SUMMARY
Group 2 innate lymphoid cells (ILC2) are innate lymphocytes that confer protective type 2 immunity during helminth infection and are also involved in allergic airway inflammation. Here we report that ILC2 development required T cell factor 1 (TCF-1, the product of the Tcf7 gene), a transcription factor also implicated in T cell lineage specification. Tcf7−/− mice lack ILC2, and were unable to mount ILC2-mediated innate type 2 immune responses. Forced expression of TCF-1 in bone marrow progenitors partially bypassed the requirement for Notch signaling in the generation of ILC2 in vivo. TCF-1 acted through both GATA-3-dependent and GATA-3-independent pathways to promote the generation of ILC2. These results are reminiscent of the critical roles of TCF-1 in early T cell development. Hence, transcription factors that underlie early steps of T cell development are also implicated in the development of innate lymphoid cells.
INTRODUCTION
Recent studies have identified innate lymphocytes that lack antigen receptors but functionally match subsets of T helper cells (Spits and Cupedo, 2012). Group 2 innate lymphoid cells (ILC2) produce T helper type 2 (Th2) cell-associated cytokines interleukin-5 (IL-5) and IL-13 and mediate innate type 2 immunity at airway and gut mucosa (Barlow et al., 2012; Chang et al., 2011; Halim et al., 2012a; Mjösberg et al., 2011; Monticelli et al., 2011; Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Wilhelm et al., 2011). ILC2 provide protective immunity to helminth infection because they are the predominant early source of type 2 cytokines (Moro et al., 2010; Neill et al., 2010; Price et al., 2010). ILC2 have also been implicated in allergic airway inflammation and in lung epithelial repair (Barlow et al., 2012; Chang et al., 2011; Halim et al., 2012a; Monticelli et al., 2011; Wilhelm et al., 2011). The development of ILC2 requires the transcription factors Id2, RORα, and GATA-3 (Halim et al., 2012b; Hoyler et al., 2012; Moro et al., 2010; Wong et al., 2012). However, other transcription factors implicated in the generation and function of ILC2 remain to be identified.
ILC2 share many similarities with T cells. ILC2 derive from lymphoid progenitors and phenotypically resemble double-negative 3 (DN3) cells that are committed to the T cell lineage (Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Wong et al., 2012; Yang et al., 2011). They also produce Th2 cell-associated cytokines and express genes characteristic of T cells (Moro et al., 2010; Wong et al., 2012). However, they lack the antigen-specific T cell receptor (TCR) or pre-TCR and develop normally in athymic mice (Moro et al., 2010). Nevertheless, the similarities between T cells and ILC2 suggest that similar transcription factors may underlie the development of these two lineages.
Here, we report that the development of ILC2 required TCF-1, a transcription factor earlier shown to be important for T cell-fate specification (Germar et al., 2011; Okamura et al., 1998; Verbeek et al., 1995; Weber et al., 2011). Tcf7−/− mice lacked ILC2 at many anatomic sites and were compromised in immune responses that required ILC2, including responses to the nematode Nippostrongylus brasiliensis and protease allergen. We found that upstream of TCF-1, Notch signaling is required for the efficient generation of ILC2 in vivo. We further show that forced expression of TCF-1 in hematopoietic progenitors partially bypassed the requirement for Notch signaling in ILC2 development. Forced expression of TCF-1 also resulted in upregulated expression of ILC2 cytokine receptors through both GATA-3-dependent and GATA-3-independent mechanisms. Thus, transcriptional regulatory events that underlie early T cell development also direct the generation of ILC2, suggesting a close developmental relationship between T cells and innate lymphoid cells.
RESULTS
ILC2 Express T Lineage Genes
During early T cell development, bone marrow lymphoid progenitors home to the thymus, gradually lose alternative lineage potentials and express T cell genes (Love and Bhandoola, 2011). Once progenitors enter the thymus, T cell-fate specification and commitment are initiated by intrathymic Notch signals that upregulate expression of transcription factors such as TCF-1 and GATA-3 (Rothenberg, 2012). Interestingly, many phenotypic and gene expression similarities between ILC2 and T cells have been noted (Moro et al., 2010; Spits and Cupedo, 2012; Wong et al., 2012). For example, ILC2 from mesenteric fat-associated lymphoid clusters (FALC) express genes characteristic of T cells and T cell progenitors such as Gata3 and Lck (Moro et al., 2010). Also, ILC2 generated in vitro with OP9-DL1 coculture express Tcf7 (the gene for TCF-1) and Bcl11b (Wong et al., 2012). Therefore, we compared the amount of key T lineage transcription factors and also T cell structural genes in ILC2, DN3 cells, and bone marrow multipotent progenitors. For these experiments, we used ILC2 obtained from the lungs of Rag1−/− mice to eliminate potential contamination with T cells. We confirmed the expression of Tcf7, Gata3, Bcl11b, and Lck by ILC2 (see Figure S1 available online). We found that lung ILC2 also expressed many other genes that are highly expressed by T-lineage-committed DN3 progenitors. These molecules included the Notch target genes Hes1 and Dtx1 and the TCR signaling molecule Lat (Figure S1). ILC2 and DN3 cells also expressed comparable amounts of several cytokine receptors, including Il7ra, Il17rb, and Il2ra. These results suggest that the transcription factors implicated in T cell-fate specification might also be involved in the generation or function of ILC2.
ILC2 Development Requires TCF-1
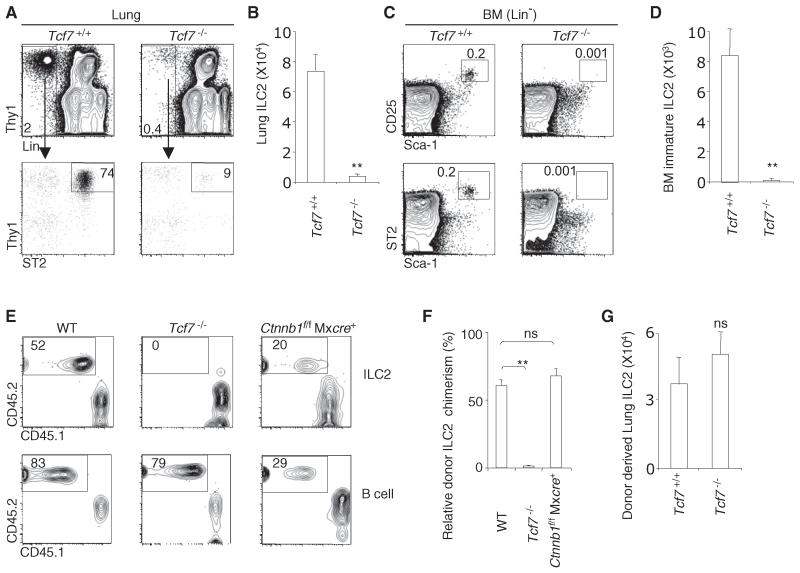
TCF-1, a T cell-specific high-mobility group (HMG) box transcription factor, is necessary for normal T lineage specification (Germar et al., 2011; Okamura et al., 1998; Verbeek et al., 1995; Weber et al., 2011). Forced expression of TCF-1 in bone marrow multipotent progenitors upregulates the expression of many T lineage genes and cytokine receptors that are also expressed by ILC2 (Weber et al., 2011). We thus asked whether TCF-1 is required for ILC2 cell development. The number of ILC2 in the lungs of Tcf7−/− mice was reduced to approximately 5% of the number in wild-type (WT) mice (Figures 1A and 1B). The recently described bone marrow immature ILC2 (Halim et al., 2012b; Hoyler et al., 2012) were nearly absent in Tcf7−/− mice (Figures 1C and 1D). These data indicate that TCF-1 is required for the generation of ILC2.
Figure 1. TCF-1 Is Required for ILC2 Development.
(A) Flow plots show a representative analysis of ILC2 in the lungs of Tcf7+/+ and Tcf7−/− mice. Plots were gated on CD45+ lung cells.
(B) Bar graph shows cellularity of lung ILC2 in Tcf7+/+ and Tcf7−/− mice. Data are from eight mice per group (error bars are SEM).
(C) Flow plots show a representative analysis of bone marrow immature ILC2 in Tcf7+/+ and Tcf7−/− mice. Plots were gated on Lin− cells.
(D) Bar graph shows cellularity of bone marrow immature ILC2. Data are from three or four mice per group (error bars are SEM).
(E) We mixed 5,000 LSK cells from WT, Tcf7−/− and poly(I:C)-treated Ctnnb1(encoding β-catenin)f/f Mxcre mice (CD45.2) with 2,500 WT competitor LSK cells (CD45.1) and together injected them into lethally irradiated (950 rads) WT recipients (CD45.1). Reconstitution of lung ILC2 and splenic B cells was examined at 4 weeks posttransplant. Deletion of Ctnnb1 in LSK cells was confirmed by PCR (data not shown).
(F) Bar graph shows relative ILC chimerism normalized to splenic B cells. Results are from two independent experiments; five mice per group (error bars are SEM).
(G) We intravenously transferred 5,000 LSK cells from WT mice (CD45.1) into lethally irradiated Tcf7+/+ and Tcf7−/− mice (CD45.2). Development of lung ILC2 was examined at 4 weeks posttransplant. Data are from two independent experiments; three mice per group (error bars are SEM; ns, not significant). **p < 0.05. See also Figure S1.
We next investigated whether the ILC2 developmental defect in Tcf7−/− mice was intrinsic to hematopoietic cells. Tcf7−/− hematopoietic progenitors failed to give rise to ILC2 in irradiated WT mice (Figures 1E and 1F), whereas WT bone marrow progenitors developed normally into ILC2 in irradiated Tcf7−/− recipient mice (Figure 1G). TCF-1 can function with β-catenin to mediate canonical Wnt signaling; however, TCF-1 drives early T cell development independently of β-catenin (Cobas et al., 2004; Jeannet et al., 2008; Weber et al., 2011). Similarly, we found that β-catenin was dispensable for the generation of ILC2 (Figures 1E and 1F). Thus, as in early T cell development, TCF-1 is required for ILC2 development in a hematopoietic cell-intrinsic and β-catenin-independent manner.
The innate lymphoid cell family includes at least three subsets: ILC2, natural killer (NK) cells, and RORγt+ innate lymphoid cells (Spits and Cupedo, 2012). TCF-1 was previously implicated in NK cell development (Held et al., 2003). We found that the number of RORγt+ innate lymphoid cells in Tcf7−/− mice was also reduced, to 20%–50% of the number in WT mice (data not shown). Thus, TCF-1 is involved in development of several innate lymphocyte subsets. However, because TCF-1 appeared most strictly required in the generation of ILC2, we focused on understanding the role of TCF-1 in ILC2 development in the present study.
ILC2-Mediated Immune Responses Are Compromised in Tcf7−/− Mice
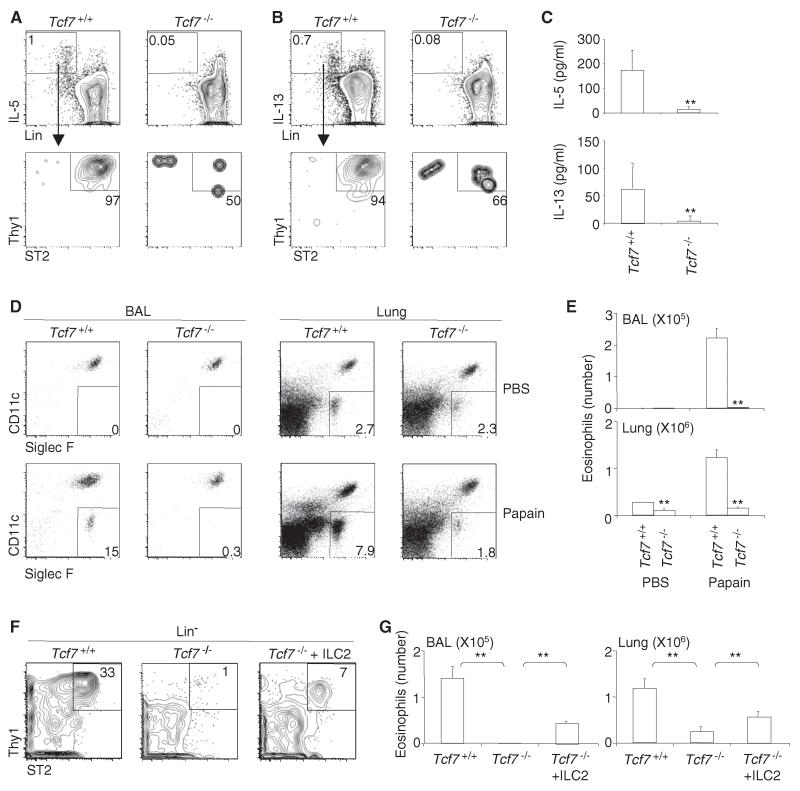
We wished to establish whether immune responses that require ILC2 are compromised in Tcf7−/− mice. Recent studies suggest a critical role for ILC2 in mediating protease-induced airway inflammation (Halim et al., 2012a; Oboki et al., 2010; Wilhelm et al., 2011). We examined the responses of Tcf7−/− and control WT mice to intranasal challenge with papain, which has been shown to induce rapid IL-5 production and eosinophilic infiltration in the airway (Halim et al., 2012a; Oboki et al., 2010; Wilhelm et al., 2011). Consistent with previous reports (Halim et al., 2012a; Oboki et al., 2010), intranasal challenge with papain induced eosinophilic infiltration in the bronchoalveolar lavage (BAL) and lungs of WT mice at 60 hr, and ILC2 produced IL-5 and IL-13 in the lung (Figures 2A and 2B). Tcf7−/− mice, however, had very few IL-5 and IL-13 producing ILC2 (Figures 2A and 2B). The few remaining Tcf7−/− ILC2 were ineffective in producing IL-5 and IL-13 (Figures S2A and S2B). The amounts of BAL IL-5 and IL-13 were significantly reduced in Tcf7−/− mice (Figure 2C), and airway eosinophilic infiltration was not observed (Figures 2D and 2E). TCF-1 may also promote the differentiation of Th2 cells and NK T cells (Ohteki et al., 1996; Yu et al., 2009), which are capable of producing Th2 cell-associated cytokines (Stock et al., 2009; Terashima et al., 2008). However, eosinophilic infiltration was observed in papain-challenged Rag2−/− mice lacking NKT cells and T cells (Halim et al., 2012a; Oboki et al., 2010). Thus, defects in TCR-negative cells, such as ILC2, are likely responsible for the complete absence of airway eosinophilic infiltration in Tcf7−/− mice. We next transferred WT ILC2 into Tcf7−/− mice. We transferred 105 WT ILC2 that were expanded in vivo by IL-33 treatment into each Tcf7−/− mouse. This manipulation resulted in partial restoration of ILC2 cell numbers in the lung (Figure 2F). Consistently, transfer of WT ILC2 partially restored eosinophilic infiltration in the BAL and lungs of Tcf7−/− mice in responses to papain challenge (Figure 2G). Together, these data establish that Tcf7−/− mice lack functional ILC2 in the lung.
Figure 2. Tcf7−/− Mice Lack Functional ILC2 in the Lung.
(A and B) Tcf7+/+ and Tcf7−/− mice were intranasally challenged with papain (10 μg in 40 μl of PBS) every 24 hr on day 0, day 1, and day 2. Mice were sacrificed 12 hr after the last challenge. Lung hematopoietic cells were isolated and stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) in the presence of monensin for 3 hr. Cells were stained for ILC2 followed by intracellular staining of IL-5 and IL-13.
(C) IL-5 and IL-13 concentrations in the BAL fluid from papain-treated mice were measured by ELISA. Data are from four mice (error bars are SEM). **p < 0.05.
(D and E) Eosinophils in BAL and lungs were examined by flow cytometry analysis. Flow plots were gated on CD45+ cells. We used the combination of CD11c and Siglec-F to distinguish airway macrophages and eosinophils as described (Dyer et al., 2011). The CD11c−Siglec-F+ cells were further confirmed to be SSChigh cells (data not shown). Data are from three to five mice per group (error bars are SEM).
(F) We intravenously transferred 105 WT ILC2 into Tcf7−/− mice 8 hr before the first challenge with papain. The restoration of lung ILC2 was examined by flow cytometry analysis 12 hr after the final challenge. Flow plots were gated on Lin− cells.
(G) The number of eosinophils in papain challenged Tcf7+/+ mice, Tcf7−/− mice, and Tcf7−/− mice with adoptive transfer of WT ILC2 is shown. Data are from three to four mice per group (error bars are SEM). **p < 0.05. See also Figure S2.
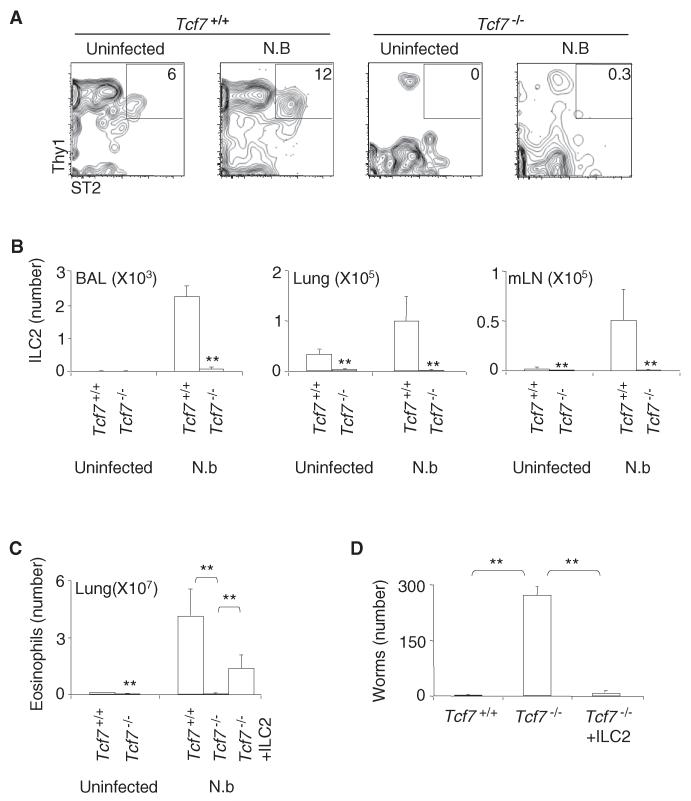
ILC2 are systemically dispersed cells (Price et al., 2010). Previous studies have demonstrated that ILC2 are the predominant early source of type 2 cytokines and can promote protective immunity to helminth infection with Nippostrongylus brasiliensis (Moro et al., 2010; Neill et al., 2010; Price et al., 2010). Therefore, we examined the responses of Tcf7−/− mice to N. brasiliensis infection. N. brasiliensis infection induced the expansion of ILC2 in multiple sites including the BAL, lungs, and mesenteric lymph nodes of WT mice at 10 days postinfection (Figures 3A and 3B). Tcf7−/− mice, however, lacked ILC2 at all of these sites (Figures 3A and 3B). Eosinophilic infiltration was observed in the lungs of WT mice in responses to N. brasiliensis infection, whereas Tcf7−/− mice lacked these responses (Figure 3C). Tcf7−/− mice were also inefficient at expelling worms from the intestine (Figure 3D). Adoptive transfer of WT ILC2 from IL-33 treated mice restored worm clearance and lung eosinophilic infiltration in N. brasiliensis infected Tcf7−/− mice (Figures 3C and 3D). The purpose of IL-33 treatment is to obtain sufficient numbers of ILC2 for adoptive transfer; whether transfer of unexpanded ILC2 from naive mice may confer similar protective immunity to helminth infection in Tcf7−/− mice requires further studies. Nevertheless, these results indicate that Tcf7−/− mice lack functional ILC2 at multiple sites and are unable to establish protective immunity to helminth infection.
Figure 3. Tcf7−/− Mice Lack ILC2-Mediated Immune Responses to Helminth Infection.
Tcf7+/+ and Tcf7−/− mice were infected with 500 L3 stage N. brasiliensis worms by subcutaneous transfer and sacrificed at 10 days after infection.
(A) Flow plots show a representative analysis of ILC2 in the mesenteric lymph nodes (mLN) of TCF-1+/+ and Tcf7−/− mice. Plots were gated on Lin− CD45+ cells.
(B) Bar graphs show the numbers of ILC2 in the BAL, lungs, and mLN. Data are from eight mice per group (error bars are SEM). **p < 0.05.
(C) We transferred 105 WT ILC2 into Tcf7−/− mice at day 0 and again at day 5 after infection. The number of eosinophils in the lungs in Tcf7+/+ mice,Tcf7−/− mice, and Tcf7−/− mice with adoptive transfer of WT ILC2 is shown. Data are from four to six mice per group (error bars are SEM). **p < 0.05.
(D) The number of intestinal worms in N. brasiliensis infected mice was assessed. Data are from four to six mice per group (error bars are SEM). **p < 0.05. See also Figure S3.
A small number of ILC2 remained evident in the lungs of Tcf7−/− mice (Figures 1A and 1B). To understand the development and function of these Tcf7−/− ILC2, we isolated these cells for gene expression analysis. We examined the expression of Lef1, another member of the TCF and LEF transcription factor family. LEF-1 shares structural features with TCF-1 (Hurlstone and Clevers, 2002), and may compensate for TCF-1 deficiency in NK and T cell development (Held et al., 2003; Okamura et al., 1998; Yu et al., 2012). Expression of Lef1 was elevated approximately 4.5-fold in Tcf7−/− ILC2 compared to WT ILC2, suggesting compensatory upregulation (Figure S7A). Tcf7−/− ILC2 expressed Gata3 and II7r, but the amounts were reduced to approximately 30% to 50% of the amounts in WT ILC2 (Figure S3), suggesting that TCF-1 might normally regulate Gata3 and II7r expression in ILC2. WT and Tcf7−/− ILC2 exhibited a comparable cell-cycle profile and Annexin V staining (Figure S7B-S7E). Haploinsufficiency of GATA-3 or overexpression of LEF-1 has been shown to impair the expression of type 2 cytokines, such as Il4 and Il5 (Hebenstreit et al., 2008; Hosoya et al., 2009; Zhu et al., 2004). Indeed, Tcf7−/− ILC2 expressed less mRNA for type 2 cytokines Il5 and Il13 (Figure S3), and the amounts remained lower than those of WT cells in responses to papain challenge (Figure S2). Hence, the few ILC2 that develop in the absence of TCF-1 are functionally compromised.
Notch Promotes ILC2 Development
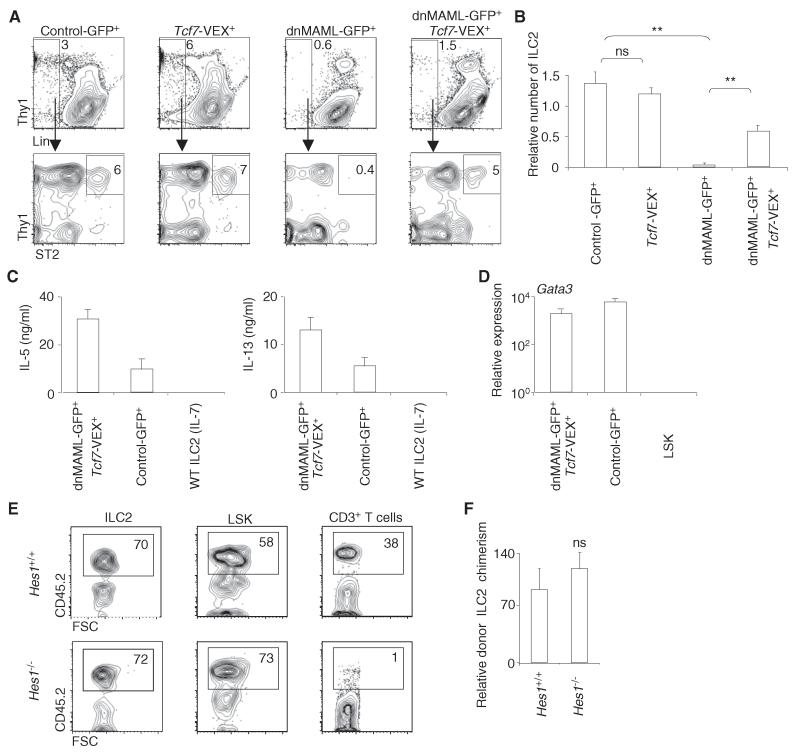
We investigated the upstream signals that elicit TCF-1 expression during ILC2 development. Notch signals directly upregulate TCF-1 expression during early T cell development (Germar et al., 2011; Weber et al., 2011). Notch also promotes the generation of ILC2 in vitro (Wong et al., 2012); however, a role for Notch in ILC2 development in vivo remains to be established. By using retroviral dominant-negative Mastermind like-1 (dnMAML), a pan-Notch inhibitor (Maillard et al., 2006), we confirmed that Notch signaling is required for ILC2 generation in vivo. DnMAML-expressing multipotent bone marrow progenitors (Lin−Sca-1+Kit+ or LSK cells) failed to efficiently give rise to ILC2 in vivo (Figures 4A and 4B). Cotransduction of Tcf7 retrovirus partially restored the generation of ILC2 from dnMAML-expressing LSK cells (Figures 4A and 4B). To determine whether the ILC2 generated from Tcf7 and dnMAML cotransduced progenitors were functional, we cultured them with the cytokines IL-2, IL-7, and IL-33, which were reported to induce the production of type-2 cytokines by ILC2 (Moro et al., 2010). These cells produced IL-5 and IL-13 (Figure 4C) and expressed Gata3 (Figure 4D), suggesting they were functional ILC2. Together, these data indicate that TCF-1 acts downstream of Notch signaling during ILC2 development.
Figure 4. Notch Promotes ILC2 Development.
(A and B) Bone marrow LSK cells were transduced with control GFP, dnMAML-GFP, Tcf7-VEX, or cotransduced with dnMAML-GFP and Tcf7-VEX retrovirus. We injected 105 cells into lethally irradiated (950 rads) recipients. Generation of lung ILC2 was examined at three weeks posttransplant. The number of donor-derived ILC2 was normalized to bone marrow LSK cells. Data are from two independent experiments; three mice per group (error bars are SEM). **p < 0.05; ns, not significant.
(C) An equal number of ILC2 that were derived from dnMAML-GFP and Tcf7-VEX retrovirus cotransduced LSK cells and from control-GFP retrovirus-transduced LSK cells were cultured with 10 ng/ml of IL-2, IL-7, and IL-33 for 12 days. The concentrations of cytokines in the culture supernatant were measured by ELISA. WT ILC2 cultured with IL-7 alone were used as a negative control. Data are from two independent experiments; three mice per group (error bars are SEM).
(D) The cultured ILC2 derived from dnMAML-GFP and Tcf7-VEX retrovirus cotransduced LSK cells and from control-GFP retrovirus transduced LSK cells were collected. The amount of Gata3 mRNA was measured by quantitative reverse-transcription PCR.
(E and F) Hes1+/+ and Hes1−/− fetal liver cells (CD45.2) were mixed with competitor wild-type bone marrow cells (CD45.1) at 1:3 ratio, and together injected into lethally irradiated wild-type recipient mice (CD45.1). The reconstitution of the indicated populations in the lungs of recipient mice was examined at 8 weeks posttransplant. Data represent 2 independent experiments; 3 mice per group (error bars are SEM). ns, not significant. See also Figure S4.
Like Tcf7, Hes1 is another direct Notch target that is critically involved in early T cell development (Tomita et al., 1999). The expression of Hes1 during early T cell development does not require TCF-1 (Weber et al., 2011). Because Hes1 is expressed by ILC2 (Figure S1), we examined whether HES-1 is involved in ILC2 development in vivo. Hes1−/− fetal liver progenitors failed to efficiently develop into T cells as expected, but they differentiated into ILC2 normally in the lungs of irradiated recipients (Figures 4E and 4F). Taken together, these data indicate that Notch signaling regulates ILC2 development through TCF-1 dependent but HES-1 independent pathways.
TCF-1 Promotes ILC2 Development through GATA-3-Dependent and GATA-3-Independent Pathways
We investigated the mechanisms through which TCF-1 promotes ILC2 development. TCF-1 promotes the survival of thymocytes by maintaining the expression of Bclxl (Goux et al., 2005); however, ectopic expression of BclXL or Bcl2 does not rescue the early T cell development defects in Tcf7−/− mice (Goux et al., 2005; Wang et al., 2011; Weber et al., 2011). Similarly, ectopic expression of BclXL did not rescue the generation of ILC2 from Tcf7−/− bone marrow progenitors in vivo (Figure S4), suggesting that TCF-1 is not exclusively required for the survival of ILC2.
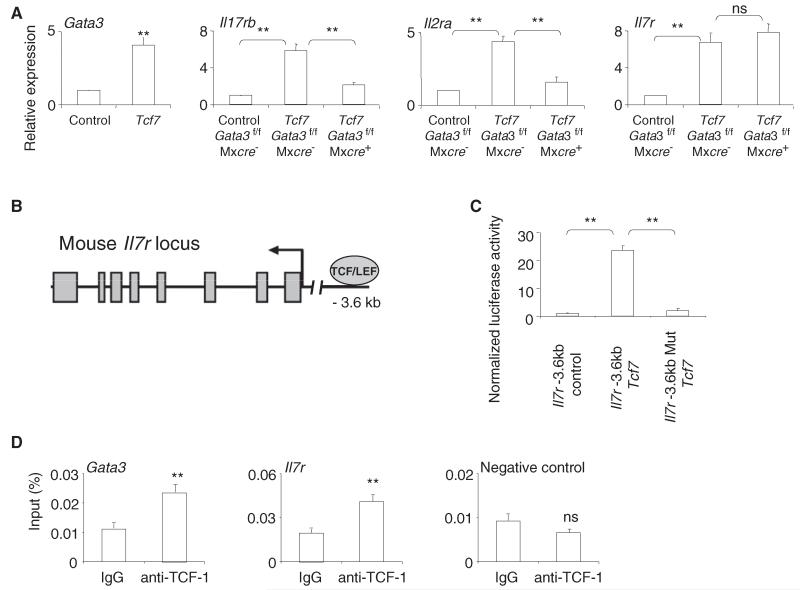
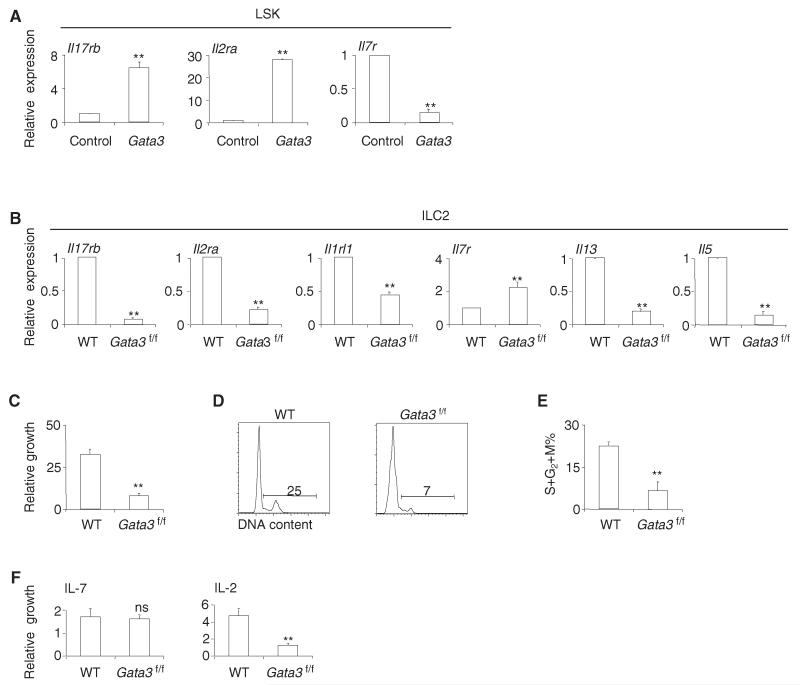
To understand how enforced expression of TCF-1 in bone marrow progenitors can bypass the Notch requirement for ILC2 development, we examined the expression of ILC2 genes in these progenitors. Forced expression of TCF-1 in multipotent bone marrow LSK cells rapidly upregulated the expression of several ILC2 cytokine receptors (Il7r, Il17rb, and Il2ra) as well as transcription factor Gata3 (Figure 5A). These cytokine receptors were previously reported to promote the generation or function of ILC2 (Moro et al., 2010; Neill et al., 2010; Wilhelm et al., 2011). GATA-3 is similarly essential for the generation and maintenance of ILC2 (Hoyler et al., 2012; Liang et al., 2012; Mjösberg et al., 2012). These data indicate that TCF-1 may direct ILC2 development by upregulating Gata3 and important ILC2 cytokine receptor genes.
Figure 5. TCF-1 Directly Regulates Expression of II7r.
(A) Tcf7-VEX or control-VEX retrovirus was transduced into LSK cells from poly (I:C) treated Gata3f/f Mxcre− or Gata3f/f Mxcre+ mice. VEX+ Cells were sorted at 60 hr post-transduction, and the amounts of mRNA were examined by qRT-PCR. Data are from two independent experiments; three or four mice per group (error bars are SEM). **p < 0.05; ns, not significant.
(B) A conversed TCF binding site was identified at a known upstream regulatory region of II7r gene locus.
(C) We cotransfected 293T cells with Tcf7 (or control) retroviral vectors and a luciferase vector containing the −3.6 kb upstream regulatory region of murine II7r gene. TCF-1 (Tcf7) was able to upregulate luciferase reporter activity. The luciferase reporter activity was ablated when the TCF-1 binding site in the −3.6 kb upstream regulatory region was mutated. Data are from two independent experiments. Error bars are SEM from triplicate samples.
(D) ILC2 from lungs of naive mice were expanded in vitro with IL-2, IL-7, and IL-33. ChIP showed that TCF-1 binds to the Gata3-1b promoter and the −3.6 kb upstream regulatory region of II7r locus in ex vivo expanded ILC2. DNA region lacking TCF-1 binding site was used as a negative control. Data are from two independent experiments. Error bars are SEM from triplicate samples. **p < 0.05; ns, not significant. See also Figure S5.
Because ectopic expression of GATA-3 is highly detrimental to the long-term survival and lymphoid-lineage differentiation of hematopoietic progenitors (Taghon et al., 2007), we could not examine the effects of ectopic expression of GATA-3 in vivo. To further understand the mechanisms by which TCF-1 promotes ILC2 development, we examined whether GATA-3 is required for TCF-1 to upregulate the expression of ILC2 genes. We first deleted GATA-3 from LSK cells by polyinosine-polycytidylic acid [poly(I:C)] treatment of Gata3f/f Mxcre+ mice. GATA-3-deficient LSK cells failed to develop into lung ILC2 when transferred into irradiated recipient mice (Figure S5). These results confirmed that GATA-3 is required for ILC2 development in vivo (Hoyler et al., 2012). To test whether GATA-3 is required for TCF-1-mediated ILC2 gene upregulation, we transduced GATA-3 deficient LSK cells with TCF-1 retrovirus. In the absence of GATA-3, ectopic expression of TCF-1 failed to upregulate expression of Il17rb and Il2ra but was sufficient for the upregulation of II7r expression (Figure 5A). Thus, TCF-1 likely upregulates the expression of ILC2 cytokine receptors through both GATA-3-dependent (for Il17rb and Il2ra) and GATA-3-independent (for II7r) mechanisms.
We further investigated the regulation of II7r by TCF-1. Like TCF-1, IL-7Rα is involved in the generation of ILC2, RORγt+ innate lymphoid cells and T cell progenitors (von Freeden-Jeffry et al., 1995; Vonarbourg et al., 2010). We therefore asked whether TCF-1 may directly regulate II7r expression. We identified a conserved TCF-1 binding site at −3.6 kb upstream of the transcription initiation site of II7r (Figure 5B), within a region previously described to be important for the regulation of II7r expression (Lee et al., 2005). We cotransfected 293T cells with Tcf7 retroviral vector and a luciferase reporter vector containing the −3.6 kb upstream regulatory domain. We found that coexpression with Tcf7 increased luciferase reporter activity 25-fold (Figure 5C). Consistently, mutation of the TCF-1 binding site at the −3.6 kb upstream regulatory domain abolished reporter activity (Figure 5C), indicating that TCF-1 may control II7r expression through this binding site. Chromatin immunoprecipitation (ChIP) on ex vivo expanded ILC2 established that TCF-1 binds to this regulatory region in ILC2 (Figure 5D). TCF-1 also binds to a previously reported Gata3-1b regulatory region (Yu et al., 2009) in ILC2 (Figure 5D). These results suggest TCF-1 may directly regulate II7r expression in ILC2.
Because GATA-3 is required for TCF-1-mediated upregulation of Il17rb and Il2ra, we investigated whether GATA-3 may upregulate or maintain the expression of these cytokine receptors during ILC2 development. We ectopically expressed GATA-3 in LSK cells. Because ectopic expression of GATA-3 is detrimental to hematopoietic progenitors (Taghon et al., 2007), we used short-term culture in which we observed minimal death of progenitors. Forced expression of GATA-3 in LSK cells induced the expression of Il2ra and Il17rb, but not II7r (Figure 6A). We next cultured lung ILC2 in vitro with IL-2, IL-33, and IL-7, and deleted Gata3 by using MSCV-GFP-cre retrovirus. We confirmed efficient deletion of Gata3 by using real-time PCR primers that amplified the deleted exons (Figure S6A). Deletion of Gata3 in ILC2 resulted in reduced expression of Il2ra, Il17rb, and to a lesser extent Il1rl1 (encoding IL-33R) (Figure 6B). Expression of II7r, however, was not reduced after Gata3 deletion (Figure 6B). Consistent with previous reports (Hoyler et al., 2012; Liang et al., 2012; Mjösberg et al., 2012), deletion of Gata3 in ILC2 also greatly decreased expression of Il5 and Il13 (Figure 6B). Because we used IL-33 for efficient retroviral transduction of ILC2, our results establish roles for GATA-3 in IL-33 activated ILC2. These results are consistent with the gene expression profile of ILC2 directly isolated from naive Tcf7−/− mice, in which we observed reduced expression of Gata3 as well as Il5 and Il13 (Figure S7A), suggesting that these GATA-3-mediated gene regulatory events are already established in ILC2 of naive mice in vivo. GATA-3 deficient ILC2 grew inefficiently in vitro in the presence of IL-2, IL-7, and IL-33 (Figure 6C). Cell-cycle analysis showed a decreased percentage of actively cycling cells (Figures 6D and 6E). Annexin V staining did not reveal a defect in survival (Figures S6B and S6C). To determine whether the growth defects of Gata3−/− ILC2 were associated with reduced expression of II7r or Il2ra, we next cultured WT and GATA-3-deficient ILC2 in the presence of IL-7 or IL-2 alone. Consistent with the reduced expression of Il2ra but not II7r after GATA-3 deletion, GATA-3-deficient ILC2 grew normally in the presence of IL-7 alone, but they expanded poorly in the presence of IL-2 alone (Figure 6F). These results indicate that GATA-3 upregulates and maintains the expression of Il2ra, but not II7r, during ILC2 development. Together, our data support a model of ILC2 development wherein TCF-1 acts downstream of Notch signaling to control the expression of ILC2 cytokine receptors Il17rb, Il2ra, and II7r via both GATA-3-dependent and GATA-3-independent mechanisms.
Figure 6. GATA-3 Is Required to Maintain the Expression of Cytokine Receptors and Cytokines by ILC2.
(A) Gata3 or empty control retrovirus was transduced into LSK cells from WT mice. GFP+ cells were sorted at 60 hr posttransduction, and the amounts of mRNA were examined by qRT-PCR. Data are from two independent experiments; three or four mice per group (error bars are SEM).
(B) WT and Gata3f/f ILC2 were cultured with the cytokines IL-7, IL-2, and IL-33. Cells were transduced with MSCV-GFP-cre retrovirus. Five days after transduction, GFP+ cells were sorted and the amounts of mRNA for the indicated genes were measured. Data are from two independent experiments; three or four mice per group (error bars are SEM).
(C) WT and Gata3f/f ILC2 were transduced with MSCV-GFP-cre retrovirus. GFP+ cells were sorted at 48 hr posttransduction and grown for a further 7 days in the presence of IL-7, IL-2, and IL-33. The relative growth of ILC2 was measured. Data are from two independent experiments; three mice per group (error bars are SEM).
(D and E) The cell-cycle status of WT and GATA-3-deficient ILC2 was examined by Hoechst 33342 staining. Data are from two independent experiments; three mice per group (error bars are SEM).
(F) WT and Gata3f/f ILC2 were transduced with MSCV-GFP-cre retrovirus. GFP+ cells were sorted at 48 hr posttransduction and grown for a further 7 days with IL-7 alone or IL-2 alone. Growth of ILC2 was measured. Data are from two independent experiments; three or four mice per group (error bars are SEM). **p < 0.05; ns, not significant. See also Figure S6.
DISCUSSION
We have identified a requirement for TCF-1 in the generation of ILC2 and further established roles for Notch and GATA-3 in this process. Our data suggest a striking similarity between early T cell and ILC2 development. Like early T cell development, Notch signaling is required for ILC2 development. Forced expression of TCF-1 in bone marrow progenitors bypasses the requirement for Notch signaling in ILC2 development, and TCF-1 acts through both GATA-3-dependent and GATA-3-independent mechanisms. Hence, the transcriptional regulatory events that specify T cell fate are similarly implicated in the development of some innate lymphoid cells.
During early T cell development, Notch signaling within the thymus upregulates expression of TCF-1 in early thymic progenitors (ETP), and TCF-1 subsequently drives T lineage specification (Germar et al., 2011; Weber et al., 2011). Strikingly, Notch signaling is also required for ILC2 development. A previous study showed that Notch signaling promotes ILC2 generation in vitro (Wong et al., 2012), and the present study establishes that a requirement for Notch signaling in ILC2 development is also evident in vivo. Unlike T cells, ILC2 do not require the thymus for their development (Moro et al., 2010). The anatomic site(s) of ILC2 maturation, and the specific Notch ligands that regulate ILC2 development, are interesting questions for future studies. Notch signaling is also required for the proper development of IL-22 producing innate lymphoid cells in the small intestine lamina propria, but not those in the Peyer’s Patches (Lee et al., 2012). Notch signaling is however dispensable for NK cell development (Lee et al., 2012; Ribeiro et al., 2010). In contrast, TCF-1 promotes efficient generation of both RORγt+ innate lymphoid cells and NK cells at distinct anatomic sites. RORγt+ ILC were reduced in small intestine lamina propria, mesenteric lymph nodes, and spleens of Tcf7−/− mice, and NK cells were reduced in bone marrow and spleens of Tcf7−/− mice (Held et al., 2003; data not shown). These results indicate that the expression of TCF-1 is likely controlled by Notch-independent mechanisms during the development of these innate lymphoid cell subsets. The nature of these mechanisms warrants future studies.
Although ILC2 share many developmental, molecular, and functional similarities with T cells, they are also clearly distinct from T cells. Unlike T cells, ILC2 lack antigen receptors and develop normally in nude mice that only possess a rudimentary thymus (Moro et al., 2010). How ILC2 cell fate is molecularly distinct from T cell fate remains unresolved. Our results indicate that ILC2 develop in a HES-1-independent manner, in contrast to T cells, which require HES-1 downstream of Notch signaling. In addition, like other innate lymphoid cells, ILC2 express high amounts of Id2 and require Id2 for their development (Moro et al., 2010). However, overexpression of Id2 in thymic progenitors is detrimental to T cell development, suggesting unique requirements for E-proteins in these lineages (Morrow et al., 1999). Also, ILC2 development requires the orphan nuclear receptor RORα, but not the thymocytes-specific isoform RORγt (Halim et al., 2012b; Moro et al., 2010; Wong et al., 2012). The overlapping and yet distinct mechanisms underlying the development of T cells versus ILC2, as well as other innate lymphoid cells, require further investigation.
The transcriptional regulators that control both T cell and ILC2 development are reminiscent of similar genes expressed by subsets of lymphocytes in lampreys. Jawless vertebrates lack RAG and TCR genes and use a distinct molecular mechanism to generate diversity among adaptive lymphocyte populations that bear clonally distributed variable lymphocyte receptors (VLR) (Guo et al., 2009). Lamprey lymphocytes include T-like “VLR-A” cells that express genes similar to the T-lineage genes expressed by ILC2 and T cells in mice. Genes expressed by lamprey T-like cells include Gata3, Bcl11b, and Notch1 (Guo et al., 2009); whether orthologs of Tcf7 are expressed by lamprey lymphocytes is unknown. Our study establishes roles for Notch, TCF-1, and GATA-3 in the development of ILC2 in mice. We speculate that transcriptional regulatory programs that establish a T cell-like fate originated prior to RAG, TCR, and VLR during evolution, in innate lymphoid cells. Our results indicate that such regulatory programs continue to underwrite the development of some innate lymphoid cells in addition to T cells in vertebrates.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 (CD45.2) and B6-Ly5.2 (CD45.1) mice were purchased from National Institutes of Health or the Jackson Laboratory. Other mice used include Tcf7−/− mice (Verbeek et al., 1995), Ctnnb1f/f (encoding β-catenin) (Brault et al., 2001), and Gata3f/f mice (Pai et al., 2003). All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee policies at the University of Pennsylvania.
Administration of Poly(I:C)
Gata3f/f Mxcre and Ctnnb1f/f Mxcre mice were treated with 0.2 mg poly(I:C) (Sigma) intraperitoneally (i.p.) every other day for 5 times for in vivo deletion of Gata3 or Ctnnb1. The mice rested for 10 days, and bone marrow LSK cells were sorted by flow cytometry cell sorting.
Potease Allergen Challenge, Helminth Infection, and Adoptive Transfer of ILC2
For protease challenge, 10 μg papain (in 40 μl of PBS) was intranasally administrated into WT or Tcf7−/− mice every 24 hr on day 0, day 1, and day 2. Mice were sacrificed at 12 hr after the last challenge. BAL fluid and lungs were analyzed.
For Nippostrongylus brasiliensis infections, WT or Tcf7−/− mice were infected subcutaneously with 500 infective third-stage larvae (L3). Mice were sacrificed at 10 days after infection for analysis of ILC2 responses. BAL fluid, lungs, and mesenteric lymph nodes were analyzed, and worm counts were performed.
For adoptive transfer of ILC2, lung ILC2 were isolated from WT mice receiving 300 ng of recombinant mIL-33 (eBioscience) i.p. daily for 7 days. We transferred 105 ILC2 into Tcf7−/− mice on the same day of papain challenge or on day 0 and day 5 of N. Brasiliensis infection.
Isolation of Lung Hematopoietic Cells
Mice were exsanguinated and lungs were perfused by injecting 10 ml PBS into the right ventricle of the heart. Lungs were carefully cut into small fragments and digested in Hank’s balanced salt solution containing 0.025 mg/ml Liberase D (Roche Diagnostics) and 10 U/ml DNase I (Roche Diagnostics). Cells were filtered by using a cell strainer.
Flow Cytometry and Cell Sorting
All antibodies (Abs) used were purchased from eBioscience unless specified otherwise. Abs in the lineage (Lin) mixture included anti-FcεR (MAR-1), anti-B200 (RA3-6B2), anti-CD19 (ID3), anti-Mac-1 (M1/70), anti-Gr-1 (8C5), anti-CD11c (HL3), anti-NK1.1 (PK 136), anti-Ter-119 (Ter-119), anti-CD3 (2C11), anti-CD8b (53.5.8), anti-TCRb (H57), and anti-gdTCR (GL-3). Additional Abs used included anti-CD45.2 (104), anti-CD45.1 (A20), anti-Kit (2B8), anti-Sca-1 (D7), anti-CD4 (GK1.5), anti-Thy1.2 (53-2.1), anti-IL-5 (TRFK5), anti-IL-13 (eBio13A), anti-Siglec F (E50-2440; BD Biosciences), and anti-ST2 (DJ8; MD Bioproducts). Cell sorting was performed on a FACSAria II (BD Biosciences), and flow cytometric analysis was performed on a LSR-II (BD Biosciences). Intracellular staining was performed by using Cytofix/Cytoperm Kit (BD Biosciences) according to the manufacturer’s instructions.
Bone Marrow Transplantation
For competitive bone marrow chimeras, 5,000 sorted LSK cells from donor mice (CD45.2) were mixed with 2,500 competitor WT LSK cells (CD45.1 or CD45.1+CD45.2+) and intravenously transferred into lethally irradiated (950 rads) WT recipient mice (CD45.1). For transplantation for fetal liver cells, fetal liver cells (CD45.2) were mixed with competitor WT bone marrow cells (CD45.1) at 1:3 ratio, and together transferred into lethally irradiated WT recipient mice (CD45.1). Reconstitution of ILC2 in the recipient mice was examined at the indicated time points.
For noncompetitive bone marrow chimeras, 5,000 sorted LSK cells from donor mice were transferred into lethally irradiated WT or TCF-1−/− recipients. In some experiments, donor LSK cells were transduced with retrovirus, and 105 cells were transferred into lethally irradiated recipients at 24 hr posttransduction.
ChIP and Luciferase Reporter Assays
For ChIP, ILC2 were sorted from the lungs of naive mice and expanded in vitro with 10 ng/ml IL-2, IL-7, and IL-33 for 2 weeks. ChIP was performed as we previously described (Weber et al., 2011). Primers to detect the TCF-1 binding site in the −3.6 kb upstream regulatory region of II7r locus are as follows: 5′-GGTACAGCATGTGCTAGTTTGCT-3′; and 5′-TGATGTCCTTTCAGCATCTCC-3′. Primers to detect the TCF-1 binding site in the Gata3-1b promoter were described previously (Yu et al., 2009).
For luciferase reporter assays, 293T cells were cotransfected with TCF-1 (or control) retroviral vector and a previously described luciferase reporter vector containing the −3.6 kb upstream regulatory region of II7r locus (Lee et al., 2005). Renilla was added at 50 ng/well for normalization of transfection efficiency. Cells were harvested 48 hr after transfection and analyzed with a Dual Assay Reporter Kit (Promega).
Cell Culture and Retroviral Transduction
Bone marrow LSK cells were cultured in DMEM medium with 15% fetal calf serum (FCS), 100 ng/ml SCF, 6 ng/ml IL-3, and 5 ng/ml IL-6. Lung ILC2 were cultured in MEM-α medium with 20% FCS and 10 ng/ml of IL-7, IL-2, and IL-33. Cytokines were purchased from Peprotech.
Retroviral supernatant preparation and retroviral transduction of LSKs were performed as described (Weber et al., 2011). For retroviral transduction of ILC2, sorted ILC2 from naive mice were cultured overnight with 10 ng/ml of IL-7, IL-2, and IL-33. The addition of IL-33 is important because we obtained very poor retroviral transduction efficiency with IL-2 and IL-7 only (data not shown). The following day, cells were resuspended in retroviral supernatant supplemented with the above cytokines, centrifuged at 1,400 g for 2 hr, and cultured at 37 °C for a further 6 hr. Cells were then washed with PBS and cultured in fresh medium with cytokines. GFP+ cells were sorted at the indicated time points after retroviral transduction.
Elisa
The concentrations of IL-5 and IL-13 were measured by standard sandwich Elisa (eBioscience) according to the manufacturer’s protocol.
Cell-Cycle Analysis and Apoptosis Assay
For cell-cycle analysis, cells were incubated with 10 mg/ml Hoechst 33342 (Invitrogen) at 37°C for 45 min. Cells were washed with PBS, and flow cytometry analyses were performed within 30 min. For apoptosis assays, cells were stained with Annexin V (BD PharMingen) according to the manufacturer’s protocol.
Statistics
Intergroup comparison was performed using Student’s t test. P values lower than 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank I.-C. Ho for permission to use GATA-3f/f mice and S. Zhang and C. Harly for critical comments. This work was supported by National Institutes of Health Grants AI059621 and AI098428 (A.B.), 5T32CA009140-38 (J.L.B.), T32CA-009140 (M.E.O), DP5OD012116 (G.F.S.), R01AI047833 (W.S.P.), T32HD007516 and 1F31CA165813 (W.B.), AI095466, AI095608, and AI097333 (D.A.), and T32-AI007532 (L.A.M.).
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2012.12.003.
REFERENCES
- Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, McKenzie AN. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 2012;129:191–198. e191–194. doi: 10.1016/j.jaci.2011.09.041. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the betacatenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobas M, Wilson A, Ernst B, Mancini SJ, MacDonald HR, Kemler R, Radtke F. Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J. Exp. Med. 2004;199:221–229. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KD, Garcia-Crespo KE, Killoran KE, Rosenberg HF. Antigen profiles for the quantitative assessment of eosinophils in mouse tissues by flow cytometry. J. Immunol. Methods. 2011;369:91–97. doi: 10.1016/j.jim.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, Gounari F. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl. Acad. Sci. USA. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux D, Coudert JD, Maurice D, Scarpellino L, Jeannet G, Piccolo S, Weston K, Huelsken J, Held W. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood. 2005;106:1726–1733. doi: 10.1182/blood-2005-01-0337. [DOI] [PubMed] [Google Scholar]
- Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergeninduced airway inflammation. Immunity. 2012a;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012b;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Giaisi M, Treiber MK, Zhang XB, Mi HF, Horejs-Hoeck J, Andersen KG, Krammer PH, Duschl A, Li-Weber M. LEF-1 negatively controls interleukin-4 expression through a proximal promoter regulatory element. J. Biol. Chem. 2008;283:22490–22497. doi: 10.1074/jbc.M804096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W, Clevers H, Grosschedl R. Redundant functions of TCF-1 and LEF-1 during T and NK cell development, but unique role of TCF-1 for Ly49 NK cell receptor acquisition. Eur. J. Immunol. 2003;33:1393–1398. doi: 10.1002/eji.200323840. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- Lee HC, Shibata H, Ogawa S, Maki K, Ikuta K. Transcriptional regulation of the mouse IL-7 receptor alpha promoter by glucocorticoid receptor. J. Immunol. 2005;174:7800–7806. doi: 10.4049/jimmunol.174.12.7800. [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- Mjösberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, te Velde AA, Fokkens WJ, van Drunen CM, Spits H. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Morrow MA, Mayer EW, Perez CA, Adlam M, Siu G. Overexpression of the Helix-Loop-Helix protein Id2 blocks T cell development at multiple stages. Mol. Immunol. 1999;36:491–503. doi: 10.1016/s0161-5890(99)00071-1. [DOI] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboki K, Ohno T, Kajiwara N, Arae K, Morita H, Ishii A, Nambu A, Abe T, Kiyonari H, Matsumoto K, et al. IL-33 is a crucial amplifier of innate rather than acquired immunity. Proc. Natl. Acad. Sci. USA. 2010;107:18581–18586. doi: 10.1073/pnas.1003059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohteki T, Wilson A, Verbeek S, MacDonald HR, Clevers H. Selectively impaired development of intestinal T cell receptor gamma delta+ cells and liver CD4+ NK1+ T cell receptor alpha beta+ cells in T cell factor-1-deficient mice. Eur. J. Immunol. 1996;26:351–355. doi: 10.1002/eji.1830260213. [DOI] [PubMed] [Google Scholar]
- Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro VS, Hasan M, Wilson A, Boucontet L, Pereira P, Lesjean-Pottier S, Satoh-Takayama N, Di Santo JP, Vosshenrich CA. Cutting edge: Thymic NK cells develop independently from T cell precursors. J. Immunol. 2010;185:4993–4997. doi: 10.4049/jimmunol.1002273. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Curr. Opin. Immunol. 2012;24:132–138. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- Stock P, Lombardi V, Kohlrautz V, Akbari O. Induction of airway hyperreactivity by IL-25 is dependent on a subset of invariant NKT cells expressing IL-17RB. J. Immunol. 2009;182:5116–5122. doi: 10.4049/jimmunol.0804213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, Iwamura C, Nakajima H, Nakayama T, Taniguchi M. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J. Exp. Med. 2008;205:2727–2733. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Hölscher C, et al. Regulated expression of nuclear receptor RORgt confers distinct functional fates to NK cell receptor-expressing RORgt(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xie H, Huang Z, Ma J, Fang X, Ding Y, Sun Z. T cell factor 1 regulates thymocyte survival via a RORgt-dependent pathway. J. Immunol. 2011;187:5964–5973. doi: 10.4049/jimmunol.1101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Hirota K, Stieglitz B, Van Snick J, Tolaini M, Lahl K, Sparwasser T, Helmby H, Stockinger B. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 2011;12:1071–1077. doi: 10.1038/ni.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, et al. Transcription factor RORa is critical for nuocyte development. Nat. Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A. Cutting edge: Natural helper cells derive from lymphoid progenitors. J. Immunol. 2011;187:5505–5509. doi: 10.4049/jimmunol.1102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat. Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr., Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.