Abstract
Canine osteosarcoma (OS) is an aggressive malignancy associated with poor outcomes. Therapeutic improvements are likely to develop from an improved understanding of signaling pathways contributing to osteosarcoma development and progression. The Wnt signaling pathway is of interest for its role in osteoblast differentiation, its dysregulation in numerous cancer types, and the relative frequency of cytoplasmic accumulation of β-catenin in canine OS. This study aimed to determine the biological impact of inhibiting canonical Wnt signaling in canine OS, by utilizing either β-catenin siRNA or a dominant-negative TCF construct. There were no consistent, significant changes in cell line behavior with either method compared to parental cell lines. Interestingly, β-catenin transcriptional activity was 3-fold higher in normal canine primary osteoblasts compared to canine osteosarcoma cell lines. These results suggest canonical Wnt signaling is minimally active in canine osteosarcoma and its targeted inhibition is not a relevant therapeutic strategy.
INTRODUCTION
Osteosarcoma (OS), the most common primary bone malignancy in humans, is a notably aggressive disease.1 Relapse and/or metastasis occur in 80% of cases, and five-year survival rates are approximately 30% when metastatic lesions are present.1,2 Naturally occurring canine OS is an accepted, clinically relevant animal model of human OS. The diseases is indistinguishable grossly and biochemically between the two species.3,4,5 Similar to the human disease, OS accounts for 85–98% of all primary bone malignancies in the dog.6,7 There has been minimal improvement in treatment outcomes during the past 15–20 years for both humans and dogs.8,9 With the advent of targeted therapeutics, there is renewed optimism that an enhanced understanding of the molecules and pathways contributing to the pathogenesis of OS will allow for the development of more targeted, efficacious therapies.
Wnt signaling is a pathway that has come under scrutiny for its potential involvement in the pathogenesis of OS, due to its necessity in osteoblast differentiation and role in normal bone development and homeostasis.10–17 Beyond bone, Wnt signaling plays a role in regulating the growth, movement, and survival of numerous normal and neoplastic cell types, further distinguishing it for investigation.18 Wnt signaling is traditionally divided into two distinct pathways: canonical and non-canonical. The keystone protein of canonical Wnt signaling is β-catenin. In the Wnt-off state (non-activated, ligand absent), free β-catenin (and thus available for transcriptional activity) is minimized through interaction with a degradation complex consisting of axin, adenomatous polyposis coli (APC), casein kinase-1α (CK1α), and glycogen synthase kinase-3β (GSK3β). Briefly, axin scaffolds with APC to direct phosphorylation of β-catenin by CK1α and subsequently GSK3β. The phosphorylation of serine/threonine amino acids promotes β-catenin ubiquitination and degradation. In the Wnt-on state (activated, ligand present), Wnt ligands complex with frizzled proteins (Fz) and low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6) to activate the receptor complex, which prevents the binding of the degradation complex to β-catenin. This unbound β-catenin is available to translocate to the nucleus, where it associates with members of the t-cell factor (TCF)/lymphoid enhancer-binding factor (LEF) transcription factor family, and promotes transcription of genes associated with differentiation and proliferation.11
The aim of this study was to determine the impact of β-catenin knockdown on OS behavior. This was achieved by either transient transfection with a β-catenin siRNA construct, or stable incorporation of a dominant-negative TCF4 (dnTCF4) construct. The latter reduces β-catenin-driven transcriptional activity by competitively binding β-catenin. Surprisingly, siRNA knockdown did not result in any consistent biological alterations, nor did it decrease β-catenin transcriptional activity; however, incorporation of the dnTCF4 construct resulted in decreased cellular metabolic activity (a surrogate for viability/proliferation) and susceptibility to doxorubicin. There were no changes in apoptosis, migration, invasion, or sensitivity to carboplatin chemotherapy associated with reduced β-catenin transcriptional activity in OS cell lines. The minimal magnitude of effects following dnTCF4 incorporation, and the lack of transcriptional reduction following significant RNA and protein knockdown with siRNA treatment, suggest that endogenous β-catenin transcriptional activity is minimal in these canine OS cell lines. This is supported by the observation that β-catenin transcriptional activity is three-fold higher in normal primary canine osteoblasts than canine OS cell lines.
MATERIALS & METHODS
Cell Culture and Reagents
Canine osteosarcoma cell lines D17 (ATCC CCL-183, originally isolated from a pulmonary metastatic OS lesion) and Abrams (originally isolated from a pulmonary metastatic OS lesion), were gifts from Dr. David Vail, and maintained in complete minimum essential medium (CMEM): minimum essential medium Eagle (MEM) supplemented with 10% cosmic calf serum, sodium pyruvate, l-glutamine, MEM vitamins, non-essential amino acids, and 1% Pen/Strep (all products from Fisher Scientific, Fair Lawn, NJ). Primary canine osteoblasts (k9Ob) were maintained in Canine Osteoblast Growth Medium, cultured using a Subculture Reagent Kit (cells and reagents from Cell Applications, Inc., San Diego, CA). All cells were maintained at 37°C in a humidified incubator with 5% CO2.
siRNA Transient Transfections
Four siRNA sequences targeting canine β-catenin were identified using the siRNA Converter and RNA Oligo Calculator (Ambion, Life Technologies, Carlsbad CA). The two sequences with the most dramatic RNA inhibition as measured by qPCR and western blot analysis were siRNA1 (sense: 5′-GCACACCAUACAACGGUUUUU-3′ antisense: 5′-AAACCGUUGUAUGGUGUGCUU-3′) and siRNA4 (sense: 5′-AGUUGUUGUAACCUGCUGUUU-3′ antisense: 5′-ACAGCAGGUUACAACAACUUU-3′). Sense and antisense oligonucleotides were obtained (Eurofins MWG Operon, Huntsville, AL) and siRNA were synthesized using Silencer siRNA Construction Kit (Ambion), according to the manufacturer’s instructions. Silencer Select negative control #1 siRNA was used as experimental control (Ambion). Transient transfections were performed with cells plated to 30–50% confluency, using Lipofectamine 2000 (Invitrogen, Carlsbad CA). In experiments plated in a 6-well plate, 100pmol siRNA was transfected with 5μL Lipofectamine diluted in 500μL EMEM per well. In experiments plated in a 96-well plate, 5pmol siRNA was transfected with 0.25μL Lipofectamine diluted in 50μL EMEM. RNA and protein lysates were isolated 48 hours following transfection. All other experiments were performed as detailed below.
dnTCF4 Stable Cell Lines
The pcDNA/dnTCF4 plasmid, originally generated by B Vogelstein, was kindly provided by V Spiegelman.19 D17 cells were plated in a six-well plate and grown to ~80% confluency, and then transfected with 2.5μg of pcDNA/dnTCF4 plasmid + 0.5μg of empty pcDNA3.1 vector using 15μL Lipofectamine LTX and 5μL PLUS reagent (Invitrogen, Carlsbad CA) in 500μL EMEM. After 24 hours, transfected cells were selected using G-418 (Invitrogen, Carlsbad, CA) at a concentration of 600μg/mL. Single-cell clones were isolated and assayed for β-catenin-transcriptional activity by TCF-responsive luciferase reporter assay. The two clones with most dramatic knockdown were propagated and maintained in CMEM supplemented with the appropriate concentration of G-418 (D17-DN1 and D17-DN6).
Quantitative PCR (qPCR)
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA), and purified by PureLink RNA Mini Kit (Ambion, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. cDNA was produced from 250ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies, Carlsbad CA) according to the manufacturer’s protocol. qPCR was performed using TaqMan Gene Expression Master Mix with the canine β-catenin/CTNNB1 (Cf02667771_m1) TaqMan Gene Expression Assay (Applied Biosystems) according to the manufacture’s protocol, on a Bio-Rad iQ5 Multicolor Real-Time PCR Detection System with Bio-Rad iCycler machine and iQ5 software. Ct values were normalized to 18S expression (4352930E, Applied Biosystems). Relative differences in mRNA expression of transfected/treated cells were compared to untreated cells using the ΔΔCt method.20 Gene expression of samples was measured in triplicate.
Western Blotting Analysis
Forty micrograms of protein lysate were separated by electrophoresis on a 7.5% Mini-PROTEAN TGX Gel (BioRad, Hercules, CA) at 150V for ~45min, transferred to nitrocellulose membranes (Whatman, Dassel, Germany) at 100V for 1h, then blocked with tris-buffered saline/0.05% Tween20 (TBST) containing 5% non-fat dry milk and 1% bovine serum albumin for 1h (all reagents from Fisher Scientific). The membranes were probed for 1h at room temperature with either (a) mouse anti-β-catenin antibody (#610154, BD Biosciences; validated for canine21) diluted 1:1000 in blocking solution or (b) mouse anti-HDAC2 (sc-56685, Santa Cruz Biotechnology, Santa Cruz, CA; validated for canine by the company) diluted 1:1000 in blocking solution. Excess primary antibody was removed by washing three times for 5 min with TBST. Membranes were incubated with 50ng/mL horseradish peroxidase-conjugated anti-mouse secondary antibody (Thermo Scientific) diluted in blocking solution for 1h at room temperature, then washed three times for 5 min at TBST, and treated with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Blots were exposed to film, developed, and then imaged using a Gel Logic 100 Imaging System (Kodak, Rochester, NY). Densitometry was performed using ImageJ (found at http://rsb.info.nih.gov/ij/index.html) according to the instructions located at http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/.
TCF-responsive Luciferase Reporter Assay
Twenty-four hours following transfection with siRNA, or plating for stable cell lines, cells were transiently transfected using Lipofectamine LTX and PLUS reagents (Invitrogen, Carlsbad CA) to introduce either TOPflash or FOPflash reporter plasmid, and TK-Renilla control plasmid at a 5:1 (Flash:Renilla) ratio (all plasmids: TCF Reporter Plasmid Kit, Millipore, Temecula, CA). For each well in a 96-well plate, cells were transfected with 60ng total DNA using 0.1μL PLUS reagent and 0.06μL LTX; Transfections were maximized using the Lipofectamine LTX optimization protocol. The TOPflash luciferase reporter plasmid contains TCF4 binding sites upstream of the luciferase gene, resulting in luciferase activity in the presence of active Wnt/β-catenin signaling, whereas the FOPflash reporter plasmid contains mutated TCF4 binding sites. The TK-Renilla plasmid serves as a control for transfection efficiency. Forty-eight hours after transfection of luciferase plasmids, cells were harvested and luciferase and Renilla luminescence were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) on a BioTek Synergy HT Multi-mode Microplate Reader, using Gen5 software (BioTek Instruments, Winooski, VT). The relative luciferase units for each transfection were adjusted by Renilla activity in the same sample, and each corrected TOPflash luciferase value was normalized to the corresponding corrected FOPflash value. Three independent transfections were performed, with each transfection assayed in triplicate.
Proliferation and Chemotoxicity Assays
Cells were plated in 96-well plates at a density such that untreated/control cells would reach an absorbance of approximately 1.0 at the conclusion of the experiment. Twenty-four hours following either plating of stable cell lines or siRNA transfection, media was aspirated off and replaced with fresh CMEM media, either with or without carboplatin (Hospira, Inc., Lake Forest, IL) or doxorubicin (Pfizer Labs, New York, NY). Cells treated with siRNA were assessed at 24 or 72 hours following media replacement (as noted in text); stable cell lines were assessed 48 hours following media replacement. Cell viability was measured by adding 20μL CellTiter96 Aqueous One Solution Cell Proliferation Assay solution (Promega, Madison, WI) per 100μL media to each well, incubated for a sufficient period, and absorbance measured at 490 nm on a VersaMax tunable microplate reader using SoftMax Pro 4.7 software (Molecular Devices, Sunnyvale, CA). Proliferation was assessed using the non-chemotherapy treated cells for modified cells vs. control cells; survival curves were generated by dividing the absorbance reading for chemotherapy-treated cells at each concentration by the absorbance for the non-chemotherapy treated cells, with the resultant modified-cell curve compared to the control-cell curve. Assays were plated in quadruplicate, and repeated in triplicate.
Apoptosis Assay
Cells were plated in 96-well plates at the same density as used in the proliferation/chemotoxicity assays. Twenty-four hours following either plating of stable cell lines or siRNA transfection, media was replaced with fresh CMEM. Forty-eight hours following media replacement, 100μL Caspase-Glo 3/7 Assay System (Promega, Madison, WI) solution was added per 100μL media in each well, incubated for 30min at room temperature and away from light, and luciferase was measured on a BioTek Synergy HT Multi-mode Microplate Reader, using Gen5 software. Measured luciferase activity (relative luciferase units) is proportional to caspase 3/7 activity, a marker of apoptosis. Assays were plated in triplicate and repeated in triplicate.
Migration & Invasion Assays
Migration and invasion assays were performed using BD BioCoat control inserts and BD BioCoat Matrigel Invasion Chambers (BD Biosciences, Bedford, MA), respectively. Cells were plated at a density of 2.5×104 cells/insert in EMEM media containing all supplements except serum in triplicate control or matrigel-coated chambers. For siRNA experiments, cells were transfected 24 hours prior to plating. CMEM media was used as a chemoattractant. Inserts were incubated 24 hours at 37°C in 5% CO2, then migrating/invading cells were fixed and stained with Diff-Quik Stain Set (Jorgensen Laboratories, Loveland, CO) and mounted onto microscope slides. Cells were imaged on an inverted microscope at 100× magnification and counted in two fields per insert. Migration was assessed by comparing the average number of migrating cells (counted cells) observed on the three un-coated control inserts per high-powered field (HPF) for untreated vs. treated/transfected cells. Invasion was assessed by comparing the ratios of invading cells for untreated vs. treated/transfected cells; this ratio was calculated by dividing the average number of cells present on three matrigel-coated inserts by the average number of cells present on three uncoated control inserts. Assays (each with three control and three matrigel-coated inserts) were repeated in duplicate.
Statistical Analyses
All graphs were generated and statistical analyses were performed using Prism 6 for Mac OS X (GraphPad Software, Inc., San Diego, CA). All data is presented as mean ± standard deviation (SD). A two-way ANOVA test with Sidak post-test was performed on all siRNA experiments, as well as dnTCF4 chemotoxicity experiments. A one-way ANOVA with Dunnett’s post-test was performed on all other experiments. Values of p<0.05 were considered significant, and are indicated by (*), values <0.01 by (**), values <0.001 by (***), and values <0.0001 by (****).
RESULTS
Knockdown of β-catenin expression using siRNA
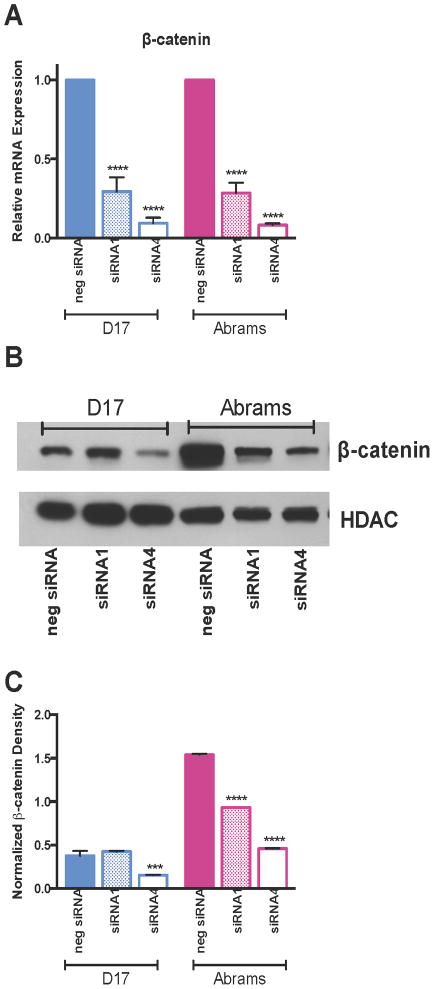
To determine whether knockdown of β-catenin expression altered canine OS behavior, D17 and Abrams canine OS cells were transiently transfected with two siRNA constructs (siRNA1 and siRNA4) targeting β-catenin, or a control negative siRNA construct (neg siRNA). In the D17 cell line, siRNA1 produced a β-catenin mRNA knockdown of 70.6% (p<0.0001) compared to the neg siRNA transfected, and siRNA4 produced a knockdown of 90.7% (p<0.0001). In the Abrams cell line, siRNA1 produced a β-catenin mRNA knockdown of 71.6% (p<0.0001) compared to the neg siRNA transfected, and siRNA4 produced a knockdown of 91.8% (p<0.0001) (Fig 1A). β-catenin protein expression was also decreased in both cell lines following siRNA transfection (Fig 1B–C), with a 58% reduction in D17 cells following siRNA4 transfection (p=0.0002; neg siRNA: 0.376±0.056, siRNA1: 0.426±0.008, siRNA4: 0.155±0.004), and a 39% reduction following siRNA1 (p<0.0001) and 70% reduction following siRNA4 (p<0.001) transfection in Abrams cells (neg siRNA: 1.539±0.013, siRNA1: 0.933±0.000, siRNA4: 0.459±0.005).
Figure 1.
Figure 1A–C. Evaluation of siRNA Efficacy in targeting β-catenin. Significant reductions in (A) β-catenin mRNA expression, (B) β-catenin protein expression, and (C) β-catenin protein as measured by densitometry were achieved following treatment with siRNA1 or siRNA4 compared to a negative control siRNA. HDAC was used as a loading control for the western blot and densitometry measurements. All calculations are a 2-way ANOVA with Sidak’s multiple comparison post-test.
Effects of β-catenin expression knockdown on cell behavior
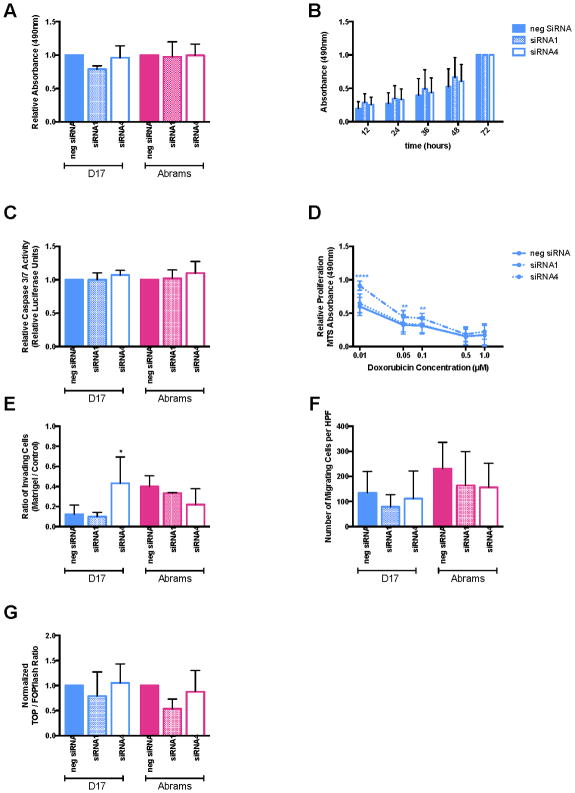
To determine whether biological behaviors were altered subsequent to knockdown of β-catenin expression, several in vitro assays were performed. These included cellular proliferation assays (a single 24 hour time-point in Fig 2A, and a 72 hour curve for D17 in Fig 2B), apoptosis assays (Fig 2C), chemoresistance to multiple doxorubicin concentrations measured at 72 hours in D17 cells (Fig 2D), invasion (Fig 2E) and migration (Fig 2F) assays. Surprisingly, cell behavior was not consistently significantly altered in any of these assessments; the only statistically significant differences observed were in siRNA1 treated D17 cells at the lower concentrations of doxorubicin, and siRNA4 treated D17 cells in the invasion assay.
Figure 2.
Figure 2A–G. Evaluation of siRNA targeting β-catenin behavioral effects. There were no consistent differences between siRNA-treated cells and negative control siRNA-treated cells for (A) proliferation at 24 hours, (B) proliferation over a 72-hour time course, (C) apoptosis, (D) doxorubicin sensitivity, (E) invasion, or (F) migration. There was also no change in β-catenin transcriptional activity as measured by TCF-responsive luciferase reporter assay (G). All calculations are a 2-way ANOVA with Sidak’s post-test.
Given the significant reduction in mRNA expression, marked reduction in protein expression, yet minimal behavioral alterations, the transcriptional activity of β-catenin following siRNA treatment was assessed using a TCF-responsive luciferase reporter assay. The normalized TOP/FOPflash ratio was not significantly reduced for either siRNA in either of the cell lines compared to the negative siRNA (Fig 2G).
Knockdown of β-catenin transcriptional activity using dominant-negative TCF4 (dnTCF4) expressing stable cell lines
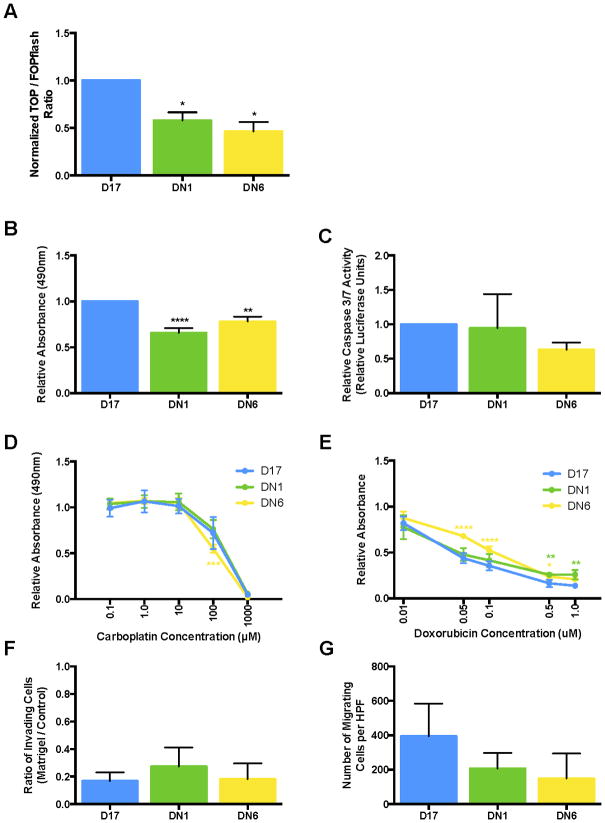
To specifically target and knockdown β-catenin transcriptional activity, and determine any subsequent effect on canine OS cell line behavior, cell lines that stably and constitutively express a dominant negative TCF4 vector were developed. The dnTCF4 construct competitively binds β-catenin, preventing β-catenin from associating with wild-type TCF4 to promote target gene transcription. The TCF-responsive luciferase reporter assay was used to confirm knockdown of β-catenin transcriptional activity. The clones DN1 and DN6 were identified as having the most robust reduction in β-catenin transcriptional activity compared to the parental D17 cell line. The normalized TOP/FOPflash ratios were 0.58±0.09 (p=0.042) and 0.46±0.10 (p=0.013) for clone DN1 and DN6, respectively (Fig 3A).
Figure 3.
Figure 3A–G. Evaluation of D17 cells stably expressing dnTCF4. (A) Following incorporation of the dnTCF4 construct, both clones exhibited reduced β-catenin transcriptional activity by TCF-responsive luciferase reporter assay. This was matched with reductions in proliferation (B) and doxorubicin sensitivity (E), but not alterations in apoptosis (C), carboplatin sensitivity (D), invasion (F), or migration (G). All calculations are a 1-way ANOVA with Dunnett’s post-test, except D and E, which are 2-way ANOVA with Sidak’s post-test.
Effects of β-catenin transcriptional activity knockdown on cell behavior
To determine whether cell behavior was altered subsequent to knockdown of β-catenin transcriptional activity, in vitro assays were performed as above. Cellular proliferation was decreased in the dnTCF4 cells compared to the parental D17 cell line by: 34% in clone DN1 cells and 36% in clone DN6 cells (1.07±0.13 vs. DN1: 0.70±0.10, p<0.0001; DN6: 0.84±0.11, p=0.0048; Fig 3B). This was not matched with alterations in apoptosis (Fig 3C) or carboplatin resistance (Fig 3D), but was paired with a corresponding reduction in sensitivity to doxorubicin (Fig 3E). Finally, no differences in invasion (Fig 3F) or migration (Fig 3G) were observed.
β-catenin transcriptional activity in canine OS compared to normal canine osteoblasts
To compare β-catenin transcriptional activity in canine OS cell lines to normal canine osteoblasts, the TCF-responsive luciferase assay was performed on two canine OS cell lines (D17 and Abrams) and normal primary canine osteoblasts (Fig 4). The normalized TOP/FOP ratio for canine normal primary osteoblasts was three-fold higher than seen in the canine OS cell lines (k9Ob: 1.28±0.310; D17: 0.48±0.103, p=0.0055; Abrams: 0.45±0.304, p=0.0042).
Figure 4.
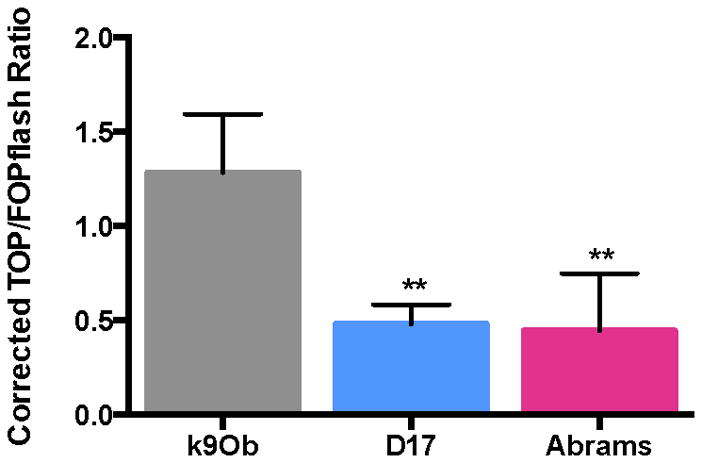
Evaluation of β-catenin transcriptional activity in canine normal primary osteoblasts compared to canine OS cells. Canine normal primary osteoblasts (k9Ob) exhibit a 2.7-fold higher β-catenin transcription activation than D17 cells (p=0.0055), and 2.9-fold higher than Abrams cells (p=0.0042). Calculation is a 1-way ANOVA with Dunnett’s post-test.
DISCUSSION
Treatment options and survival rates for both human and canine OS have essentially stagnated the last 15–20 years, prompting the investigation of various molecules and signaling pathways that could be targeted for therapy in hopes of improving outcomes.8,9 Wnt signaling is a pathway of interest for its role in osteoblast differentiation, its dysregulation in numerous cancer types, and the frequency of cytoplasmic accumulation in canine and human OS.12,13,18,21–23 While in epithelial-based cancers – such as hepatocellular, colorectal, and mammary carcinomas – β-catenin activation is considered a pro-oncogenic alteration, the role of β-catenin in mesenchymal-based tumors such as melanoma and OS is less well-defined.23–30 Some authors have reported that the Wnt/β-catenin pathway is active in OS, as indicated by nuclear and cytoplasmic β-catenin staining in human OS cells, increased cytoplasmic and/or nuclear β-catenin expression in tissue from xenogeneic murine pulmonary metastasis, and decreased tumorigenesis by inhibition of Wnt receptors; while others have reported that activation of the Wnt/β-catenin pathway is selected against in OS development and that inhibition of Wnt/β-catenin signaling transforms human mesenchymal stem cells into sarcoma cells in vitro.21, 28, 30–34 Therefore, the aim within this study was to determine whether β-catenin knockdown would result in alterations of OS cell line behavior. The overall hypothesis was that β-catenin knockdown would result in a less biologically aggressive phenotype – i.e. decreased proliferation, migration and invasion, and increased apoptosis and sensitivity to chemotherapeutics.
To achieve this aim, two approaches were utilized: (1) β-catenin expression knockdown via two siRNA constructs; and (2) β-catenin transcriptional activity reduction via the integration of a dnTCF4 construct. No significant consistent alterations were found in the tested biological behaviors following significant and dramatic knockdown in mRNA and protein expression following siRNA treatment, including chemoresistance to carboplatin (data not shown). While two differences in behavior were noted to be statistically significant (siRNA1 and doxorubicin sensitivity; siRNA4 and invasion), these changes were not uniform across both siRNA treatments, nor between cell lines, thus are most likely due to off-target effects of the siRNA.35 The lack of biological effects was paired with a surprising lack of reduction in transcriptional activity. It is possible that a reduction in β-catenin expression alone, without corresponding transcriptional reductions, is insufficient to alter in vitro cellular behavior as β-catenin’s role in the canonical Wnt signaling pathway is that of transcriptional co-factor, and thus affects cellular activity through the activation of downstream transcriptional targets.11,18 To achieve a reduction in β-catenin’s transcriptional activity, stable cell line clones expressing a dnTCF4 construct were generated. The roughly 50% decrease in β-catenin transcriptional activity was associated with a minimal, yet statistically significant, reduction in proliferation and decreased sensitivity to doxorubicin.
β-catenin knockdown in the human OS cell line MG63 resulted in no change in cell proliferation, similar to the canine OS cell lines tested here.36 Additionally, no change in apoptosis was noted, consistent the results of Wu et al using shRNA in both MG63 and SaOS2, and with our results in canine OS cell lines.37,38 However, other groups have observed increases in apoptosis following β-catenin knockdown: Xia et al used a lentiviral siRNA construct in MG63 cells; Leow et al used PKF118-310, a chemical inhibitor that disrupts the β-catenin/TCF4 complex, on U2OS cells; and Ma et al treated SaOS2 cells with either a β-catenin shRNA construct or CCT036477, a chemical inhibitor that blocks β-catenin transcription without altering β-catenin expression.38,39 One potential reason for differences in apoptosis results is that the assays used by each group to quantify apoptosis are different, and may thus be picking up mechanisms of apoptosis that are not mediated by Caspase 3/7, which our apoptosis assay detects. Another potential reason for the lack of observed behavioral changes with the siRNA is, of course, the lack of knock down in transcriptional activity. However, barring the decrease in proliferation and sensitivity to doxorubicin, the lack of behavioral changes associated with β-catenin knock down were consistent between siRNA and dnTCF4 inhibition.
In the current study, reduction of β-catenin’s transcriptional activity was associated with decreased sensitivity to doxorubicin treatment in the dnTCF4 clones compared to the parental line. Zhang et al observed a similar decreased sensitivity to doxorubicin in MG63 human OS cells.36 While the decreased sensitivity to doxorubicin may be due, at least in part, to the reduction in proliferation, Zhang et al. did not see a reduction in proliferation accompanying changes to doxorubicin sensitivity. It is interesting to note that reducing β-catenin’s transcriptional activity had no impact on sensitivity to carboplatin treatment. This would suggest the reduction in cell proliferation is not the only factor in altering doxorubicin sensitivity, as similar changes in sensitivity would be expected for carboplatin as well. Others have identified increased sensitivity to chemotherapeutics following β-catenin inhibition, with Wu et al observing enhanced sensitivity to cisplatin and Ma et al observing increased sensitivity to methotrexate treatment.37,40 Finally, we observed no differences in invasion and migration; this finding is in contrast to the human OS studies of Zhang et al and Leow et al.35,38
Despite an attempt to compare the results of our current study to those of previous studies, caution must be exercised due to the differences in the analyses employed to assess β-catenin inhibition. Most studies have relied upon Western blot analysis to demonstrate β-catenin expression knockdown, with no assessment of alterations in transcriptional activity (beyond the biological assays themselves). As the current study demonstrates, it is possible to have significant knockdown in mRNA and protein without alterations in transcriptional activity. Without a method to directly compare transcriptional activity, it is possible that β-catenin knockdown in the previous studies yielded transcriptional alterations greater than the approximately 50% decrease produced here. Thus, it is conceivable that differences in apoptosis and response to chemotherapeutics may have been noted with further reductions in transcriptional activity. This type of dose-dependent response to knockdown of transcriptional activity may underlie the variable results observed and published, however such a comparison is impossible to perform without transcriptional activity data.
Given the minimal magnitude of effects following dnTCF4 incorporation, and the lack of transcriptional reduction following significant RNA and protein knockdown with siRNA treatment, the hypothesis that endogenous β-catenin transcriptional activity is minimal in canine OS cell lines would logically follow. To test this, β-catenin transcriptional activity was compared between normal primary canine osteoblasts and canine OS cell lines. β-catenin transcriptional activity was found to be three-fold higher in the normal primary canine osteoblasts relative to these canine OS cell lines. This assessment is the first of its kind for canine cell lines, but is corroborated by the results of four other studies. Bongiovanni et al found increased β-catenin intensity and nuclear localization in canine osteoblasts compared to canine OS clinical samples.41 Similarly, Cai and colleagues identified strong membranous and nuclear β-catenin localization in human osteoblastomas without nuclear localization in OS samples.28 In a study utilizing rat cell lines there was decreased expression of β-catenin and Lef1, a downstream target of active β-catenin transcription, in rat OS cells compared to rat mesenchymal stem cells (MSCs) on microarray analysis.41 Finally, Cleton-Janson et al reported on decreased upstream mediators of Wnt signaling in human OS compared to osteoblastoma, MSC and MSCs differentiated into osteoblasts.29 Contrary to these results, other groups have observed some OS cell lines to have relatively increased β-catenin transcriptional activity relative to human MSCs, and increased expression of β-catenin and Lef1 mRNA in human OS compared to human fetal osteoblasts, paired with increased active β-catenin protein by western blot.40,42
The inconsistencies of relative β-catenin transcriptional activity in MSCs, osteoblasts and OS cells are both a function and a clear representation of the lack of precise knowledge concerning the cell-of-origin for OS. OS is a disease of multiple histological sub-types, suggesting either that tumor-originating cells are multipotent, or that there are multiple points along the differentiation spectrum where cells are able to achieve neoplastic growth.43 This differentiation spectrum – which comprises of mesenchymal progenitors on the least differentiated side, moving through pre-osteoblastic states defined by the acquisition or loss of particular transcription factors, until a state of mature osteoblast is reached – is driven by five signaling pathways, including Wnt signaling.44 In particular, β-catenin is required for differentiation from mid-pre-osteoblastic states through mature osteoblasts.43 The flux of Wnt signaling controlling osteoblast-lineage differentiation, combined with the uncertainty of the cell-of-origin for OS, are additional complications in the effort to determine transcriptional activation of β-catenin in OS. A final point that should be considered in the interpretation of these results is the limited number of cell lines that have been assessed. It is possible the utilization of additional cell lines may have yielded different results. Further confounding the results of any in vitro study of OS is the relatively high degree of disease heterogeneity, which is further exacerbated by selecting for a clonal population of cells when generating cell lines. However, the basal endogenous Wnt signaling activity in the cell lines reported here is similar to that of other human, murine, and canine OS cell lines we have assessed (unpublished data), as well as the results of Cai et al with human OS cell lines, and the gene-expression profiling of osteosarcoma tissue, osteoblastoma and mesenchymal stem cells by Cleton-Jansen et al.28,30
The determination of β-catenin’s sub-cellular localization of clinical samples through immunohistochemistry is commonly used to assess the activation status of canonical Wnt signaling. Often, the presence of cytoplasmic and/or nuclear localization of β-catenin is used as surrogate for pathway activation. While both humans and canines exhibit frequent cytoplasmic β-catenin staining, there is minimal nuclear β-catenin staining, with 10% (5/52) of human OS samples and 2% (1/56) to 10% (3/30) of canine OS samples displaying positive nuclear β-catenin.22,23,28 Contrary to these low numbers, Bongiovanni et al observed 47% (8/17) of canine OS samples to display nuclear staining; however in nuclear-positive samples, less than 10% of nuclei/cells within the sample were positive, indicating that these positive nuclei were very weakly positive.41 The results of these studies are similar to those evaluating β-catenin in human and canine malignant melanoma, in which the presence of cytoplasmic β-catenin is frequent, though nuclear β-catenin is rare and there is low transcriptional activity in melanoma cell lines.45,46 Interestingly, Kuphal and Bosserhoff found serine 33 and 37 residues in β-catenin to be hypo-phosphorylated, likely preventing β-catenin from being targeted for ubiquitin-mediated degradation. However, β-catenin was concurrently phosphorylated at threonine 41 and serine 45 residues, thereby preventing its transcriptional activity.45 The current results suggest a similar scenario may exist in OS.
In summary, our data suggests that β-catenin transcriptional activation is minimal in canine OS cell lines. This conclusion is based on minimal clinical OS β-catenin transcriptional activation, paired with this study’s findings of: (a) a lack of transcriptional activity knockdown concurrent with robust mRNA and protein expression reduction following siRNA treatment, and (b) minimal behavioral alterations subsequent to transcriptional inhibition of roughly 50%. This conclusion is supported by the observation that β-catenin transcriptional activity in canine osteoblasts is three-fold more than in canine OS cells. These findings support further study to determine whether transcriptional activation of β-catenin is selected against in the development and/or progression of both human and canine OS.
Acknowledgments
This project was supported by a Chase Away K9 Cancer Grant through the American College of Veterinary Internal Medicine Foundation (#10-08)(TJS), the Clinical and Translational Science Award (CTSA) program, previously through the National Center for Research Resources (NCRR) grant 1UL1RR025011, and now by the National Center for Advancing Translational Sciences (NCATS) grants 9U54TR000021 (TJS), T32RR17503 (CMP), and 1UL1RR025011 (CMP).
Footnotes
Portions of this material were presented at the Veterinary Cancer Society Annual Meeting, Las Vegas, NV, 2012.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. International Journal of Cancer. 2009;125:229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorlick R, Khanna C. Osteosarcoma. Journal of Bone and Mineral Research. 2010;25:683–691. doi: 10.1002/jbmr.77. [DOI] [PubMed] [Google Scholar]
- 4.Morello E, Martano M, Buracco P. Biology, diagnosis and treatment of canine appendicular osteosarcoma: Similarities and differences with human osteosarcoma. The Veterinary Journal. 2011;189:268–277. doi: 10.1016/j.tvjl.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Paoloni M, Davis S, Lana S, Withrow S, Sangiorgi L, Picci P, Hewitt S, Triche T, Meltzer P, Khanna C. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liptak JM, Dernell WS, Ehrhart NP, Withrow SJ. Canine appendicular osteosarcoma: diagnosis and palliative treatment. Compendium of Continuing Education for the Practicing Veterinarian. 2004;26:172–182. [Google Scholar]
- 7.Dernell WS, Ehrhart NP, Straw RC, Vail DM. In: Withrow and MacEwen’s Small Animal Oncology. Withrow SJ, Vail DM, editors. St. Louis, MO: Saunders, Elsevier; 2007. pp. 540–582. [Google Scholar]
- 8.O’Day K, Gorlick R. Novel therapeutic agents for osteosarcoma. Expert Reviews in Anticancer Therapy. 2009;9:511–523. doi: 10.1586/era.09.7. [DOI] [PubMed] [Google Scholar]
- 9.Phillips B, Powers BE, Dernell WS, Straw RC, Khanna C, Hogge GS, Vail DM. Use of single-agent carboplatin as adjuvant or neoadjuvant therapy in conjunction with amputation for appendicular osteosarcoma in dogs. Journal of the American Animal Hospital Association. 2009;45:33–38. doi: 10.5326/0450033. [DOI] [PubMed] [Google Scholar]
- 10.McQueen P, Ghaffar S, Guo Y, Rubin EM, Zi X, Hoang BH. The Wnt signaling pathway: implications for therapy in osteosarcoma. Expert Reviews in Anticancer Therapy. 2011;11:1223–1232. doi: 10.1586/era.11.94. [DOI] [PubMed] [Google Scholar]
- 11.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes and Development. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaes BL, Dechering KJ, van Someren EP, Hendriks JM, van de Ven CJ, Feijen A, Mummery CL, Reinders MJ, Olijve W, van Zoelen EJ, Steegenga WT. Microarray analysis reveals expression regulation of Wnt antagonists in differentiating osteoblasts. Bone. 2005;36:803–811. doi: 10.1016/j.bone.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends in Cell Biology. 2006;16:151–158. doi: 10.1016/j.tcb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 15.Parr BA, McMahon AP. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature. 1995;374:350–353. doi: 10.1038/374350a0. [DOI] [PubMed] [Google Scholar]
- 16.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. Journal of Cell Bioliology. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Jüppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2011;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Haydon RC, Deyrup A, Ishikawa A, Heck R, Jiang W, Zhou L, Feng T, King D, Cheng H, Breyer B, Peabody T, Simon MA, Montag AG, He TC. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. International Journal of Cancer. 2002;102:338–342. doi: 10.1002/ijc.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein TJ, Holmes KE, Muthuswamy A, Thompson V, Huelsmeyer MK. Characterization of β-catenin expression in canine osteosarcoma. Veterinary and Comparative Oncology. 2011;9:65–73. doi: 10.1111/j.1476-5829.2010.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piskun CM, Muthuswamy A, Huelsmeyer MK, Thompson V, Stein TJ. Wnt/β-catenin expression does not correlate with serum alkaline phosphatase concentration in canine osteosarcoma patients. PLoS One. 2011;6:e26106. doi: 10.1371/journal.pone.0026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 25.White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142:219–232. doi: 10.1053/j.gastro.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: β-catenin. Breast Cancer Research. 2010;12:213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucero OM, Dawson DW, Moon RT, Chien AJ. A re-evaluation of the “oncogenic” nature of Wnt/beta-catenin signaling in melanoma and other cancers. Current Oncology Reports. 2010;12:314–318. doi: 10.1007/s11912-010-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, Mohseny AB, Karperien M, Hogendoorn PC, Zhou G, Cleton-Jansen AM. Inactive Wnt/beta-catenin pathway in conventional high-grade osteosarcoma. Journal of Pathology. 2010;220:24–33. doi: 10.1002/path.2628. [DOI] [PubMed] [Google Scholar]
- 29.Honoki K, Fujii H, Tohma Y, Tsujiuchi T, Kido A, Tsukamoto S, Mori T, Tanaka Y. Comparison of gene expression profiling in sarcomas and mesenchymal stem cells identifies tumorigenic pathways in chemically induced rat sarcoma model. International Scholarly Research Network Oncology. 2012 doi: 10.5402/2012/909453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleton-Jansen AM, Anninga JK, Briaire-de Bruijn IH, Romeo S, Oosting J, Egeler RM, Gelderblom H, Taminiau AH, Hogendoorn PC. Profiling of high-grade central osteosarcoma and its putative progenitor cells identifies tumourigenic pathways. British Journal of Cancer. 2009;101:1909–1918. doi: 10.1038/sj.bjc.6605405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwaya K, Ogawa H, Kuroda M, Izumi M, Ishida T, Mukai K. Cytoplasmic and/or nuclear staining of beta-catenin is associated with lung metastasis. Clinical Experimental Metastasis. 2003;20:525–529. doi: 10.1023/a:1025821229013. [DOI] [PubMed] [Google Scholar]
- 32.Rubin EM, Guo Y, Tu K, Xie J, Zi X, Hoang BH. Wnt inhibitory factor 1 decreases tumorigenesis and metastasis in osteosarcoma. Molecular Cancer Therapeutics. 2010;9:731–741. doi: 10.1158/1535-7163.MCT-09-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Rubin EM, Xie J, Zi X, Hoang BH. Dominant negative LRP5 decreases tumorigenicity and metastasis of osteosarcoma in an animal model. Clinical Orthopaedics and Related Research. 2008;466:2039–2045. doi: 10.1007/s11999-008-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA, Singer S, Maki RG, Cordon-Cardo C. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. Journal of Clinical Investigation. 2007;117:3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Chen A, Chen J, Yu T, Guo F. SiRNA-mediated silencing of beta-catenin suppresses invasion and chemosensitivity to doxorubicin in MG-63 osteosarcoma cells. Asian Pacific Journal of Cancer Prevention. 2011;12:239–245. [PubMed] [Google Scholar]
- 37.Wu J, Liao Q, He H, Zhong D, Yin K. TWIST interacts with β-catenin signaling on osteosarcoma cell survival against cisplatin. Molecular Carcinogenesis. 2012 doi: 10.1002/mc.21991. [DOI] [PubMed] [Google Scholar]
- 38.Xia JJ, Pei LB, Zhuang JP, Ji Y, Xu GP, Zhang ZP, Li N, Yan JL. Celecoxib inhibits β-catenin-dependent survival of the human osteosarcoma MG-63 cell line. Journal of Internal Medicine Research. 2010;38:1294–1304. doi: 10.1177/147323001003800411. [DOI] [PubMed] [Google Scholar]
- 39.Leow PC, Tian Q, Ong ZY, Yang Z, Ee PL. Antitumor activity of natural compounds, curcumin and PKF118-310, as Wnt/β-catenin antagonists against human osteosarcoma cells. Investigational New Drugs. 2010;28:766–82. doi: 10.1007/s10637-009-9311-z. [DOI] [PubMed] [Google Scholar]
- 40.Ma Y, Ren Y, Han EQ, Li H, Chen D, Jacobs JJ, Gitelis S, O’Keefe RJ, Konttinen YT, Yin G, Li TF. Inhibition of the Wnt-β-catenin and Notch signaling pathways sensitizes osteosarcoma cells to chemotherapy. Biochemical and Biophysical Research Communications. 2013;43:274–279. doi: 10.1016/j.bbrc.2012.12.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bongiovanni L, Mazzocchetti F, Malatesta D, Romanucci M, Ciccarelli A, Buracco P, De Maria R, Palmieri C, Martano M, Morello E, Maniscalco L, Della Salda L. Immunohistochemical investigation of cell cycle and apoptosis regulators (survivin, β-catenin, p53, caspase 3) in canine appendicular osteosarcoma. BMC Veterinary Research. 2012;8:78. doi: 10.1186/1746-6148-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayakumar S, Liu G, Rus IA, Yao S, Chen Y, Akiri G, Grumolato L, Aaronson SA. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/β-catenin target gene, CDC25A. Cancer Cell. 2011;19:601–612. doi: 10.1016/j.ccr.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohseny AB, Hogendoorn PC. Concise review: mesenchymal tumors: when stem cells go mad. Stem Cells. 2011;29:397–403. doi: 10.1002/stem.596. [DOI] [PubMed] [Google Scholar]
- 44.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nature Reviews Molecular and Cellular Biology. 2011;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 45.Kuphal S, Bosserhoff A. Phosphorylation of β-catenin results in lack of β-catenin signaling in melanoma. International Journal of Oncology. 2011;39:235–243. doi: 10.3892/ijo.2011.1028. [DOI] [PubMed] [Google Scholar]
- 46.Chon E, Thompson V, Schmid S, Stein TJ. Activation of the canonical Wnt/β-catenin signaling pathway is rare in canine malignant melanoma tissue and cell lines. Journal of Comparative Pathology. 2013;148:178–187. doi: 10.1016/j.jcpa.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]