively).
Table 3.
HCV genotypes in the study population of 286 HIV/HCV co-infected patients
| |
Genotype 1 |
Genotype 2 |
Genotype 3 |
Genotype 4 |
Mixed* |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
N (%) |
N (%) |
N (%) |
N (%) |
N(%) |
|||||||||||||||
| Gender | Total N | Genotyped N (%) | Total | NS | a | b | c | a/b | Total | NS | a | a/c | Total | NS | a | Total | NS | a | |
| Unknown |
13 |
10 |
6 |
1 |
3 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
1 |
1 |
2 |
2 |
0 |
0 |
| (76.9) |
(60.0) |
(-) |
(20.0) |
(20.0) |
(-) |
||||||||||||||
| Female |
74 |
59 |
33 |
9 |
13 |
7 |
4 |
0 |
0 |
0 |
0 |
0 |
19 |
2 |
17 |
7 |
4 |
3 |
0 |
| (79.7) |
(55.9) |
(-) |
(25.7) |
(11.9) |
(-) |
||||||||||||||
| Male |
199 |
176 |
78 |
18 |
25 |
18 |
15 |
3 |
3 |
1 |
1 |
1 |
58 |
8 |
50 |
35 |
27 |
8 |
2 |
| (88.4) |
(44.3) |
(1.7) |
(33.0) |
(19.9) |
(1.1) |
||||||||||||||
|
Total |
286 |
245 |
117 |
28 |
41 |
27 |
19 |
2 |
3 |
1 |
1 |
1 |
79 |
11 |
68 |
44 |
33 |
11 |
2 |
| (85.7) | (47.8) | (1.2) | (32.2) | (18.0) | (0.8) | ||||||||||||||
N, number; NS, not subtyped; The two mixed cases (*) were infected by genotype 1a and 3 and genotype 1a and 3a. respectively. Forty-one genotypes (14.3%) were not available due to very low levels of HCV-RNA.
Figure 2.
Distribution of HCV-RNA (A) and HCVAg (B) by HCV genotype. Significant differences between median values were observed only between genotypes 1 and 3 for both HCV-RNA (p = 0.028) and HCVAg (p = 0.0098).
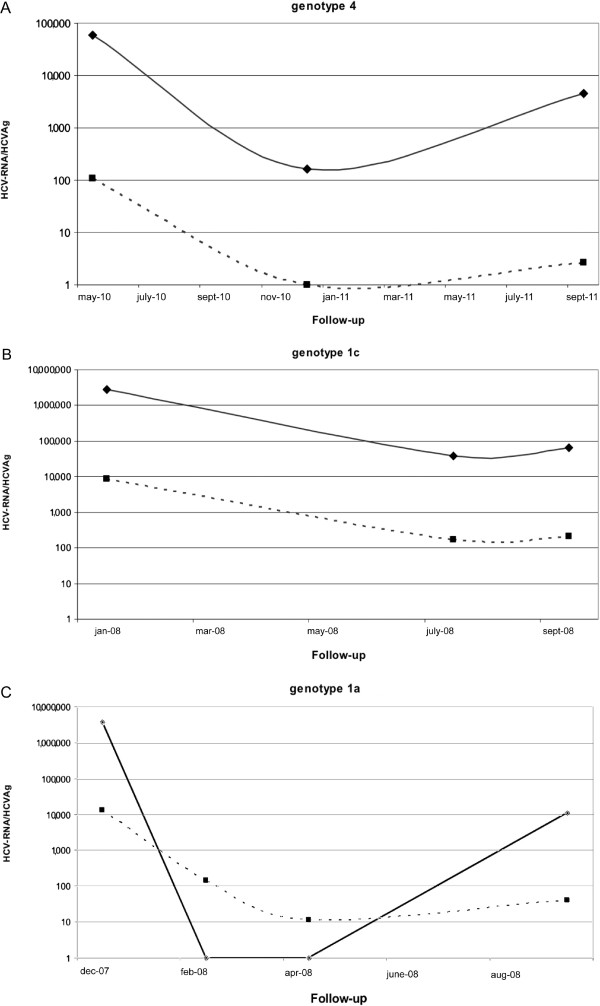
In 17 of the 18 patients sampled 3 or more times during PEG-IFN + Ribavirin therapy the behaviour of HCV-RNA and HCVAg was coincident, as 12 patients showed a sustained decrease, 2 patients a less pronounced decrease and 3 patients no significant variations: two examples are reported in Figure 3A and B. For one patient a significant discrepancy was observed (Figure 3C): both HCV-RNA and HCVAg declined initially, but while the former was undetectable on two consecutive draws, taken 2 months apart, HCVAg was still positive at 139.96 and 11.21 fmol/L. On the last draw, both HCV-RNA and HCVAg were detected (11,050 IU/mL and 40.20 fmol/L, respectively).
Figure 3.
Monitoring profiles from three patients co-infected by HCV and HIV. The values of HCV-RNA and HCVAg are expressed on a logarithmic scale as IU/mL and fmol/L. respectively. The trends of the two parameters in the two patients described in Figure 3A and B were almost identical whereas the patient in Figure 3C showed a persistence of HCVAg positivity and a virological relapse on the following draw. Dotted line: HCVAg; solid line: HCV-RNA.
Discussion
HCVAg has been initially proposed as a surrogate marker of HCV-RNA for blood screening [5] and soon afterwards it was suggested that the quantitative determination of this parameter could surrogate RNA testing also in the monitoring of chronic HCV infections and possibly also in patients under therapy [24-27]. The availability of a fully automated immunoassay with on-board sample pretreatment for the quantitative determination of HCVAg has brought up a series of new studies. Most of those have confirmed the original data presented by the manufacturer [11], reporting a good specificity [12,13,17] and a sensitivity in the range of 500–3,000 IU/mL of HCV-RNA, according to the HCV genotype and possibly to other factors. Indeed, the theoretical and experimental equivalency between the two parameters was investigated by Schuettler et al. [28], who established that according to the HCV structure 1 pg of HCVAg corresponds to 43,000 IU of HCV-RNA, whereas on clinical samples this ratio was much lower (1 pg = about 7,900 IU of HCV-RNA). This has been confirmed in subsequent studies based on different HCV RNA quantification systems [13-16,18]; in particular, very recently Descamps et al. [14] found a very significant correlation of HCVAg to HCV-RNA, measured by TaqMan amplification (r = 0.90; p < 0.001) and an equivalency of 1 pg of HCVAg = 9,755 IU of HCV-RNA, that equals about 600 IU/mL at the HCVAg assay threshold of 3 fmol/L. From those data, confirmed also by our observation, only a portion of the circulating HCVAg seems to be associated with complete virions, implying that a variable fraction of circulating HCVAg may be secreted by the infected cells. Additionally the HCVAg measured by the immunoassay derives also from antigen-antibody complexes that are disrupted during sample pre-treatment. Furthermore, it has been observed that also HCV, like the hepatitis B virus, may be present in the bloodstream as complete virions and defective particles, presumably with low or no infectivity [29] especially in the late phases of infection. The discrepancy between HCV-RNA and HCVAg production may be large enough in some instances to justify the huge differences observed in 1.4% of samples in our observation as well as in other studies [12,13,15] and the combined action of all the above mentioned factors may help explain why the equivalency and correlation between viral RNA and core antigen differs among the various studies.Viral variability may also be implicated in generating these discrepancies, such as the finding of one specimen with >1 million IU/mL of HCV-RNA and negative for HCVAg. For instance, it has been shown that a variability in the core gene products is able to affect the recognition of circulating HCVAg because polymorphisms at amino acids 47 to 49 of the core Ag were responsible for underestimation [30] and that the levels of HCVAg and ratio of HCV-RNA to HCVAg may be affected by the polymorphism of the IL28B genotype [31].
In our experience the correlation (r = 0.818) was weaker than reported by others, as previous studies with the same HCVAg assay reported r values up to 0.90 or even higher [14,32]. This does not seem to be ascribed to the relatively high rate (30%) of samples from treated patients in our selection, since the relationship between these two indexes of active HCV infections appears stronger in treated than in untreated patients. Recently, Kuo et al. [32] assayed 405 subjects identified by a community screening for HCVAg and found a high degree of correlation with HCV-RNA (R2 = 0.94), concluding that the combined use of anti-HCV levels and of HCVAg would have a very high predictive value for viremia, helping to identify individuals with active HCV infection in the screening of an asymptomatic population. Indeed, the currently available HCVAg assay is suitable for the purpose of identifying the presence of active HCV infection in anti-HCV positive individuals [16,17,33], bearing also a significant prognostic value [34].
We analyzed the potential effect of HCV-RNA levels on the RNA/Ag Log ratio separately in samples from treated and untreated patients. We did not observe any significant differences between the two groups, neither between groups with HCV-RNA levels lower or higher than 10 IU/mL of HCV-RNA.
The target population of our study was different from the majority of the other studies on HCVAg, as it was entirely composed of patients co-infected by HCV and HIV. The double infection is rather frequent, due to common routes of transmission [2,35-37], and liver disease is the second leading cause of death in patients infected by HIV [38].
In Italy HCV infection has a higher prevalence than in other Western countries [39], and IDUs have represented the highest risk group for HIV for at least 2 decades, thus the number of co-infected individuals should be quite high [39,40]. The HCV genotype distribution gives an indirect confirmation to this hypothesis: while in Italian population genotypes 1 and 2 appear the most represented [39], in our study genotype 2 was almost absent and genotype 4, mainly transmitted by parenteral routes and linked to drug injection [39,40], is rare in other settings, while it was observed in 18% of patients from the present study.
HCV replication appears to be higher in HIV co-infected than in HCV monoinfected patients as indicated by previous studies [41,42]. Thus far, only 3 very recent studies have explored the behaviour of HCV antigen in HIV/HCV co-infections. Xu et al. [43] have shown a higher variability of the HCV Core gene sequences and a more diversified HCV quasispecies population in association with the progressive decrease in the immune pressure caused by long-term HIV infection, but HIV co-infection did not seem to influence the correlation between HCV-RNA and HCVAg. In residents from a rural village in China Shen et al. [42] have found a high correlation both in 129 HCV-only infected (r = 0.808) and in 98 patients with the double infection (r = 0.952). The results of Mederacke et al. [21] were substantially similar, with a 96% concordance and a very high correlation (r = 0.97) on 71 samples from 58 co-infected individuals. Our results were different, since the concordance (91.5%) and correlation (r = 0.816) were both lower. It is also of note that ratio of HCV-RNA/HCVAg log10 levels reported in 16 Chinese co-infected patients [43] were lower and more homogeneous (1.78 ± 0.16; median 1.74) than in the present study (2.25 ± 0.88; median 1.98) and not different from those found in HCV-monoinfected subjects.
Several factors may help explain these discrepancies: first, our population was numerically much more consistent (355 samples from 292 patients, i.e. almost twice as many as in the 3 other studies combined). Furthermore no information on the HCV genotype was available from the latter study while in the former study [21] most samples were collected from patients infected by genotype 1 (41/71) or genotype 2 (23/71) and these Authors found a very high correlation for both, while in the present study the correlation in genotype 3 infection was weaker. Finally, a different proportion of patients undergoing treatment for HCV and host factors related to the different populations may account for the observed discrepancies observed between this and previous studies.
The kinetics of HCV-RNA and HCVAg in a subgroup of 18 co-infected patients under treatment for chronic hepatitis C was also evaluated and confirms the substantial concordance and similar temporal trends already observed in other studies [12,25,27], though in some cases the decrease of HCVAg may be slower as we found samples reactive for HCVAg and negative or with very low levels of HCV-RNA only in samples obtained during antiviral treatment for hepatitis C.We also describe one notable exception, in which the persistence of HCVAg after the disappearance of HCV-RNA from the blood stream anticipated a virological relapse. While the complete mechanisms of inhibition of viral replication and of the production of viral antigens in patients undergoing treatment for chronic hepatitis C are still unclear, testing for HCVAg during either conventional or DAA-based treatment, as recently suggested [26,27], will bring only a marginal additional cost and may enable to identify some cases that will benefit from a closer monitoring of HCV-RNA levels in order to anticipate viral rebound.
Conclusions
Testing for HCVAg may represent a suitable option for assessing the presence and degree of active HCV infection also in patients co-infected by HIV. As in other viral infections, the adoption of standardized serological testing on high-volume automated systems will guarantee a faster turnaround time. This is particularly useful for the close monitoring requested in the new therapeutic options with direct antiviral acting drugs (DAA). In addition, this approach can help reducing the cost, especially in terms of personnel, for the initial screening and routine patient assessment and will allow to focus the time and skills on more demanding and cumbersome virological assays.
Ethical approval
This study was exempt from ethical review, since it was based on a retrospective chart review, and analyses were performed on an anonymous database. In this respect, the local policy was employed in accordance with the recently reviewed international policy [44], and with current Italian legislation (see Methods section).
Competing interests
Dr. Claudio Galli is currently employed by Abbott Diagnostics, Italy, as the Scientific Affairs Manager. The other Authors have no conflicts of interest to declare.
Authors’ contributions
ARG, CG, MrC, PB conceptualised the study. Samples were collected and prepared by RS,MF,AM. The assays were carried out by RS, MF, AM, PB. ARG, CG interpreted the results, and edited the manuscript. MRC, PB participated in critical review of the paper. All authors read and approved the final version of the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Anna Rosa Garbuglia, Email: argarbuglia@iol.it.
Alessia Monachetti, Email: alexraff@infinito.it.
Claudio Galli, Email: Claudio.galli@abbott.com.
Rosella Sabatini, Email: Rosella.sabatini@inmi.it.
Monica Lucia Ferreri, Email: ml.ferreri@ospedaliriuniti.marche.it.
Maria Rosaria Capobianchi, Email: maria.capobianchi@inmi.it.
Patrizia Bagnarelli, Email: p.bagnarelli@univpm.it.
Acknowledgements
This work was supported by Italian Ministry of Health, funds Ricerca Corrente. The HCVAg reagents employed were supplied free of charge from Abbott Diagnostics.
References
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;14:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;14:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Hatzakis A, Wait S, Bruix J, Buti M, Carballo M, Cavaleri M, Colombo M, Delarocque-Astagneau E, Dusheiko G, Esmat G, Esteban R, Goldberg D, Gore C, Lok ASF, Manns M, Marcellin P, Papatheodoridis G, Peterle A, Prati D, Piorkowsky N, Rizzetto M, Roudot-Thoraval F, Soriano V, Thomas HC, Thursz M, Valla D, van Damme P, Veldhuijzen IK, Wedemeyer H, Wiessing L, Zanetti AR, Janssen HLA. et al. The state of hepatitis B and C in Europe: report from the hepatitis B and C summit conference. J Viral Hepat. 2011;14(suppl. 1):1–16. doi: 10.1111/j.1365-2893.2011.01499.x. [DOI] [PubMed] [Google Scholar]
- European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatitis C virus infection. J Hepatol. 2011;14:245–264. doi: 10.1016/j.jhep.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Hofmann WP, Sarrazin C, Zeuzem S. Current standards in the treatment of chronic hepatitis C. Dtsch Arztebl Int. 2012;14:352–358. doi: 10.3238/arztebl.2012.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon P, Bourlière M, Pénaranda G, Khiri H, Ouzan D. Real-time PCR assays for hepatitis C virus (HCV) RNA quantitation are adequate for clinical management of patients with chronic HCV infection. J Clin Microbiol. 2006;14:2507–2511. doi: 10.1128/JCM.00163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne MT, Konnick EQ, Phansalkar A, Hillyard DR. Evaluation of the Abbott investigational use only RealTime hepatitis C virus (HCV) assay and comparison to the Roche TaqMan HCV analyte-specific reagent assay. J Clin Microbiol. 2009;14:2872–2878. doi: 10.1128/JCM.02329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoletto G, Campagnolo D, Mirandola S, Comastri G, Severini L, Pulvirenti FR, Alberti A. Comparable performance of TMA and Real-Time PCR in detecting minimal residual hepatitis C viraemia at the end of antiviral therapy. J Clin Virol. 2011;14:217–220. doi: 10.1016/j.jcv.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Bossler A, Gunsolly C, Pyne MT, Rendo A, Rachel J, Mills R, Miller M, Sipley J, Hillyard D, Jenkins S, Essmyer C, Young S, Lewinski M, Rennert H. AmpliPrep/COBAS TaqMan® automated system for hepatitis C virus (HCV) quantification in a multi-center comparison. J Clin Virol. 2011;14:100–103. doi: 10.1016/j.jcv.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Chevaliez S. Virological tools to diagnose and monitor hepatitis C virus infection. Clin Microbiol Infect. 2011;14:116–121. doi: 10.1111/j.1469-0691.2010.03418.x. [DOI] [PubMed] [Google Scholar]
- Morota K, Fujinami R, Kinukawa H, Machida T, Ohno K, Saegusa H, Takeda K. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods. 2009;14:8–14. doi: 10.1016/j.jviromet.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Ross RS, Viazov S, Salloum S, Hilgard P, Gerken G, Roggendorf M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;14:1161–1168. doi: 10.1128/JCM.01640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici MC, Furlini G, Rodella A, Fuertes A, Monachetti A, Calderaro A, Galli S, Terlenghi L, Olivares M, Bagnarelli P, Costantini A, De Conto F, Sainz M, Galli C, Manca N, Landini MP, Dettori G, Chezzi C. Hepatitis C virus core antigen: analytical performances, correlation with viremia and potential applications of a quantitative, automated immunoassay. J Clin Virol. 2011;14:260–265. doi: 10.1016/j.jcv.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Descamps V, Op de Beck A, Plassart C, Brochot E, Francois C, Helle F, Adler M, Bourgeois N, Degré D, Duverlie G, Castelain S. Strong correlation between liver and serum levels of hepatitis C virus core antigen and RNA in chronically infected patients. J Clin Microbiol. 2012;14:465–468. doi: 10.1128/JCM.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I, Wedemeyer H, Ciesek S, Steinmann E, Raupach R, Wursthorn K, Manns MP, Tillmann HL. Performance and clinical utility of a novel fully automated quantitative HCV-core antigen assay. J Clin Virol. 2009;14:210–215. doi: 10.1016/j.jcv.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Kesly R, Polat H, Terzi Y, Kurtoglu MG, Uyar Y. Comparison of a newly developed automated and quantitative hepatitis C virus (HCV) core antigen test with the HCV RNA assay for clinical usefulness in confirming anti-HCV results. J Clin Microbiol. 2011;14:4089–4093. doi: 10.1128/JCM.05292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamili S, Drobeniuk J, Araujo AC, Hayden TM. Laboratory diagnostics for hepatitis C virus infection. Clin Infect Dis. 2012;14(S1):S43–S48. doi: 10.1093/cid/cis368. [DOI] [PubMed] [Google Scholar]
- Miedouge M, Saune K, Kamar N, Rieu M, Rostaing L, Izopet J. Analytical evaluation of HCV core antigen and interest for HCV screening in haemodialysis patients. J Clin Virol. 2010;14:18–21. doi: 10.1016/j.jcv.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Icardi G, Ansaldi F, Bruzzone BM, Durando P, Lee S, de Luigi C, Crovari P. Novel approach to reduce the hepatitis C virus (HCV) window period: clinical evaluation of a new enzyme-linked immunosorbent assay for HCV Core antigen. J Clin Microbiol. 2001;14:3110–3114. doi: 10.1128/JCM.39.9.3110-3114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary TP, Gutierrez RA, Muerhoff AS, Birkenmeyer LG, Desai SM, Dawson GJ. A chemiluminescent, magnetic particle-based immunoassay for the detection of hepatitis C virus core antigen in human serum or plasma. J Med Virol. 2006;14:1436–1440. doi: 10.1002/jmv.20716. [DOI] [PubMed] [Google Scholar]
- Mederacke I, Potthoff A, Meyer-Olson D, Meier M, Raupach R, Manns MP, Wedemeyer H, Tillmann HL. HCV core antigen testing in HIV- and HBV-coinfected patients, and in HCV-infected patients on hemodialysis. J Clin Virol. 2012;14:110–115. doi: 10.1016/j.jcv.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Verbeeck J, Stanley MJ, Shieh J, Celis L, Huyck E, Wollants E, Morimoto J, Farrior A, Sablon E, Jankowski-Hennig M, Schaper C, Johnson P, Van Ranst M, Van Brussel M. Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0. J Clin Microbiol. 2008;14:1901–1906. doi: 10.1128/JCM.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martró E, González V, Buckton AJ, Saludes V, Fernández G, Matas L, Planas R, Ausina V. Evaluation of a new assay in comparison with reverse hybridization and sequencing methods for hepatitis C virus genotyping targeting both 5′ noncoding and nonstructural 5b genomic regions. J Clin Microbiol. 2008;14:192–197. doi: 10.1128/JCM.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti AR, Romanò L, Brunetto M, Colombo M, Bellati G, Tackney C. Total HCV core antigen assay: a new marker of hepatitis C viremia for monitoring the progress of therapy. J Med Virol. 2003;14:27–30. doi: 10.1002/jmv.10355. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Saito H, Higashimoto M, Atsukawa K, Ishii H. Benefit of hepatitis C virus core antigen assay in prediction of therapeutic response to interferon and ribavirin combination therapy. J Clin Microbiol. 2005;14:186–191. doi: 10.1128/JCM.43.1.186-191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermehren J, Susser S, Berger A, Perner D, Peiffer KH, Allwinn R, Zeuzem S, Sarrazin C. Clinical utility of the ARCHITECT HCV Ag assay for early treatment monitoring in patients with chronic hepatitis C genotype 1 infection. J Clin Virol. 2012;14:17–22. doi: 10.1016/j.jcv.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Loggi E, Cursaro C, Scuteri A, Grandini E, Martello Panno A, Galli S, Furlini G, Bernardi M, Galli C, Andreone P. Patterns of HCV-RNA and HCV Core antigen in the early monitoring of standard treatment for chronic hepatitis C. J Clin Virol. 2013;14:291–295. doi: 10.1016/j.jcv.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Schuettler CG, Thomas C, Discher T, Friese G, Lohmeyer J, Schuster R, Schaefer S, Gerlich WH. Variable ratio of hepatitis C virus RNA to viral Core antigen in patient sera. J Clin Microbiol. 2005;14:1977–1981. doi: 10.1128/JCM.42.5.1977-1981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;14:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Sugiyama N, Watashi K, Masaki T, Suzuki R, Aizaki H, Mizuochi T, Wakita T, Kato T. Japanesereferencepanel of bloodspecimens for evaluation of hepatitis C virus RNA and core antigen quantitative assays. J Clin Microbiol. 2012;14:1943–1949. doi: 10.1128/JCM.00487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante-Mangoni E, Vallefuoco L, Sorrentino R, Lossa D, Perna E, Molaro R, Braschi U, Zampino R, Sodano G, Adinolfi LE, Utili R, Portella G. Clinico-pathological significance of hepatitis C virus core antigen levels in chronic infection. J Med Virol. 2013;14:1913–1918. doi: 10.1002/jmv.23672. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Chang KC, Wang JH, Tsai PS, Hung SF, Hung CH, Chen CH, Lu SN. Is hepatitis C virus core antigen an adequate marker for community screening? J Clin Microbiol. 2012;14:1989–1993. doi: 10.1128/JCM.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergunay K, Sener B, Alp A, Karakaya J, Hascelick G. Utility of a commercial quantitative hepatitis C virus core antigen assay in a diagnostic laboratory setting. Diagn Microbiol Infect Dis. 2011;14:486–491. doi: 10.1016/j.diagmicrobio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Ohshawa M, Kato K, Tanno K, Ikai K, Fujishima Y, Okayama A, Turin TC, Onoda T, Suzuki. Seropositivity for anti-HCV core antigen is independently associated with increased all-cause, cardiovascular, and liver disease-related mortality in hemodialysis patients. J Epidemiol. 2011;14:491–499. doi: 10.2188/jea.JE20100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Bansi L, Gilson R, Gazzard B, Walsh J, Pillay D, Orkin C, Phillips A, Easterbrook P, Johnson M, Porter K, Schwenk A, Hill T, Leen C, Anderson J, Fisher M, Sabin C. UK Collaborative HIV Cohort (UK CHIC) Study. The prevalence of hepatitis C virus (HCV) infection in HIV-positive individuals in the UK – trends in HCV testing and the impact of HCV on HIV treatment outcomes. J Viral Hepat. 2010;14:569–577. doi: 10.1111/j.1365-2893.2009.01215.x. [DOI] [PubMed] [Google Scholar]
- Taylor LE, Swan T, Mayer KH. HIV coinfection with hepatitis C virus: evolving epidemiology and treatment paradigms. Clin Infect Dis. 2012;14(S1):s33–s42. doi: 10.1093/cid/cis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggorowati N, Yano Y, Heriyanto DS, Hinonce HT, Utsumi T, Mulya DP, Subronto YW, Hayashi Y. Clinical and virological characteristics of hepatitis B or C virus co-infection with HIV in Indonesian patients. J Med Virol. 2012;14:857–865. doi: 10.1002/jmv.23293. [DOI] [PubMed] [Google Scholar]
- Righi E, Beltrame A, Bassetti M, Lindstrom V, Mazzarello G, Dentone C, Di Biagio A, Ratto S, Viscoli C. Therapeutical aspects and outcome of HIV/HCV coinfectedpatients treated with pegylated interferon plus ribavirin in an Italian cohort. Infection. 2008;14:358–361. doi: 10.1007/s15010-008-7319-5. [DOI] [PubMed] [Google Scholar]
- Cornberg M, Razavi HA, Alberti A, Bernasconi E, Buti M, Cooper C, Dalgard O, Dillion JF, Flisiak R, Forns X, Frankova S, Goldis A, Goulis I, Halota W, Hunyady B, Lagging M, Largen A, Makara M, Manolakopoulos S, Marcellin P, Marinho RT, Pol S, Poynard T, Puoti M, Sagalova O, Sibbel S, Simon K, Wallace C, Young K, Yurdaydin C. et al. A systematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Internat. 2011;14(suppl. 2):30–60. doi: 10.1111/j.1478-3231.2011.02539.x. [DOI] [PubMed] [Google Scholar]
- Stroffolini T, D’Egidio PF, Aceti A, Filippini P, Puoti M, Leonardi C, Almasio PL. and the DAVIS (Drug Addicted, HCV Prevalence in Italy) participating Centers. An epidemiological, observational, cross-sectional, multicenter study: Hepatitis C virus infection among drug addicts in Italy. J Med Virol. 2012;14:1608–1612. doi: 10.1002/jmv.23370. [DOI] [PubMed] [Google Scholar]
- Danta M, Semmo N, Fabris P, Brown D, Pybus OG, Sabin CA, Bhagani S, Emery VC, Dusheiko GM, Klenerman P. Impact of HIV on host-virus interactions during early hepatitis C virus infection. J Infect Dis. 2008;14:1558–1566. doi: 10.1086/587843. [DOI] [PubMed] [Google Scholar]
- Shen T, Chen X, Zhang W, Xi Y, Cao G, Zhi Y, Wang S, Xu C, Wei L, Lu F, Zhuang H. A higher correlation of HCV core antigen with CD4+ T cell counts compared with HCV RNA in HCV/HIV-1 coinfected patients. PLoS ONE. 2011;14:e23550. doi: 10.1371/journal.pone.0023550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C-H, Shen T, Zheng J-J, Tu J, Zhang W-D, Lu F-M. Higher dN/dS ratios in the HCV core gene, but not in the E1/HVR1 gene, are associated with human immunodeficiency virus-associated immunosuppression. Arch Virol. 2012;14:2153–2162. doi: 10.1007/s00705-012-1390-z. [DOI] [PubMed] [Google Scholar]
- Millum J, Menikoff J. Streamlining ethical review. Ann Intern Med. 2010;14(153):655–657. doi: 10.7326/0003-4819-153-10-201011160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]