Abstract
Long-term, in vitro propagation of tumor-specific endothelial cells (TEC) allows for functional studies and genome-wide expression profiling of clonally-derived, well-characterized subpopulations. Using a genetically engineered mouse model (GEMM) of mammary adenocarcinoma, we have optimized an isolation procedure and defined growth conditions for long-term propagation of mammary TEC. The isolated TEC maintain their endothelial specification and phenotype in culture. Furthermore, gene expression profiling of multiple TEC subpopulations revealed striking, persistent overexpression of several candidate genes including Irx2 and Zfp503 (transcription factors), Alcam and Cd133 (cell surface markers), Ccl4 and neurotensin (Nts) (angiocrine factors), and Gpr182 and Cnr2 (G protein-coupled receptors, GPCRs). Taken together, we have developed an effective method for isolating and culture-expanding mammary TEC, and uncovered several new TEC-selective genes whose overexpression persists even after long-term in vitro culture. These results suggest that the tumor microenvironment may induce changes in vascular endothelium in vivo that are stably transmittable in vitro.
Keywords: Tumor angiogenesis, tumor endothelial cells, endothelial heterogeneity, tumor microenvironment, angiocrine, endothelial cell plasticity, breast cancer
Introduction
Vascular endothelial cells (EC) are highly specialized in order to meet the metabolic demands of specific tissues. For example, a recent study demonstrated that isolated, organ-specific EC express combinations of factors unique to each organ [1]. Moreover, transplanted “generic” EC acquire specific features of the organ in which they occupy [1]. These studies suggest that EC are remarkably malleable and “tuned” to conditions found in the surrounding microenvironment. In tumors and other pathophysiological states, the endothelium may also be modified or educated by specific microenvironmental cues. Indeed, most tumors are typified by hypoxia, acidosis, and the presence of inflammatory cytokines to which the endothelium must adapt. Multiple studies have described unique gene expression patterns in freshly isolated TEC from different tumor types which may reflect either a transient adaptive response or stable reprogramming due to epigenetic modifications [2, 3].
A challenge to better understanding the biology of the tumor vasculature is that it is difficult to isolate and maintain a homogeneous population of TEC for long-term, in vitro expansion. But as we have shown, clonally-derived TEC may be propagated in culture and further characterized using functional in vitro assays [4–6]. On one hand, ex vivo propagated TEC may no longer resemble TEC in situ once they are removed from influences of the tumor-microenvironment. For example, it is well-known that tissue-specific EC undergo genetic drift in culture and lose expression of key factors [7–10]. On the other hand, TEC which are immediately processed following isolation cannot be further characterized in vitro nor can individual subpopulations of TEC be studied [11, 12]. Moreover, it is difficult to ensure that TEC isolated by methodologies such as laser capture micro-dissection or fluorescence-activated cell sorting are entirely free of contaminating tumor cells or perivascular cells which can confound genome-wide expression profiling studies.
Using a GEMM of mammary adenocarcinoma (C3-TAg) we have optimized an isolation procedure and defined an ideal growth medium for maintaining highly-enriched, clonally-derived TEC populations (from the same tumor) for long-term culture. For comparison, we have isolated normal mammary gland endothelial cells (NEC) from age-matched littermates and carried out genome-wide mRNA expression profiling of these different populations under identical culture conditions. Overall, we show that clonally-derived populations of NEC and TEC can be maintained free of tumor cell/stromal cell contamination, and they retain their endothelial specification. In addition, we have identified several new candidate genes, some of unknown function in EC, that are persistently up regulated in TEC cultures.
Methods
Mice
C3-TAg (FVB/N C31-TAg) mice were provided by the Mouse Phase 1 Unit from the Lineberger Comprehensive Cancer Center at UNC Chapel Hill. Tumors were harvested under sterile conditions when mice were ~ 5 months of age. Normal mammary tissue was obtained from age-matched, wildtype littermates.
Cell culture and media
Isolated EC were maintained in 1 g/L D-glucose DMEM (LG-DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 10% Nu-Serum IV (BD), and Lonza EGM™-2 SingleQuots including hFGF, VEGF, hEGF, R3-IGF-1, and heparin. Mammary tumor cells isolated from C3-TAg tumors were cultured in DMEM with 4.5 g/L D-glucose (Gibco) and 10% FBS. Mouse monocytes (M1) were purchased from ATCC and were grown in RPMI containing 10% FBS. Mouse mesenchymal stem cells (MSC) were purchased from Gibco and were maintained in DMEM/F-12 (Gibco) with 10% MSC-qualified FBS (Gibco). All media were supplemented with antibiotic-antimycotic (Gibco).
Endothelial cell isolation
The EC isolation method was modified from previously published procedures [5]. Briefly, freshly-resected tumors were minced into ~ 3 mm pieces and transferred to LG-DMEM containing 1 mg/mL collagenase type II (Worthington), 100 μg/mL deoxyribonuclease (Worthington), and 0.25 U/mL neutral protease (Worthington) at an approximate ratio of 1 tumor volume to 3–5 volumes of digestion solution. The sample was homogenized on the Miltenyi Tissue Dissociator (Miltenyi Biotec) before being placed on a shaker at 37°C for 75 min. The tumor digests were then filtered with a 100 μm cell strainer to obtain a single-cell suspension before pelleting at 1200 rpm for 10 min. If blood was visible in the cell pellet, 10 mL of 1X PharmLyse B (BD Pharmingen) was used to lyse the red blood cells, and the sample was immediately centrifuged at 1200 rpm for 5 min. Next, the cells were resuspended in 10 mL MACS buffer (de-gassed phosphate-buffered saline [PBS] containing 2 mM EDTA and 0.5% BSA) and counted. Cells were pelleted and then resuspended in MACS buffer at 1.0 × 107 cells/100 μL. FcR Blocking Reagent (Miltenyi) (10 μL/100 μL cell suspension) was added to the sample, which was then incubated on ice for 15 min. The subsequent two-step antibody incubation includes first adding a PE-rat anti-mouse CD31 antibody (BD Pharmingen, 553373) and then anti-PE microbeads (Miltenyi). Each antibody incubation step was performed on ice for 15 min. After each antibody incubation, the cells were washed twice with MACS buffer, centrifuged, and resuspended in 500 μL MACS buffer. The final cell suspension was filtered through a 35 μm cell strainer and then passed through the magnetic Miltenyi LS Column pre-equilibrated with MACS buffer. The column was washed three times with 2–3 mL MACS buffer to remove unbound cells. Bound cells were eluted with 2–3 mL MACS buffer three times. The eluted cells were washed with growth medium once and plated at a density of ~ 1.0 × 106 cells/10 mL in the same medium onto 10 cm tissue-culture dishes coated with 0.5% gelatin (Sigma-Aldrich). The medium was changed every 2–3 days and Dil-Ac-LDL (Biomedical Technologies) was added to the plates to monitor the size of the EC colonies. When EC colonies grew to ~ 3–5 mm in diameter, LDL-negative cells surrounding the EC clones were lightly scraped off using a 200-μL pipette tip. Cloning rings of appropriate sizes were glued onto the plate using Vetbond (3M) to trap EC clones which were then washed once in PBS, detached with 25 μL Accutase (Sigma), and transferred into individual wells of a gelatin-coated 96-well plate. Cells were slowly expanded over the next 3–4 weeks into larger wells and then 10 cm dishes.
Polymerase chain reaction (PCR) and real-time quantitative PCR (qPCR)
Primers were designed using either Invitrogen Primer Perfect Design Software or NCBI-Primer Blast. Total RNA was isolated using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. cDNA synthesis was completed using an iScript cDNA Synthesis Kit (Bio-Rad). End-point PCR was carried out using a Taq PCR Kit (NEB) with the products resolved on agarose gels. qPCR was run in triplicate with Maxima SYBR Green (ThermoFisher) on an Applied Biosystems Step One Plus analyzer.
Flow cytometry
Cells were analyzed by flow cytometry as previously described using a BD Accuri® C6 Flow Cytometer [5, 6]. Data were post-analyzed using FloJo (Version X).
Immunofluorescence (IF)
IF was carried out as previously described [5, 6]. Antibodies include: 1:100 rat anti-mouse CD31 antibody (BD, 550274), 1:200 Alexa Flour® 488 goat anti-rat antibody (Invitrogen, A11006), and 1:500 monoclonal mouse anti-α-smooth muscle actin (αSMA) Cy3 antibody (Sigma, C6198). Slides were mounted with Vectashield Hardset Mounting Medium with DAPI (Vector Labs) and imaged on a Leica DM IRB inverted microscope.
Gene expression microarrays and bioinformatics analysis
All EC and tumor cells were profiled using mouse oligo gene expression microarrays (Agilent Technologies, Santa Clara, CA, USA) as previously described [13]. Microarray data are available at UNC Microarray Database (https://genome.unc.edu) and have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE50555. Heat maps were generated using the Gene-E software package (http://www.broadinstitute.org/cancer/software/GENE-E/). Statistical analysis for the microarrays was carried out with WinSTAT, R v2.15.1, Cluster v3.0, and Prism. The probes were filtered by requiring the Lowess normalized intensity values in both sample and control to be > 10. All probes for each gene were averaged. The normalized log2 ratios (Cy5 sample/Cy3 control) of probes mapping to the same gene (Entrez ID as defined by the manufacturer) were averaged to generate independent expression estimates for each gene. For each cell type the “vascular content” gene expression signature value was identified by averaging the normalized log2 ratio value for each gene within the signature [14]. Sixty-six of 74 vascular content signature genes were present in this dataset. For the heat map contrasting array data from TEC and mammary tumor cells, the normalized log2 ratio values were utilized for selected genes known to be expressed in vascular cells. A two-class significance analysis of microarrays was utilized to identify NEC and TEC-specific genes (FDR<5), which were then median centered and hierarchically clustered.
Results
Endothelial cell isolation and characterization
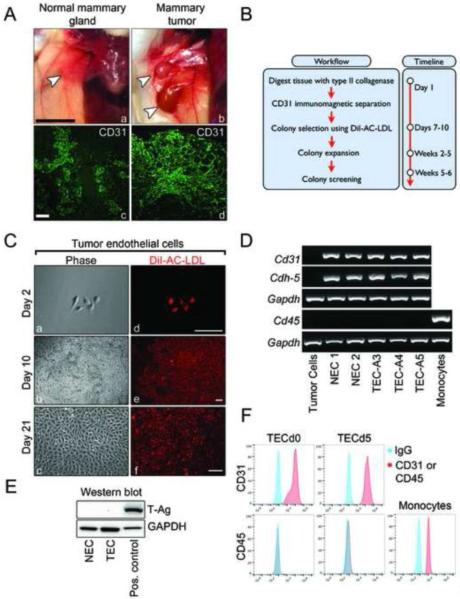
C3-TAg female mice develop spontaneous mammary intra-epithelial neoplasia at ~ 12 weeks of age which resembles human ductal carcinoma in situ, and at ~ 16 weeks of age tumors progress to palpable, well-vascularized, invasive carcinoma [15]. Comparison of normal mammary gland of wild-type mice and mammary tumors of C3-TAg mice are shown in Figure 1A (a–b). CD31 immunohistochemistry of normal mammary gland and mammary tumors shows numerous CD31+ blood vessels (Figure 1A, c–d). To isolate CD31+ EC, we used a protocol developed by us, with minor modifications [5]. The workflow and timeline are summarized in Figure 1B. At day 7, cultures were incubated with DiI-AC-LDL to identify EC colonies [16]. Twenty to thirty colonies were captured using cloning rings, transferred to 96-well plates, and then expanded. Characteristic DiI-AC-LDL+ TEC cultures are shown at day 2 after the isolation, day 10 when cloning rings were applied, and day 21 when the clone expansion was completed (Figure 1C, a–f). Cells remained 100% DiI-AC-LDL positive even after several passages in culture (not shown), demonstrating the high efficiency of the cloning ring method. When cells reached approximately 6–8 passages, RNA was isolated, reverse transcribed, and subjected to PCR. The results showed that several representative colonies of NEC and TEC expressed Cd31 and Cdh-5 mRNAs, whereas mammary tumor cells derived from C3-Tag mice did not express these markers (Figure 1D). None of the EC expressed the pan leukocyte marker Cd45 (Figure 1D), or the SV40 large T-antigen (T-Ag) carried by C3-TAg mice (Figure 1E), thus ruling out a tumor cell of origin for TEC. Cell surface CD31 expression was retained in all EC culture, even after repeated passages, whereas CD45 was entirely absent (Figure 1F).
Figure 1. Endothelial cell isolation and characterization.
(A) Normal mammary glands from wild-type mice and mammary tumors from C3-TAg mice (a–b) and their corresponding CD31 staining (c–d) reveals vascular structures. White arrowheads indicate the tissues resected for EC isolation. Black scale bar is 10 mm; white scale bar is 10 μm. (B) Workflow summary and timeline of the EC isolation procedure. (C) Representative phase contrast and fluorescent images of TEC clones incubated with DiI-AC-LDL at indicated time points. Scale bars are 10 μm. (D) RT-PCR analysis showing expression of EC markers Cd31 and Cdh-5 and absence of the monocyte/macrophage marker Cd45 in NEC and TEC clones. (E) Western blot of SV40 large T-antigen carried by C3-TAg mice indicating the absence of T-antigen protein in NEC and TEC cultures. (E) Flow cytometry for CD31 and CD45 expression in cells at different time points. Cells were split 3–4 times between day 0 (d0) and day 5 (d5).
Isolated TEC maintain the expression of endothelial-selective genes and are free of mesenchymal cells and tumor cells
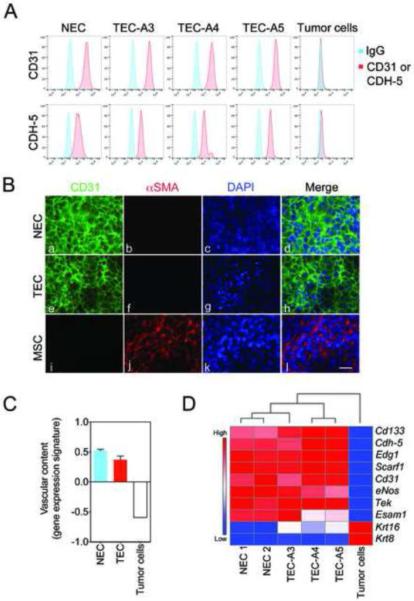
Using flow cytometry, we found that both NEC and TEC continued to express CD31 and CDH-5 during prolonged culturing (greater than 6–8 passages) (Figure 2A). Immunocytochemistry confirmed that ~ 100% of the cultured cells were CD31+ and did not express the mesenchymal marker αSMA (Figure 2B, a–l). CD31 was localized at cell-cell junctions in NEC and TEC indicating that all primary EC maintained their endothelial features in vitro. To examine the gene expression profiles of EC clones and C3-TAg tumor cells, we performed genome-wide mRNA expression microarrays and generated a “vascular content” genetic signature recently described by us [14]. As predicted, both NEC and TEC were enriched for endothelial-selective genes as defined by the “vascular content” genetic signature and when compared to C3-TAg tumor cells (p=0.00073) (Figure 2C). Hierarchical clustering and analysis of candidate, endothelial-selective factors demonstrate the relative difference between NEC, TEC, and mammary tumor cells derived from C3-Tag mice (Figure 2D). Furthermore, the expected vascular-specific factors including Cd133, Cdh-5, Edg1, Scarf1, Cd31, eNos, Tek, and Esam1 were highly enriched in all TEC clones, whereas the levels of tumor cell markers, including several cytokeratins (Krt), were essentially absent.
Figure 2. Isolated TEC maintain the expression of endothelial-selective genes and are free of mesenchymal cells and tumor cells.
(A) Flow cytometry plots demonstrating expression of CD31 and CDH-5 by NEC and TEC. (B) Representative images of CD31 (green) and αSMA (red) expression in NEC and TEC. MSC were used as a positive control for αSMA. Nuclei were counter-stained with DAPI (blue). Scale bar represents 10 μm. (C) Vascular content gene expression signature scores in NEC, TEC, and the mammary tumor cell line. (D) Hierarchical clustering and gene expression heat map identifies enrichment of EC-selective markers and absence of tumor cell markers (Krt) in different TEC clones.
Genome-wide expression profiling reveals a distinct molecular signature in mammary TEC
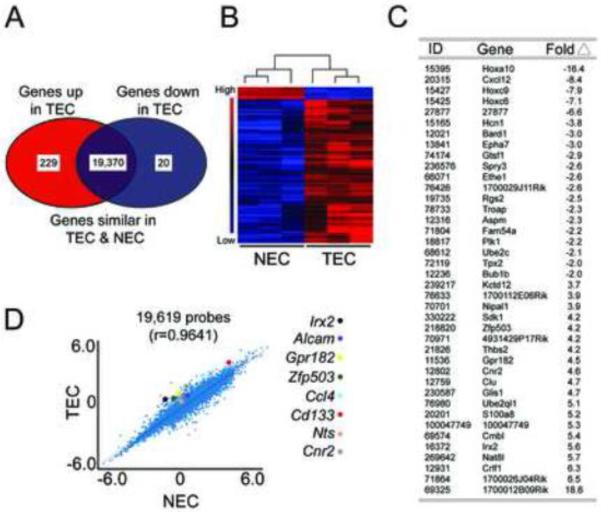
We next compared the gene expression profiles of TEC and NEC, represented as a Venn diagram, to identify differentially expressed genes (Figure 3A). Hierarchical clustering of 249 differentially-expressed genes demonstrated a consistent expression pattern in NEC and TEC populations as indicated by the dendrogram (Figure 3B). The entire microarray dataset is available for further query (https://genome.unc.edu) and a gene list of the 20 most down-regulated and up-regulated genes in TEC relative to NEC is shown (Figure 3C). A regression analysis using multiple NEC and TEC clones (19,619 genes) showed an overall striking congruency in gene expression (r=0.9641) in TEC versus NEC but multiple genes remained either up- or down-regulated in TEC cultures (Figure 3D). We used qPCR to validate eight up-regulated candidate genes from four different functional groups identified by our microarray. On average, the expression of each of these genes displayed the same trend as revealed by the microarray (Figure 4A). The most dramatically up-regulated genes in TEC clones included Irx2, Grp182, and Ccl4, with ~ average increases of 160, 28, and 12 fold, respectively (when compared to NEC). However, mRNA expression of individual clones for each gene was variable. For example, the relative expression of Cnr2 for the three TEC clones ranged from 0.6 – 6.5 fold, whereas Gpr182 expression ranged from 8 – 50 fold, indicating that TEC from the same tumor, while retaining similar gene expression profiles, may be heterogeneous in the expression of different factors.
Figure 3. Genome-wide expression profiling reveals a distinct molecular signature in mammary TEC.
(A) Venn diagram depicting up-regulated, down-regulated, and similarly-expressed genes in TEC versus NEC. (B) Supervised hierarchical clustering and gene expression heat map of 249 genes differently expressed in TEC versus NEC. (C) The 20 most down-regulated and up-regulated genes in TEC relative to NEC. The selected genes have an FDR<5. The ID is the mouse Entrez Gene ID. (D) Scatter plot comparing NEC and TEC gene expression for 19,619 genes. Each point represents the mean expression level of each gene in NEC and TEC. Colored dots identify 8 selected candidate genes that were up-regulated > 2-fold in TEC versus NEC.
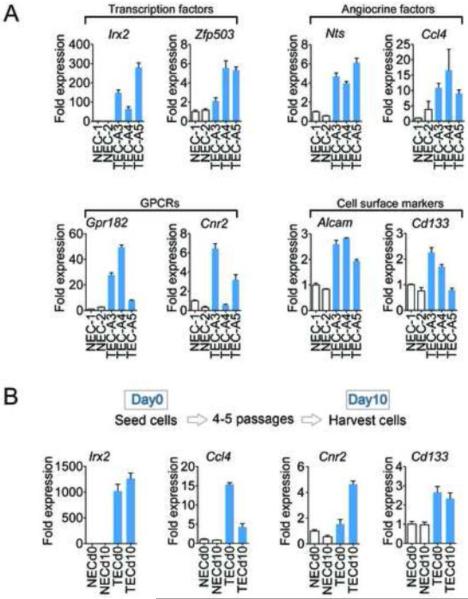
Figure 4. Aberrant expression of TEC-selective genes persists in vitro.
(A) qPCR confirmation of genes identified/selected from the microarray screen using different NEC and TEC clones. (B) qPCR analysis of selected genes at different time points. Depending on the cell type, cells were split 4–5 times between day 0 (d0) and day 10 (d10).
Aberrant expression of TEC-selective genes persists in vitro
Finally, we chose one gene from each category, and then carried out qPCR in cells at two time points separated by 4–5 sequential passages depending on the cell type. The results showed that Irx2, Ccl4, Cnr2, and Cd133 were on average persistently up-regulated in TEC relative to NEC, even after prolonged culturing and repeated passages (Figure 4B). Taken together, these results suggest that even after their removal from the tumor microenvironment, TEC retain over-expression of several candidate genes relative to their normal counterparts.
Discussion
A congruent TEC-specific genetic profile in TEC from different tumor types, or in TEC from the same tumor, has not been established [2]. This inconsistency likely arises from the heterogeneous nature of EC found in different organs (and tumors) as well as limitations of TEC isolation methodologies which include laser capture micro-dissection and positive selection by magnetic columns or cell sorting. Both methods can produce highly enriched, but not entirely pure TEC populations. Our use of magnetic columns followed by cloning rings to capture and expand individual TEC colonies has proven effective for deriving long-term cultures that are free of perivascular cells and tumor cells.
Because the microarray revealed aberrant and persistent expression of multiple factors in TEC cultures, we propose that stable, transmittable changes are induced in TEC during tumor progression that are retained ex vivo. However, it will be important to confirm the expression of these factors in tumors in vivo, either by in situ hybridization or immunohistochemistry in tumors during different stages of development. We stratified the microarray analysis into four categories (transcription factors, angiocrine factors, cell surface markers, and GPCRs) and further confirmed the over-expression of two candidate genes from each category by qPCR. For example, the transcription factors Irx2 and Zfp503 (Nolz-1), both highly enriched in TEC, regulate neural patterning and differentiation during development [17, 18]. Proliferating EC and neural precursors rely on common factors for network formation during development, and many of the same signaling pathways that regulate axon guidance overlap with those that control vessel sprouting [19]. Thus, activated TEC may co-opt neuronal cues during tumor angiogenesis. We also identified two secreted angiocrine factors (Nts and Ccl4) that are up-regulated in TEC. These factors are pro-inflammatory and may stimulate leukocyte proliferation and mobilization that create a permissive environment for tumor growth [20]. Thus, in addition to forming blood vessels, TEC may have perfusion-independent functions in the tumor microenvironment by orchestrating chronic inflammatory responses through secreted factors [21].
Other genes identified by our microarray include cell surface markers and GPCRs. For example, Cd133 was up-regulated in mammary TEC and in isolated prostate TEC as previously reported by us [5]. Though its function is unclear, CD133 was long-considered a marker of endothelial progenitor cells but more recent evidence supports a role for CD133 as a marker of proangiogenic hematopoietic precursors [22]. Similarly, ALCAM (CD166) is typically up-regulated on activated hematopoietic cells but is also expressed in inflamed/activated blood vessels where it regulates leukocyte capture and trafficking [23]. Expression of the GPCRs Gpr182 and Cnr2 were both markedly increased in TEC. The orphan receptor GPR182, once thought to be the adrenomedulin receptor, has no known function in EC but is enriched in embryonic vasculature [24]. On the other hand, CNR2 expression is well-characterized in vascular cells where it appears to play a role in dampening inflammatory responses and endothelial activation upon receptor stimulation [25, 26].
Folkman proposed that anti-angiogenic therapies could be used to attack tumor blood vessels and shrink solid tumors [27]. Although anti-angiogenic therapies are highly effective in curbing pathological neovascularization in non-malignant diseases such as wet macular degeneration, these same therapies have not produced a durable benefit in patients with different cancers [28, 29]. Thus, it is possible that TEC are more diverse and complex than previously recognized and may not respond predictably to angiogenesis inhibition [30]. Further characterization of TEC from different tumor types, or perhaps multiple subpopulations of TEC as we have done here, may reveal new vascular-selective targets for anti-angiogenic therapy [31].
Acknowledgements
ACD is supported by grants from the National Institute of Health (R00-CA140708) and the University Cancer Research Fund at UNC Chapel Hill. LX is a scholar in the HHMI-funded translational medicine program at UNC Chapel Hill. We would like to thank Mimi Kim and Clayton Davis for their excellent technical assistance.
References
- 1.Nolan DJ, Ginsberg M, Israely E, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Developmental Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aird WC. Molecular heterogeneity of tumor endothelium. Cell Tissue Res. 2009;335:271–281. doi: 10.1007/s00441-008-0672-y. doi: 10.1007/s00441-008-0672-y. [DOI] [PubMed] [Google Scholar]
- 3.Dudley AC. Tumor endothelial cells. Cold Spring Harb Perspect Med. 2012;2:a006536. doi: 10.1101/cshperspect.a006536. doi: 10.1101/cshperspect.a006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh K, Thodeti CK, Dudley AC, et al. Tumor-derived endothelial cells exhibit aberrant Rho-mediated mechanosensing and abnormal angiogenesis in vitro. Proc Natl Acad Sci. 2008;105:11305–11310. doi: 10.1073/pnas.0800835105. doi: 10.1073/pnas.0800835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley AC, Khan ZA, Shih S-C, et al. Calcification of multipotent prostate tumor endothelium. Cancer Cell. 2008;14:201–211. doi: 10.1016/j.ccr.2008.06.017. doi: 10.1016/j.ccr.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudley AC, Udagawa T, Melero-Martin JM, et al. Bone marrow is a reservoir for proangiogenic myelomonocytic cells but not endothelial cells in spontaneous tumors. Blood. 2010;116:3367–3371. doi: 10.1182/blood-2010-02-271122. doi: 10.1182/blood-2010-02-271122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. doi: 10.1101/cshperspect.a006429. doi: 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacorre D-A, Baekkevold ES, Garrido I, et al. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Burridge KA, Friedman MH. In vivo differences between endothelial transcriptional profiles of coronary and iliac arteries revealed by microarray analysis. Am J Physiol Heart Circ Physiol. 2008;295:H1556–61. doi: 10.1152/ajpheart.00540.2008. doi: 10.1152/ajpheart.00540.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burridge KA, Friedman MH. Environment and vascular bed origin influence differences in endothelial transcriptional profiles of coronary and iliac arteries. Am J Physiol Heart Circ Physiol. 2010;299:H837–46. doi: 10.1152/ajpheart.00002.2010. doi: 10.1152/ajpheart.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St Croix B, Rago C, Velculescu V, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 12.Seaman S, Stevens J, Yang MY, et al. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–554. doi: 10.1016/j.ccr.2007.04.017. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrell JC, Pfefferle AD, Zalles N, et al. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clin Exp Metastasis. 2013 doi: 10.1007/s10585-013-9607-4. doi: 10.1007/s10585-013-9607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green JE, Shibata MA, Yoshidome K, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–1027. doi: 10.1038/sj.onc.1203280. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 16.Voyta JC, Via DP, Butterfield CE, Zetter BR. Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. J Cell Biol. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto K, Nishihara S, Kamimura M, et al. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci. 2004;7:605–612. doi: 10.1038/nn1249. doi: 10.1038/nn1249. [DOI] [PubMed] [Google Scholar]
- 18.Chang C-W, Tsai C-W, Wang H-F, et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci USA. 2004;101:2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klagsbrun M, Eichmann A. A role for axon guidance receptors and ligands in blood vessel development and tumor angiogenesis. Cytokine Growth Factor Rev. 2005;16:535–548. doi: 10.1016/j.cytogfr.2005.05.002. doi: 10.1016/j.cytogfr.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 21.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson MR, Yoder MC. Endothelial progenitor cells: Quo Vadis? J Mol Cell Cardiol. 2010 doi: 10.1016/j.yjmcc.2010.07.009. doi: 10.1016/j.yjmcc.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cayrol R, Wosik K, Berard JL, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat Immunol. 2008;9:137–145. doi: 10.1038/ni1551. doi: 10.1038/ni1551. [DOI] [PubMed] [Google Scholar]
- 24.Takase H, Matsumoto K, Yamadera R, et al. Genome-wide identification of endothelial cell-enriched genes in the mouse embryo. Blood. 2012;120:914–923. doi: 10.1182/blood-2011-12-398156. doi: 10.1182/blood-2011-12-398156. [DOI] [PubMed] [Google Scholar]
- 25.Molica F, Matter CM, Burger F, et al. Cannabinoid receptor CB2 protects against balloon-induced neointima formation. Am J Physiol Heart Circ Physiol. 2012;302:H1064–74. doi: 10.1152/ajpheart.00444.2011. doi: 10.1152/ajpheart.00444.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajesh M, Mukhopadhyay P, Bátkai S, et al. CB2-receptor stimulation attenuates TNF-alpha-induced human endothelial cell activation, transendothelial migration of monocytes, and monocyte-endothelial adhesion. Am J Physiol Heart Circ Physiol. 2007;293:H2210–8. doi: 10.1152/ajpheart.00688.2007. doi: 10.1152/ajpheart.00688.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folkman J. Anti-angiogenesis: new concept for therapy of solid tumors. Ann Surg. 1972;175:409–416. doi: 10.1097/00000658-197203000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim LA, D'Amore PA. A Brief History of Anti-VEGF for the Treatment of Ocular Angiogenesis. Am J Pathol. 2012;181:376–379. doi: 10.1016/j.ajpath.2012.06.006. doi: 10.1016/j.ajpath.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res. 2012;72:1909–1914. doi: 10.1158/0008-5472.CAN-11-3406. doi: 10.1158/0008-5472.CAN-11-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhary A, Hilton MB, Seaman S, et al. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell. 2012;21:212–226. doi: 10.1016/j.ccr.2012.01.004. doi: 10.1016/j.ccr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]