Abstract
Aspergillus flavus has received much attention owing to its severe impact on agriculture and fermented products induced by aflatoxin. Sclerotia morphogenesis is an important process related to A. flavus reproduction and aflatoxin biosynthesis. In order to obtain an extensive transcriptome profile of A. flavus and provide a comprehensive understanding of these physiological processes, the isolated mRNA of A. flavus CA43 cultures was subjected to high-throughput strand-specific RNA sequencing (ssRNA-seq). Our ssRNA-seq data profiled widespread transcription across the A. flavus genome, quantified vast transcripts (73% of total genes) and annotated precise transcript structures, including untranslated regions, upstream open reading frames (ORFs), alternative splicing variants and novel transcripts. We propose natural antisense transcripts in A. flavus might regulate gene expression mainly on the post-transcriptional level. This regulation might be relevant to tune biological processes such as aflatoxin biosynthesis and sclerotia development. Gene Ontology annotation of differentially expressed genes between the mycelia and sclerotia cultures indicated sclerotia development was related closely to A. flavus reproduction. Additionally, we have established the transcriptional profile of aflatoxin biosynthesis and its regulation model. We identified potential genes linking sclerotia development and aflatoxin biosynthesis. These genes could be used as targets for controlled regulation of aflatoxigenic strains of A. flavus.
Introduction
Aspergillus flavus is a ubiquitous pathogenic fungus that infects plants and animals. Recently, studies of A. flavus gained tremendous attention owing to its health impact on agricultural commodities and related fermented products induced by aflatoxin contamination. Initially, studies using A. flavus as a model organism have been focused mainly on the aflatoxin biosynthesis pathway and on mechanisms of regulation of aflatoxin formation [1]–[5]. The highly negative impact of infection of agriculturally relevant plants by A. flavus caused gradual expansion of studies into the related fundamental areas of biology of this fungus, including the A. flavus secondary metabolism, sclerotia morphogenesis and propagation [2], [4], [6]–[9]. The recently revealed genomic sequence of A. flavus NRRL3357 provided a powerful tool for detailed analysis of biology of this fungus [10].
Sclerotia morphogenesis is an physiological process important for A. flavus propagation and involves various secondary metabolism pathways, including aflatoxin biosynthesis. Sclerotia are pigmented, specialized structures composed of compact A. flavus mycelia, which makes A. flavus resistant to harsh environmental conditions. Sclerotia likely derive from cleistothecia and might represent a vestige of sexual ascospore production [6]. According to the size of sclerotia, A. flavus could be divided into two groups: the L strain has sclerotia that are >400 µm in diameter and the S strain has sclerotia that are <400 µm in diameter [11], [12]. The S strains produce greater amounts of sclerotia and aflatoxin compared to the L strains under the same conditions of medium and cultivation [13], suggesting sclerotia morphogenesis and aflatoxin biosynthesis are closely related. To interpret this correlation, Chang and colleagues proposed the “substrate (acetate) competition” hypothesis. In their proposal, the increased production of aflatoxin results in a progressive decrease in sclerotial size, alterations in sclerotial shape and causes weakening of the sclerotial structure [6]. Recent efforts provided insights into the regulation of sclerotia morphogenesis. Ammonium, light, oxidative stress, temperature, organic acids and endogenous levels of cAMP might influence sclerotia formation and maintenance [14]–[19]. It was reported that A. flavus laeA and veA mutants are not able to form sclerotia [20], [21]. Cary and colleagues used DNA array to analyze sclerotia-related genes by comparing the A. flavus wild type strain and a veA mutant strain [22]; however, a detailed comprehensive description of sclerotia morphogenesis is lacking.
Natural antisense transcripts (NATs), a subset of non-coding RNAs (ncRNAs), are endogenous RNA molecules transcribed from the opposite DNA strand and can be complementary to the sense RNA through base pairing [23]–[25]. Expressed sequence tag (EST) sequencing, tiling microarrays, SAGE libraries, asymmetric strand-specific analysis of gene expression and global run-on sequencing (GRO-seq) could be used to identify NATs [26]. NATs are involved in transcriptional and post-transcriptional gene regulation by RNA interference (RNAi) [27], [28], chromatin-level gene silencing [23], [26], [29]–[31], chromatin remodeling [28], [32] and local chromatin modifications [33]–[35]. Global analysis of NATs has been done for mammals [23], insects [36], worms [37] and plants [38], [39]. It was reported that a large portion of the mammalian genome could produce transcripts from both strands [23], [40], [41]. For instance, >70% of mouse transcripts contain NATs and, owing to their prevalence, NATs are treated as pervasive features of mammalian genomes [26]. Aspergillus spp. have beenused as model organisms to investigate many molecular processes that govern the life of eukaryotic cells. Moreover, Aspergillus spp. have been used extensively to study fungal-specific pathways. NATs have been found in a divergent group of fungi, including the ascomycetes Saccharomyces cerevisiae, Candida albicans, A. flavus, Magnaporthe oryzae, Tuber melanosporum and Schizosaccharomyces pombe, and the basidiomycetes Cryptococcus neoformans, Ustilago maydis and Schizophyllum commune [28]. There are two distinct drawbacks in the study of A. flavus NATs, however; first, there were only 352 NATs (2.8% of the total open reading frames (ORFs)) found in A. flavus by analyzing ∼23,000 A. flavus cDNAs from cells grown under different nutritional conditions [25], while this number soars to 16.7–85.2% in comprehensively analyzed transcriptomes such as S. cerevisiae (16.7%), C. albicans (40.0%) and S. pombe (85.2%) [28]. Second, the biological function of NATs in A. flavus remained elusive due to the limited amount of NATs detected [42].
Owing to the importance of transcriptional regulation in the development of fungi, transcriptome profiling could be a valuable tool for establishing an in-depth understanding of A. flavus biology. The transcriptome of A. flavus has been studied by several groups, but these studies were not focused on sclerotia development or secondary metabolism. ESTs and microarrays have been used to identify genes involved in aflatoxin production using 7218 unique ESTs and microarrays containing more than 5000 unique A. flavus gene amplicons [1]–[3]. However, only 263 genes were expressed differentially and only 20 of the 29 aflatoxin pathway genes were identified owing to the limitation of microarrays in detecting genes with low levels of expression. Even an important transcriptional factor in the aflatoxin biosynthesis pathway, aflR, has not been detected [1]. Chang et al identified 22 features common to the aflatoxigenic/non-aflatoxigenic pairs by cross comparison. Possible roles of these identified genes have been discussed in relation to the regulation of aflatoxin biosynthesis [3]. Yu et al profiled the transcriptome of A. flavus under various temperature conditions in order to understand the effect of temperature on mycotoxin biosynthesis using RNA-seq technology. Only 23–29% of the total reads were mapped to genes, however, and >50% of the reads were mapped to rRNA genes and mitochondria and, therefore, were regarded as useless data [4].
We used strand-specific RNA sequencing (ssRNA-seq) to obtain an extensive transcriptome profile of A. flavus that might facilitate comprehensive understanding of the physiological processes of A. flavus. ssRNA-seq technology has all of the advantages of conventional RNA-seq technology, including low detection background, high dynamic detection range, high reproducibility and precise definition of transcript structure [43]. Furthermore, in contrast to conventional RNA-seq technology, ssRNA-seq data contain RNA polarity information and could decode a complex eukaryote transcriptome, including genome annotation, de novo transcriptome assembly and accurate digital gene expression analysis [44], [45]. ssRNA-seq technology has been used to parse the transcriptome of many organisms, including S. cerevisiae [46], Mycoplasma pneumonia [47], Mus musculus [47] and Oryza sativa [48]. By using ssRNA-seq data of the A. flavus mycelia and sclerotia cultures, we profiled genome-scale transcription, annotated precise transcript structure and provided in-depth depiction of A. flavus NATs. Analysis of differentially expressed genes might contribute to explicating sclerotia development and aflatoxin biosynthesis. In summary, our ssRNA-seq-based annotation provided a more extensive depiction of the A. flavus transcriptome and might contribute to the reduction of detrimental effects of A. flavus infection on agriculture.
Materials and Methods
Strain and Culture Conditions
A. flavus CA43 was kindly provided by Professor Perng-Kuang Chang (Southern Regional Research Center, Agricultural Research Service, U.S. Department of Agriculture, Washington, DC, USA). A. flavus CA43 belongs to the S strain isolates, which can produce numerous sclerotia and high amounts of aflatoxin [13].
Potato dextrose agar (PDA) medium (20 g of dextrose, 15 g of agar and the infusion from 200 g of potatoes per 1 L of medium, pH = 6.0) was used for A. flavus cultivation. For harvesting the A. flavus culture, a layer of cellophane was placed over the PDA medium plate (PDA-cellophane plate). A. flavus sclerotia (1.2×106) were inoculated onto each PDA-cellophane plate and cultivated at 30°C in darkness. For RNA sequencing, the A. flavus mycelia were harvested after 48 h of cultivation (Aflavus_CA43_M) and sclerotia were collected after 7 days of cultivation (Aflavus_CA43_S).
Strand-specific RNA-seq Library and Sequencing
The total RNA of each sample was extracted using RNAiso™ Plus (TaKaRa, Japan), treated with RNase-free DNase I (TaKaRa, Japan) and purified with a NucleoSpin® RNA Clean-up Kit (Macherey-Nagel, Germany). RNA integrity was analyzed by an Agilent Technologies 2100 Bioanalyzer. All samples had an RNA integrity number (RIN) >7.
The strand-specific RNA-seq library was prepared essentially as described but with some modifications [49]. Sera-mag magnetic oligo(dT) beads were used to isolate poly(A) mRNA from 20 µg of purified total RNA. mRNA was eluted by 10 mmol/L Tris buffer and fragmented into small pieces in the range of 100–500 bp by treatment with divalent cations. Taking these short fragments as templates, random hexamer primers were used to synthesize the first-strand cDNA using SuperScript II, RNaseH and DNA polymerase I. The first-strand cDNA was purified using a QIAquick PCR Purification Kit and used as the template for synthesizing dUTP-containing second-strand cDNA by adding buffer, dNTPs with dTTP replaced by dUTP, RNAseH and DNA polymerase I. Double-stranded cDNA fragments were purified with a QIAquick PCR Purification Kit and eluted with elution buffer for end-repair, phosphorylation and 3′-adenylation. After that, cDNA fragments were purified with a Qiagen MinElute PCR Purification Kit. Illumina sequencing adapters were ligated to the 3′-adenylated cDNA fragments followed by purification using a MinElute PCR Purification Kit. Fragments of 100–500 bp were purified using a QIAquick Gel Extraction Kit from the modified double-stranded cDNA fragments described above separated by TAE-agarose gel electrophoresis (2% (w/v) agarose; Certified Low-Range Ultra Agarose, Biorad). Uracil-DNA glycosylase (UNG) was added to digest dUTP-containing second-strand cDNA in an alkalescent medium at high temperature. The remaining first-strand cDNA was purified using a MinElute PCR Purification Kit and enriched by 15 rounds of PCR amplification using primers P7 and P5, which were homologous to the Illumina sequencing adapters. After purification using a QIAquick Gel Extraction Kit, fragments of 100–500 bp (average 200 bp) were selected as a strand-specific RNA-seq cDNA library. Finally, after quantification on an Agilent Technologies 2100 Bioanalyzer, the cDNA library was sequenced on the Illumina HiSeq2000 platform by a 90 bp paired-end sequencing strategy (using primers P7 and P5). The raw Illumina sequencing data for the A. flavus CA43 transcriptome was deposited in the SRA (http://www.ncbi.nlm.nih.gov/sra/) with ID no.SRP018670.
Primary Splitting of Raw Strand-specific Sequencing Data
In the course of strand-specific paired-end (PE) sequencing, reads consistent with the reverse strand of a transcript were first obtained by primer P5-triggered sequencing (read1), and reads consistent with the forward strand of the same transcript were then obtained by primer P7-triggered sequencing (read2). Read1 and read2 are PE reads.
According to strand-specific sequencing features, read1 mapped to the forward strand of a transcript and read2 reversely mapped to the forward strand of the transcript could not reflect the real transcription and was discarded. The remaining mapping reads were used to analyze gene expression and genes expressed differentially.
As the forward strand of a gene might be located in either the forward or the reverse strand of the genome, we classified read1 reversely mapped to the forward strand of the genome and read2 mapped to the forward strand of the genome into one group representing transcription from the forward strand of the genome. We classified read1 mapped to the forward strand of the genome and read2 reversely mapped to the forward strand of the genome into a group representing transcription from the reverse strand of the genome. The mapping result with genome sequence was used for the following analysis, including novel transcript predication, untranslated region (UTR) analysis and alternative splicing (AS) events. This splitting strategy was designed according to the BGI analysis pipeline (http://www.genomics.cn/index).
Read Mapping and Normalization of Gene Expression
The reference genome (A. flavus NRRL3357) was downloaded from GenBank (accession no. AAIH00000000.2) and the gene annotation data were downloaded from the AspGD website (http://www.aspergillusgenome.org/download/sequence/A_flavus_NRRL_3357/current/). After removing the reads containing sequencing adapters and low-quality reads (reads containing Ns >9), the remaining 90 bp clean reads were aligned to the A. flavus reference genome and annotated genes using SOAP2 software [50], allowing up to five base mismatches for genome alignment and three base mismatches for gene alignment. Reads that failed to be mapped were first trimmed off 1 base from the 5′-end. If there was still no match, these reads were trimmed off progressively two bases each time from the 3′-end and mapped to the genome until a match was found (unless the read was trimmed <48 bases for genome alignment or <28 bases for gene alignment). The insert between PE reads was set to 1 bp-10 kb, allowing them to span introns or the intergenic regions of various size in the genome. When PE reads were mapped to non-redundant genes, the insert was set to 1 bp-1 kb. The coverage per base in the genome and the reads count covering each transcript could be calculated by reads mapping.
We used the popular RPKM method to normalize the transcript level, which was expressed as the number of reads per kilobase of exon region per million mapped reads (RPKM) [51], [52]. The cutoff value for gene transcriptional activity was determined on the basis of the 95% confidence interval (CI) for all RPKM values. GO annotation of the A. flavus genes was done by Blast2GO (version 2.6.3) software [53] and visualized by WEGO software [54].
Non-redundant EST Sequence
Non-redundant ESTs were obtained by mapping all A. flavus EST sequences (http://compbio.dfci.harvard.edu/cgi-bin/tgi/est_ann.pl?gudb=a_flavus) to A. flavus annotated genes using BLAT software [55], [56]. ESTs that could not be mapped to annotated genes (http://www.aspergillusgenome.org/download/sequence/A_flavus_NRRL_3357/current/) were selected as non-redundant ESTs.
UTR and Upstream Open Reading Frame Analysis
UTRs were defined as regions flanking a gene coding sequence, with contiguous expression of each base supported by at least two uniquely mapped reads. Transcripts whose ends overlapped with each other were discarded. The length of 5′- or 3′-UTRs was limited to a maximum length of 1000 bp. The upstream ORFs (uORFs) were searched for in the 5′-UTRs of genes. The length of uORFs was set to 9–150 bp, and the distance between a uORF and a gene start codon should be <500 bp.
Detection of Novel Transcripts
Novel transcriptionally active regions (TARs) were determined in the intergenic regions by contiguous expression of each base supported by at least two uniquely mapped reads and length >35 bp [43]. A novel TAR should be separated by at least 200 bp from the upstream or downstream region of the transcript. A novel transcript is composed of novel TARs connected by at least one paired read. Novel transcripts of length >150 bp were selected for further analysis.
Alternative Splicing Events in A. flavus
Potential junction sites were detected using the TopHat program [57]. Reads used to identify junction sites could not map to the genome but were able to map to the genome after several terminal bases were trimmed off. An acknowledged junction site should be supported by at least two mapped reads with different mapping positions within the junction site region and with a minimum of 5 bp mapping on each side of the junction site and a tolerance of 2 bp mismatch [43]. A novel junction site should be supported by at least four such reads. Using the method described by Wang et al [58], seven types of AS events were analyzed in A. flavus CA43, including skipped exons (SE), retained introns (RI), alternative 5′-splice sites (A5SS), alternative 3′-splice sites (A3SS), mutually exclusive exons (MXE), alternative first exons (AFE) and alternative last exons (ALE).
Natural Antisense Transcript
Putative antisense transcripts were detected for A. flavus annotated genes with RPKM >33. For a certain annotated transcript, the corresponding antisense transcript was denoted as a complementary overlapping region on the opposite strand with contiguous expression of each base supported by at least one uniquely mapped read and length >200 bp. In order to obtain reliable results, the detected antisense transcripts should have an average coverage depth >2. To eliminate the influence of an intronic region, antisense transcripts were analyzed in the transcriptional exons and UTRs.
Analysis of Differentially Expressed Genes between A. flavus Mycelia and Sclerotia States
The DEGseq R package was used to analyze differentially expressed genes (DEGs) by reads count covering each gene on the basis of the Random Sampling model [59]. Functional enrichment analysis of certain DEGs was performed with Blast2GO software using Fisher’s exact test with robust false discovery rate (FDR) correction of 0.05.
Results and Discussion
Sequencing Summary
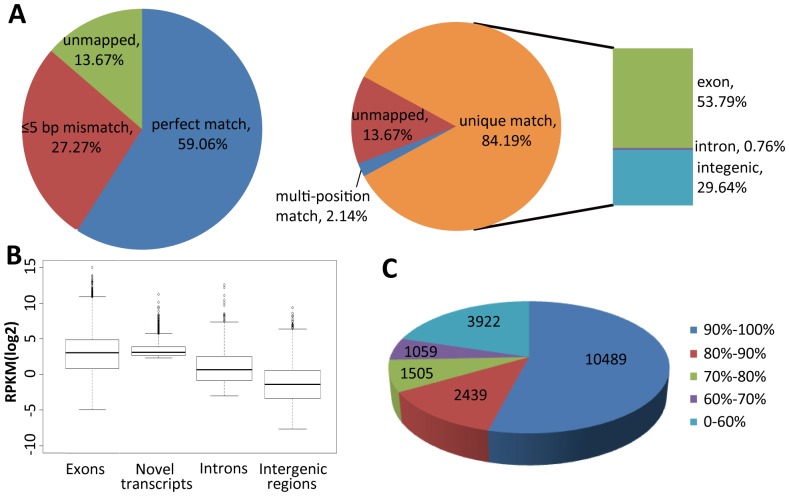
To obtain an elaborate transcriptome profile of A. flavus, the isolated mRNA of A. flavus CA43 mycelia (Aflavus_CA43_M) and sclerotia (Aflavus_CA43_S) were subjected to high-throughput sequencing by the strand-specific paired-end sequencing strategy. In total, 26.9 and 27.7 million of 90 bp Illumina reads were obtained for Aflavus_CA43_M and Aflavus_CA43_S, reaching an average genome coverage depth of 60-fold and 62-fold, respectively. About 85% of all reads were mapped uniquely to the A. flavus genome with a tolerance of 5 bp mismatch (84.48% for Aflavus_CA43_M and 83.91% for Aflavus_CA43_S; Fig. 1A and Table S1). Approximately 55% of all reads were mapped uniquely to annotated A. flavus genes (Table S1), which represents much greater accuracy compared to earlier A. flavus transcriptome data (23–29%) [4]. The 30% of reads mapped uniquely that could be mapped to the genome but could not be mapped to the annotated genes might represent novel transcripts hidden in the intergenic regions. Massive non-redundant ESTs in the A. flavus genome (Table S2) suggest the presence of a large number of novel transcripts. For all A. flavus transcripts, 38.34 million reads were mapped to their sense strand but only 0.65 million reads (1.7%) were mapped to their opposite strand, suggesting our RNA-seq library is strand specific. The reads mapped to the opposite strand might represent natural antisense transcripts that occur normally in eukaryotes. Moreover, the low ratio of reads mapped to introns suggests that our RNA-seq data could depict the transcription of A. flavus genomic loci precisely.
Figure 1. Summary of RNA sequencing of A. flavus CA43.
(A) Matching summary of ssRNA-seq reads to the A. flavus genome. (B) Box and whisker plots of log2-transformed RPKM for exons, novel transcripts, introns and intergenic regions. Horizontal lines in boxes, the first, median and third quartile. Other horizontal lines, the inner boundaries. Diamonds, data outside the inner boundaries. (C) Mapping coverage of the transcribed A. flavus genes.
Extensive Depiction of A. flavus Transcriptome by ssRNA-seq Data
The global transcriptional profile of the A. flavus mycelia culture (Aflavus_CA43_M) is shown in Fig. S1. About 67.55% of the A. flavus genome (39.91 Mb) was expressed as ssRNA-seq reads, quantifying the transcriptional abundance for 9871 A. flavus genes (73% of 13,487 genes) with a 95% CI (Table S2). However, the EST-based annotation assembled only 3749 tentative consensus sequences [2]. The genome coverage was comparable to that of A. oryzae RIB40 (76.66%) [43], and >50% of the expressed genes had a sequencing coverage of >90% ( Fig. 1C ). GO annotation showed that 7652 of the 9871 transcribed genes were assigned to GO categories (Table S2), providing rich information for the investigation of gene function. The expression levels of exons were much higher compared to introns and intergenic regions ( Fig. 1B ), and only very few introns (1621 of total 27,783 introns) were mapped by ssRNA-seq reads. Therefore, ssRNA-seq based annotation provided a more extensive depiction of the A. flavus transcriptome.
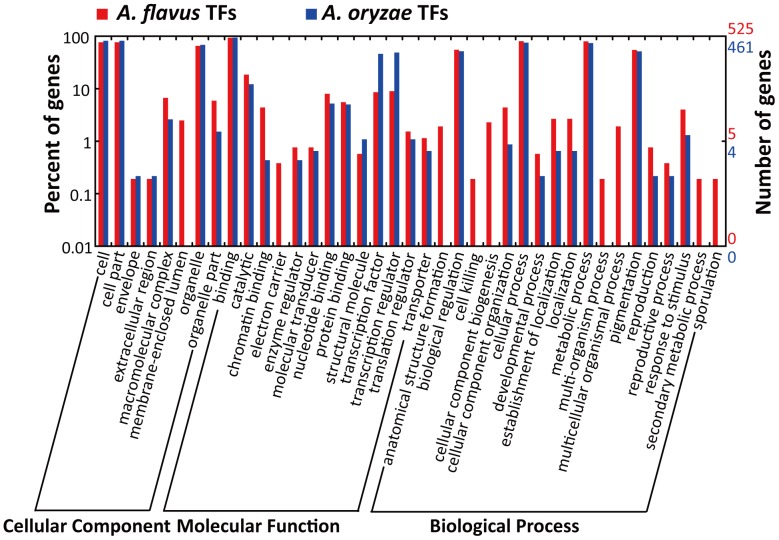
We analyzed the transcriptional activity of A. flavus transcriptional factors (TFs) to address extensive transcription of the A. flavus CA43 genome. Fungal TF information was downloaded from the web (http://ftfd.snu.ac.kr/tf.php?a=summary0&o=o). The A. flavus genome contains 667 TFs and 617 of them were expressed as RNA-seq reads, while A. oryzae contains 603 TFs and 571 of them were transcribed (Table S3; the data for A. oryzae TFs were obtained from our earlier work [43]). GO analysis showed A. flavus CA43 TFs were enriched mainly in the GO categories of binding, catalytic, biological regulation, nucleotide binding, cellular process and metabolic process ( Fig. 2 ). Particularly, A. flavus CA43 contains transcribed TFs involved in secondary metabolic process and sporulation.
Figure 2. WEGO comparison of A. flavus and A. oryzae transcribed transcriptional factors.
In total, 525 of the 617 A. flavus transcribed TFs and 461 of the 571 A. oryzae transcribed TFs were assigned to GO categories.
The non-homologous end joining (NHEJ) pathway is important for DNA repair and gene recombination. NHEJ-deficient strains could be used efficiently for gene targeting and homologous gene recombination [60]. ku70, ku80 and LigD are the essential genes in the NHEJ pathway of Aspergillus spp. [61]. For A. oryzae RIB40, the transcriptional level of ku80 (AO090026000842) reached RPKM 82.82 in the solid-state culture, and ku70 (AO090701000906) and LigD (AO090113000208) were not expressed [43]. For A. flavus CA43, ku70 (AFL2G_06276), ku80 (AFL2G_06694) and LigD (AFL2G_08329) were all transcribed under both mycelia and sclerotia cultures (RPKM 23.57, 18.84 and 14.28 for mycelia culture, and 150.82, 31.34 and 231.45 for sclerotia culture) (Table S2), suggesting A. flavus CA43 possessed highly efficient non-homologous gene targeting.
Improved Annotation of A. flavus Gene Models
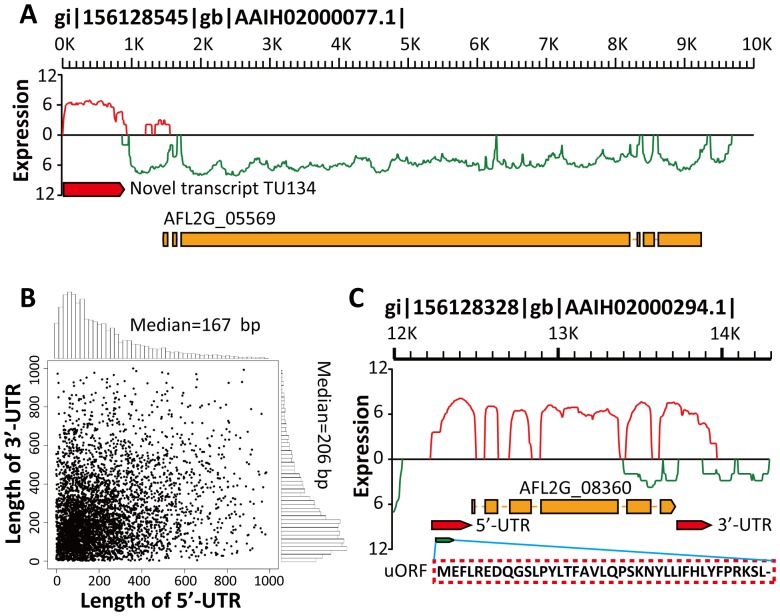
Besides the reads mapped to the 13,487 annotated A. flavus genes, 30% of reads mapped uniquely were located in the intergenic regions. Extensive mapping and clustering of these intergenic reads revealed 939 and 1196 previously unrecognized novel transcripts for mycelia and sclerotia samples, respectively (Table S4). The levels of expression of these novel transcripts were almost as high compared to exons ( Fig. 1B ) and 62.62% of the identified novel transcripts were longer than 500 bp (Table S4). This ratio is much higher compared to A. oryzae RIB40 [43]. Most of the identified novel transcripts were non-coding RNAs (ncRNAs) and only 25 (1.17%) novel transcripts were predicted to be potential protein-coding genes with an ORF length of ≥150 bp. The vast majority of transcribed ncRNAs might act as NATs to regulate gene expression. One of the identified novel transcripts (TU134) in mycelia culture is illustrated by Fig. 3A , with a length of 848 bp and an average sequencing depth of 11.61.
Figure 3. Detailed annotation of the A. flavus gene model.
(A) A novel transcript (TU134) identified in contig gi|156128545|gb|AAIH02000077.1| in the forward orientation. Red bar, the novel transcript TU134 identified in the mycelia sample. Red curve, expression level (log2-transformed reads count) of the transcripts located in the forward strand. Green curve, the expression level of the transcripts located in the reverse strand. (B) Scatterplot and histograms showing the length distribution of the 5′- and 3′-UTRs of A. flavus CA43 genes. (C) UTR and uORF illustration for an annotated A. flavus gene (AFLG2_08360).
The transcribed genome sequence assembled by our ssRNA-seq reads could be used to define or extend untranslated regions (UTRs) of the gene, which have important roles in post-transcriptional regulation [62]. The 5′-UTRs for 5994 transcripts and the 3′-UTRs for 6407 transcripts were determined in this study (Table S5). Most (88.87%) of these identified UTRs were <500 bp with a median length of 167 bp for 5′-UTRs and 206 bp for 3′-UTRs ( Fig. 3B ). The median length of UTRs in A. flavus CA43 was longer compared to A. oryzae RIB40 (107 bp for 5′-UTRs and 156 bp for 3′-UTRs) [43] and Schizosaccharomyces pombe (152 bp for 5′-UTRs and 169 bp for 3′-UTRs) [63]. According to the study reported by Lackner et al suggesting the most stable transcripts have short 5′-UTRs and the least stable transcripts have long 5′-UTRs [64], A. flavus CA43 has less stable transcripts and a much higher RNA turnover rate compared to A. oryzae and S. pombe. It was reported that ORFs in 5′-UTRs upstream of annotated start codons (uORFs) might constitute important regulatory factors for gene expression [65], [66]. We have predicted 2600 uORFs for A. flavus annotated genes (19.28%; Table S5), which is much higher compared to A. oryzae RIB40 (11.14%) [43] and S. cerevisiae (6%) [67]. An example of a uORF and the corresponding 5′-UTR is shown in Fig. 3C . Genes containing uORFs are enriched specifically for GO terms of cellular protein modification process, protein serine/threonine kinase activity, Ras GTPase activator activity, positive regulation of Ras GTPase activity, phosphorylation, autophagy, phospholipid binding and protein transport (FDR-adjusted p<0.05; Fig. S2 and Table S5).
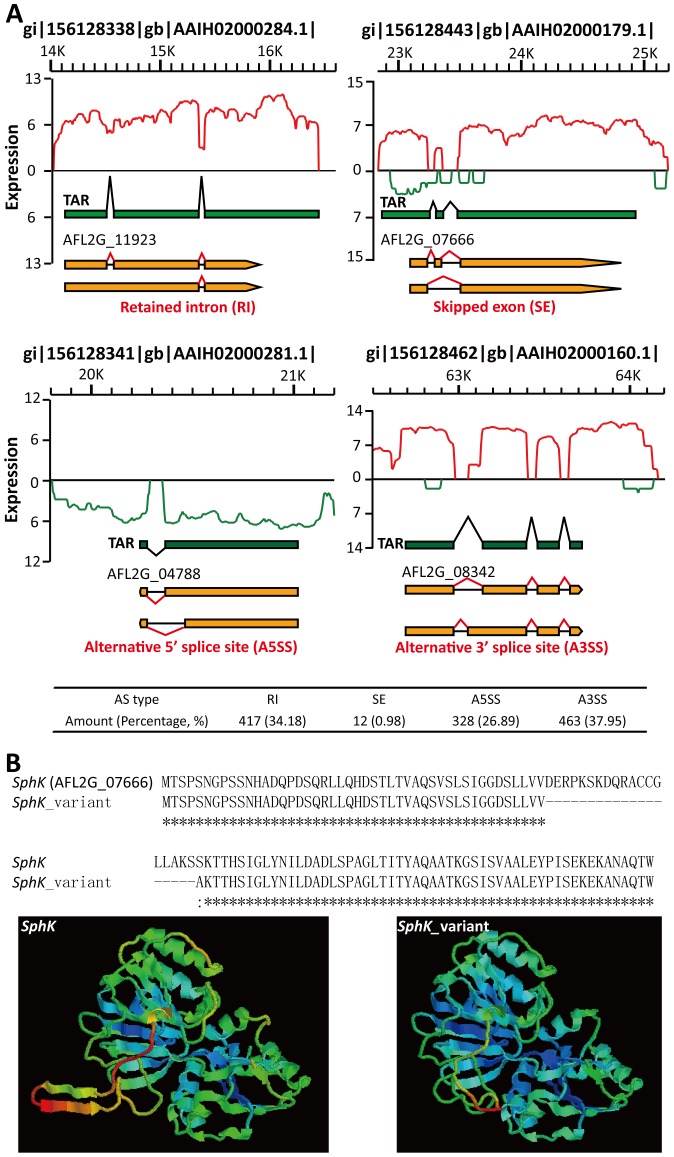
AS events contribute to producing multiple proteins from genes with two or more exons in fungi. This could enrich the proteomic diversity of fungi and provide the ability to survive in a hazardous environment. AS events were analyzed in 9545 A. flavus multi-exon genes (70.77% of all A. flavus genes) using the method described by Wang et al [58]. A total of 1220 AS events took place in 941 A. flavus genes (Table S6 and Fig. 4A ), including retained introns (RIs), skipped exons (SEs), alternative 5′-splice sites (A5SSs) and alternative 3′-splice sites (A3SSs). About 12.78% of the A. flavus multi-exon genes produced AS isoforms, similar to A. oryzae RIB40 (11.10%) [43] and many more than Pichia pastoris (4.78%) [68]. This is in agreement with the conclusion that the frequency of AS events is proportional to the ratio of multi-exon genes in a genome (76.98% for A. oryzae and 11.91% for P. pastoris) [69]. AS events might alter the amino acid composition and the structure of the target protein. For example, AFL2G_07666 ( Fig. 4A ), encodes sphingosine kinase (SphK), which participates in the sphingosine 1-phosphate (S1P) metabolism. Together with S1P phosphatase (S1PP) and S1P lyase (SPL), AFL2G_07666 controls the intracellular S1P level and has important roles in the regulation of cell migration, survival, differentiation, angiogenesis and development through an extracellular signaling pathway mediated by a family of specific G protein-coupled receptors. We performed homologous modeling using SWISS-MODEL Workspace [70], [71] and constructed a 3D model of the AFL2G_07666 transcript and its AS variant ( Fig. 4B ). The skipped exon alters the 3D structure of AFL2G_07666 and might influence its biological function. To investigate the mechanism of AS events, we calculated the ratio of the amount of RIs to cassette exons (CEs, including SE, AFE, ALE and MXE). The high RI/CE ratio (34.75) indicates A. flavus might recognize splicing sites and produce AS events mainly by the intron definition (ID) mechanism, according to the study reported by McGuire et al [69].
Figure 4. Illustration of AS events (RI, SE, A5SS and A3SS) identified in the A. flavus transcriptome.
(A) Green bar, the transcriptional active region (TAR). Orange bar, A. flavus annotated genes. Knuckle lines (black or red), the relation with TARs or exons. The types and amounts of AS events are shown. (B) Amino acid alignment and homologous 3D modeling of AFL2G_07666 (SphK) and its AS variant (SphK _variant). Broken line, the skipped exon of SphK.
Natural Antisense Transcript
The “G-value paradox”, that the amount of protein-coding genes does not correlate with the complexity of an organism, suggests that so-called junk DNA in a genome contains rich regulatory information exerting via transcribed natural antisense transcripts (NATs) [23], [28]. So far, little is known about the role of NATs in Aspergillus spp. It was suggested that NATs might have an important role in differential expression of genes involved in secondary metabolism in A. oryzae [42]. According to an earlier study, the most prominent NAT type is the non-protein-coding antisense RNA partner of a protein-coding gene [26]. We searched for NATs in our data for all A. flavus protein-coding genes with the RPKM>33 using stringent criteria. In all, 1124 and 839 NATs were identified in the AflavusCA43_M and AflavusCA43_S samples, respectively (Table S7). This is many more compared to A. flavus NATs on the basis of EST data (352, 2.8%) and Cryptococcus neoformans NATs (53, 0.8%) [25]. The number of RNA-seq-based A. flavus NATs is in the same range as S. cerevisiae (1103, 16.7%) [46]. However, this frequency is much lower compared to Candida albicans (2458, 40%) and M. oryzae (4215, 32.8%) [28].
There are more NATs located inside coding regions (768, 39.14%) compared to UTR regions (466, 23.76%, Fig. 5A ). This means NATs in A. flavus might regulate gene expression mainly at the post-transcriptional level, which is consistent with the conclusion reported by Donaldson et al [28]. The inside-biased NAT distribution in A. flavus, however, was different from the 3′-biased antisense transcription in A. nidulans [72] and the mammalian NAT distribution, where NATs are usually enriched in the region of the 250 bp upstream sequence and the 1.5 kb downstream sequence [26]. The transcriptional level of A. flavus genes with NATs (average RPKM 253.59) was much higher compared to genes without NATs (average RPKM 54.50; Fig. 5B ). This is consistent with the earlier study by Katayama et al [23], strongly denying the simple hypothesis that NATs are just negative regulatory elements and artifacts of leaky bidirectional transcription [26], [28], [73].
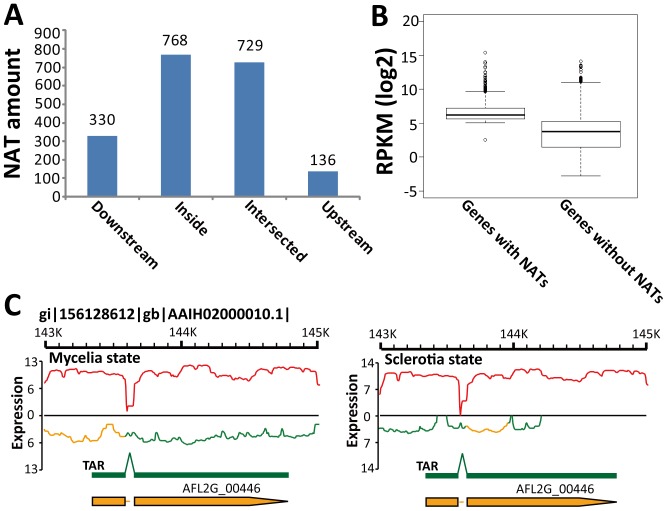
Figure 5. NAT analysis in A. flavus CA43.
(A) The relationship between the location of NAT and its corresponding genes. Downstream, Upstream and Inside, NATs located in the downstream, upstream or inside of their corresponding genes, respectively. Intersected, NAT located across one gene and its 5′- or 3′- flanking region. (B) Box and whisker plots of log2-transformed RPKM for A. flavus annotated genes with or without NATs. (C) Occurrence of NAT in the SdeA gene (AFL2G_00446). Orange curves, the NAT in the SdeA gene.
Among the 352 EST-based NATs in A. flavus, only 19 were verified by our RNA-seq data (Table S7). One of the EST-based NATs is that of the aflatoxin biosynthetic regulator gene (aflR, AFL2G_07224), which was identified in our study when the RPKM cutoff for NAT analysis was set at 5 (Table S7). The existence of a NAT in the transcriptional factor aflR suggests the interaction between a NAT and its sense transcript can occur in the nucleus, consistent with the conclusion from studies in mammals [23], [26]. Another example is SdeA (AFL2G_00446), which is involved in the regulation of morphology under temperature change and in the production of multicellular developmental structures (conidiophores, cleistothecia and sclerotia) [25]. Changes in the expression of SdeA and its NAT in the mycelia and sclerotia states suggests NAT participates in the regulation of SdeA and thus biological processes controlled by this gene ( Fig. 5C ).
GO enrichment analysis demonstrated genes with NATs were enriched specifically in GO terms of protein complex, protein binding, RNA binding, translation, ribosome, intracellular protein transport, cellular amino acid metabolic process, vesicle-mediated transport, hexose catabolic process and biological regulation (FDR-adjusted p<0.05; Fig. S3 and Table S7). Thus, these NAT-containing genes are closely related to protein expression, secretion and energy production in A. flavus. For example, the A. flavus Set1 gene (AFL2G_02936) has a NAT located inside its coding sequence (Table S7). According to the report that the corresponding NAT could prevent Set1-mediated transcription initiation in S. cerevisiae [28], NAT might influence the Set1-mediated transcription initiation in A. flavus. The discussion about NAT function in fungi was focused mainly on the NAT-mediated alteration of physiological processes in response to environmental nutrient starvation [28] and nitrogen metabolism [72]. Our analysis is the first global investigation of NAT function in A. flavus.
Sclerotia Development and Reproduction
Sclerotia are considered to derive from cleistothecia, which is the sexual reproductive organ in Aspergillus spp. [6]. Genes involved in sexual reproduction and the balance of sexuality and asexuality of Aspergillus spp. are given in Table S8 [74]–[76]. To identify genes involved in A. flavus sclerotia development, differentially expressed genes (DEGs) between the A. flavus mycelia and sclerotia cultures were detected. For the 13,487 A. flavus genes, 9871 (73.19%) were transcribed under both conditions. A total of 661 genes were expressed specifically in the mycelia state and 343 genes were specific for the sclerotia state. These genes might represent factors critical for the physiological development of A. flavus. There were 7609 DEGs (56.42%) between the A. flavus mycelia and sclerotia states, with 1821 up-regulated genes and 5788 down-regulated genes in the sclerotia state (p<0.001; Table S9). DEGs between the A. flavus mycelia and sclerotia states are much more abundant compared to A. flavus cultivated at different temperatures (2709) [25], indicating different developmental stages are accompanied by a high level of diversity in gene expression.
To identify genes closely related to the developmental stages, DEGs with change >2-fold were selected for further analysis: 1149 up-regulated genes and 4492 down-regulated genes were detected for the sclerotia state. WEGO illustration showed GO terms of the reproductive cellular process, the reproductive process, sexual reproduction and sporulation contained more up-regulated genes in the sclerotia state compared to the mycelia state ( Fig. 6A ), indicating sclerotia is closely related to A. flavus reproduction instead of being only a sexual vestige. Additionally, the abundance of residual mating process genes in A. flavus suggests it might be capable of sexual development.
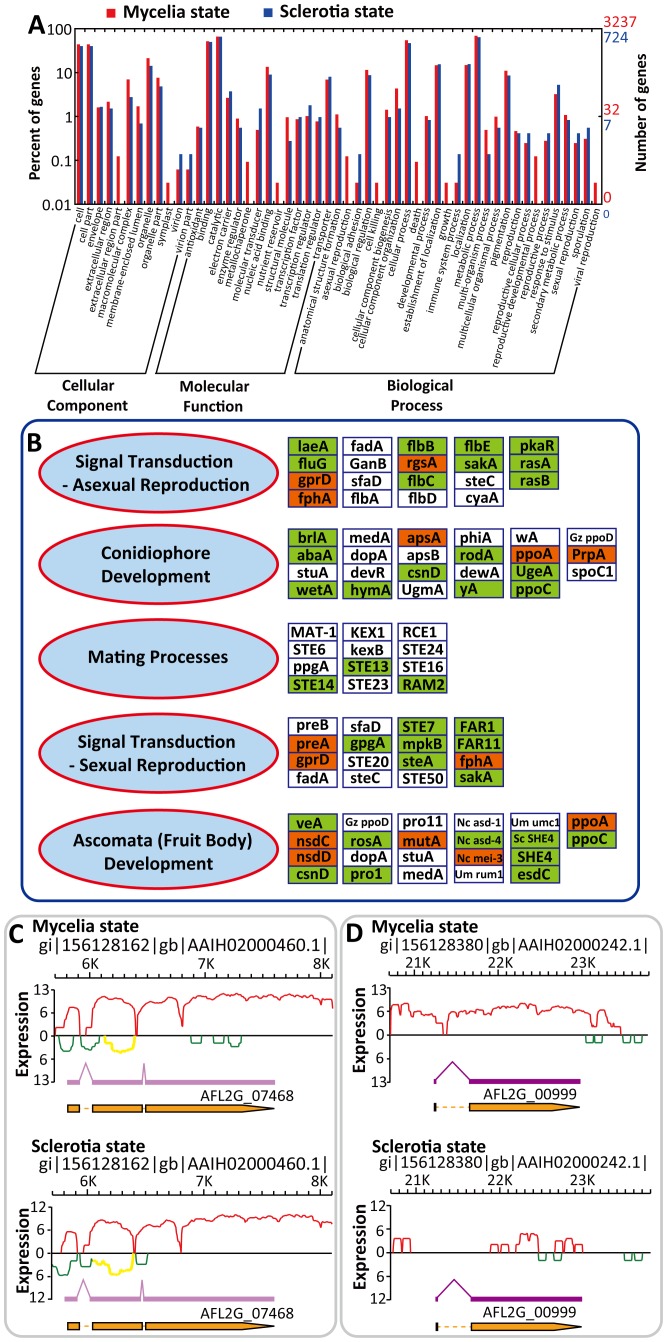
Figure 6. Genes expressed differentially between A. flavus mycelia and sclerotia states.
(A) WEGO illustration of the up-regulated genes in the A. flavus mycelia and sclerotia states. In total, 3237 of the 4492 up-regulated genes in the mycelia state and 724 of 1149 up-regulated genes in the sclerotia state are assigned to GO categories. (B) The model of regulation of A. flavus reproduction-related genes in the mycelia and sclerotia states. Boxes, DEGs (p<0.001 and change >2-fold) between the mycelia and sclerotia states, denoted by the names of their homologs in A. nidulans, S. cerevisiae (Sc) and Neurospora crassa (Nc). Red and green boxes, the up-regulated and the down-regulated genes in the sclerotia state, respectively. White boxes, genes with no significant difference between the mycelia and the sclerotia states. (C) Occurrence of NAT in the veA gene (AFL2G_07468). Yellow curve, NAT in the veA gene. (D) Transcriptional status of the conidial transcriptional factor brlA (AFL2G_00999).
DEGs related to reproduction are shown in Fig. 6B , with 14 up-regulated genes and 37 down-regulated genes in the sclerotia state. The zinc finger protein-encoding gene brlA (AFL2G_00999) is the primary activator of asexual conidiation reproduction [77], [78]. Down-regulated brlA in the sclerotia state ( Fig. 6D ), together with a series of conidiation-related genes (abaA, wetA, flbB, flbC and flbE; Fig. 6B ), suggests conidial development is repressed during sclerotia development. These findings might contribute to understanding the sexual and asexual balance of A. flavus.
Secondary Metabolism
The secondary metabolic (SM) pathways in fungi consist of many genes encoding polyketide synthetases (PKS), fatty acid synthases, dehydrogenases, reductases, oxidoreductases, epoxide hydrolases, cytochrome P450 monooxygenases and methyltransferases [1]. However, it is difficult to determine whether such genes are involved in SM pathways, because vast numbers of genes are categorized as belonging to the gene clusters mentioned above. On the basis of the web tool SMURF [9], 55 putative SM pathways were identified in A. flavus, including 22 PKS and 27 non-ribosomal peptide synthetase (NRPS) pathways. It is important to study secondary metabolism in A. flavus because it was reported that secondary metabolism was often related to sporulation and sclerotia development [2], [76]. In our RNA-seq data, backbones of 38 SM pathways were transcribed in both the mycelia and sclerotia cultures (Table S10), including the aflatoxin (AF) biosynthetic pathway (cluster #54).
The AF biosynthesis pathway is the best studied SM pathway in A. flavus, containing at least 23 enzymatic reactions and 29 genes in a 75 kb cluster on chromosome III (Table S10) [79]. Only eight transcribed genes in the AF pathway were scored by microarray technology [1]. Our RNA-seq data provided precise information about AF biosynthesis. The transcriptional factor aflR (AFL2G_07224) was transcribed under both the mycelia and sclerotia conditions and the NAT of aflR was transcribed under the sclerotia condition, suggesting AF biosynthesis might be down-regulated by NAT-mediated RNAi during sclerotia development (Table S10). This is consistent with the fact that the transcripts of all AF pathway genes were detected in the mycelia state but three genes (aflD, aflCa and aflP) were not detected in the sclerotia state (Table S10). Additionally, changes in expression of the global SM transcriptional factor veA (AFL2G_07468) during sclerotia development suggest it has roles in linking sclerotia development with secondary metabolism ( Fig. 6C and Table S8) [80]. These findings are in agreement with the transcriptional profile of AF biosynthesis ( Fig. 7A ) and the down-regulation of most AF biosynthetic structural genes in the sclerotia state ( Fig. 7B ).
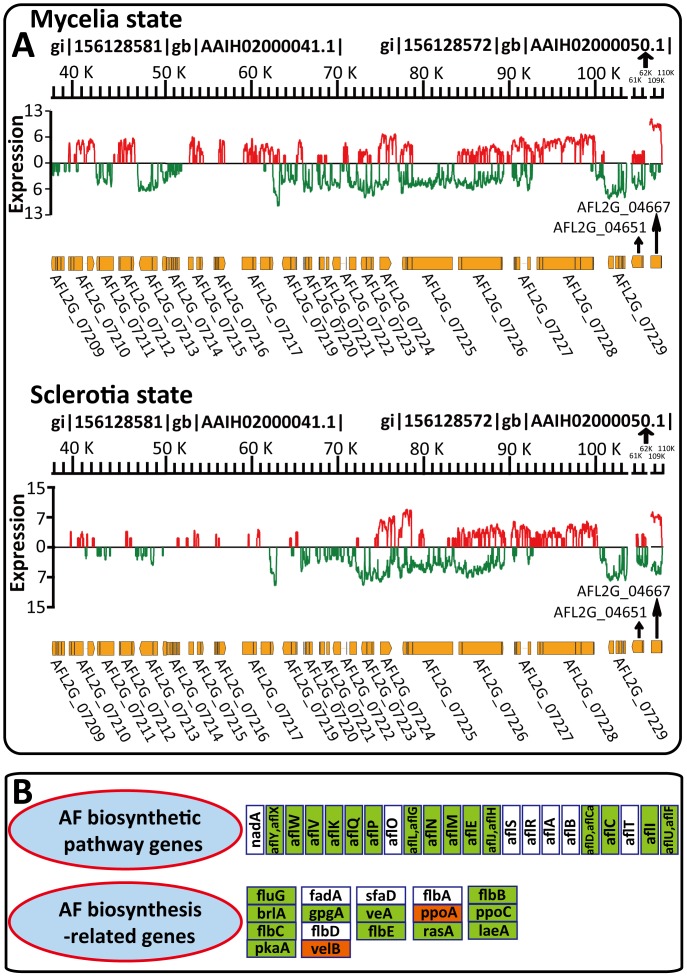
Figure 7. Differential transcription of A. flavus aflatoxin biosynthetic pathway.
(A) The expression status of AF biosynthesis in the mycelia and sclerotia states. (B) The regulation model of AF biosynthesis-related genes in the mycelia and sclerotia states. The orthologs of the AF biosynthetic pathway genes are in the same order as in (A). Boxes, DEGs (p<0.001 and change >2-fold) between mycelia and sclerotia states, denoted by the names of their homologs in A. nidulans. Red and green boxes, up-regulated and down-regulated genes in the sclerotia state, respectively. White boxes, genes with no significant change between the mycelia and sclerotia states.
Genes participating in the AF metabolic pathway contain biosynthesis genes, signal transduction genes, regulatory genes and genes involved in the stress response [2]. Although most of the AF structural genes are in a single cluster, this cluster might be regulated by genes spread throughout the A. flavus genome [81]. Among the AF-related genes given in Table S10, 11 were down-regulated in the sclerotia state and two were up-regulated ( Fig. 7B ). According to our analysis of sclerotia development (Table S8 and Fig. 6B ), these AF genes were also related to sclerotia development, suggesting they link sclerotia development and aflatoxin biosynthesis. Therefore, high throughput RNA-seq data brought new insights into A. flavus mycotoxin production and other secondary metabolism pathways in this fungus. AF contamination of crops is a heavy economic burden on farmers and it is important, therefore, that the target genes identified by RNA-seq data can be used in the biocontrol of aflatoxigenic strains by genetic manipulation.
Conclusions
The A. flavus transcriptome has been studied intensively by several research groups in recent years. Despite these efforts, however, the mechanism underlying the regulation of A. flavus physiology is unknown. Our data profiled the A. flavus transcriptome on the genomic scale and annotated transcript structures precisely. UTR annotation revealed A. flavus might have a much higher RNA turnover rate compared to A. oryzae or S. pombe. A. flavus might recognize the splicing sites and produce AS events mainly by the intron definition (ID) mechanism owing to the high RI/CE ratio (34.75). Among the novel transcripts we were able to identify, the vast majority of transcribed ncRNAs might act as NATs to regulate gene expression at the post-transcriptional level. The transcriptional activity of A. flavus genes with NATs was much higher compared to genes without NATs, suggesting NATs are true transcripts rather than artifacts of leaky bidirectional transcription. Our analysis is the first global investigation of NAT function in A. flavus. As for DEGs, it is quite likely that the 14 up-regulated and 37 down-regulated reproduction-related genes in the sclerotia state link A. flavus reproduction and sclerotia development. In our ssRNA-seq data, the backbones of 38 SM pathways were transcribed in both mycelia and sclerotia states, and we identified genes linking sclerotia development and aflatoxin biosynthesis. Our data could be used to develop strategies to control aflatoxin synthesis by aflatoxigenic strains. Therefore, ssRNA-seq data provided in this study could expand our understanding of A. flavus.
Supporting Information
Global transcriptional profile of A. flavus CA43 in the mycelia state, denoted by log2-transformed reads count. Coverage, the percentage of the genomic region covered by ssRNA-seq reads with a window size of 5 kb. Gene, the number of A. flavus genes within a window size of 5 kb. The chromosome number is shown at the left.
(TIF)
GO functional enrichment analysis of A. flavus uORF-containing genes. The abscissa is the percentage of genes in each GO term. The ordinate is in GO terms.
(TIF)
GO functional enrichment analysis of A. flavus genes with NATs. The abscissa is the percentage of genes in each GO term. The ordinate is in GO terms.
(TIF)
Mapping summary of ssRNA-seq reads.
(XLS)
Expression level of A. flavus genes and non-redundant ESTs at different development stages revealed by ssRNA-seq.
(XLS)
The transcription status of transcriptional factors in A. flavus CA43 and A. oryzae RIB40.
(XLS)
Novel transcripts in A. flavus CA43.
(XLS)
UTRs and uORFs identified for A. flavus annotated genes.
(XLS)
Alternative splicing events in A. flavus CA43 mycelia state.
(XLS)
Antisense transcripts of A. flavus in the mycelia and sclerotia states.
(XLS)
Genes related to asexual and sexual reproduction in A. flavus.
(XLS)
Genes expressed differentially between A. flavus mycelia and sclerotia states.
(XLS)
Transcriptional status of A. flavus secondary metabolism pathways.
(XLS)
Acknowledgments
We are grateful to Dr Perng-Kuang Chang (Southern Regional Research Center, Agricultural Research Service, U.S. Department of Agriculture, Washington, DC) for providing A. flavus strains and for helpful suggestions concerning experimental design.
Funding Statement
The authors wish to acknowledge the financial support by National Natural Science Foundation of China (Project 31100026), the 863 (Hi-tech research and development program of China) program (contract NO. 2012AA022108), Guangdong Provincial Department of Science and Technology Research Project (NO. 2012B010900028 and 2012A080800013). Part of this work was supported by Guangdong Provincial Natural Science Foundation of China (No. S2011040003020) and the Fundamental Research Funds for the Central Universities (NO. 2012ZM0080). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yu J, Cleveland TE, Wilkinson JR, Campbell BC, Kim JH, et al. (2006) Aspergillus flavus expressed sequence tags and microarray as tools in understanding aflatoxin biosynthesis. Mycotoxin Res 22: 16–21. [DOI] [PubMed] [Google Scholar]
- 2. Yu J, Whitelaw CA, Nierman WC, Bhatnagar D, Cleveland TE (2004) Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiol Lett 237: 333–340. [DOI] [PubMed] [Google Scholar]
- 3. Chang PK, Wilkinson JR, Horn BW, Yu J, Bhatnagar D, et al. (2007) Genes differentially expressed by Aspergillus flavus strains after loss of aflatoxin production by serial transfers. Appl Microbiol Biotechnol 77: 917–925. [DOI] [PubMed] [Google Scholar]
- 4. Yu J, Fedorova ND, Montalbano BG, Bhatnagar D, Cleveland TE, et al. (2011) Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol Lett 322: 145–149. [DOI] [PubMed] [Google Scholar]
- 5. Wilkinson JR, Yu J, Bland JM, Nierman WC, Bhatnagar D, et al. (2007) Amino acid supplementation reveals differential regulation of aflatoxin biosynthesis in Aspergillus flavus NRRL 3357 and Aspergillus parasiticus SRRC 143. Appl Microbiol Biotechnol 74: 1308–1319. [DOI] [PubMed] [Google Scholar]
- 6. Chang PK, Bennett JW, Cotty PJ (2002) Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus . Mycopathologia 153: 41–48. [DOI] [PubMed] [Google Scholar]
- 7. Cotty PJ (1988) Aflatoxin and sclerotical production by Aspergillus flavus: influence of pH. Phytopathology 78: 1250–1253. [Google Scholar]
- 8. Geiser DM, Timberlake WE, Arnold ML (1996) Loss of meiosis in Aspergillus . Mol Biol Evol 13: 809–817. [DOI] [PubMed] [Google Scholar]
- 9. Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, et al. (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47: 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, et al. (2006) Whole genome comparison of Aspergillus flavus and A. oryzae . Med Mycol 44: 9–11. [DOI] [PubMed] [Google Scholar]
- 11. Horn BW, Dorner JW (1999) Regional differences in production of aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Appl Environ Microbiol 65: 1444–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotty P (1989) Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79: 808–814. [Google Scholar]
- 13. Bayman P, Cotty PJ (1993) Genetic diversity in Aspergillus flavus: association with aflatoxin production and morphology. Can J Bot 71: 23–31. [Google Scholar]
- 14. Bennett J, Fernholz F, Lee L (1978) Effect of light on aflatoxins, anthraquinones, and sclerotia in Aspergillus flavus and A. parasiticus . Mycologia 70: 104–116. [PubMed] [Google Scholar]
- 15. Littley E, Rahe J (1992) Sclerotial morphogenesis in Sclerotium cepivorum in vitro. Can J Bot 70: 772–778. [Google Scholar]
- 16. Willetts H, Bullock S (1992) Developmental biology of sclerotia. Mycol Res 96: 801–816. [Google Scholar]
- 17. Rollins JA, Dickman MB (1998) Increase in endogenous and exogenous cyclic AMP levels inhibits sclerotial development in Sclerotinia sclerotiorum . Appl Environ Microbiol 64: 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Georgiou CD, Patsoukis N, Papapostolou I, Zervoudakis G (2006) Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integr Comp Biol 46: 691–712. [DOI] [PubMed] [Google Scholar]
- 19. Chang P-K, Hua S (2007) Molasses supplementation promotes conidiation but suppresses aflatoxin production by small sclerotial Aspergillus flavus . Lett Appl Microbiol 44: 131–137. [DOI] [PubMed] [Google Scholar]
- 20. Calvo AM, Bok J, Brooks W, Keller NP (2004) veA is required for toxin and sclerotial production in Aspergillus parasiticus . Appl Environ Microbiol 70: 4733–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amaike S, Keller NP (2009) Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus . Eukaryot Cell 8: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cary J, OBrian G, Nielsen D, Nierman W, Harris-Coward P, et al. (2007) Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl Microbiol Biotechnol 76: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 23. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566. [DOI] [PubMed] [Google Scholar]
- 24. Zhoug S, Wen SM, Zhang DF, Wang QL, Wang SQ, et al. (1998) Sequencing of PCR amplified HBV DNA pre-c and c regions in the 2.2.15 cells and antiviral action by targeted antisense oligonucleotide directed against sequence. World J Gastroenterol 4: 434–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith CA, Robertson D, Yates B, Nielsen DM, Brown D, et al. (2008) The effect of temperature on Natural Antisense Transcript (NAT) expression in Aspergillus flavus . Curr Genet 54: 241–269. [DOI] [PubMed] [Google Scholar]
- 26. Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 28. Donaldson ME, Saville BJ (2012) Natural antisense transcripts in fungi. Mol Microbiol 85: 405–417. [DOI] [PubMed] [Google Scholar]
- 29. Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35. [DOI] [PubMed] [Google Scholar]
- 30. Kawasaki H, Taira K (2004) Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211–217. [DOI] [PubMed] [Google Scholar]
- 31. Morris KV, Chan SW, Jacobsen SE, Looney DJ (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292. [DOI] [PubMed] [Google Scholar]
- 32. Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, et al. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6: 784–791. [DOI] [PubMed] [Google Scholar]
- 33. Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, et al. (2004) Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun 322: 593–600. [DOI] [PubMed] [Google Scholar]
- 34. Murrell A, Heeson S, Reik W (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893. [DOI] [PubMed] [Google Scholar]
- 35. Andersen AA, Panning B (2003) Epigenetic gene regulation by noncoding RNAs. Curr Opin Cell Biol 15: 281–289. [DOI] [PubMed] [Google Scholar]
- 36. Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, et al. (2002) Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol 3: RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen N, Stein LD (2006) Conservation and functional significance of gene topology in the genome of Caenorhabditis elegans . Genome Res 16: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, et al. (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana . Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J, Sun M, Kent WJ, Huang X, Xie H, et al. (2004) Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- 42.Yu J, Nierman WC, Fedorova ND, Bhatnagar D, Cleveland TE, et al. (2011) What can the Aspergillus flavus genome offer to mycotoxin research. Mycology 2 218–236.
- 43. Wang B, Guo G, Wang C, Lin Y, Wang X, et al. (2010) Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res 38: 5075–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, et al. (2011) High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949. [DOI] [PubMed] [Google Scholar]
- 45. Mamanova L, Turner DJ (2011) Low-bias, strand-specific transcriptome Illumina sequencing by on-flowcell reverse transcription (FRT-seq). Nat Protoc 6: 1736–1747. [DOI] [PubMed] [Google Scholar]
- 46. Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, et al. (2010) Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol 11: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vivancos AP, Guell M, Dohm JC, Serrano L, Himmelbauer H (2010) Strand-specific deep sequencing of the transcriptome. Genome Res 20: 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang L, Si Y, Dedow LK, Shao Y, Liu P, et al. (2011) A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One 6: e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, et al. (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 51. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 52. Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, et al. (2012) A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. doi: 10.1093/bib/bbs046 [DOI] [PubMed] [Google Scholar]
- 53. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 54. Ye J, Fang L, Zheng H, Zhang Y, Chen J, et al. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34: W293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kent WJ (2002) BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yavatkar AS, Lin Y, Ross J, Fann Y, Brody T, et al. (2008) Rapid detection and curation of conserved DNA via enhanced-BLAT and EvoPrinterHD analysis. BMC Genomics 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, et al. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang L, Feng Z, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
- 60. Arentshorst M, Ram AF, Meyer V (2012) Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Methods Mol Biol 835: 133–150. [DOI] [PubMed] [Google Scholar]
- 61. Machida M, Yamada O, Gomi K (2008) Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res 15: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3: REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, et al. (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- 64. Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, et al. (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ruiz-Echevarria MJ, Peltz SW (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101: 741–751. [DOI] [PubMed] [Google Scholar]
- 66. Vilela C, McCarthy JE (2003) Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol Microbiol 49: 859–867. [DOI] [PubMed] [Google Scholar]
- 67. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liang S, Wang B, Pan L, Ye Y, He M, et al. (2012) Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics 13: 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McGuire AM, Pearson MD, Neafsey DE, Galagan JE (2008) Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol 9: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 71. Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sibthorp C, Wu H, Cowley G, Wong PW, Palaima P, et al. (2013) Transcriptome analysis of the filamentous fungus Aspergillus nidulans directed to the global identification of promoters. BMC Genomics 14: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, et al. (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. [DOI] [PubMed] [Google Scholar]
- 74. Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231. [DOI] [PubMed] [Google Scholar]
- 75. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, et al. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- 76. Jorgensen TR, Nielsen KF, Arentshorst M, Park J, van den Hondel CA, et al. (2011) Submerged conidiation and product formation by Aspergillus niger at low specific growth rates are affected in aerial developmental mutants. Appl Environ Microbiol 77: 5270–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yamada O, Lee BR, Gomi K, Iimura Y (1999) Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J Biosci Bioeng 87: 424–429. [DOI] [PubMed] [Google Scholar]
- 78. Wieser J, Adams TH (1995) flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev 9: 491–502. [DOI] [PubMed] [Google Scholar]
- 79. Yu J, Cleveland TE, Nierman WC, Bennett JW (2005) Aspergillus flavus genomics: gateway to human and animal health, food safety, and crop resistance to diseases. Rev Iberoam Micol 22: 194–202. [DOI] [PubMed] [Google Scholar]
- 80. Calvo AM (2008) The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol 45: 1053–1061. [DOI] [PubMed] [Google Scholar]
- 81. Georgianna DR, Payne GA (2009) Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet Biol 46: 113–125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global transcriptional profile of A. flavus CA43 in the mycelia state, denoted by log2-transformed reads count. Coverage, the percentage of the genomic region covered by ssRNA-seq reads with a window size of 5 kb. Gene, the number of A. flavus genes within a window size of 5 kb. The chromosome number is shown at the left.
(TIF)
GO functional enrichment analysis of A. flavus uORF-containing genes. The abscissa is the percentage of genes in each GO term. The ordinate is in GO terms.
(TIF)
GO functional enrichment analysis of A. flavus genes with NATs. The abscissa is the percentage of genes in each GO term. The ordinate is in GO terms.
(TIF)
Mapping summary of ssRNA-seq reads.
(XLS)
Expression level of A. flavus genes and non-redundant ESTs at different development stages revealed by ssRNA-seq.
(XLS)
The transcription status of transcriptional factors in A. flavus CA43 and A. oryzae RIB40.
(XLS)
Novel transcripts in A. flavus CA43.
(XLS)
UTRs and uORFs identified for A. flavus annotated genes.
(XLS)
Alternative splicing events in A. flavus CA43 mycelia state.
(XLS)
Antisense transcripts of A. flavus in the mycelia and sclerotia states.
(XLS)
Genes related to asexual and sexual reproduction in A. flavus.
(XLS)
Genes expressed differentially between A. flavus mycelia and sclerotia states.
(XLS)
Transcriptional status of A. flavus secondary metabolism pathways.
(XLS)