Abstract
HLA-DM (DM) functions as a peptide editor that mediates the exchange of peptides loaded onto MHCII molecules by accelerating peptide dissociation and association kinetics. The relative DM-susceptibility of peptides bound to MHCII molecules correlates with antigen presentation and immunodominance hierarchy, and measurement of DM-susceptibility has been a key effort in this field. Current assays of DM-susceptibility, based on differential peptide dissociation rates measured for individually labeled peptides over a long time base, are difficult and cumbersome. Here, we present a novel method to measure DM-susceptibility based on peptide binding competition assays performed in the presence and absence of DM, reported as a delta-IC50 (change in 50% inhibition concentration) value. We simulated binding competition reactions of peptides with various intrinsic and DM-catalyzed kinetic parameters and found that under a wide range of conditions the delta-IC50 value is highly correlated with DM-susceptibility as measured in off-rate assay. We confirmed experimentally that DM-susceptibility measured by delta-IC50 is comparable to that measured by traditional off-rate assay for peptides with known DM-susceptibility hierarchy. The major advantage of this method is that it allows simple, fast and high throughput measurement of DM-susceptibility for a large set of unlabeled peptides in studies of the mechanism of DM action and for identification of CD4+ T cell epitopes.
Keywords: MHCII-peptide complex, DM-susceptibility, IC50, fluorescence polarization, binding competition, off-rate
INTRODUCTION
HLA-DM (DM)4 is a nonclassical MHCII molecule that serves as a peptide editor by mediating the exchange of peptides loading onto MHCII during antigen presentation. DM-mediated peptide exchange has been shown to play a key role in CD4+ T cell epitope selection (Hartman et al., 2010; Lazarski et al., 2006; Lich et al., 2003; Lovitch et al., 2003; Sant et al., 2005; Yin et al., 2012). Measurements of differential DM-susceptibility of various sets of peptides have been crucial in understanding the molecular mechanism of DM-mediated peptide exchange, in identifying CD4+ T cell epitopes, and in improving the efficiency of CD4+ T cell epitope prediction algorithms.
Previously, we and others have measured DM-susceptibility by determining dissociation kinetics of a labeled peptide in the presence of different concentrations of DM, and calculated DM-susceptibility as the slope of the off-rate versus DM concentration curve (Belmares et al., 2002; Hou et al., 2011; Painter et al., 2011; Weber et al., 1996; Yin et al., 2012). This method allows for a direct measurement of peptide off-rate and DM-susceptibility. However, the assay is cumbersome in that each test peptide has to be individually labeled for detection, and difficult in that multiple time points have to be collected for reliable off-rate determination, over a time base that can extend to >10 days for stable peptide complexes as often observed for epitope peptides. These factors limit the application of this assay for measuring DM-susceptibility of a large set of peptides. Moreover, DM has been shown to catalyze peptide association to MHCII molecules, and different peptides might be differentially susceptible to DM-accelerated peptide association, even though the detailed mechanism is still in debate (Grotenbreg et al., 2007; Guce et al., 2013; Pashine et al., 2003; Zarutskie et al., 2001). Nevertheless, the traditional measurement of DM-susceptibility by off-rate has not taken into account of impact of DM on peptide association.
In this study, we developed a novel method to measure DM-susceptibility by assessing the difference of IC50 in the absence and presence of DM in conventional peptide competition binding assays. The resultant ΔIC50 value correlates with conventional off-rate based measures of DM-susceptibility but also takes into account the effect of DM on both peptide association and dissociation reactions. The method described here allows for a reliable and high throughput measurement of DM-susceptibility for a large set of peptides.
MATERIALS AND METHODS
Peptide synthesis and labeling
Influenza hemagglutinin peptide HA306–318 (PKYVKQNTLKLAT), class II-associated invariant chain peptide CLIP (VSKMRMATPLLMQ), human MHCI A2104–117 (GSDWRFLRGYHQYA) and its P1 pocket residue substituted mutants (W1I and W1T) were synthesized for IC50 assay (21st Century Biochemicals, Marlboro, MA). N-terminally acetylated HA306–318 analog peptide (Ac-PRFVKQNTLRLAT) was labeled with Alexa488 tetrafluorophenyl ester (Invitrogen, Eugene, OR) through primary amine of K5 to be used as the probe peptide in the IC50 assay. N-terminal biotin-labeled MHCI A2104–117 and its P1 pocket residue substituted mutants, Alexa488-labeled HA306–318 and Alexa488-labeled CLIP were used in the dissociation kinetics assay. Labeled peptides were purified by Jupiter C18 reverse-phase chromatography (Phenomenex, Torrance, CA).
DR1 and DM expression and purification
Soluble recombinant MHCII molecules DR1 (HLA-DRA*01:01;DRB1*01:01) and DM were expressed in Drosophila S2 cells and purified by immunoaffinity chromatography followed by Superdex200 (GE Healthcare) size exclusion chromatography as described previously (Busch et al., 1998; Frayser et al., 1999; Stern and Wiley, 1992).
KinTek simulation of peptide association, dissociation, and binding competition reactions
Peptide dissociation, association and binding competition reactions in the absence or presence of DM were simulated with KinTek Explorer (Johnson, 2009; Johnson et al., 2009). Peptides with various intrinsic and DM-catalyzed kinetic parameters were set as the test peptides. The model used to simulate the reactions was described as:
| (Eq.1) |
| (Eq.2) |
| (Eq.3) |
For intrinsic peptide association and dissociation reactions, Eq. 1 was used. For peptide binding competition reactions, two versions of Eq. 1 were used, with different parameters for each peptide. For DM-catalyzed reactions, Eqs. 2 and 3 also were included. Values used in the simulation for each kinetic parameter for various peptides were: kass = 0.048–0.114 μM−1min−1, kass,DM = 0.114–0.912 μM−1min−1, kdis = 0.00017–0.0108 min−1, kdis,DM = 0.00043–0.052 min−1, k+DM = 0.0216 μM−1min−1 and k−DM = 0.216 min−1 (detailed values for test peptides shown in Table I). For calculation of IC50, the binding competition simulations using a series of test peptide concentrations were exported and IC50 was fitted from the concentration-dependent inhibition curve (using the equation described in the following IC50 assay) for each peptide. For calculation of intrinsic (koff) and DM-catalyzed off-rate (koff,DM), dissociation simulations of 0.1 μM DR-peptide complex were exported and off-rate was fitted from the dissociation curve using one-phase exponential decay for each peptide.
Table I.
Kinetic parameters and calculated IC50 and off-rate for peptides used in the simulations
| Peptide name | kass (μM−1min−1) | kass,DM (μM−1min−1) | kdis (min−1) | kdis,DM (min−1) | koff a (min−1) | koff,DM (min−1) | IC50 b (μM) | IC50,DM (μM) |
|---|---|---|---|---|---|---|---|---|
| Probe peptide | 0.048 | 0.192 | 0.00017 | 0.00043 | 0.0002 | 0.0003 | 0.0308 | 0.0336 |
| Peptide 1 | 0.114 | 0.228 | 0.00027 | 0.013 | 0.0003 | 0.0035 | 0.0224 | 0.1919 |
| Peptide 2 | 0.048 | 0.192 | 0.00017 | 0.00043 | 0.0002 | 0.0003 | 0.0308 | 0.0336 |
| Peptide 3 | 0.114 | 0.228 | 0.0027 | 0.0027 | 0.0027 | 0.0033 | 0.1803 | 0.1645 |
| Peptide 4 | 0.114 | 0.228 | 0.0027 | 0.0054 | 0.0027 | 0.0040 | 0.1803 | 0.2011 |
| Peptide 5 | 0.114 | 0.228 | 0.0027 | 0.013 | 0.0027 | 0.0059 | 0.1803 | 0.3123 |
| Peptide 6 | 0.114 | 0.228 | 0.0027 | 0.026 | 0.0027 | 0.0091 | 0.1803 | 0.5255 |
| Peptide 7 | 0.114 | 0.228 | 0.0027 | 0.013 | 0.0027 | 0.0059 | 0.1803 | 0.3123 |
| Peptide 8 | 0.114 | 0.228 | 0.0054 | 0.026 | 0.0053 | 0.0117 | 0.3531 | 0.6585 |
| Peptide 9 | 0.114 | 0.228 | 0.0081 | 0.039 | 0.0080 | 0.0176 | 0.5255 | 1.021 |
| Peptide 10 | 0.114 | 0.228 | 0.0108 | 0.052 | 0.0106 | 0.0234 | 0.6959 | 1.399 |
| Peptide 11 | 0.114 | 0.228 | 0.0027 | 0.013 | 0.0027 | 0.0059 | 0.1803 | 0.3123 |
| Peptide 12 | 0.114 | 0.456 | 0.0027 | 0.013 | 0.0027 | 0.0059 | 0.1803 | 0.2713 |
| Peptide 13 | 0.114 | 0.684 | 0.0027 | 0.013 | 0.0027 | 0.0058 | 0.1803 | 0.2455 |
| Peptide 14 | 0.114 | 0.912 | 0.0027 | 0.013 | 0.0027 | 0.0058 | 0.1803 | 0.2273 |
| Peptide 15 | 0.114 | 0.114 | 0.0027 | 0.0027 | 0.0027 | 0.0033 | 0.1803 | 0.1706 |
| Peptide 16 | 0.114 | 0.114 | 0.0027 | 0.0054 | 0.0027 | 0.0040 | 0.1803 | 0.2111 |
| Peptide 17 | 0.114 | 0.114 | 0.0027 | 0.013 | 0.0027 | 0.0059 | 0.1803 | 0.3402 |
| Peptide 18 | 0.114 | 0.114 | 0.0027 | 0.026 | 0.0027 | 0.0091 | 0.1803 | 0.6069 |
Intrinsic (koff, without DM) and DM-catalyzed (koff,DM, with 0.25 μM DM) off-rates were calculated by simulating dissociation reactions of 0.1 μM DR-peptide complex for each peptide and fitting the dissociation curves with a one-phase exponential decay equation. Input values for kass, kass,DM, kdis and kdis,DM rate constants for simulation of each peptide dissociation reaction are shown. Values for k+DM and k−DM were kept constant at 0.0216 μM−1min−1 and 0.216 min−1 respectively for all the peptides.
IC50 was calculated at equilibrium time point of 19200 minutes.
IC50 assay and calculation of DM-susceptibility by IC50
A fluorescence polarization (FP) assay was used to measure the IC50 of each peptide, using Alexa488-HA306–318 as probe peptide as previously described(Yin et al., 2012). 100 nM DR1 was incubated with 25 nM probe peptide Alexa488-HA306–318, together with a series dilution of test peptides, starting with 20 μM with a diluting factor of 5. DR1 concentration is set by titrating DR1 against fixed labeled peptide concentration (25 nM), with the selected DR1 concentration which allows for ~50–75% of the maximum binding as judged by FP assay. The starting concentration and dilution factor for serial dilution of target peptides can be changed depending on specific assay configuration. Competition of each test peptide with binding of probe Alexa488-HA306–318 to DR1 was measured by FP. FP values were converted to % bound as [(FP_sample-FP_free) / (FP_no_comp – FP_free)] ×100, where FP_sample is the FP values for sample well; FP_free is the FP values for free Alexa488-HA306–318; FP_no_comp is the FP values for wells without competitor peptides. Typically FP_free was 70 mP, and FP_no_comp varied with experimental conditions with a maximum of 350 mP for fully bound peptide. We plotted % bound versus concentration of test peptide, and fit the curve to equation y=1/(1+[pep]/IC50), where [pep] is the concentration of test peptide, y is the % of probe peptide bound at that concentration of test peptide and IC50 is the 50% inhibition concentration of the test peptide. To measure DM-susceptibility, IC50,DM was obtained by including DM in the binding competition assay and ΔIC50 was calculated (IC50,DM-IC50). DM-susceptibility measured by IC50 was calculated as ΔIC50/[DM], where [DM] is the concentration of DM.
Peptide dissociation assay and calculation of DM-susceptibility by off-rate
Peptide dissociation kinetics were measured by europium time-resolved fluorescence after addition of excess unlabeled peptide to purified DR-biotinylated test peptide complexes using an antibody capture assay with streptavidin-Europium detection as previously described (Tompkins et al., 1993; Yin et al., 2012), or measured by FP if the test peptide was labeled with Alexa488 (HA306–318 and CLIP in this study) as previously described (Anders et al., 2011; Ferrante et al., 2008; Guce et al., 2013; Painter et al., 2011). DM-susceptibility measured by off-rate was calculated as the slope of off-rate versus DM concentration curve, or as (koff,DM-koff)/[DM] when only one DM concentration was included, where koff,DM and koff are the off-rates in the presence or absence of DM respectively; and [DM] is the concentration of DM.
RESULTS
Influence of DM on MHCII-peptide binding reactions
DM is required for efficient MHCII-peptide loading in antigen presenting cells (Albert et al., 1998; Morris et al., 1994). In vitro DM catalyzes peptide association, dissociation, and exchange reactions (Kropshofer et al., 1996; Morris et al., 1994; Sloan et al., 1995; Weber et al., 1996). Different peptides are differentially susceptible to the action of DM (Belmares et al., 2002; Kropshofer et al., 1996; Weber et al., 1996). The DM susceptibility of a MHCII-peptide complex, usually is measured in a DM-dependent dissociation assay, and characterized as the slope of the linear portion of the off-rate versus DM concentration curve (Yin et al., 2012). DM-dependent peptide dissociation plots and off-rate vs. DM concentration plots are shown in Fig. 1A–C for DR1 complexes of two peptides with different DM-susceptibilities: influenza hemagglutinin derived HA306–318 (HA306–318) and class II-associated invariant chain Ii105–117 peptide (CLIP). HA306–318 is a well-characterized immunodominant epitope with high affinity to DR1 (Roche and Cresswell, 1990a). The DR1-HA306–318 complex has extremely low DM-susceptibility (Ferrante et al., 2008; Ferrante and Gorski, 2010; Joshi et al., 2000; Narayan et al., 2007; Roche and Cresswell, 1990a; Stern et al., 1994; Yin et al., 2012; Zhou et al., 2009). CLIP is the naturally processed remnant of the class II-associated invariant chain chaperone that stabilizes nascent MHCII molecules, with CLIP exchanged for antigenic peptides during epitope selection in antigen presenting cells (Denzin and Cresswell, 1995; Kropshofer et al., 1996; Roche and Cresswell, 1990b; Xu et al., 1995). Although CLIP exhibits similar binding affinity as HA306–318, it has a much higher DM-susceptibility (Anders et al., 2011; Bakke and Dobberstein, 1990; Painter et al., 2011; Roche and Cresswell, 1990b). Consistent with previous studies, HA306–318 displayed slower dissociation kinetics compared with CLIP (koff of 0.00026 vs 0.20 hr−1, Fig. 1A and 1B) and lower DM-susceptibility (0.0013 vs 1.43 hr−1μM−1, Fig. 1C). In general faster dissociating peptides are more susceptible to DM. In early studies it appeared that the ratio between the slope of the DM-susceptibility curve and intrinsic dissociation rate would be constant, however the relationship is now believed to hold only approximately with many outliers (Belmares et al., 2002; Painter et al., 2011; Stratikos et al., 2004; Weber et al., 1996)
FIGURE 1. DM-susceptibility measured by off-rate and influence of DM on IC50.
(A, B) Dissociation kinetics of 0.1 μM (A) DR1-HA306–318 or (B) DR1-CLIP were measured in the absence or presence of various concentrations of DM. (C) The off-rate versus DM concentration of HA306–318 and CLIP bound to DR1 were plotted. The linear range of this plot was amplified and DM-susceptibility measured by off-rate was calculated as the slope. (D–G) IC50 values in the absence or presence of 0.25 μM DM were measured for (D) HA306–318, and (E) CLIP after incubation at 37 °C for 24 hours, and (F) HA306–318, and (G) CLIP after incubation at 37 °C for 72 hours. Alexa488-HA306–318 was used as the probe peptide. These data represent at least three independent experiments with two replicates each. (H, I) Binding competition assay in the presence of a series concentration of DM for (H) HA306–318, and (I) CLIP after incubation at 37 °C for 72 hours. (J) IC50 versus DM concentration was plotted for HA306–318 and CLIP.
In the experiments shown in Fig. 1A–C, peptides were labeled with the fluorophore Alexa488 and dissociation of MHCII-peptide complexes was measured by fluorescence polarization. In previous studies of the dissociation kinetics of these peptides, a variety of fluorophore, biotin, or radioactive labels were used, with dissociation tracked in situ by fluorescence polarization or fluorescence resonance energy transfer (FRET) assay, or after separation of bound and free peptide with fluorescence, gamma radiation, scintillation counting, or enzyme-linked assays (De Wall et al., 2006; Kim et al., 2013; Nicholson et al., 2006; Rothbard and Busch, 2001; Sidney et al., 2013; Tompkins et al., 1993; Vollers and Stern, 2008). Peptide association kinetics have been measured using similar techniques (Call et al., 2009; Ferrante et al., 2008; Guce et al., 2013; Joshi et al., 2000; Kropshofer et al., 1996; Nicholson et al., 2006; Painter et al., 2011). In every case, the test peptides need to be individually labeled in order to detect the MHCII-peptide complex, and samples at multiple time points have to be collected to plot the dissociation kinetics curve.
In contrast to studies of MHCII-peptide association and dissociation kinetics, studies of MHCII-peptide affinity often have employed competition assays. In these assays unlabeled test peptides compete for MHCII binding with a labeled probe peptide. FP-based competition assays have been used widely to measure peptide binding affinities to HLA-DR molecules (De Wall et al., 2006; Ferrante and Gorski, 2010; Ferrante and Gorski, 2012; Guce et al., 2013; Nastke et al., 2012; Nicholson et al., 2006; Pos et al., 2012; Yin et al., 2012; Zhou et al., 2009). The major advantage of this method is that only the probe peptide has to be labeled, which makes the measurement for a large set of unlabeled target peptides possible. The relative binding affinities of peptides as measured in competition assays usually are reported as IC50 values, i.e. the concentration of test peptide needed to inhibit 50% of the binding of probe peptide. Under certain conditions IC50 values can be related to equilibrium binding affinities (KD) using a simple relationship KD,test= IC50,test/(1+ [probe]/KD,probe (Cheng and Prusoff, 1973), but under the conditions typically used for FP assays and constraints of MHCII-peptide association/dissociation kinetics the relationship is considerably more complicated (McFarland and Beeson, 2002; Munson and Rodbard, 1988; Nikolovska-Coleska et al., 2004).
We expected that an IC50 assay might be useful in measurements of DM-susceptibility, with DM influencing the IC50 value more for DM-susceptible peptides than for DM-resistant peptides. DM previously has been shown to influence IC50 of peptides based their sensitivities to DM, in studies using size-exclusion chromatography (Kropshofer et al., 1996). We measured IC50 for unlabeled HA306–318 and CLIP in the absence or presence of DM after 24 hours of reaction using Alexa488-labeled HA306–318 as the probe in a FP assay (Fig. 1D–E). DM had little impact on the IC50 of HA306–318 (0.074 and 0.079 μM without or with DM respectively, Fig. 1D). In the absence of DM the IC50 value for CLIP was 0.046 μM, similar to that of HA306–318. However, in the presence of DM the IC50 value for CLIP was substantially increased to 0.15 μM, for a ΔIC50 of 0.1 μM (Fig. 1E). Considering the long half-life of HA306–318 bound to DR1, we also performed the binding competition assay after 72 hours incubation. Consistently, we observed that DM had little effect on the IC50 of HA306–318 (Fig. 1F), but a ΔIC50 of 0.54 μM was observed for CLIP (Fig. 1G). DM catalyzes peptide dissociation in a dose-dependent manner (Fig. 1A–1C). We evaluated whether DM also influences IC50 in a dose-dependent manner by including a series of concentrations of DM in the binding competition assay (Fig. 1H–1I). We found that similar to the effect of DM on peptide dissociation, DM increased IC50 in a dose-dependent manner, with CLIP having a much greater slope (1.57) in the IC50 versus DM concentration curve compared with HA306–318 (0.06) (Fig. 1J). Thus, our data suggested that DM influences IC50 and ΔIC50 could be used to measure DM-susceptibility. DM-susceptibility measured by off-rate is characterized as the slope of the linear portion of the off-rate versus DM concentration curve. In this linear portion (usually DM concentration less than 1 μM, i.e. Fig. 1C), this slope can be simplified as (koff,DM-koff)/[DM] when only one DM concentration was included, where koff,DM and koff are the off-rates in the presence or absence of DM respectively; and [DM] is the concentration of DM. This simplification is used in previous studies of DM kinetics and epitope selection (Belmares et al., 2002; Ferrante and Gorski, 2010; Hall et al., 2002; Kropshofer et al., 1996; Narayan et al., 2007; Schafer et al., 1996; Sloan et al., 1995; Yin et al., 2012; Zarutskie et al., 2001). We will also use this simplification in this study for Fig. 3–6. For DM-susceptibility measured by IC50 assay, as we demonstrated in Fig. 1J, IC50 versus DM concentration curve was also linear in the range of DM concentrations tested. Therefore, like using (koff,DM-koff)/[DM], ΔIC50/[DM] is a reasonable simplification for the slope of IC50 versus DM concentration curve when only one specific DM concentration is performed as a measure for DM-susceptibility.
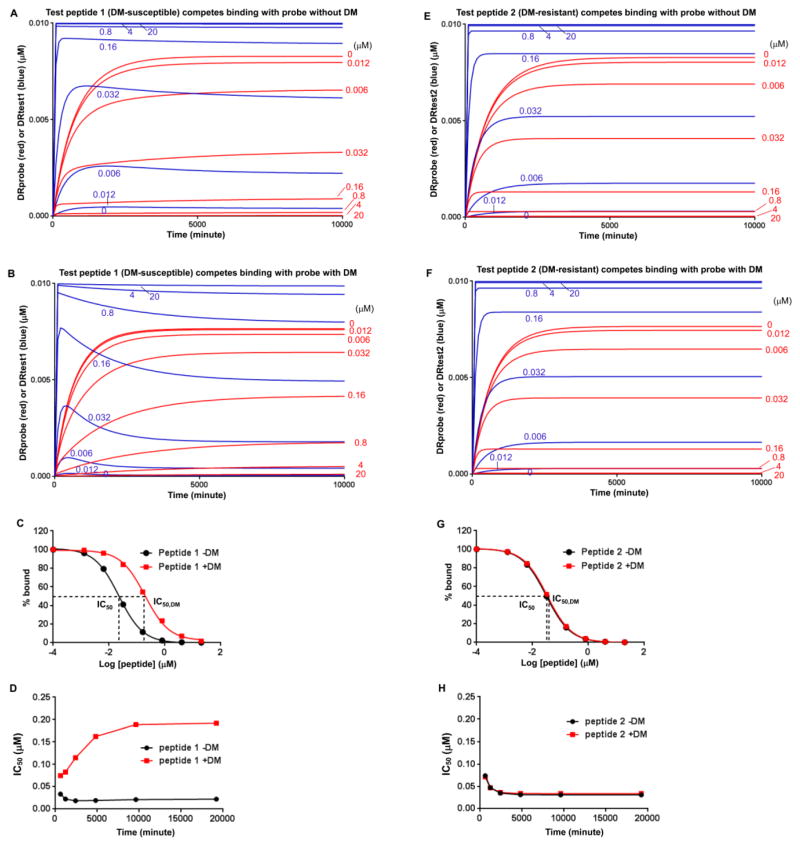
FIGURE 3. Significant difference in IC50 value (ΔIC50) is observed for DM-susceptible peptide but not for DM-resistant peptide.
(A, B) Competition of test peptide 1 (DM-susceptible) with 0.025 μM probe peptide for binding of 0.01 μM DR (A) without DM, or (B) with 0.25 μM DM. Test peptide 1 was included as a 5-fold dilution series from 20 to 0.012 μM. Concentration of complex formed with probe peptide (DRprobe, red) and test peptide 1 (DRtest1, blue) at different initial concentrations of test peptide 1 are shown. (C) Concentration-dependent inhibition plots for test peptide 1 without or with 0.25 μM DM using 19200 minute data from panels (A) and (C). (D) IC50 without or with 0.25 μM DM shown for various time points. (E–H) Competition of test peptide 2 (DM-resistant).
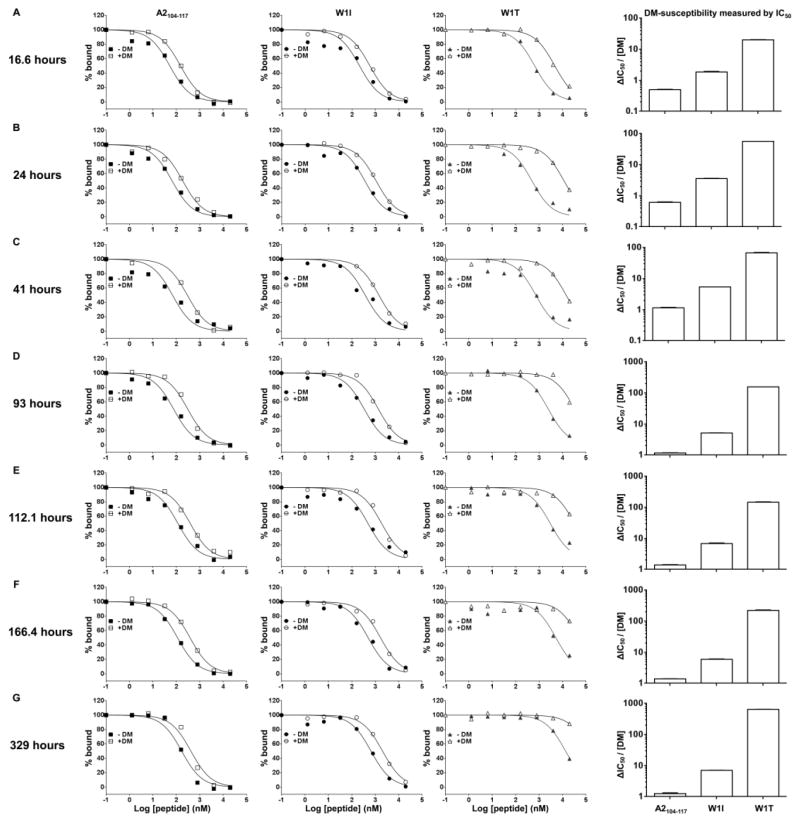
FIGURE 6. Hierarchy of DM-susceptibility measured by ΔIC50 is independent of time of detection over a long period.
IC50 in the absence or presence of 0.2 μM DM were measured for A2104–117, W1I, and W1T, and DM-susceptibility by ΔIC50 was calculated for each peptide. The IC50 assay was read at (A) 16.6 hours, (B) 24 hours, (C) 41 hours, (D) 93 hours, (E) 112.1 hours, (F) 166.4 hours, and (G) 329 hours.
Numerical simulation of binding reactions
We used a computational approach to help understand the relationship between DM susceptibility and ΔIC50 values, We simulated intrinsic and DM-catalyzed peptide dissociation and association reactions in the KinTek modeling program (Johnson, 2009; Johnson et al., 2009) using a simplified reaction scheme (see Materials and Methods, Fig. 2A–D) and values for intrinsic association (kass=0.114 μM−1min−1 ) and dissociation (kdis=0.0027 min−1 ) rate constants. We simulated a binding competition reaction (Fig. 2E–F) as two concurrent binding reactions, with kass of 0.048 and 0.114 μM−1min−1, and kdis of 0.00017 and 0.0027 min−1 for probe and test peptide, respectively. Previous studies of peptide binding and dissociation kinetics have provided values in this range. For peptide dissociation reactions, half-times of ~5 hours for CLIP and ~200 hours for HA306–318 have been reported (Belmares et al., 2002; Ferrante et al., 2008; Ferrante and Gorski, 2010; Joshi et al., 2000; Kropshofer et al., 1996; Narayan et al., 2007; Pashine et al., 2003; Sloan et al., 1995; Stratikos et al., 2004; Weber et al., 1996; Zarutskie et al., 2001; Zhou et al., 2009) corresponding to kdis values of 0.002 min−1 and 0.00006 min−1, respectively. For peptide association reactions, the situation is more complicated because multiple intermediates are involved, including peptide-receptive and peptide-averse MHCII species (Natarajan et al., 1999; Rabinowitz et al., 1998) and open and closed states of the MHCII-peptide complex (Joshi et al., 2000). By combining rate constants for individual steps and estimates of the peptide-receptive fraction we can estimate kass values for HA306–318 and CLIP as ~0.05 and ~0.1 μM−1min−1 respectively (Joshi et al., 2000; Zarutskie et al., 2001). Another study determined kass as 0.1 μM−1min−1 for a fluorescence-labeled myelin basic protein-derived peptide by computationally fitting experimental data into a kinetic model for peptide exchange (Grotenbreg et al., 2007).
FIGURE 2. Simulation of peptide dissociation, peptide association, and peptide binding competition in the absence or presence of DM.
(A, B) Simulation of 0.01 μM DRpeptide dissociation (A) without, or (B) with 0.25 μM DM. (C, D) Simulation of 0.025 μM peptide association with 0.01 μM DR (C) without or (D) with 0.25 μM DM. (E, F) Simulation of competition of 0.025 μM test peptide with 0.025 μM probe peptide for the binding to 0.01 μM DR (E) without DM or (F) with 0.25 μM DM. The equations for each reaction have been indicated, and the simulated values for peptide-bound and peptide-free DR species in each reaction have been plotted.
Fig. 2 shows calculated concentrations of peptide-free DR, DRpeptide, DRprobe and DRtest for simulated dissociation (Fig. 2A), association (Fig. 2C), and competition binding (Fig. 2E) reactions. We modeled the effect of DM by including additional reaction intermediates: a DM-bound form of the DR-peptide complex with increased peptide dissociation (kdis,DM), and a DM-bound form of DR with increased peptide association (kass,DM) (Fig. 2B and 2D) (Pashine et al., 2003; Zarutskie et al., 2001). In the peptide binding competition simulation, we used values for kass,DM of 0.192 and 0.228, kdis,DM of 0.00043 and 0.013 for probe and test peptide, respectively. In the simulations, DM increased the rates of peptide binding, release, and exchange (Fig. 2B, 2D and 2F). Notably, DM changed the competition profile in terms of how much DRprobe relative to DRtest was formed (Fig. 2F). Similar curve shapes were observed when intrinsic and DM-dependent kinetic values were varied in a reasonable range (data not shown).
Relationship of ΔIC50 and DM susceptibility
To obtain IC50 values from the simulations, multiple concentrations of test peptide were included in competition binding reactions, and DRprobe was plotted against concentration of test peptide with IC50 value determined by curve fitting (Fig. 3). To test our hypothesis that ΔIC50 could be used to measure DM-susceptibility, we simulated binding competition reaction for a DM-susceptible test peptide 1 (kass=0.114 μM−1min−1, kass,DM=0.228 μM−1min−1, kdis=0.00027 min−1, kdis,DM=0.013 min−1, and calculated koff,DM-koff equals 0.0032 min−1, Table I) and a DM-resistant peptide 2 (kass=0.048 μM−1min−1, kass,DM=0.192 μM−1min−1, kdis=0.00017 min−1, kdis,DM=0.00043 min−1, and calculated koff,DM-koff equals 0.0001 min−1, Table I), with peptides 1 and 2 having similar KD (kdis/kass ~ 3 nM). We simulated the binding competition reactions with or without DM and calculated IC50 values. In the absence of DM, peptide 1 competed binding with probe peptide in a concentration-dependent manner and remained bound due its slow koff (Fig. 3A and 3C). In the presence of DM, the competition capacity of peptide 1 was weakened due to its high DM-susceptibility, and a large fraction of peptide 1 dissociated from DR1 resulting in increased binding of probe peptide and higher IC50 (Fig. 3B and 3C). The influence of DM on IC50 for peptide 1 was observed at different time points (Fig. 3D). In sharp contrast, for the DM-resistant peptide 2 no difference for IC50 was observed with or without DM (Fig. 3E–H). These data indicated that ΔIC50 could be used to measure DM-susceptibility.
ΔIC50 reports the DM-susceptibility for peptides with a wide range of kinetic parameters
To further test the hypothesis that ΔIC50 values could be used to monitor DM susceptibility, we simulated the reactions for a set of peptides (peptides 3, 4, 5 and 6), which had equal kass, kass,DM and kdis, but various kdis,DM (Table I). Intrinsic (koff) and DM-catalyzed off-rates (koff,DM) were calculated by simulating dissociation reactions of 0.1 μM DR-peptide complex using the input kass, kass,DM, kdis and kdis,DM rate constants for each peptide in the absence (koff) or presence of 0.25 μM DM (koff,DM) and fitting the dissociation curves with one-phase exponential decay (Fig. 4A). Again, we found that ΔIC50 correlated with koff,DM-koff (Fig. 4B). For a set of peptides (peptides 7, 8, 9 and 10) with same kass and kass,DM, but different kdis and kdis,DM (Fig. 4C, Table I), the correlation between ΔIC50 and koff,DM-koff still held (Fig. 4D). Taken together, these simulations demonstrate that ΔIC50 could be a reliable measure of DM-susceptibility for peptides with various kinetic parameters.
FIGURE 4. ΔIC50 is a reliable measure of DM-susceptibility for peptides with various kinetic parameters.
(A) Intrinsic (koff) and DM-catalyzed off-rates (koff,DM) were calculated by simulating dissociation reactions of 0.1 μM DR-peptide complex using the input kass, kass,DM, kdis and kdis,DM rate constants for each peptide in the absence (koff) or presence of 0.25 μM DM (koff,DM) and fitting the dissociation curves with one-phase exponential decay. Simulated reactions for peptides 3, 4, 5 and 6 used the same kass, kass,DM, kdis, but various kdis,DM. (B) Calculated ΔIC50 correlated with calculated koff,DM-koff of peptides 3, 4, 5 and 6. (C) Simulated reactions for peptides 7, 8, 9 and 10 used the same kass, kass,DM, various kdis and kdis,DM. (D) Calculated ΔIC50 correlated with calculated koff,DM-koff of peptides 7, 8, 9 and 10. (E) Simulated reactions for peptides 11, 12, 13 and 14 used the same kass, kdis, kdis,DM, but various kass,DM. Calculated ΔIC50 negatively correlated with kass,DM for these peptides. (F) Set 1 (peptides 3, 4, 5 and 6, black circle) had the same kass, kass,DM, and kdis, but various kdis,DM. Set 2 (peptides 15, 16, 17 and 18, red square) had the same kass, kass,DM, and kdis, but various kdis,DM. Set 1 had a kass,DM of 0.228 μM−1min−1, while set 2 had a kass,DM of 0.114 μM−1min−1. Calculated ΔIC50 correlated with calculated koff,DM-koff of set 1 and set 2 peptides, but ΔIC50 of set 2 was systemically higher than that of set 1.
Although most studies of DM have focused on DM-mediated peptide dissociation, several studies have clearly demonstrated a role of DM on catalyzing peptide loading onto MHCII molecules during antigen presentation and epitope selection (Ferrante et al., 2008; Grotenbreg et al., 2007; Guce et al., 2013; Nicholson et al., 2006; Zarutskie et al., 2001). Therefore, we examined whether ΔIC50 could capture the effect of DM on peptide association. We first simulated the binding competition reactions with or without DM for a set of peptides (peptides 11, 12, 13 and 14) with same kass, kdis and kdis,DM, but different kass,DM (Table I). As shown, ΔIC50 was negatively correlated with kass,DM (Fig. 4E). To test the effect of DM on both peptide dissociation and association, we simulated the binding competition reactions for two additional sets of peptides. Set 1 (peptides 3, 4, 5 and 6) had the same kass,DM of 0.228 μM−1min−1, but various koff,DM-koff (same in Fig. 4A–B). Set 2 (peptides 15, 16, 17 and 18) had the same kass,DM of 0.114 μM−1min−1, but various koff,DM-koff (Table I). Interestingly, we found that ΔIC50 correlates with koff,DM-koff for both sets of peptides, but ΔIC50 for set 2 is systemically higher than that of set 1 due to its lower kass,DM (Fig. 4F). These simulation data demonstrate that ΔIC50 represents a measurement of DM-susceptibility of peptides that takes into account the catalytic effect of DM on both peptide association and dissociation reactions.
DM-susceptibility measured by IC50 assay correlates with that measured by dissociation assay
To experimentally test the hypothesis that ΔIC50 is a reliable measure of DM-susceptibility, we measured DM-susceptibility by IC50 for a set of peptides based on the immunodominant alloantigen A2104–117 (derived from HLA-A2) and compared these values to DM-susceptibility measured conventionally by off-rate analysis. To generate peptides with increased DM-susceptibility, we substituted the P1 pocket residue of A2104–117 from tryptophan to isoleucine (W1I) or to threonine (W1T), because the P1 pocket residue is one of the major anchor residues (Murthy and Stern, 1997; Stern et al., 1994) and has been implicated in determining DM-susceptibility (Anders et al., 2011; Narayan et al., 2009; Pos et al., 2012; Schulze and Wucherpfennig, 2012; Yin and Stern, 2013). As expected, A2104–117, W1I and W1T all showed a concentration-dependent inhibition on the binding of probe peptide Alexa488-HA306–318 to DR1, with A2104–117 having the highest affinity and W1T the lowest (Fig. 5A–C). Notably, DM exerted an influence on the IC50 of each peptide, and DM-susceptibility calculated by IC50 of A2104–117, W1I and W1T exhibited the expected hierarchy (Fig. 5D). To confirm the accuracy of this IC50-based method, we also measured the dissociation kinetics for each peptide bound to DR1 (Fig. 5E–G), and calculated the DM-susceptibility by off-rate, which showed consistent hierarchy as that measured by IC50 (Fig. 5H). The correlation coefficient of DM-susceptibility measured by IC50 and off-rate was 0.99 (data not shown).
FIGURE 5. DM-susceptibility measured by ΔIC50 correlates with that calculated by off-rate.
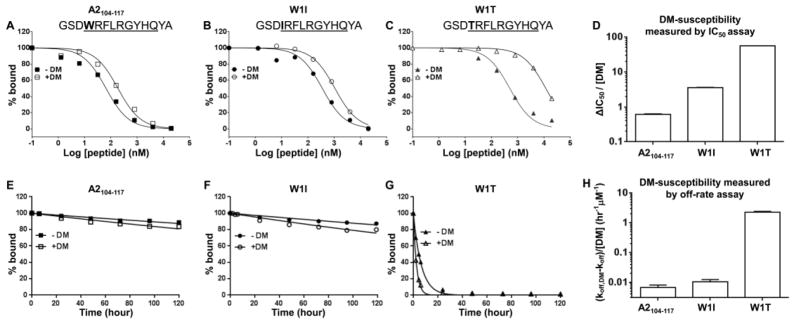
(A–C) IC50 measurements for in the absence or presence of 0.2 μM DM measured for (A) A2104–117, and its pocket 1 substituted variants (B) W1I, and (C) W1T. The sequence of each peptide is indicated with peptide binding motif underlined and P1 pocket residue highlighted in bold. This assay was read at 24 hours after incubation at 37 °C. Alexa488-HA306–318 was used as the probe peptide. (D) DM-susceptibility by IC50 for each peptide was calculated as ΔIC50 divided by DM concentration. (E–G) Dissociation kinetics measurements for 0.1 μM DR1 bound with (E) A2104–117, (F) W1I, and (G) W1T measured in the absence or presence of 0.1 μM DM. (H) DM-susceptibility by off-rate was calculated for each peptide, using function (koff,DM-koff)/[DM]. These data are representative of two independent experiments with at least two replicates each.
Hierarchy of DM-susceptibility measured by IC50 is independent of time of detection over a wide range of assay times
To be noted, IC50 measurement both in the absence or presence of DM is dependent on the time of detection, especially for peptides with higher DM-susceptibility and early detection times (Fig. 3D and 3H). This is due to the fact that probe peptide and test peptide have different association and dissociation kinetics. To test the influence of time of detection on DM-susceptibility measured by IC50, we read the FP at different time points and calculated IC50 and DM-susceptibility (Fig. 6). As shown, although IC50 in the absence or presence of DM changed during the time course of over 300 hours, the hierarchy of DM-susceptibility measured by IC50 of these tested peptides remained the same.
DISCUSSION
DM-mediated peptide exchange plays a key role in MHCII antigen presentation (Amria et al., 2008; Lazarski et al., 2006) and CD4+ T cell epitope selection (Hartman et al., 2010; Kremer et al., 2012; Lazarski et al., 2005; Sant et al., 2005; Yin et al., 2012). In this study, we developed a novel IC50-based method to measure DM-susceptibility. The underlying principle for this measurement is that the DM-susceptibility of a test peptide would be reflected by its differential ability to inhibit binding of probe peptide to MHCII in the presence or absence of DM. In the FP-based binding competition assay, labeled probe peptide and unlabeled test peptide compete for binding to MHCII. If test peptide bound to MHCII is more susceptible to DM, it will dissociate more easily and allow for more labeled probe peptide to bind. Instead, if test peptide bound to MHCII is resistant to DM, it will stay bound and prevents more labeled probe peptide to bind. This difference is reflected in ΔIC50 in the absence or presence of DM. Using both numerical simulation and experimental data, we showed that DM-susceptibility measured by IC50 was comparable with that measured by traditional kinetic off-rate analysis.
In this study we used a simplified reaction scheme to simulate peptide association, dissociation and binding competition reactions (Fig. 2–4). Some previous studies have suggested that these reactions may be more complex than modeled. For instance, it has been demonstrated that MHCII molecules undergo a reversible isomerization between peptide-receptive and peptide-averse states, and DM may catalyze peptide association by accelerating the transition of peptide-averse to peptide-receptive conformation, or stabilizing the peptide-receptive form (Grotenbreg et al., 2007; Natarajan et al., 1999; Rabinowitz et al., 1998). Moreover, multiple intermediates formed between peptide, MHCII and DM during peptide association, dissociation and exchange have been proposed although the detailed mechanisms are still in debate (Anders et al., 2011; Ferrante et al., 2008; Grotenbreg et al., 2007; Narayan et al., 2009; Pashine et al., 2003; Pos et al., 2012; Zarutskie et al., 2001). It is possible that reaction steps not included in our simulation reaction scheme might put some constraints on the interpretation on the IC50 data. Nevertheless, simulations using the reasonably simplified reactions demonstrated that DM-susceptibility could be reliably measured by ΔIC50, which is confirmed by our experimental evaluations.
Measuring IC50 by a FP-based method has been a standard protocol in determining MHCII-peptide interactions (De Wall et al., 2006; Ferrante and Gorski, 2012; Guce et al., 2013; Pos et al., 2012; Yin et al., 2012; Zhou et al., 2009). By incorporating DM into the binding competition reaction, we developed a novel IC50-based method to measure DM-susceptibility. Compared with the previous used kinetic method, the major advantage of the method described in this study is only a single probe peptide needs to be labeled and only a single final read is needed, allowing for measurement of DM-susceptibility of many peptides at the same time. The protocol we described here uses 96-well plate, but should be easily converted to 384-well or 1536-well format for screening of peptides with higher or lower DM-susceptibility, because the basic FP measurement is concentration- and volume-independent. Another advantage of this method to measure DM-susceptibility is that it considers the catalyzing effect of DM on both peptide association and dissociation, which is important in antigen presentation and epitope selection. In this protocol, we used a well-characterized and widely-used HA306–318 derived peptide as the probe peptide, which has high affinity, high kinetic stability, and low DM-susceptibility (koff,DM-koff =0.0001 min−1). However, when we changed the koff,DM-koff of probe peptide to 0.1 min−1, the correlation between ΔIC50 and koff,DM-koff of test peptides still held, which indicated that having a probe peptide with low DM-susceptibility is not necessary for using ΔIC50 as a measure of DM-susceptibility. This might be important for applying this method to other MHCII alleles, for which a probe peptide with low DM-susceptibility may not be established. One potential disadvantage of this protocol is that it only measures relative DM-susceptibility from binding competition with the probe peptide, instead of absolute DM-susceptibility calculated from dissociation kinetics. However, in most cases, we are interested in screening out peptides with lowest DM-susceptibility to identify epitopes, and measurement of the relative DM-susceptibility is sufficient for this goal.
Epitope prediction algorithms have been widely used to help identify CD4 T cell epitopes from various pathogens (Borras-Cuesta et al., 2000; Calvo-Calle et al., 2007; Doolan et al., 2003; Wang et al., 2010). Most current CD4+ T cell epitope prediction algorithms are based explicitly on measurements of peptide binding affinity to MHCII (Hammer et al., 1992; Peters et al., 2005), with no consideration of the effect of DM. We have previously demonstrated that DM susceptibility is a strong and independent factor governing peptide immunogenicity and epitope selection (Yin et al., 2012). Therefore, the method developed here capable of measuring DM-susceptibility for a large set of peptides might be useful in training the prediction algorithms to account for DM effects, and should be directly useful in predicting epitopes based on differential DM-susceptibilities of tested peptides.
In summary, in this study we describes a novel IC50-based protocol for reliable, fast, simple and high throughput measurement of DM-susceptibility of MHCII-peptide complexes, which will facilitate our understanding of the mechanism of DM-mediated peptide exchange and improve our ability to screen and predict CD4+ T cell epitopes.
Highlights.
ΔIC50 is a fast, reliable and high-throughput measure of MHCII DM-susceptibility
ΔIC50 correlates with DM-susceptibility measured conventionally by off-rate
ΔIC50 accounts for the effect of DM in both peptide association and dissociation
Acknowledgments
We would like to thank Liying Lu and Loretta Lee for soluble DR1 and DM expression and purification. This work was supported by NIH grant AI-38996 and AI-57319.
Footnotes
Abbreviations: DM, HLA-DM; MHCII, major histocompatibility complex class II molecules; IC50, concentration required for 50% inhibition; IC50,DM, IC50 measured in the presence of DM; koff,DM and koff, DM-catalyzed and intrinsic off-rate respectively; kass,DM and kass, DM-catalyzed and intrinsic association rate constant respectively; kdis,DM and kdis, DM-catalyzed and intrinsic dissociation rate constant respectively; FP, fluorescence polarization
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFENCES
- Albert LJ, Denzin LK, Ghumman B, Bangia N, Cresswell P, Watts TH. Quantitative defect in staphylococcal enterotoxin A binding and presentation by HLA-DM-deficient T2.Ak cells corrected by transfection of HLA-DM genes. Cellular immunology. 1998;183:42–51. doi: 10.1006/cimm.1997.1236. [DOI] [PubMed] [Google Scholar]
- Amria S, Hajiaghamohseni LM, Harbeson C, Zhao D, Goldstein O, Blum JS, Haque A. HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur J Immunol. 2008;38:1961–1970. doi: 10.1002/eji.200738100. [DOI] [PubMed] [Google Scholar]
- Anders AK, Call MJ, Schulze MS, Fowler KD, Schubert DA, Seth NP, Sundberg EJ, Wucherpfennig KW. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nat Immunol. 2011;12:54–61. doi: 10.1038/ni.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- Borras-Cuesta F, Golvano J, Garcia-Granero M, Sarobe P, Riezu-Boj J, Huarte E, Lasarte J. Specific and general HLA-DR binding motifs: comparison of algorithms. Hum Immunol. 2000;61:266–278. doi: 10.1016/s0198-8859(99)00153-6. [DOI] [PubMed] [Google Scholar]
- Busch R, Doebele RC, von Scheven E, Fahrni J, Mellins ED. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J Immunol. 1998;160:734–743. [PubMed] [Google Scholar]
- Call MJ, Xing X, Cuny GD, Seth NP, Altmann DM, Fugger L, Krogsgaard M, Stein RL, Wucherpfennig KW. In vivo enhancement of peptide display by MHC class II molecules with small molecule catalysts of peptide exchange. J Immunol. 2009;182:6342–6352. doi: 10.4049/jimmunol.0803464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- De Wall SL, Painter C, Stone JD, Bandaranayake R, Wiley DC, Mitchison TJ, Stern LJ, DeDecker BS. Noble metals strip peptides from class II MHC proteins. Nature chemical biology. 2006;2:197–201. doi: 10.1038/nchembio773. [DOI] [PubMed] [Google Scholar]
- Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR, 3rd, Carucci DJ, Sette A. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante A, Anderson MW, Klug CS, Gorski J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS One. 2008;3:e3722. doi: 10.1371/journal.pone.0003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante A, Gorski J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J Immunol. 2010;184:1153–1158. doi: 10.4049/jimmunol.0902878. [DOI] [PubMed] [Google Scholar]
- Ferrante A, Gorski J. A Peptide/MHCII conformer generated in the presence of exchange peptide is substrate for HLA-DM editing. Sci Rep. 2012;2:386. doi: 10.1038/srep00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayser M, Sato AK, Xu L, Stern LJ. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr Purif. 1999;15:105–114. doi: 10.1006/prep.1998.0987. [DOI] [PubMed] [Google Scholar]
- Grotenbreg GM, Nicholson MJ, Fowler KD, Wilbuer K, Octavio L, Yang M, Chakraborty AK, Ploegh HL, Wucherpfennig KW. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, Stern LJ. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol. 2013;20:90–98. doi: 10.1038/nsmb.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall FC, Rabinowitz JD, Busch R, Visconti KC, Belmares M, Patil NS, Cope AP, Patel S, McConnell HM, Mellins ED, Sonderstrup G. Relationship between kinetic stability and immunogenicity of HLA-DR4/peptide complexes. Eur J Immunol. 2002;32:662–670. doi: 10.1002/1521-4141(200203)32:3<662::AID-IMMU662>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Hammer J, Takacs B, Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992;176:1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, Cole RN, Sadegh-Nasseri S. A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat Med. 2010;16:1333–1340. doi: 10.1038/nm.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Macmillan H, Chen Z, Keech CL, Jin X, Sidney J, Strohman M, Yoon T, Mellins ED. An insertion mutant in DQA1*0501 restores susceptibility to HLA-DM: implications for disease associations. J Immunol. 2011;187:2442–2452. doi: 10.4049/jimmunol.1100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods in enzymology. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Analytical biochemistry. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Joshi RV, Zarutskie JA, Stern LJ. A three-step kinetic mechanism for peptide binding to MHC class II proteins. Biochemistry. 2000;39:3751–3762. doi: 10.1021/bi9923656. [DOI] [PubMed] [Google Scholar]
- Kim A, Ishizuka I, Hartman I, Poluektov Y, Narayan K, Sadegh-Nasseri S. Studying MHC class II peptide loading and editing in vitro. Methods in molecular biology. 2013;960:447–459. doi: 10.1007/978-1-62703-218-6_33. [DOI] [PubMed] [Google Scholar]
- Kremer AN, van der Meijden ED, Honders MW, Goeman JJ, Wiertz EJ, Falkenburg JH, Griffioen M. Endogenous HLA class II epitopes that are immunogenic in vivo show distinct behavior toward HLA-DM and its natural inhibitor HLA-DO. Blood. 2012;120:3246–3255. doi: 10.1182/blood-2011-12-399311. [DOI] [PubMed] [Google Scholar]
- Kropshofer H, Vogt AB, Moldenhauer G, Hammer J, Blum JS, Hammerling GJ. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lich JD, Jayne JA, Zhou D, Elliott JF, Blum JS. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J Immunol. 2003;171:853–859. doi: 10.4049/jimmunol.171.2.853. [DOI] [PubMed] [Google Scholar]
- Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Medicinal research reviews. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- Munson PJ, Rodbard D. An exact correction to the “Cheng-Prusoff” correction. Journal of receptor research. 1988;8:533–546. doi: 10.3109/10799898809049010. [DOI] [PubMed] [Google Scholar]
- Murthy VL, Stern LJ. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of the specificity of peptide binding. Structure. 1997;5:1385–1396. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM mediates peptide exchange by interacting transiently and repeatedly with HLA-DR1. Mol Immunol. 2009;46:3157–3162. doi: 10.1016/j.molimm.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastke MD, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, Calvo-Calle JM. Human CD4+ T cell response to human herpesvirus 6. J Virol. 2012;86:4776–4792. doi: 10.1128/JVI.06573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999;162:4030–4036. [PubMed] [Google Scholar]
- Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, Wucherpfennig KW. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Analytical biochemistry. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Painter CA, Negroni MP, Kellersberger KA, Zavala-Ruiz Z, Evans JE, Stern LJ. Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proc Natl Acad Sci U S A. 2011;108:19329–19334. doi: 10.1073/pnas.1108074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger S, Stewart S, Surko P, Way S, Wilson S, Sette A. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, Wucherpfennig KW. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, Davis MM, McConnell HM. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J Immunol. 1990a;144:1849–1856. [PubMed] [Google Scholar]
- Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990b;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- Rothbard JB, Busch R. Binding of biotinylated peptides to MHC class II proteins on cell surfaces. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 18. Chapter 18. 2001. p. 11. [DOI] [PubMed] [Google Scholar]
- Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- Schafer PH, Green JM, Malapati S, Gu L, Pierce SK. HLA-DM is present in one-fifth the amount of HLA-DR in the class II peptide-loading compartment where it associates with leupeptin-induced peptide (LIP)-HLA-DR complexes. J Immunol. 1996;157:5487–5495. [PubMed] [Google Scholar]
- Schulze MS, Wucherpfennig KW. The mechanism of HLA-DM induced peptide exchange in the MHC class II antigen presentation pathway. Curr Opin Immunol. 2012;24:105–111. doi: 10.1016/j.coi.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, Sette A. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 18. Chapter 18. 2013. p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- Stratikos E, Wiley DC, Stern LJ. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II alpha-chain. J Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- Tompkins SM, Rota PA, Moore JC, Jensen PE. A europium fluoroimmunoassay for measuring binding of antigen to class II MHC glycoproteins. J Immunol Methods. 1993;163:209–216. doi: 10.1016/0022-1759(93)90124-p. [DOI] [PubMed] [Google Scholar]
- Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123:305–313. doi: 10.1111/j.1365-2567.2007.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- Xu X, Song W, Cho H, Qiu Y, Pierce SK. Intracellular transport of invariant chain-MHC class II complexes to the peptide-loading compartment. J Immunol. 1995;155:2984–2992. [PubMed] [Google Scholar]
- Yin L, Calvo-Calle JM, Dominguez-Amorocho O, Stern LJ. HLA-DM constrains epitope selection in the human CD4 T cell response to vaccinia virus by favoring the presentation of peptides with longer HLA-DM-mediated half-lives. J Immunol. 2012;189:3983–3994. doi: 10.4049/jimmunol.1200626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Stern LJ. HLA-DM Focuses on Conformational Flexibility Around P1 Pocket to Catalyze Peptide Exchange. Frontiers in immunology. 2013;4:336. doi: 10.3389/fimmu.2013.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc Natl Acad Sci U S A. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]